Abstract
Polyunsaturated fatty acids (PUFAs) are normal constituents of the diet, but have properties different from other fatty acids (e.g., through generation of signaling molecules). N-3 PUFAs reduce cancer cell growth, but no unified mechanism has been identified. We show that docosahexaenoic acid (DHA; 22:6 n-3) causes extensive changes in gene expression patterns at mRNA level in the colon cancer cell line SW620. Early changes include unfolded protein response (UPR) and increased levels of phosphorylated eIF2α as verified at protein level. The latter is considered a hallmark of endoplasmic reticulum (ER) stress and is abundantly present already after 3 h. It may coordinate many of the downstream changes observed, including signaling pathways for cell cycle arrest/apoptosis, calcium homeostasis, cholesterol metabolism, ubiquitination, and proteasomal degradation. Also, eicosapentaenoic acid (EPA), but not oleic acid (OA), induced key mediators of ER stress and UPR at protein level. Accumulation of esterified cholesterol was not compensated for by increased total levels of cholesterol, and mRNAs for cholesterol biosynthesis as well as de novo synthesis of cholesterol were reduced. These results suggest that cytotoxic effects of DHA are associated with signaling pathways involving lipid metabolism and ER stress.
Keywords: gene expression, phosphorylated eIF2α, antioxidant response, heat shock response, cytosolic free Ca2+, cell cycle, total cholesterol level, cholesterol synthesis
Long-chain polyunsaturated fatty acids (PUFAs) of the n-3 type are important dietary components that may prevent or alleviate coronary heart disease and inflammatory conditions (1, 2). Even though epidemiological studies on the association between fish consumption and cancer risk are not consistent, evidence from animal- and cell-culture studies demonstrate that PUFAs inhibit cancer-cell growth, induce apoptosis, and increase the efficiency of chemotherapeutic drugs (3–5). Several mechanisms have been proposed for the antiproliferative effect of n-3 PUFAs; among these are alterations in eicosanoid formation (6), lipid peroxidation initiated by free radicals (7–9), accumulation of cytotoxic lipid droplets (10), and specific changes in gene expression patterns (11, 12). This could be mediated directly by PUFAs as ligands of transcription factors, or indirectly through metabolites of PUFAs or other secondary events. There is evidence indicating that fatty acids may regulate gene expression directly. The activity and abundance of several nuclear transcription factors, like peroxisome proliferator-activated receptors (PPARα/δ/γ), liver X receptors (LXRα/β), and sterol regulatory element-binding proteins (SREBP1/2), have been shown to be regulated by dietary PUFAs and their metabolites (11, 13).
Cellular stress from cytotoxic agents may result in adaptive mechanisms in several cellular compartments, including endoplasmic reticulum (ER). ER has three main functions: 1) folding, glycosylation, and sorting of proteins to their proper destination; 2) synthesizing cholesterol and other lipids; and 3) maintenance of Ca2+ homeostasis. Disruption of any of these processes causes ER stress and activates the unfolded protein response (UPR). The UPR up-regulates genes that support adaptation to and recovery from ER stress as well as initiating apoptotic pathways when damage is severe. Three transmembrane proteins mediate the UPR signal across the ER membrane: inositol-requiring enzyme 1 (IRE1), eukaryotic translation initiation factor 2α (eIF2α) kinase 3 (EIF2AK3/PKR-like ER kinase [PERK]), and activating transcription factor 6 (ATF6). PERK belongs to a family of eIF2α kinases that regulates the translational control during the UPR. Phosphorylation of eIF2α by PERK leads to attenuation of global protein synthesis, but promotes translation of certain mRNAs, like activating transcription factor 4 (ATF4) mRNA (14). Downstream targets of ATF4 are CHOP, GADD34, ATF3, and genes involved in amino acid metabolism, glutathione biosynthesis, resistance to oxidative stress, and protein secretion. Loss of cyclin D1 during ER stress leads to G1 arrest and provides the cell with an opportunity to restore cell homeostasis (15). However, prolonged ER stress may cause cell death. ER stress-induced apoptosis may be mediated by caspase-12, caspase-9, and caspase-7 (16).
Several links exist between signaling pathways controlling the UPR and lipogenesis. Activation of the transcription factors ATF6 as well as the SREBPs that control cholesterol and lipid synthesis requires translocation from the ER to the Golgi followed by cleavage by site-1 protease (S1P) and site-2 protease (S2P) (17). Also, ER stress may activate expression of genes involved in cholesterol biosynthesis (18, 19). Both elevated levels of cholesterol as well as depletion have been shown to induce ER stress (20, 21).
We have previously shown that the human colon cancer cell lines SW480 and SW620, derived from a primary and a secondary tumor of the same patient, respectively, were strongly growth inhibited by docosahexaenoic acid (DHA) (5). DHA enhanced lipid peroxidation in both cell lines significantly, measured as accumulation of the end product malondialdehyde. The antioxidant vitamin E (α-tocopherol) completely abolished the increase in malondialdehyde, without restoring cell growth, demonstrating that the cells were resistant to lipid peroxidation products. DHA accumulated mainly as triglyceride and cholesteryl ester-enriched lipid droplets in SW480 and SW620, respectively. The protein level of the nuclear form of SREBP1 (nSREBP1) decreased in both cell lines, indicating a possible relationship between disturbances in lipid homeostasis and cell-cycle arrest. We demonstrate that DHA-treatment of SW620 cells results in extensive changes in gene expression patterns at the mRNA level. Early changes include induction of ER stress, as evident from the abundant presence of phosphorylated eIF2α (eIF2α-P); increase in cytosolic Ca2+; and disturbances in lipid metabolism. Downstream signaling subsequently results in growth arrest and protein degradation. Key mediators of ER stress, eIF2α-P, as well as ATF4 were also induced by eicosapentaenoic acid (EPA), but not by oleic acid (OA) treatment.
MATERIALS AND METHODS
Cell culture and fatty acid treatment
Human colon adenocarcinoma cell line, SW620, was obtained from American Type Culture Collection (Rockville, MD). Cells were cultured in Leibovitz's L-15 medium (Cambrex, BioWhittaker, Walkersville, MD) supplemented with L-glutamine (2 mM), FBS (10%), and gentamicin (45 mg/l) (complete growth medium) and maintained in a humidified atmosphere of 5% CO2: 95% air at 37°C. Stock solutions of DHA, EPA, and OA in ethanol (Cayman Chemical, Ann Arbor, MI) were stored at −20°C and diluted in complete growth medium before experiments (final concentration of ethanol < 0.025% v/v).
RNA isolation
Seeded in 75 cm2 flasks were 1.5 × 106 cells. After 8 h, complete growth medium supplemented with DHA (70 μM) or an equal volume of ethanol (control) was added, and cells were incubated for 12, 24, and 48 h. Cells were harvested by scraping in ice-cold phosphated buffered saline (PBS) and stored at −80°C. Total RNA was isolated using the High Pure RNA Isolation Kit (Roche, Mannheim, Germany) according to instruction manual. RNase inhibitor rRNasin (40U/μl, 1 μl) (Promega, Madison, WI) was added, and RNA was up-concentrated on a speed vac and resuspended in RNase free distilled H2O. RNA concentration and quality were determined using the NanoDrop1000 (NanoDrop Technologies, Wilmington, DE) and agarose gel electrophoresis.
Gene expression profiling
Five micrograms total RNA was used for cDNA and cRNA synthesis according to the eukaryote expression manual (Affymetrix, Santa Clara, CA). Detailed gene expression profiling procedure can be found in supplementary data. cRNA was hybridized to the Human Genome Focus Array (Affymetrix). Washing and staining were performed using the Fluidics Station 400 (Midi-Euk2v3 protocol). The arrays were scanned using an Affymetrix GeneChip GA2500 Scanner, controlled by GeneChip® Operating Software 1.2 (GCOS, Affymetrix). Expression profiling was performed in triplicates at all time points using RNA from independent biological replicates. All experiments have been submitted to ArrayExpress with accession number E-MEXP-1014.
Statistical analysis of gene expression data
Statistical analysis was performed based on summary expression measures for each probe set of the GeneChips, using the raw data (CEL) files and a linear statistical model for background-corrected, quantile normalized, and log-transformed perfect match values, performed by the robust multiarray average (RMA) method (22, 23).
For each transcript, a linear regression model including parameters representing treatment effects and time effects for the treatment group was fitted to the RMA expression measures. Based on the estimated effects, tests for significant differential expression due to DHA treatment were performed using moderated t-tests, in which gene-specific variance estimates are replaced by variance estimates found by borrowing strength from data on the remaining genes (24).
To account for multiple testing, adjusted P values controlling the false discovery rate were calculated (25) by inserting the estimated value of the proportion of nondifferentially expressed genes (26). Differentially expressed genes were selected based on a threshold of 0.05 on the adjusted P values.
Time effect in the control groups were considered negligible and omitted from the model. Statistical analysis was performed in R (27), using the packages Limma and affy from Bioconductor (28). Differentially expressed genes were annotated using the NetAffx Analysis Centre (http://www.Affymetrix.com) and NMC Annotation Tool/eGOn V2.0 (http://www.GeneTools.no).
Immunoblot analysis
DHA treatment of cells and preparation of total protein extracts were performed as described previously (5). Nuclear extracts for detection of ATF4 were prepared using a Nuclear extract kit (Active Motif, Belgium) according to manufacturer's instructions. To detect phosphorylated eIF2α, cells were washed and scraped in ice-cold PBS-1 mM EDTA and pelleted by centrifugation. Pellets were lysed in 2 × packed cell volumes of lysis buffer on ice for 10 min. Equal amounts of protein were separated on 10% precast denaturing NuPAGE gels (Invitrogen, Carlsbad, CA) and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). The blots were incubated with the indicated primary and horse radish peroxidase-conjugated secondary antibodies (DAKO, Carpinteria, CA) and detected by chemiluminescense using Super Signal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL) and visualized by Kodak Image Station 4000R (Eastman Kodak Co., Rochester, NY). Quantification was performed using Kodak Molecular Imaging Software (version 4.0.1). Details about buffers and primary antibodies can be found in the supplementary data.
Measurement of cytosolic calcium in single cells
Cytosolic free Ca2+ in single cells was determined as previously described (29). In short, cells were incubated for 40 min at 37°C with a solution of 5 μM fura-2, 0.25% DMSO, and 0.025% Pluronic F-127 (TefLab, Austin, TX) in HEPES-buffered salt solution (HSS). Cells were then washed once and incubated in 400 μl HSS. Applications to cells were done by injecting 100 μl of agonist into the well. Ca2+ imaging and registration software has been developed by Rotnes and Iversen (30). Cytosolic Ca2+ concentration was calculated using the equation: [Ca2+] = Kdβ(R − Rmin)/(Rmax − R) (31). Fluorescence data were analyzed using the program LICS (32).
Analysis of total cholesterol levels
Seeded in 175 cm2 flasks were 4 × 106 cells. The following day, medium was replaced with complete growth medium supplemented with DHA (70 μM) or medium with equal volume of ethanol and harvested after 3, 6, 12, 24, and 48 h incubation. Cells were harvested by trypsination and resuspended in PBS together with floating cells collected by centrifugation. The cell suspension was counted using a Coulter Counter (Beckman Coulter, Fullerton, CA) and an aliquot of 4 × 106 cells was collected by centrifugation. Lipids were extracted from the cell pellet (33) using chloroform-methanol-water (1:2:0.8 v/v/v). Total cholesterol levels (cholesteryl ester and free cholesterol) in the lipid extracts were determined using the Amplex Red Cholesterol Assay Kit (Invitrogen) according to the instruction manual. Data were expressed as μg cholesterol/mg protein.
Analysis of cholesterol and cholesteryl ester synthesis
Seeded in 175 cm2 flasks were 4 × 106 cells. The following day, medium was replaced with complete growth medium supplemented with DHA (70 μM) or with equal volume of ethanol (control) and incubated for 24 h. Cells were then incubated with complete growth medium containing 14C-acetate (1.2 μCi/ml) and DHA (70 μM) or ethanol (control cells) for 4 and 6 h. Cells (12 × 106) were harvested by trypsination, and cellular lipids were extracted with chloroform/methanol according to a method modified after Bligh and Dyer (33). Lipids were separated by thin layer chromatography using hexane/diethyl ether/acetic acid (70:30:1, v/v/v) and lipids were visualized using iodine vapor. Lipid fractions were solubilized in Insta-gel plus (Perkin Elmer) before counting. Data were expressed as incorporation of 14C-acetate into cholesterol/cholesteryl ester (cpm/mg protein).
RESULTS
Growth inhibition by DHA through ER stress and growth arrest signaling
The human colon cancer cell line SW620 is strongly growth-inhibited by DHA. Between 72 h and 144 h, essentially no growth could be observed after treatment with 70 μM DHA (5). Although growth retardation was modest after 24 h, [3H]thymidine incorporation was reduced 30–40% in SW620 cells already by 12 h, while no effect was observed after 6 h (data not shown).
We demonstrate that complex gene networks and cell signaling pathways are affected at the mRNA level after DHA treatment in SW620 cells (Fig. 1 and Table 1). The number of transcripts differentially expressed increased from 12 to 24 h of DHA treatment [up-regulated: 839 (12 h) vs. 1157 (24 h); down-regulated: 1066 (12 h) vs. 1222 (24 h)], while the number decreased at 48 h (up-regulated: 288; down-regulated: 267). The fold change of transcript levels after DHA treatment ranged from 1.2 to 24, the majority of which being toward the lower end. Transcripts could be classified into several functional categories after annotation as shown in Table 1 and outlined in later discussion. More comprehensive information on changes in gene expression is found in the supplementary Table I. Early changes include induction of ER stress and UPR.
Fig. 1.
Docosahexaenoic acid (DHA) induces endoplasmic reticulum (ER) stress in SW620 cells. Diagram showing transcripts found to be affected by DHA treatment in SW620 cells by gene expression analysis (up-regulated, pink; down-regulated, blue) in the main pathways of ER stress signaling. Three transmembrane proteins mediate the unfolded protein response (UPR) across the ER membrane after dissociation from BiP, activating transcription factor 6 (ATF6), PERK, and inositol-requiring enzyme 1 (IRE1). Each of these proteins represents distinct pathways of the ER stress response.
TABLE 1.
Functional categories of differentially expressed transcripts affected in SW620 cells treated with docosahexaenoic acid (DHA) (70 μM) at time points indicated
| SW620 Fold Change |
||||||
|---|---|---|---|---|---|---|
| Gene Symbol | Affymetrix ID | Refseq NCBI ID | Description | 12 h | 24 h | 48 h |
| ER Stress Response | ||||||
| ATF3 | 202672_s_at | NM_001030287 | Activating transcription factor 3 | 3.7 | 4.1 | 3.1 |
| NM_001040619 | ||||||
| NM_001674 | ||||||
| NM_004024 | ||||||
| ATF4 | 200779_at | NM_001675 | Activating transcription factor 4 | 2.1 | 2.0 | 1.6 |
| NM_182810 | ||||||
| ATF6 | 203952_at | NM_007348 | Activating transcription factor 6 | 1.3 | 1.2 | — |
| EIF2S1 | 201142_at | NM_004094 | Eukaryotic translation initiation factor 2-α | NC | NC | NC |
| GADD34 | 37028_at | NM_014330 | Growth arrest and DNA-damage-inducible 34 | 6.3 | 3.9 | — |
| NRF2 | 201146_at | NM_006164 | Nuclear factor E2-related factor | 2.0 | 1.8 | — |
| PERK | 218696_at | NM_004836 | PKR-like ER kinase | 1.4 | 2.0 | — |
| VCP | 208649_s_at | NM_007126 | Valocin containing protein | 1.9 | 1.6 | — |
| XBP1 | 200670_at | NM_001079539 | X-box binding protein 1 | 2.0 | 1.8 | — |
| NM_005080 | ||||||
| Chaperones/Protein Folding/UPR Response | ||||||
| DNAJB1 | 200666_s_at | NM_006145 | DnaJ homolog, subfamily B, member 1 | 8.0 | 4.1 | — |
| HMOX1 | 203665_at | NM_002133 | Heme oxygenase (decycling) 1 | 24.0 | 10.6 | 5.7 |
| HSPA1A/B | 200800_s_at | NM_005345 | Heat shock 70 kDa protein 1A/B | 17.8 | 9.8 | 5.1 |
| NM_005346 | ||||||
| HSPA1B | 202581_at | NM_005346 | Heat shock 70 kDa protein 1B | 9.8 | 6.5 | 3.1 |
| HSP47 | 207714_s_at | NM_001235 | Heat shock protein 47 | 4.4 | 1.8 | — |
| Ubiquitine/Proteasome | ||||||
| PSMD1/RPN2 | 211198_s_at | NM_002807 | Proteasome 26S subunit, non-ATPase, 1 | 2.2 | 2.2 | — |
| SQSTM1 | 213112_s_at | NM_003900 | Sequestosome 1 | 7.7 | 6.7 | 5.0 |
| SQSTM1 | 201471_s_at | NM_003900 | Sequestosome 1 | 6.7 | 7.3 | 3.9 |
| Ca2+ Homeostasis | ||||||
| CAMLG | 203538_at | NM_001745 | Calcium modulating ligand | 1.9 | 1.9 | — |
| CAPN2 | 208683_at | NM_001748 | Calpain 2, large subunit | 1.3 | 1.8 | 1.4 |
| CAPN7 | 203356_at | NM_014296 | Calpain 7 | — | 1.5 | — |
| IP3R1 | 203710_at | NM_002222 | Inositol 1,4,5-triphosphate receptor, type 1 | 1.5 | 2.2 | 1.4 |
| IP3R3 | 201189_s_at | NM_002224 | Inositol 1,4,5-triphosphate receptor, type 3 | — | — | 1.3 |
| Antioxidants/Oxidative Stress | ||||||
| CAT | 201432_at | NM_001752 | Catalase | — | −1.4 | — |
| GCLC | 202922_at | NM_001498 | Glutamate-cysteine ligase, catalytic subunit | 1.6 | 1.3 | — |
| GCLM | 203925_at | NM_002061 | Glutamate-cysteine ligase, modifier subunit | 3.7 | 3.5 | 2.0 |
| HMOX1 | 203665_at | NM_002133 | Heme oxygenase (decycling) 1 | 24.0 | 10.6 | 5.7 |
| SOD1 | 200642_at | NM_000454 | Superoxide dismutase 1 | 1.5 | 1.6 | — |
| TXNRD1 | 201266_at | NM_003330 | Thioredoxin reductase 1 | 3.2 | 2.9 | 1.9 |
| NM_182729 | ||||||
| NM_182742 | ||||||
| NM_182743 | ||||||
| Cell Cycle/Apoptosis | ||||||
| BAG3 | 217911_s_at | NM_004281 | BCL2-associated athanogene 3 | 9.9 | 5.4 | — |
| CASP4 | 209310_s_at | NM_001225 | Caspase 4 | 1.6 | 2.9 | — |
| NM_033306 | ||||||
| NM_033307 | ||||||
| CASP7 | 207181_s_at | NM_001227 | Caspase 7 | 1.6 | 2.1 | — |
| NM_033338 | ||||||
| NM_033339 | ||||||
| NM_033340 | ||||||
| CCND1 | 208712_at | NM_053056 | Cyclin D1 | −1.7 | −2.0 | — |
| TRIB3 | 218145_at | NM_021158 | Tribbles homolog 3 (Drosophila) | 7.4 | 6.5 | 3.3 |
| Cholesterol Biosynthesis, Uptake, Metabolism, and Transport | ||||||
| CAV1 | 203065_s_at | NM_001753 | Caveolin 1, caveolae protein, 22 kDa | −1.5 | −1.4 | — |
| DHCR24 | 200862_at | NM_014762 | 24-dehydrocholesterol reductase | −1.6 | −1.7 | — |
| DHCR7 | 201791_s_at | NM_001360 | 7-dehydrocholesterol reductase | −1.6 | −1.5 | — |
| FDPS | 201275_at | NM_002004 | Farnesyl diphosphate synthase | −1.3 | −1.2 | — |
| HMGCR | 202539_s_at | NM_000859 | 3-hydroxy-3-methylglutaryl-CoA reductase | NC | NC | NC |
| LDLR | 202068_s_at | NM_000527 | Low density lipoprotein receptor | 2.4 | 2.4 | — |
| LSS | 202245_at | NM_002340 | Lanosterol synthase | −1.3 | — | |
| NPC1 | 202679_at | NM_000271 | Niemann-Pick disease, type C1 | 3.0 | 4.5 | 1.9 |
| NPC2 | 200701_at | NM_006432 | Niemann-Pick disease, type C2 | 1.5 | 1.5 | |
| OSBP | 201800_s_at | NM_002556 | Oxysterol binding protein | 1.4 | 1.4 | — |
| PMVK | 203515_s_at | NM_006556 | Phosphomevalonate kinase | -1.3 | −1.8 | — |
| SREBP2 | 201247_at | NM_004599 | Sterol regulatory element binding protein 2 | NC | NC | NC |
| TM7SF2 | 210130_s_at | NM_003273 | Transmembrane 7 superfamily member 2 | −1.4 | −1.9 | — |
| VLDLR | 209822_s_at | NM_001018056 | Very low density lipoprotein receptor | 1.6 | 1.6 | — |
| NM_003383 | ||||||
NC, no change; UPR, unfolded protein response.
The protein levels of selected target genes were measured in SW620 cells after DHA treatment. Importantly, we demonstrate that an abundant amount of eIF2α-P is found as early as 3 h after DHA treatment, preceding the activation of ATF4 and HMOX1 (Fig. 2A). Phosphorylation of eIF2α is considered a hallmark of the UPR and ER stress and leads to attenuation of global protein synthesis, but promotes translation of certain mRNAs, like ATF4 mRNA (15). In accordance with this, ATF4 is up-regulated here both at the mRNA and protein level (Table 1 and Fig. 2A). Downstream targets of ATF4, ATF3, and genes involved in amino acid metabolism (ASNS), glutathione biosynthesis, resistance to oxidative stress (HMOX1), and protein secretion are up-regulated at the mRNA level and (when examined) at the protein level (Fig. 2A, Table 1 and supplementary Table I). Also, ATF6, PERK, and X-box binding protein 1 (XBP1), a downstream target of ATF6, are up-regulated at mRNA level in SW620 cells (Table 1). XBP1 regulates a subset of ER-resident chaperones that are essential for protein folding, maturation, and degradation in the ER (34).
Fig. 2.
Analysis of proteins involved in ER stress signaling and UPR. A: Western blot analysis of proteins involved in ER stress signaling and UPR from total cell extracts (except for ATF4: nuclear extracts; eIF2α: cytoplasmic extracts) of SW620 cells treated with DHA for indicated time periods (h). Controls were harvested at all time points; only the 24 h control (C means control) is shown. B: Western blot analysis of cyclin D1 from total extracts of SW620 cells treated or not treated (controls) with DHA for the indicated time periods (h). C: Quantification of cyclin D1 Western blots in B [DHA treated cells (gray bars) compared with controls (black bars)]. Results were verified in three independent experiments; one representative blot is shown. β-actin (total extracts), lamin C (nuclear extacts), or total eIF2α was used as a control for equal protein loading. The blots were quantified and protein band intensities normalized relative to loading control. The adjusted band intensities from the DHA and control membranes were then normalized relative to the 24 h control band, present at all membranes, to adjust for differences in signal intensities between the membranes. The numbers under the blots represent mean fold change (SD) of DHA samples relative to control at the indicted time points for three independent experiments. * Significantly different from control (Student's t-test, P < 0.05).
Cyclin D1 is significantly down-regulated both at mRNA and protein level (Table 1, Fig. 2B, C), probably mediated by phosphorylated eIF2α that has been shown to attenuate cyclin D1 translation and cause cell-cycle arrest (G1 phase) in response to prolonged ER stress (15). mRNA for GADD34, a subunit in phosphorylated eIF2α phosphatase, is up-regulated in SW620 cells, possibly explaining in part the decrease in phosphorylated eIF2α at 12 h and later (Fig. 2A).
Induction of the UPR is initiated through dissociation of PERK from the ER-resident chaperone BiP/GRP78 that engages in numerous complexes (35). The protein level of BiP remained constant at all time points (data not shown). Treatment of SW620 cells with the ER stress inducers tunicamycin (1 μg/ml) and thapsigargin (0.2 μM) for 6 h, caused a marked down-regulation of cyclin D1 compared with control (results not shown). These results confirm that ER stress in SW620 cells leads to down-regulation of cyclin D1.
Induction of ER stress and UPR is followed by disruption of protein folding and destruction of defective proteins by ER-associated degradation. Several members of the molecular chaperone Hsp40, Hsp70, and Hsp90 families were up-regulated at the mRNA level (Table 1 and supplementary Table I). Hsp70 was also found to be up-regulated at the protein level (Fig. 2A).
Several transcripts belonging to the ubiquitin/proteasome system were up-regulated (Table 1 and supplementary Table I). The proteasome family of proteins is responsible for the degradation of damaged and short-lived proteins. In SW620, mRNA for 27 out of 34 subunits (present on the Human Genome Focus array) of the proteasome 26S were up-regulated. The proteasomal subunit proteasome 26S subunit, non-ATPase, 1 (PSMD1)/Rpn2 was significantly increased relative to control at protein level (Fig. 2A). Also, the mRNA level of sequestosome 1 (SQSTM1), which serves as a storage place for ubiquitinated proteins in the cytoplasm, was up-regulated at all time points in SW620 cells (Table 1). These results support the view that the ER stress response initiated by DHA causes extensive changes in protein homeostasis in these cells.
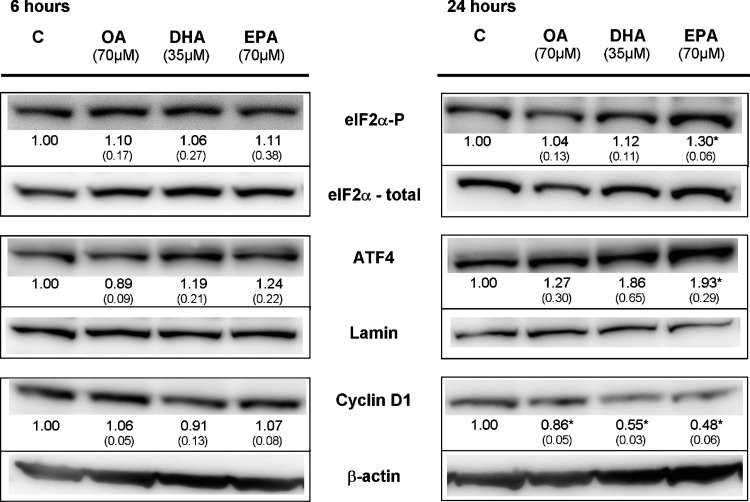
We have previously shown that EPA has an antiproliferative effect on SW620 cells, although to a lesser extent than DHA, while OA has no effect (5). Key mediators of ER stress and UPR were also induced at protein level by EPA, but not OA, at equal molar concentrations (Fig. 3). DHA-treatment (70 μM) of SW620 cells induced phosphorylation of eIF2α already after 3 h (Fig. 2A), while a weaker response is observed after treatment with EPA (70 μM) or half molar concentrations of DHA (35 μM) first at 24 h (Fig. 3). A similar time- and concentration-dependent response is also observed for the induction of ATF4 (Fig. 3). Also, EPA (70 μM) and DHA (35 and 70 μM) reduced the level of cyclin D1 after 24 h (Fig. 2B, C and Fig. 3). After 6 h, only DHA (70 μM) reduced the level of cyclin D1 significantly, whereas DHA (70 μM) and EPA (70 μM) both reduced cyclin D1 substantially and with comparable effects after 24 h. OA did not affect the proliferation of SW620 cells. However, OA reduced the level of cyclin D1 after 24 h, although to a much lesser extent (Fig. 3). This reflects the differences in the antiproliferative effect between the PUFAs observed earlier.
Fig. 3.
ER stress signaling and UPR in response to n-3 polyunsaturated fatty acids (PUFAs) and oleic acid (OA). Western blot analysis of proteins involved in ER stress signaling and UPR (ATF4: nuclear extracts; cyclin D1, eIF2α: cytoplasmic extracts) in SW620 cells treated with complete growth medium supplemented with either OA (70 μM), DHA (35 μM), eicosapentaenoic acid (EPA, 70μM), or ethanol (control media, C) for 6 and 24 h. β-actin (cytoplasmic extracts), lamin C (nuclear extracts), or total eIF2α was used as a control for equal protein loading. One representative blot is shown. The blots were quantified and intensities normalized relative to loading control. The numbers under the blots represent mean fold change (SD) relative to control for three independent experiments. * Significantly different from control (Student's t-test, P < 0.05).
DHA induces antioxidant response
PUFAs are subject to lipid peroxidation, thus causing oxidative stress. Some cell lines that have weak antioxidant defense are highly sensitive to n-3 PUFAs for this reason (36). However, SW480 and SW620 cell lines display little sensitivity to lipid peroxidation products (5). Activated PERK phosphorylates nuclear factor (erythroid-derived 2)-like 2 (Nrf2) (up-regulated in SW620) (Table 1) that promotes transcription of genes involved in redox homeostasis, which contributes to survival of ER stress induced in mammalian cells. Oxidative stress-related genes that were found to be up-regulated in SW620 cells included thioredoxin reductase 1 (TXNRD1), superoxide dismutase 1 (SOD1), HMOX1, glutamate-cysteine ligase modifier (GCLM), and glutamate-cysteine ligase catalytic subunits, (GCLC) indicating a disturbance in the redox balance (Fig. 1 and Table 1). HMOX1, GCLM, and TXNRD1 are downstream targets of Nrf2 (14). HMOX1 displays a 24-fold induction at 12 h and is also strongly induced at the protein level (Fig. 2A and Table 1). HMOX1 catabolizes cellular heme to biliverdin, which is reduced to bilirubin, both being very potent cytoprotective antioxidants (37). However, knockdown of HMOX1 by siRNA did not result in increased DHA-sensitivity in SW620 cells (data not shown), supporting our previous findings that lipid peroxidation is not the key mediator of cytotoxicity.
Effect of DHA treatment on Ca2+ homeostasis and genes involved in apoptosis
ER stress-induced depletion of Ca2+ stores or dysregulation of Ca2+ homeostasis may trigger apoptosis. We found that mRNA for a large number of genes involved in Ca2+ homeostasis was changed, mostly up-regulated, after DHA treatment in SW620 cells. Thus, transcripts for the inositol 1,4,5-triphosphate receptors (IP3R1 and 3) were up-regulated, indicating a release of Ca2+ regulated by these receptors (Table 1 and supplementary Table I). In agreement with this, treatment with DHA (70 μM) for 12–48 h resulted in an increase in cytosolic Ca2+ concentration (Fig. 4A). Cytosolic [Ca2+] is mainly regulated by means of transport across cell membranes (e.g., the plasma and ER membranes). Thapsigargin is a specific inhibitor of a Ca2+ ATPase, which pumps Ca2+ into ER. The rate of [Ca2+] increases in cytosol after addition of thapsigargin thus reflects Ca2+ turnover in ER, the main intracellular Ca2+ store. In SW620 this effect was apparent first after 48 h DHA incubation (average [Ca2+] increase 267nM vs. 187nM in control). The time course of these registrations is shown in Fig. 4B.
Fig. 4.
Cytosolic Ca2+ release after DHA treatment. Registrations of cytosolic Ca2+ in DHA-treated SW620 cells. A: DHA treatment increases the basic cytosolic Ca2+ level in SW620 cells. SW620 cells were incubated with DHA (70 μM) for various time periods as indicated. Average Ca2+ concentrations in 196–324 cells are shown. Bars indicate SEM values. The average basic cytosolic Ca2+ level from each time period was tested against time 0. Statistically significant difference from control (no treatment): ** P < 0.01. B: DHA treatment affects the thapsigargin-inhibited Ca2+ transport. SW620 cells were incubated with DHA (70 μM) for 48 h as indicated. After 30 s of [Ca2+] registration thapsigargin (5 μM) (Sigma-Aldrich) or vehicle was added (arrow). Average registrations from all cells are shown since virtually all cells responded. Cytosolic [Ca2+] at the end of the registration (180 s) in DHA-treated cells was statistically significant different from control (P < 0.05). C: ATP stimulation causes a prolonged Ca2+ signal in DHA-treated cells. SW620 cells were incubated with DHA (70 μM) for various time periods as indicated. After 30 s of [Ca2+] registration ATP (1 μM) was added (arrow). Average registrations from responding cells are shown. D: Removal of extracellular Ca2+ with ethylene glycol tetraacetic acid (EGTA) abolishes the prolonged ATP response in DHA-treated cells. SW620 cells were incubated with DHA (70 μM) for 24 h. The cells were incubated in a 10 mM HEPES buffer without Ca2+, but with 0.1 mM EGTA or in a 10 mM HEPES buffer containing 1.2 mM Ca2+ for 10 min before registration. After 30 s of [Ca2+] registration, ATP (1 μM) was added (arrow). Average registrations from responding cells are shown.
A Ca2+ signal after agonist-binding to a G protein coupled receptor often displays two phases: an initial peak response, which is due to release from intracellular stores; and a prolonged phase, which is attributable to Ca2+ entry through the plasma membrane due to emptying of intracellular Ca2+ stores (“capacitative” or store operated Ca2+ influx). This Ca2+ entry is abolished when free extracellular [Ca2+] is chelated by ethylene glycol tetraacetic acid (EGTA). We find that ATP is an agonist that releases a two-phasic Ca2+ signal in most SW620 cells. Incubation with DHA for 12 h and more seemed to accentuate the second phase Ca2+ elevation in SW620 cells (Fig. 4C, Table 2), whereas the first peak response was virtually unchanged. The time course of the registrations from ATP-stimulated SW620 cells are shown in Fig. 4C. When EGTA was added to SW620 cells treated with DHA for 24 h and then stimulated with ATP, we found that the second phase Ca2+ elevation was abolished (Fig. 4D, Table 3). The elevation in prolonged Ca2+ signal in DHA-treated cells after ATP stimulation can therefore be ascribed to Ca2+ entry, probably of the capacitative type.
TABLE 2.
Ca2+ registrations in SW620 cells pretreated with DHA (70 μM) at time points indicated: SW620 cells stimulated with ATP, 1 μM
| Pretreatment | # Cells (Responding Cells, %) | Maximal [Ca2+] Increase, nM | Decline of the Response |
|---|---|---|---|
| None (control) | 120 (56) | 190 (± 13.2) | 1.64 (± 0.06) |
| DHA 6 h | 110 (53) | 196 (± 10.8) | 1.66 (± 0.16) |
| DHA 12 h | 107 (55) | 226 (± 15.9)a | 1.39 (± 0.06)a |
| DHA 24 h | 94 (51) | 201 (± 18.4) | 1.27 (± 0.09)b |
| DHA 48 h | 97 (45) | 198 (± 21.1) | 1.23 (± 0.04)b |
After 30 s of [Ca2+] registration ATP (1μM) (Sigma-Aldrich) or vehicle was added. Maximal [Ca2+] increase is calculated as difference between baseline and peak [Ca2+] in the responding cells. The decline of the response is quantified as the ratio between peak [Ca2+] and [Ca2+] at the end of the registration (180 s). Registrations are depicted in Fig. 4C. The data are presented as means with standard errors (± SEM).
Statistically significant difference from control (P < 0.05).
Statistically significant difference from control (P < 0.01).
TABLE 3.
Ca2+ registrations in SW620 cells pretreated with DHA (70 μM) at time points indicated: SW620 cells stimulated with ATP, 1μM
| Pretreatment | # Cells | Baseline [Ca2+], nM | Maximal [Ca2+] Increase, nM | Decline of the Response |
|---|---|---|---|---|
| None (control) | 103 | 123 (± 4.1) | 247 (± 10.6) | 2.07 (± 0.09) |
| DHA 24 h | 85 | 141 (± 4.7)a | 238 (± 14.1) | 1.61 (± 0.07)a |
| 2. EGTA | 81 | 103 (± 3.7)a | 259 (± 18.2) | 2.51 (± 0.14)a |
| EGTA, DHA 24 h | 73 | 105 (± 3.8)a,b | 227 (± 17.3) | 2.11 (± 0.12)a,b |
EGTA, ethylene glycol tetraacetic acid. After 30 s of [Ca2+] registration ATP (1μM) (Sigma-Aldrich) or vehicle was added. Maximal [Ca2+] increase is calculated as difference between baseline and peak [Ca2+] in the responding cells. The decline of the response is quantified as the ratio between peak [Ca2+] and [Ca2+] at the end of the registration (180 s). Registrations are depicted in Fig. 4D. The data are presented as means with standard errors (± SEM).
Significantly different from control (P < 0.05).
Significantly different from DHA 24 h (÷ EGTA) (P < 0.001).
Disturbances in the Ca2+ pool of ER activate calpain in the cytosol, which then converts ER-localized procaspase 12 to caspase 12 (38). Calpain 7 and a large subunit of calpain 2 as well as the ER stress-related caspases (caspase 4 and 7) were up-regulated in SW620 cells (Table 1). The proapoptotic members of the Bcl-2 family, BAD and BIK, were down-regulated, while BID was up-regulated (SW620). BCL2-associated athanogene 3 (BAG3), known to participate in regulation of apoptosis, was up-regulated 9.9-fold in SW620 cells after 12 h incubation with DHA. Also, the proapoptotic factor Tribbles homolog 3 (TRIB3), known to be induced by ER stress through the PERK-ATF4-CHOP pathway, was up-regulated at all time points (Table 1) (39). The protein level of active caspase 7 was found to increase with time in SW480 cells, while not detected in SW620 cells (data not shown).
Effect of DHA on cellular cholesterol and cholesterol metabolism
We have previously shown that treatment of SW480 and SW620 with DHA leads to accumulation of numerous large lipids droplets, mainly containing triglycerides in SW480 and cholesteryl esters in SW620 (5). However, an increase in cholesteryl esters was also seen in SW480. The formation of lipid droplets is probably induced by DHA, since they are highly enriched in this PUFA. To examine whether this accumulation of esterified cholesterol was compensated for by increased total cholesterol levels, we measured cellular cholesterol content after DHA treatment. No significant differences in total cholesterol were found at 3–24 h when comparing control and DHA-treated cells (data not shown). A slight, but significant increase in total cholesterol levels in DHA-treated cells was observed at 48 h (31.28 ± 1.29 (control) vs. 38.75 ± 3.39 μg cholesterol/mg protein, P < 0.05). These results may indicate that cholesterol available for organelles is reduced due to deposition in lipid droplets.
From the gene expression data, it was apparent that several genes encoding proteins involved in cholesterol biosynthesis were down-regulated at the mRNA level in DHA-treated SW620 cells (Table 1). These include 7- and 24-dehydrocholesterol reductase (DHCR7, DHCR24), farnesyl diphosphate synthase (FDPS), phosphomevalonate kinase (PMVK), 3β-hydroxysterol Δ14-reductase (TM7SF2) (12, 24 h), and lanosterol synthase (LSS) (12 h). However, some transcripts involved in cholesterol uptake and intracellular cholesterol transport, such as low and very low density lipoprotein receptor (LDLR, VLDLR), the Niemann-Pick C1 protein (NPC1), NPC2, and the oxysterol binding protein (OSBP), were found to be up-regulated in SW620 cells after DHA-treatment (Table 1). NPC1 protein levels were in addition analyzed by Western blot and were increased in DHA-treated cells compared with control at 24 h (Fig. 5A). Since several of the differentially expressed transcripts listed above are regulated by sterol regulatory element binding protein 2 (SREBP2), the protein levels of mSREBP2 (mature) and pSREBP2 (precursor) were analyzed by Western blot. An increase in mSREPB2 levels was observed in control and DHA-treated cells over the time period assayed, but DHA-treated cells displayed higher levels of mSREBP2 compared with control cells at all time points (Fig. 5A, B). The level of pSREBP2 was unchanged in SW620 control cells at all time points, while a slight decrease was observed after 48 h treatment with DHA (data not shown). We also analyzed the level of the rate-limiting enzyme in cholesterol biosynthesis, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) by Western blot after DHA treatment. The protein levels of HMGCR in DHA-treated and control cells were similar at 3 and 6 h. At 12–48 h, the HMGCR protein levels were reduced in controls, while the protein level in DHA-treated cells increased slightly (Fig. 5A, B).
Fig. 5.
Changes in cholesterol metabolism induced by DHA. A: Western blot analysis of HMGCR, mSREBP2, and NPC1 protein levels in total protein extracts from SW620 cells treated with DHA for the indicated time periods (h). Controls were harvested at all time points; only 24 h control is shown for mSREBP2 and NPC1. For HMGCR, controls are shown for all time points. β-actin was used as a control for equal protein loading. One blot, representing three independent experiments, is shown. The blots were quantified and protein band intensities normalized relative loading control. The actin adjusted band intensities from the DHA and control membranes were further normalized relative to the 24 h control band, present at all membranes, to adjust for differences in signal intensities between the membranes. The numbers under the blots represent mean fold change (with SD) of DHA samples relative to control at indicted time points for three independent experiments. * Significantly different from control (Student's t-test, P < 0.05). B: Alterations in HMGCR and mSREBP2 protein levels in control (baseline) and DHA treated cells at the indicated time periods. The plots show the mean value of the actin adjusted band intensitites normalized relative to the 24 h control band for DHA treated cells (gray bars) and control cells (black bars). The data represent the mean and SD of three independent experiments. * Significantly different from control (Student's t-test, P < 0.05).
To investigate de novo synthesis of cholesterol in DHA-treated cells, the incorporation of 14C-acetate into cholesterol and cholesteryl esters was measured after treating the cells with DHA for 24 h. The amount of 14C-acetate incorporated into cholesterol in DHA treated cells was slightly, but significantly lower relative to control at 4 h; a similar trend, although not significant was seen after 6 h (Fig. 6). The amount of 14C-acetate incorporated into cholesteryl esters in DHA treated cells was reduced by approximately 60% relative to control at both time points (Fig. 6).
Fig. 6.
Effect of DHA on incorporation of 14C- acetate into cholesterol and cholesteryl esters. Amount of 14C-acetate (% of control) incorporated in cholesterol (black bars) and cholesteryl esters (gray bars) in SW620 cells treated with DHA for 24 h, and further coincubated with DHA and 14C-acetate for 4 and 6 h. The mean and ± SD from (4 h, n = 3; 6 h, n = 2) independent experiments is displayed. * Significantly different from control (Student's t-test, P < 0.05).
DISCUSSION
Exploring how dietary factors interact with and modulate signaling pathways to promote or counteract cancer development and progression constitutes a major challenge. The purpose of the present study was to examine whether n-3 PUFAs like DHA exert their cytotoxicity by changing gene expression patterns and signaling pathways regulating cell growth. We found that ER stress is established already after 3 h treatment with DHA, as demonstrated by increased levels of phosphorylated eIF2α, a hallmark of ER stress. Phosphorylation of eIF2α adapts cells to various conditions of stress by attenuation of protein synthesis. We found that the n-3 PUFAs DHA and EPA, but not OA, cause phosphorylation of eIF2α, thereby generally inhibiting translation initiation. This is in agreement with previous results showing that inhibition of translation initiation mediates the antiproliferative action of EPA in NIH 3T3 cells by decreased levels of cyclin D1 (40). Increased expression of genes downstream of phosphorylated eIF2α is mediated through induction of the transcription factor ATF4 (14). Genes with ATF4 binding sites are involved in restoring ER homeostasis in response to various stresses (41). Several downstream targets of ATF4 are affected at the mRNA and protein level in our study, indicating that ER stress induced by DHA in SW620 cells is mediated through the ER-localized PERK pathway. Induction of the UPR is initiated by dissociation of PERK from the ER-resident chaperone BiP. However, the protein level of BiP remained constant at all time points. Pimpl et al. (42) have reported that transcriptional induction of BiP rarely leads to increased protein levels of BiP/GRP78, this being due to increased turnover.
UPR is activated to restore cellular homeostasis and induces transcription of genes encoding proteins that mediate ER-associated degradation in response to prolonged ER stress. A large number of 20S and 26S proteasomal subunits were up-regulated in SW620 cells. The proteasome plays a central role in proteolysis of ubiquitinated proteins and are responsible for cleaving many regulatory proteins, like cyclins and members of the NFκB family (43). Prolonged ER stress may cause induction of apoptosis. We show that even though the ER stress-related caspases 4 and 7 are up-regulated in DHA-treated SW620 cells, active caspase 7 is not detectable. On the other hand, active caspase 7 was detected in SW480 cells (data not shown). Chen and Istfan (44) have studied the apoptotic response to DHA in several cell lines, among these SW480 and SW620. A DNA ladder was observed after incubation with DHA (150 μM) for 24 h in SW480, but not in SW620 cells; this is in accordance with our results. Previously, we were not able to detect apoptosis by the TUNEL-assay in either SW480 or SW620 (5). This may indicate that the survival threshold is not exceeded in these cells within the time period assayed and concentration used.
ER is the principal site for protein synthesis and folding, Ca2+ storage and signaling, as well as biosynthesis of fatty acids and cholesterol. Any perturbation that interferes with these activities promotes ER stress and initiates the UPR. We found that DHA treatment mobilizes Ca2+ from ER into the cytosol, in agreement with previous results investigating the effects of n-3 and n-6 PUFAs (40, 45–47), but the mechanism is not known. Our results indicate that calcium release induced by DHA may be linked to induction of ER stress. A redistribution of cholesterol from intracellular regulatory compartments like ER to DHA-cholesteryl ester-enriched lipid droplets (5), causing functional depletion of cholesterol in the ER could potentially lead to ER stress and Ca2+ mobilization, since total cholesterol is not increased (this work). Harding et al. (21) have shown that compounds that deplete cellular cholesterol stores activate an integrated stress response (ISR) by promoting ER stress.
The observed stabilization of HMGCR, the rate limiting enzyme in cholesterol biosynthesis, and the increased level of mSREBP2 observed in DHA-treated SW620 cells, indicate an increased cellular need for de novo synthesized cholesterol during DHA treatment. Surprisingly, both increased and decreased expression of several SREBP2 target genes is observed in SW620, despite an increase in the active transcription factor. Inhibition of transcription of SREBP2 target genes has previously been associated with ER stress-induced activation and cleavage of ATF6, and is mediated by interaction of the two transcription factors in the nucleus (48). Reduced expression of SREBP2 target genes may result in a decreased ability of the cells to synthesize new cholesterol, in spite of activated SREBP2. In line with this, we show that DHA promotes a reduced de novo synthesis of cholesterol. Surprisingly, we also find a reduced incorporation of newly synthesized cholesterol into cholesteryl esters (this work), despite the previous observed accumulation of cholesteryl esters in SW620 cells treated with DHA (5). This might possibly result from reduced turnover of DHA-enriched cholesteryl esters in droplets resulting in accumulation in spite of reduced synthesis.
Recently, a link between molecular chaperones, heat stress, and cholesterol synthesis was demonstrated (49). In this work, the chaperone DnaJA4 (DnaJ/Hsp40) was identified as a novel SREBP target gene that can be turned on under conditions of low sterol availability and heat shock. They postulated that SREBP-regulated chaperones may function as effectors linking heat-shock response and the maintenance of membrane components. Also, Lee and Ye (18) have shown that both hypotonic conditions and thapsigargin induced ER stress in CHO-7 cells leads to activation of SREBP2, while no increase in cholesterol synthesis was observed.
The reasons why SREBP2 target genes are regulated differently, and consequences thereof, remain to be investigated in our system.
Up-regulation of transcripts involved in cellular uptake and intracellular transport of cholesterol, like LDLR, VLDLR, NPC1/NPC2, and OSBP in SW620 cells treated with DHA, also suggests an increased demand for intracellular cholesterol. A recent report indicates that DHA treatment inhibits transport of exogenous cholesterol from the plasma membrane to the ER by an unknown mechanism in CaCo2 colon cancer cells (50). In addition, a study on a panel of colon carcinoma cell lines revealed a deficiency of the LDLR in SW480 cells, indicating a dependency on endogenous cholesterol biosynthesis (51). This would probably also apply to the SW620 cell line, which is established from a metastasis derived from the primary SW480 tumor. Reduced de novo cholesterol synthesis and inhibition of transport of exogenous cholesterol to the ER pool, in combination with increased cholesterol esterification as observed earlier, may lead to depletion of cholesterol in the ER. This may be an important factor contributing to the observed prolonged ER stress that may cause growth inhibition and eventually cell death.
In vitro studies suggest that pharmacological activation of the UPR can alter the sensitivity of tumor cells to chemotherapeutic agents (52). Understanding how common dietary chemicals like DHA affect gene expression and signaling pathways in tumor cells may reveal possible treatment strategies that may be targeted and possibly enhance the impact of conventional therapy.
Supplementary Material
Acknowledgments
Technical assistance from Beate Buland and Jens Erik Slagsvold is highly appreciated. We are sincerely grateful for the generous financial support given to the project in remembrance of Egon Leren.
Published, JLR Papers in Press, June 19, 2008.
Footnotes
The project was financed by The Faculty of Medicine, NTNU, The Cancer Research Fund, Trondheim University Hospital, and The Research Council of Norway through grants from the Functional Genomics Program (FUGE). Microarray experiments were performed at the microarray core facility at the Norwegian Microarray Consortium (NMC), Trondheim, which is supported by FUGE, The Norwegian Research Council. Financial support was also given by the cross-disciplinary project “BIOEMIT-Prediction and modification in functional genomics: combining bioinformatical, bioethical, biomedical, and biotechnological research,” NTNU.
The online version of this article (available at http://www.jlr.org) contains supplementary data.
References
- 1.von Schacky C., and W. S. Harris. 2007. Cardiovascular benefits of omega-3 fatty acids. Cardiovasc. Res. 73 310–315. [DOI] [PubMed] [Google Scholar]
- 2.Stamp L. K., M. J. James, and L. G. Cleland. 2005. Diet and rheumatoid arthritis: a review of the literature. Semin. Arthritis Rheum. 35 77–94. [DOI] [PubMed] [Google Scholar]
- 3.Terry P. D., T. E. Rohan, and A. Wolk. 2003. Intakes of fish and marine fatty acids and the risks of cancers of the breast and prostate and of other hormone-related cancers: a review of the epidemiologic evidence. Am. J. Clin. Nutr. 77 532–543. [DOI] [PubMed] [Google Scholar]
- 4.Kato T., R. L. Hancock, H. Mohammadpour, B. McGregor, P. Manalo, S. Khaiboullina, M. R. Hall, L. Pardini, and R. S. Pardini. 2002. Influence of omega-3 fatty acids on the growth of human colon carcinoma in nude mice. Cancer Lett. 187 169–177. [DOI] [PubMed] [Google Scholar]
- 5.Schønberg S. A., A. G. Lundemo, T. Fladvad, K. Holmgren, H. Bremseth, A. Nilsen, O. Gederaas, K. E. Tvedt, K. W. Egeberg, and H. E. Krokan. 2006. Closely related colon cancer cell lines display different sensitivity to polyunsaturated fatty acids, accumulate different lipid classes and downregulate sterol regulatory element-binding protein 1. FEBS J. 273 2749–2765. [DOI] [PubMed] [Google Scholar]
- 6.Das U. N. 1999. Essential fatty acids and their metabolites and cancer. Nutrition. 15 239–240. [DOI] [PubMed] [Google Scholar]
- 7.Das U. N. 1999. Essential fatty acids, lipid peroxidation and apoptosis. Prostaglandins Leukot. Essent. Fatty Acids. 61 157–163. [DOI] [PubMed] [Google Scholar]
- 8.Stoll B. A. 2002. N-3 fatty acids and lipid peroxidation in breast cancer inhibition. Br. J. Nutr. 87 193–198. [DOI] [PubMed] [Google Scholar]
- 9.Finstad H. S., M. C. Myhrstad, H. Heimli, J. Lomo, H. K. Blomhoff, S. O. Kolset, and C. A. Drevon. 1998. Multiplication and death-type of leukemia cell lines exposed to very long-chain polyunsaturated fatty acids. Leukemia. 12 921–929. [DOI] [PubMed] [Google Scholar]
- 10.Finstad H. S., H. Dyrendal, M. C. Myhrstad, H. Heimli, and C. A. Drevon. 2000. Uptake and activation of eicosapentaenoic acid are related to accumulation of triacylglycerol in Ramos cells dying from apoptosis. J. Lipid Res. 41 554–563. [PubMed] [Google Scholar]
- 11.Sampath H., and J. M. Ntambi. 2005. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 25 317–340. [DOI] [PubMed] [Google Scholar]
- 12.Narayanan B. A., N. K. Narayanan, B. Simi, and B. S. Reddy. 2003. Modulation of inducible nitric oxide synthase and related proinflammatory genes by the omega-3 fatty acid docosahexaenoic acid in human colon cancer cells. Cancer Res. 63 972–979. [PubMed] [Google Scholar]
- 13.Wahle K. W., D. Rotondo, and S. D. Heys. 2003. Polyunsaturated fatty acids and gene expression in mammalian systems. Proc. Nutr. Soc. 62 349–360. [DOI] [PubMed] [Google Scholar]
- 14.Schroder M. 2007. Endoplasmic reticulum stress responses. Cell. Mol. Life Sci. 65 862–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroder M., and R. J. Kaufman. 2005. ER stress and the unfolded protein response. Mutat. Res. 569 29–63. [DOI] [PubMed] [Google Scholar]
- 16.Momoi T. 2004. Caspases involved in ER stress-mediated cell death. J. Chem. Neuroanat. 28 101–105. [DOI] [PubMed] [Google Scholar]
- 17.Ye J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 6 1355–1364. [DOI] [PubMed] [Google Scholar]
- 18.Lee J. N., and J. Ye. 2004. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 279 45257–45265. [DOI] [PubMed] [Google Scholar]
- 19.Colgan S. M., D. Tang, G. H. Werstuck, and R. C. Austin. 2007. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int. J. Biochem. Cell Biol. 39 1843–1851. [DOI] [PubMed] [Google Scholar]
- 20.Feng B., P. M. Yao, Y. Li, C. M. Devlin, D. Zhang, H. P. Harding, M. Sweeney, J. X. Rong, G. Kuriakose, E. A. Fisher, et al. 2003. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat. Cell Biol. 5 781–792. [DOI] [PubMed] [Google Scholar]
- 21.Harding H. P., Y. Zhang, S. Khersonsky, S. Marciniak, D. Scheuner, R. J. Kaufman, N. Javitt, Y. T. Chang, and D. Ron. 2005. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19 185–193. [DOI] [PubMed] [Google Scholar]
- 24.Smyth G. K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 1–27. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y., and Y. Hochberg. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. Royal Statistical Soc. B. 57 289–300. [Google Scholar]
- 26.Langaas M., B. H. Lindqvist, and E. Ferkingstad. 2005. Estimating the proportion of true null hypotheses, with application to DNA microarray data. J. Royal Statistical Soc. B. 67 555–572. [Google Scholar]
- 27.R Development Core Team. 2004. R; A language and environment for statistical computing. Accessed July 28, 2008 at http://www.R-project.org. Vol. 2005.
- 28.Gentleman R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottingen J. A., T. Enden, E. Camerer, J. G. Iversen, and H. Prydz. 1995. Binding of human factor VIIa to tissue factor induces cytosolic Ca2+ signals in J82 cells, transfected COS-1 cells, Madin-Darby canine kidney cells and in human endothelial cells induced to synthesize tissue factor. J. Biol. Chem. 270 4650–4660. [DOI] [PubMed] [Google Scholar]
- 30.Rotnes J. S., and J. G. Iversen. 1992. Thapsigargin reveals evidence for fMLP-insensitive calcium pools in human leukocytes. Cell Calcium. 13 487–500. [DOI] [PubMed] [Google Scholar]
- 31.Grynkiewicz G., M. Poenie, and R. Y. Tsien. 1985. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260 3440–3450. [PubMed] [Google Scholar]
- 32.Rotnes J. S., and J. A. Rottingen. 1994. Quantitative analysis of cytosolic free calcium oscillations in neutrophils by mathematical modelling. Cell Calcium. 15 467–482. [DOI] [PubMed] [Google Scholar]
- 33.Bligh E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. [DOI] [PubMed] [Google Scholar]
- 34.Okada T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleizen B., and I. Braakman. 2004. Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 16 343–349. [DOI] [PubMed] [Google Scholar]
- 36.Schonberg S. A., P. K. Rudra, R. Noding, F. Skorpen, K. S. Bjerve, and H. E. Krokan. 1997. Evidence that changes in Se-glutathione peroxidase levels affect the sensitivity of human tumour cell lines to n-3 fatty acids. Carcinogenesis. 18 1897–1904. [DOI] [PubMed] [Google Scholar]
- 37.Stocker R. 2004. Antioxidant activities of bile pigments. Antioxid. Redox Signal. 6 841–849. [DOI] [PubMed] [Google Scholar]
- 38.Orrenius S., B. Zhivotovsky, and P. Nicotera. 2003. Regulation of cell death: the calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 4 552–565. [DOI] [PubMed] [Google Scholar]
- 39.Corcoran C. A., X. Luo, Q. He, C. Jiang, Y. Huang, and M. S. Sheikh. 2005. Genotoxic and endoplasmic reticulum stresses differentially regulate TRB3 expression. Cancer Biol. Ther. 4 1063–1067. [DOI] [PubMed] [Google Scholar]
- 40.Palakurthi S. S., R. Fluckiger, H. Aktas, A. K. Changolkar, A. Shahsafaei, S. Harneit, E. Kilic, and J. A. Halperin. 2000. Inhibition of translation initiation mediates the anticancer effect of the n-3 polyunsaturated fatty acid eicosapentaenoic acid. Cancer Res. 60 2919–2925. [PubMed] [Google Scholar]
- 41.Harding H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11 619–633. [DOI] [PubMed] [Google Scholar]
- 42.Pimpl P., J. P. Taylor, C. Snowden, S. Hillmer, D. G. Robinson, and J. Denecke. 2006. Golgi-mediated vacuolar sorting of the endoplasmic reticulum chaperone BiP may play an active role in quality control within the secretory pathway. Plant Cell. 18 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olivier S., P. Robe, and V. Bours. 2006. Can NF-kappaB be a target for novel and efficient anti-cancer agents? Biochem. Pharmacol. 72 1054–1068. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z. Y., and N. W. Istfan. 2000. Docosahexaenoic acid is a potent inducer of apoptosis in HT-29 colon cancer cells. Prostaglandins Leukot. Essent. Fatty Acids. 63 301–308. [DOI] [PubMed] [Google Scholar]
- 45.Chow S. C., and M. Jondal. 1990. Polyunsaturated free fatty acids stimulate an increase in cytosolic Ca2+ by mobilizing the inositol 1,4,5-trisphosphate-sensitive Ca2+ pool in T cells through a mechanism independent of phosphoinositide turnover. J. Biol. Chem. 265 902–907. [PubMed] [Google Scholar]
- 46.Aktas H., and J. A. Halperin. 2004. Translational regulation of gene expression by omega-3 fatty acids. J. Nutr. 134 2487S–2491S. [DOI] [PubMed] [Google Scholar]
- 47.Kolar S. S., R. Barhoumi, J. R. Lupton, and R. S. Chapkin. 2007. Docosahexaenoic acid and butyrate synergistically induce colonocyte apoptosis by enhancing mitochondrial Ca2+ accumulation. Cancer Res. 67 5561–5568. [DOI] [PubMed] [Google Scholar]
- 48.Zeng L., M. Lu, K. Mori, S. Luo, A. S. Lee, Y. Zhu, and J. Y. Shyy. 2004. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 23 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robichon C., M. Varret, X. Le Liepvre, F. Lasnier, E. Hajduch, P. Ferre, and I. Dugail. 2006. DnaJA4 is a SREBP-regulated chaperone involved in the cholesterol biosynthesis pathway. Biochim. Biophys. Acta. 1761 1107–1113. [DOI] [PubMed] [Google Scholar]
- 50.Mathur S. N., K. R. Watt, and F. J. Field. 2007. Regulation of intestinal NPC1L1 expression by dietary fish oil and docosahexaenoic acid. J. Lipid Res. 48 395–404. [DOI] [PubMed] [Google Scholar]
- 51.Fabricant M., and S. A. Broitman. 1990. Evidence for deficiency of low density lipoprotein receptor on human colonic carcinoma cell lines. Cancer Res. 50 632–636. [PubMed] [Google Scholar]
- 52.Mann M. J., and L. M. Hendershot. 2006. UPR activation alters chemosensitivity of tumor cells. Cancer Biol. Ther. 5 736–740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.