Abstract
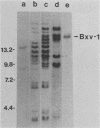
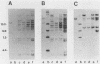
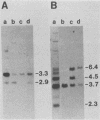
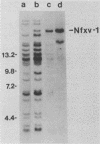
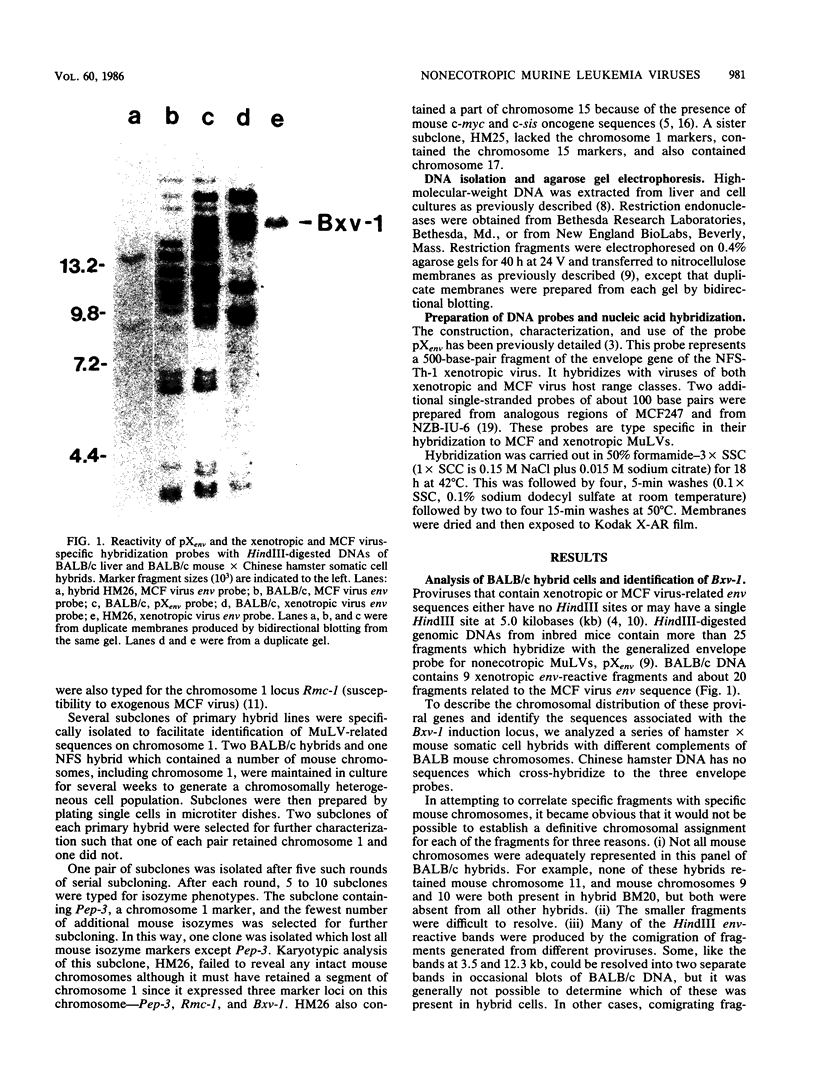
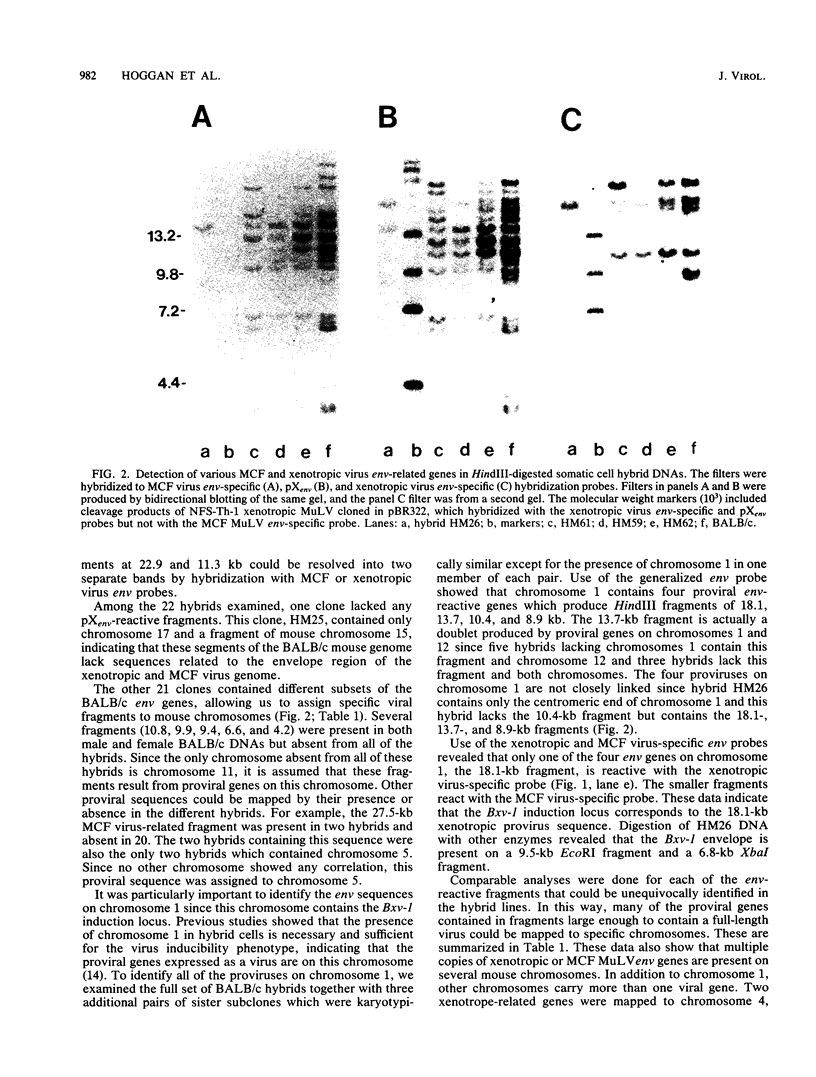
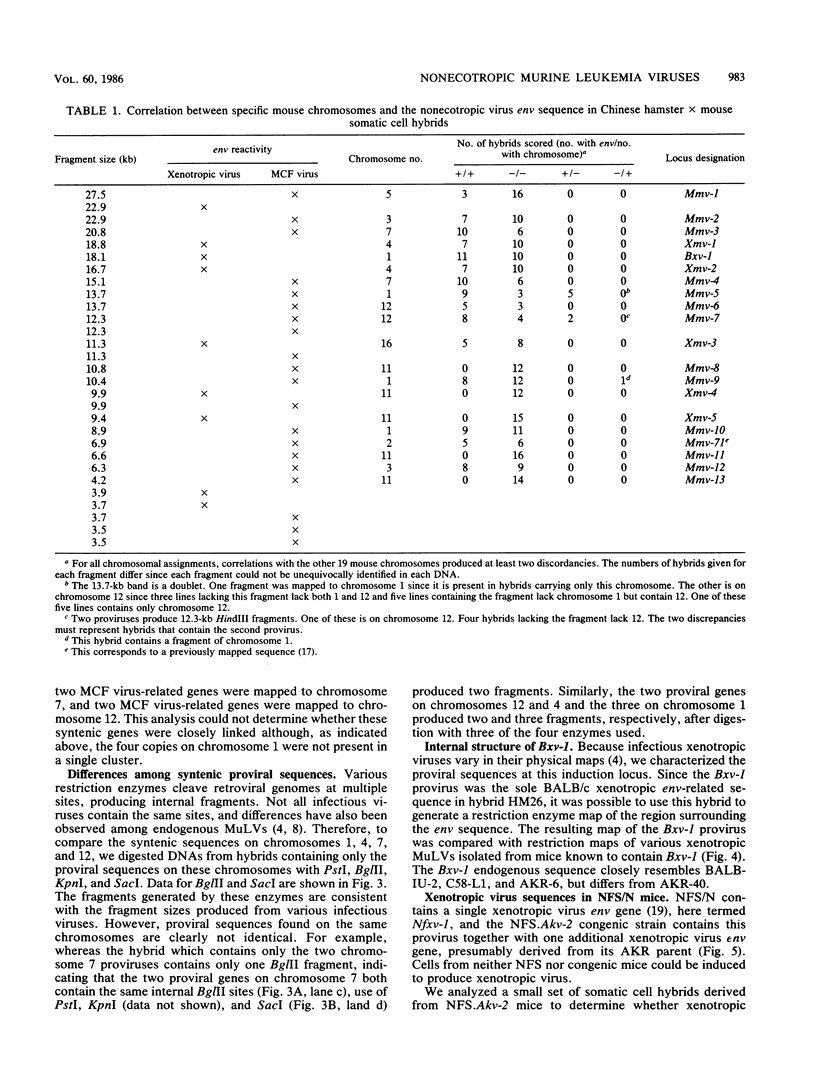
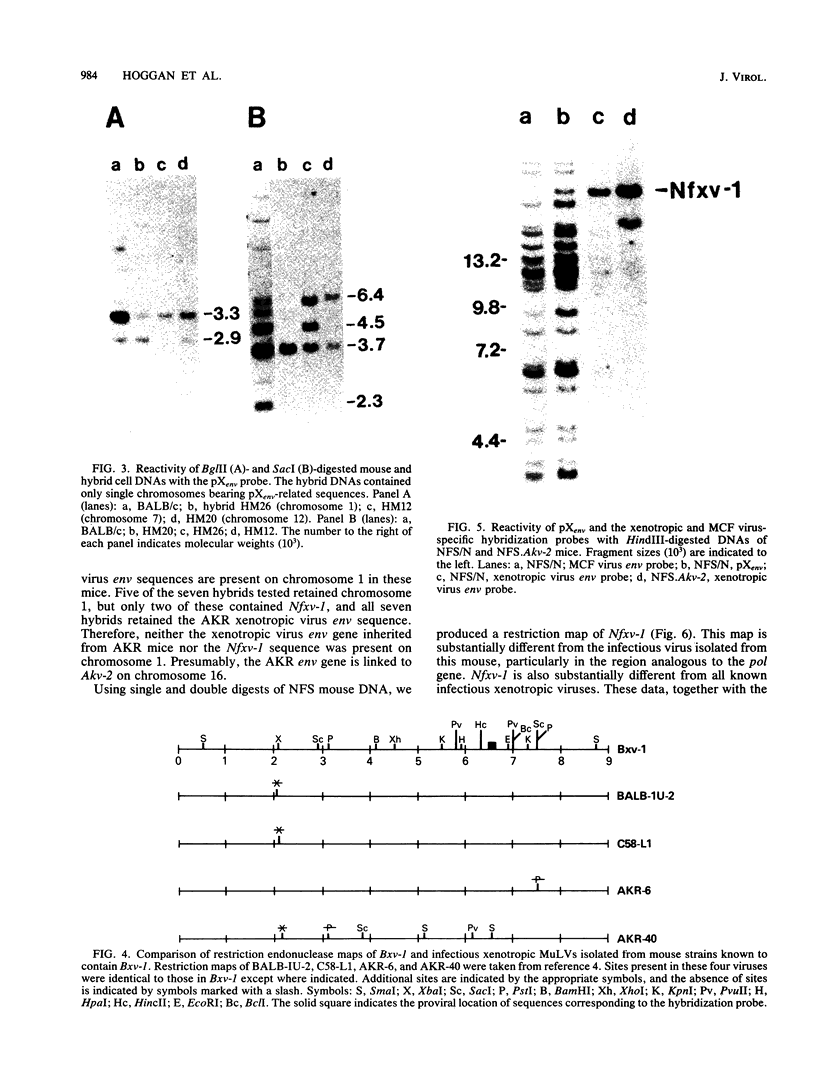
We used hybridization probes that react specifically with xenotropic and mink cell focus-forming virus envelope sequences to characterize the nonecotropic proviruses of BALB/c and NFS/N mice. Analysis of somatic cell hybrids with different BALB/c chromosomes showed that the 9 xenotropic and more than 20 MCF virus-related proviral sequences in this mouse were present on more than nine BALB/c chromosomes. Multiple copies were found on chromosomes 1, 4, 7, 12, and probably 11, and the copies found on a single chromosome were not identical by restriction enzyme mapping. We also identified and characterized the proviral sequences that give rise to infectious xenotropic virus in both BALB/c and NFS/N mice. BALB/c contains the major locus for induction of infectious virus in inbred mice, Bxv-1, which is on chromosome 1. We showed that this locus contains a single xenotropic provirus on an 18-kilobase HindIII fragment. Restriction enzyme analysis of a hybrid cell DNA that contains only the Bxv-1 xenotropic provirus showed that the Bxv-1 provirus contains restriction enzyme sites characteristic of the infectious virus induced from BALB/c fibroblasts. The Bxv-1 provirus and its flanking sequences also contain the same restriction sites as the provirus thought to contribute U3 long terminal repeat sequences to leukemogenic (class I) AKR MCF viruses. Analysis of cell hybrids made with the nonvirus-inducible strain NFS/N showed that the single xenotropic virus env gene of NFS mice, here termed Nfxv-1, is not on chromosome 1. Unlike that of Bxv-1, the restriction map of Nfxv-1 does not resemble that of any known infectious xenotropic virus including xenotropic viruses isolated from NFS mice. These data suggest that Bxv-1, but not Nfxv-1, is a full-length xenotropic provirus that can be transcribed directly to produce infectious virus.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Stephenson J. R. Independent segregation of loci for activation of biologically distinguishable RNA C-type viruses in mouse cells. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2055–2058. doi: 10.1073/pnas.70.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt C., Mileham K., Haas M., Nesbitt M. N., Harper M. E., Simon M. I. Chromosomal mapping of the mink cell focus-inducing and xenotropic env gene family in the mouse. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6298–6302. doi: 10.1073/pnas.80.20.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler C. E., Hoggan M. D., Chan H. W., Sears J. F., Khan A. S., Moore J. L., Hartley J. W., Rowe W. P., Martin M. A. Cloning and characterization of an envelope-specific probe from xenotropic murine leukemia proviral DNA. J Virol. 1982 Jan;41(1):228–236. doi: 10.1128/jvi.41.1.228-236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Lander M. R., Gupta S., Rands E., Lowy D. R. Origin of mink cytopathic focus-forming (MCF) viruses:comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981 Sep;113(2):465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- Crews S., Barth R., Hood L., Prehn J., Calame K. Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science. 1982 Dec 24;218(4579):1319–1321. doi: 10.1126/science.7146913. [DOI] [PubMed] [Google Scholar]
- Datta S. K., Schwartz R. S. Mendelian segregation of loci controlling xenotropic virus production in NZB crosses. Virology. 1977 Dec;83(2):449–452. doi: 10.1016/0042-6822(77)90193-3. [DOI] [PubMed] [Google Scholar]
- Greenberger J. S., Phillips S. M., Stephenson J. R., Aaronson S. A. Induction of mouse type-C RNA virus by lipopolysaccharide. J Immunol. 1975 Jul;115(1):317–320. [PubMed] [Google Scholar]
- Hoggan M. D., Buckler C. E., Sears J. F., Chan H. W., Rowe W. P., Martin M. A. Internal organization of endogenous proviral DNAs of xenotropic murine leukemia viruses. J Virol. 1982 Jul;43(1):8–17. doi: 10.1128/jvi.43.1.8-17.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Rowe W. P., Martin M. A. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J mice: identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J Virol. 1982 Nov;44(2):625–636. doi: 10.1128/jvi.44.2.625-636.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A. Genetic mapping of a mouse chromosomal locus required for mink cell focus-forming virus replication. J Virol. 1983 Oct;48(1):300–303. doi: 10.1128/jvi.48.1.300-303.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., Hartley J. W., Morse H. C., 3rd Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J Virol. 1984 Jul;51(1):77–80. doi: 10.1128/jvi.51.1.77-80.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., Rowe W. P. Genetic mapping of the ecotropic murine leukemia virus-inducing locus of BALB/c mouse to chromosome 5. Science. 1979 Apr 6;204(4388):69–71. doi: 10.1126/science.219475. [DOI] [PubMed] [Google Scholar]
- Kozak C. A., Rowe W. P. Genetic mapping of the ecotropic virus-inducing locus Akv-2 of the AKR mouse. J Exp Med. 1980 Nov 1;152(5):1419–1423. doi: 10.1084/jem.152.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., Rowe W. P. Genetic mapping of xenotropic murine leukemia virus-inducing loci in five mouse strains. J Exp Med. 1980 Jul 1;152(1):219–228. doi: 10.1084/jem.152.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., Sears J. F., Hoggan M. D. Genetic mapping of the mouse proto-oncogene c-sis to chromosome 15. Science. 1983 Aug 26;221(4613):867–869. doi: 10.1126/science.6308764. [DOI] [PubMed] [Google Scholar]
- Meruelo D., Rossomando A., Offer M., Buxbaum J., Pellicer A. Association of endogenous viral loci with genes encoding murine histocompatibility and lymphocyte differentiation antigens. Proc Natl Acad Sci U S A. 1983 Aug;80(16):5032–5036. doi: 10.1073/pnas.80.16.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse H. C., 3rd, Kozak C. A., Yetter R. A., Hartley J. W. Unique features of retrovirus expression in F/St mice. J Virol. 1982 Jul;43(1):1–7. doi: 10.1128/jvi.43.1.1-7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill R. R., Khan A. S., Hoggan M. D., Hartley J. W., Martin M. A., Repaske R. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J Virol. 1986 May;58(2):359–366. doi: 10.1128/jvi.58.2.359-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint W., Boelens W., van Wezenbeek P., Cuypers T., Maandag E. R., Selten G., Berns A. Generation of AKR mink cell focus-forming viruses: a conserved single-copy xenotrope-like provirus provides recombinant long terminal repeat sequences. J Virol. 1984 May;50(2):432–438. doi: 10.1128/jvi.50.2.432-438.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wejman J. C., Taylor B. A., Jenkins N. A., Copeland N. G. Endogenous xenotropic murine leukemia virus-related sequences map to chromosomal regions encoding mouse lymphocyte antigens. J Virol. 1984 Apr;50(1):237–247. doi: 10.1128/jvi.50.1.237-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]