Abstract
The ability to manipulate the genomes of herpesviruses is of eminent importance for obtaining insight into gene function and regulation of gene expression of these complex viruses. Here we report the use of in vivo overlap recombination to generate pseudorabies virus mutants. Cotransfection of up to five overlapping cloned subgenomic fragments, which together constitute the entire genomic information of pseudorabies virus, results in the efficient reconstitution of virus. This allows the efficient introduction of multiple well-defined mutations in herpesvirus genomes in a single step, without any selection or screening for a particular phenotype.
Full text
PDF
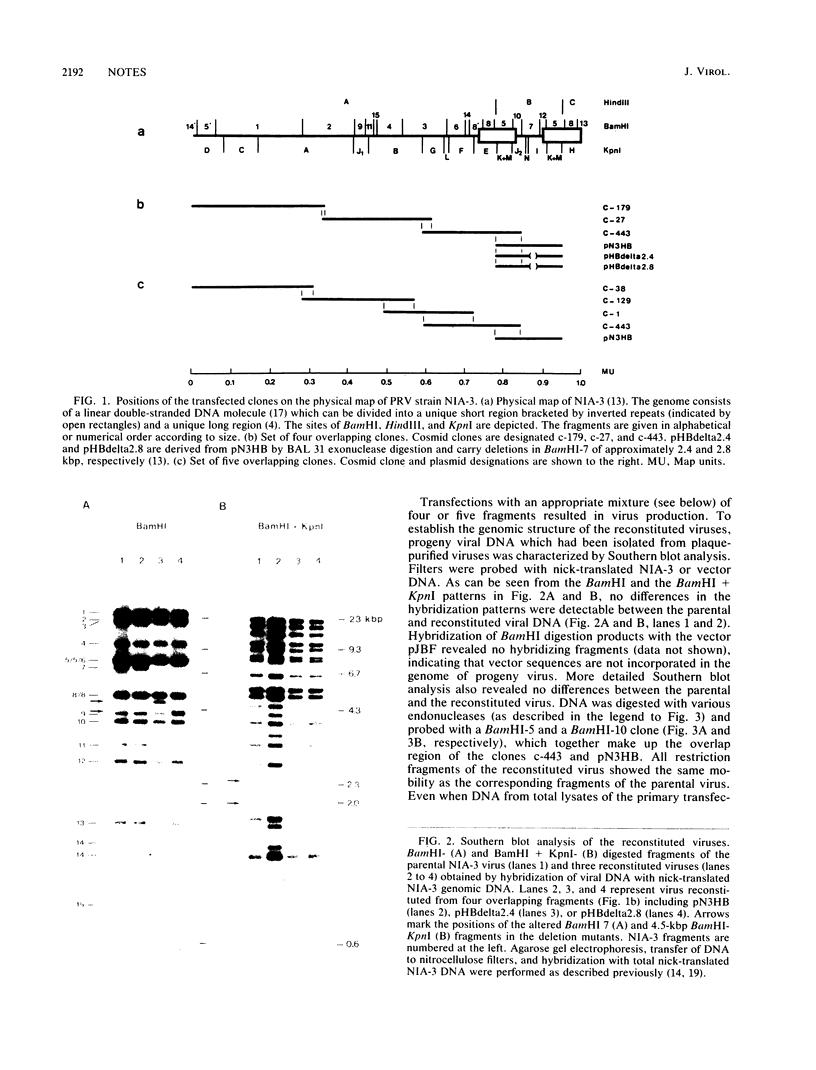



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. A., Eliason S. L. Recombination of homologous DNA fragments transfected into mammalian cells occurs predominantly by terminal pairing. Mol Cell Biol. 1986 Sep;6(9):3246–3252. doi: 10.1128/mcb.6.9.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Deatly A., Veach R. A., Blankenship M. L. Equalization of the inverted repeat sequences of the pseudorabies virus genome by intermolecular recombination. Virology. 1984 Jan 30;132(2):303–314. doi: 10.1016/0042-6822(84)90037-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J., Blankenship M. L. Analysis of the structure of the genome of pseudorabies virus. Virology. 1979 Jun;95(2):285–294. doi: 10.1016/0042-6822(79)90484-7. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Sharp P. A. Generation of adenovirus by transfection of plasmids. Nucleic Acids Res. 1983 Sep 10;11(17):6003–6020. doi: 10.1093/nar/11.17.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R., Vaessen M. J., Van der Eb A. J., Sussenbach J. S. Construction and characterization of an adenovirus type 5/adenovirus type 12 recombinant virus. Virology. 1983 Nov;131(1):30–38. doi: 10.1016/0042-6822(83)90530-5. [DOI] [PubMed] [Google Scholar]
- Berns A., van den Ouweland A., Quint W., van Oirschot J., Gielkens A. Presence of markers for virulence in the unique short region or repeat region or both of pseudorabies hybrid viruses. J Virol. 1985 Jan;53(1):89–93. doi: 10.1128/jvi.53.1.89-93.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., Wright S. H., Wolff R. K., Grzesiuk E., Maryon E. B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986 Jun;6(6):2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielkens A. L., Van Oirschot J. T., Berns A. J. Genome differences among field isolates and vaccine strains of pseudorabies virus. J Gen Virol. 1985 Jan;66(Pt 1):69–82. doi: 10.1099/0022-1317-66-1-69. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Quint W., Gielkens A., Van Oirschot J., Berns A., Cuypers H. T. Construction and characterization of deletion mutants of pseudorabies virus: a new generation of 'live' vaccines. J Gen Virol. 1987 Feb;68(Pt 2):523–534. doi: 10.1099/0022-1317-68-2-523. [DOI] [PubMed] [Google Scholar]
- Quint W., Quax W., van der Putten H., Berns A. Characterization of AKR murine leukemia virus sequences in AKR mouse substrains and structure of integrated recombinant genomes in tumor tissues. J Virol. 1981 Jul;39(1):1–10. doi: 10.1128/jvi.39.1.1-10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A. K., Whealy M. E., Watson R. J., Enquist L. W. Pseudorabies virus gene encoding glycoprotein gIII is not essential for growth in tissue culture. J Virol. 1986 Sep;59(3):635–645. doi: 10.1128/jvi.59.3.635-645.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Jacob R. J., Knipe D. M., Morse L. S., Ruyechan W. T. On the structure, functional equivalence, and replication of the four arrangements of herpes simplex virus DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):809–826. doi: 10.1101/sqb.1979.043.01.088. [DOI] [PubMed] [Google Scholar]
- Rubenstein A. S., Kaplan A. S. Electron microscopic studies of the DNA of defective and standard pseudorabies virions. Virology. 1975 Aug;66(2):385–392. doi: 10.1016/0042-6822(75)90211-1. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomsen D. R., Marchioli C. C., Yancey R. J., Jr, Post L. E. Replication and virulence of pseudorabies virus mutants lacking glycoprotein gX. J Virol. 1987 Jan;61(1):229–232. doi: 10.1128/jvi.61.1.229-232.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]