Abstract
Abnormal lipid levels are important risk factors for cardiovascular diseases. We conducted genome-wide variance component linkage analyses to search for loci influencing total cholesterol (TC), LDL, HDL and triglyceride in families residing in American Samoa and Samoa as well as in a combined sample from the two polities. We adjusted the traits for a number of environmental covariates, such as smoking, alcohol consumption, physical activity, and material lifestyle. We found suggestive univariate linkage with log of the odds (LOD) scores > 3 for LDL on 6p21-p12 (LOD 3.13) in Samoa and on 12q21-q23 (LOD 3.07) in American Samoa. Furthermore, in American Samoa on 12q21, we detected genome-wide linkage (LODeq 3.38) to the bivariate trait TC-LDL. Telomeric of this region, on 12q24, we found suggestive bivariate linkage to TC-HDL (LODeq 3.22) in the combined study sample. In addition, we detected suggestive univariate linkage (LOD 1.9–2.93) on chromosomes 4p-q, 6p, 7q, 9q, 11q, 12q 13q, 15q, 16p, 18q, 19p, 19q and Xq23 and suggestive bivariate linkage (LODeq 2.05–2.62) on chromosomes 6p, 7q, 12p, 12q, and 19p-q. In conclusion, chromosome 6p and 12q may host promising susceptibility loci influencing lipid levels; however, the low degree of overlap between the three study samples strongly encourages further studies of the lipid-related traits.
Keywords: environmental effects, genetic linkage analysis, variance component analysis, total cholesterol, low density lipoprotein, high density lipoprotein, triglyceride
Noncommunicable diseases, especially cardiovascular diseases (CVDs), have increased worldwide, and while CVDs have been a major cause of death in the established market economies for many decades, CVDs now rank in the top five causes of death globally (1). There are many risk factors involved in the development of CVD; some of the most prominent ones include abnormal blood lipid levels, obesity, and hypertension, all of which are influenced by genetic as well as environmental factors (2). In addition, environmental factors such as cigarette smoking (3) and lack of physical activity (4) are established CVD risk factors.
We have previously studied genetic influences on adiposity-related phenotypes in samples from American Samoa (5) and Samoa (6) as well as in a combined study sample from both polities (our unpublished data). The two polities belong to a single genetic population (7) that has been fairly isolated and has a common evolutionary history of ∼3,000 years. During the last several decades, the two polities have been differently influenced by economic modernization, which has resulted in increased differences in dietary intakes, physical activity, and other aspects of the social and behavioral environment (8 –11).
In this study, we investigate the serum lipid profile, including total cholesterol (TC), LDL, HDL, and triglyceride (TG), in the combined study sample and in the polity-specific study samples from the Samoan islands. We apply variance component (VC) analysis, as implemented in the software LOKI (12) and SOLAR (13, 14), to search for genetic linkage to the traits. The studied lipid traits are strongly influenced by genetic components; however, environmental factors are also of great importance (15). In an attempt to adjust for environmental influences on the traits, we include information on physical activity, consumption of alcohol and cigarettes, education, and an index of household possessions as covariates in the genetic model.
SUBJECTS AND METHODS
Study population
The study samples are from the Samoan islands of Polynesia, which consist of two polities, the United States territory of American Samoa and the independent nation of Samoa. The total population on the Samoan islands consisted of ∼235,000 habitants in 2000–2001 (16, 17). American Samoa, which has a higher level of education, a higher proportion of adults in wage and salary occupations, and higher economic and material lifestyle indicators than Samoa (8 –11), contains approximately one-fourth of the total population of the archipelago (17). The two polities have a common evolutionary history (7, 18, 19), but during the last few decades American Samoa has been influenced by economic modernization to a much greater extent than Samoa (11, 20).
Samples
The participating families were selected based on the number of adult family members available. As described previously (5, 6), participants were selected from villages throughout American Samoa and Samoa, and probands and families were not selected based on any specific trait. All participants gave their informed consent, and protocols for this study were approved by the Brown University Institutional Review Board, the American Samoan Institutional Review Board, and the Government of Samoa, Ministry of Health, Health Research Committee.
Families
Interviews were used to collect information on pedigree structure. The combined study sample include 71 pedigrees containing 3,016 individuals, age ⩾ 18 years. The American Samoa sample set and the Samoan sample set include 34 families and 46 families, respectively. Twenty of the 71 families in the combined sample set had a mixed origin, containing family members from American Samoa as well as from Samoa. The number of genotyped individuals for each population is shown in Table 1 . Each family has at least two genotyped individuals. The largest family has 246 genotyped individuals. Details about the pedigree structures can be found in supplementary Table I.
TABLE 1.
Overview of samples and characteristics of nontransformed phenotypes
| American
Samoa |
Samoa
|
Combined
|
|||||
|---|---|---|---|---|---|---|---|
| Characteristic | Males | Females | Males | Females | Males | Females | |
| Genotyped markers a | 368 (377 b ) + 14 (18) | 368 (378) + 14 (14) | 368 + 14 | ||||
| Pedigrees | 34 | 46 | 71 (20 c ) | ||||
| Genotyped individuals | 246 | 332 | 278 | 294 | 534 | 630 | |
| Phenotyped individuals | 261 | 334 | 336 | 338 | 597 | 672 | |
| Age (years) d | 43.2 (16.5) | 43 (16.1) | 41.7 (16.3) | 45.2 (17.4) | 42.4 (16.4) | 44.1 (16.8) | |
| Sex e | 44 | 56 | 50 | 50 | 47 | 53 | |
| Education (years) d | 11.7 (2.4) | 12.0 (2.4) | 9.7 (3.4) | 10.0 (3.0) | 10.6 (3.1) | 11.0 (2.9) | |
| Material lifestyle index d | 9.1 (1.8) | 9.0 (1.8) | 7.6 (2.6) | 7.7 (2.6) | 8.3 (2.4) | 8.4 (2.3) | |
| Smoking cigarettes (yes/no) e | 38/56 | 20/74 | 38/51 | 13/67 | 38/53 | 17/71 | |
| Drinking alcohol (yes/no) e | 43/48 | 8/80 | 32/54 | 3/74 | 37/51 | 6/77 | |
| Physical activity (hours per week) d | 4.1 (6.3) | 1.9 (3.6) | 8.6 (14.8) | 2.3 (5.7) | 6.7 (12.0) | 2.1 (4.8) | |
| Body mass index (kg/m2) d | 33.5 (7.6) | 36.6 (8.4) | 28.9 (5.4) | 33.0 (7.6) | 30.9 (6.9) | 34.8 (8.2) | |
| Total cholesterol (mg/dl) d | 189.4 (37.8) | 187.2 (38.2) | 198 (39.3) | 202.9 (37) | 194.2 (38.8) | 195 (38.3) | |
| LDL (mg/dl) d | 118.3 (34.5) | 119.1 (33.6) | 127.9 (36.7) | 133.4 (33.2) | 123.8 (36.1) | 126.1 (34.1) | |
| HDL (mg/dl) d | 38.6 (8.8) | 42.1 (8.4) | 47.1 (11.6) | 47.9 (10.8) | 43.3 (11.3) | 45.0 (10.0) | |
| Triglyceride (mg/dl) d | 199.1 (205.2) | 129.7 (76.6) | 115.7 (71) | 108.4 (55) | 152.7 (152.2) | 119.2 (67.6) | |
Number of autosomal + X chromosomal markers included.
Total number of markers genotyped.
Pedigrees including individuals from both polities.
Mean and SD.
Percentage of sample. The percentages do not sum to 100 due to missing values.
Phenotypes
Fasting blood samples were collected from participants after a 10 h overnight fast. Serum was separated and then stored at −40°C in the field sites. Serum was transported frozen on dry ice from the field sites to Providence, Rhode Island, for analysis. The TC and TG contents of the serum samples were determined by enzymatic assays on a Gilford Impact 400 computer-directed analyzer (Gilford Instruments, Oberlin, OH) (21, 22). Serum HDL was determined by the double precipitation methods of Gidez et al. (22). Serum LDL was calculated by the Friedewald equation (23):
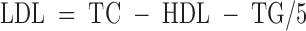 |
For samples with TG values of >400 mg/dl, the Friedewald equation is not valid (23); therefore, the LDL trait was set to missing for these individuals. Standard anthropometric techniques and measurements were used to measure stature and weight and to calculate body mass index (BMI).
Questionnaires were used to collect information on environmental factors such as education (years), physical activity (hours per week doing moderate to very hard sport activities and/or performing farm work), alcohol consumption (yes/no), smoking (yes/no), and household physical characteristics and possessions. Similar to prior Samoan studies by Galanis et al. (8), we used the information on household physical characteristics and possessions to create a household possessions or material lifestyle index (MLSI) ranging from 1 (low material lifestyle standard) to 12 (high material lifestyle standard). The MLSI is based on information regarding domestic flooring type, electricity, cooking facilities, bathroom fixtures, water supply, and possession of a refrigerator, freezer, television, video cassette recorder, stereo, portable stereo, and motor vehicle, where each of the 12 subunits of the information may contribute one point to the index.
Our current data sets do not contain information regarding treatment for nonnormal lipid levels. However, the awareness and care for risky blood lipid levels in the Samoan islands is very limited, so the use of treatments for these CVD risk factors is extremely rare.
Genotypes
We genotyped the samples from American Samoa and from Samoa with the markers in the ABI PRISM linkage mapping set v2.5 MD10 (Applied Biosystems, Inc., Foster City, CA) as described previously (5, 6). The samples from American Samoa were genotyped with an ABI PRISM 3100 genetic analyzer and with an ABI PRISM 3130XL (Applied Biosystems), while the Samoan samples were genotyped using the ABI PRISM 3130XL. Even though the same set of markers was used throughout, the numbers of successfully genotyped markers in the study samples are slightly different. American Samoa and Samoa have 368 overlapping autosomal markers and 14 overlapping X chromosomal markers. The total number of genotyped markers in each study sample is shown in Table 1. All results reported in this study use only the overlapping markers. However, to ensure that the uniquely genotyped markers did not detect any linkage signals in American Samoa or Samoa, we also performed polity-specific genome-wide scans with all available markers. No differences in log of the odds (LOD) score were observed (data not shown).
When a study sample contains pedigrees that have individuals originating from different subsets (here, the American Samoa and Samoa study samples) that have been genotyped with different instruments, it is important to ensure that the same allele label defines identical alleles, to be able to perform linkage analysis. We previously described how we merged the two sets of genotypes according to the minimal sum of differences in allele frequencies (our unpublished data).
Error checking and data handling
Phenotypes
For the TG trait, we found some very extreme outliers. Therefore, we used winsorization to bring the upper and lower 5% of the TG values closer to the trait mean (24, 25). As described by Shete et al. (25), winsorization increases the power to detect linkage and reduces the bias in estimation of the major VC. Box-Cox power transformations (26) were applied to the lipid traits and to BMI, since they were not normally distributed. Using a VC analysis when the trait is nonnormally distributed can lead to a biased estimate of the major gene effect. Also, falsely assuming normality may lead to excessive type I errors (27). To further guard against false positives due to possible nonnormality, we used the option “tdist” for multivariate t-distribution in SOLAR (13, 14).
Genotypes
In our previous studies investigating adiposity-related phenotypes in the population on the Samoan islands, we extensively checked for genotype errors and errors of pedigree structure prior to statistical analysis (5, 6) (our unpublished data). In short, to detect errors in pedigree structure, PEDSTATS (28) was used to check for internal consistency of ages, and RELPAIR v2.0.1 (29, 30) and PREST (31, 32) were used to check the accuracy of the self-reported pedigree relationships. The “set correct_errors 1” option in LOKI (12) was used to remove a minimal set of genotypes to generate Mendelianly consistent pedigrees for the autosomes. For the X chromosome, we used the option in Mega2 (33) and Pedcheck (34) to remove all genotypes within the entire pedigree for a locus where a Mendelian inconsistency was detected. Mega2 and the statistical software R (The R Project for Statistical Computing) were used interactively to set up files for the analyses performed in this study.
Multipoint linkage analysis
For the American Samoan study sample, the Samoa study sample, and the combined study sample from the two polities, we used the same genetic map based on Kosambi centimorgan (35) and applied the same statistical strategy previously used when investigating adiposity-related phenotypes in these study samples (5, 6) (our unpublished data). As previously, we estimated marker allele frequencies from our pedigree data while simultaneously estimating the identity-by-descent (IBD) sharing matrices using LOKI (12). However, in contrast to our previous studies (5, 6), in this study we generated the IBD matrices using information from all available genotyped pedigree members for all of the data regardless of originating polity. With this strategy, the IBD matrix itself will not cause any differences in the LOD score calculation regardless of whether the American Samoa, Samoa, or combined study sample is studied.
Univariate analysis
We used the multipoint VC linkage analysis as implemented in SOLAR (13, 14) to search for quantitative trait loci (QTLs) for serum lipid levels on the autosomes. Prior to the actual multipoint linkage analysis, we used the “polygenic -s” option in SOLAR to fit the VC model and screen for significance of covariates. Information from all individuals that have complete phenotype information available, from investigated traits and covariates, is used to generate the polygenic model.
In an attempt to adjust for environmental factors that might influence the traits, we screened two sets of covariates for their significant (P ⩽ 0.10) effect on the traits using SOLAR. The initial covariate set included education, physical activity, cigarette smoking, alcohol consumption, and MLSI as well as age, age2, sex, age × sex, and age2 × sex. To investigate the serum lipid-related traits independently of body composition, we used a second covariate set that included BMI in addition to all other covariates. Only significant covariates were included in the final genetic model.
For a given phenotype, a likelihood ratio test for linkage was carried out and classical LOD scores were obtained by converting the statistic into values of log10. A LOD score of ⩾3.3 was taken as evidence of significant linkage, which is equivalent to a marginal P value of 0.0001 or less. LOD scores of ⩾1.9 and ⩾1.175 were considered to show evidence of suggestive linkage and potential linkage, respectively (36). As described above, for some traits we calculated LOD scores using two different models (i.e., using two different covariate sets); thus, correction for multiple testing is appropriate. We applied a conservative correction for multiple models by subtracting log10 t from the obtained maximum LOD scores, where t is the number of models tested (37).
Since the current version of SOLAR cannot correctly carry out multipoint X-linked VC analysis, we used Mendel v6.0.1 (38), which fits a variety of X-linked VC models. We applied two different models in which X-linked QTLs, autosomal additive polygenic VCs, and random environmental VCs were always included, while X-linked additive polygenic VCs were included or left out. Mendel is not able to handle our largest pedigrees. Therefore, we broke our pedigrees into their component nuclear families using Mega2 (33) before running Mendel.
Bivariate analysis
For all chromosomes that, in the univariate analyses, obtained QTLs with LOD scores of ⩾1.5 for two or more phenotypes within the same chromosomal region, we performed bivariate multipoint linkage analysis using SOLAR (13, 14). The bivariate analysis tests for simultaneous linkage of two phenotypes to a single genetic region. We here report the bivariate LOD score transformed to 1 degree of freedom (LODeq) with the SOLAR command “loddf -default - c_rhoq,” which is comparable to a univariate LOD score. Covariate screening in bivariate models is not supported in SOLAR. Therefore, we regressed the covariates that were significant in the univariate analysis onto each trait using a linear model. The differences between the observed and the fitted values (i.e., the residuals) were transformed using Box-Cox power transformation (26) and used as the traits in the bivariate analysis.
We carried out two separate likelihood ratio tests at the location of the maximum multipoint bivariate LOD score on each chromosome, one to test whether the QTL signal is due to pleiotropy (i.e., a major gene affects both phenotypes) and the other to test whether it is due to coincident linkage (i.e., a set of clustered genes, each influencing a particular trait) (39, 40). We used a Chi-square test to evaluate the results. A P of 0.05 to reject either pleiotropy or coincident linkage was used (39).
Web resources
Web resources used were as follows: PEDSTATS (http://www.sph.umich.edu/csg/abecasis/Pedstats), RELPAIR 2.0.1(http://csg.sph.umich.edu/index.php), PREST (http://fisher.utstat.toronto.edu/sun/Software/Prest), Mega2 (http://watson.hgen.pitt.edu), R statistical software (http://www.r-project.org), LOKI (http://www.stat.washington.edu/thompson/Genepi/Loki.shtml), SOLAR (http://www.sfbr.org/solar), and Mendel v6.0.1 (http://www.genetics.ucla.edu/software).
RESULTS
Characteristics for nontransformed traits and covariates are presented in Table 1, and their distributions in American Samoa versus Samoa are shown in supplementary Figure I. Most notable is the high mean for TG (199.1 ± 205.2) observed in males from the American Samoa. However, as reflected by the SD and as mentioned above, some extreme measurements were observed for TG in this group. Also as pointed out previously (5, 6) (our unpublished data), the studied sample sets have remarkably high BMI values. According to body composition studies of Polynesians, BMI > 26 kg/m2 should be considered overweight and BMI > 32 kg/m2 should be considered obese (41). Interestingly, the frequency of heavily obese (BMI > 34 kg/m2) individuals is almost twice as high in American Samoa compared with Samoa (see supplementary Figure I, left column, bottom row). A similar skewness to the right is observed in the American Samoa for the MLSI (see supplementary Figure I, left column, top row). Cigarette smoking and alcohol consumption frequencies as well as hours of physical activity are lower in females than in males (Table 1).
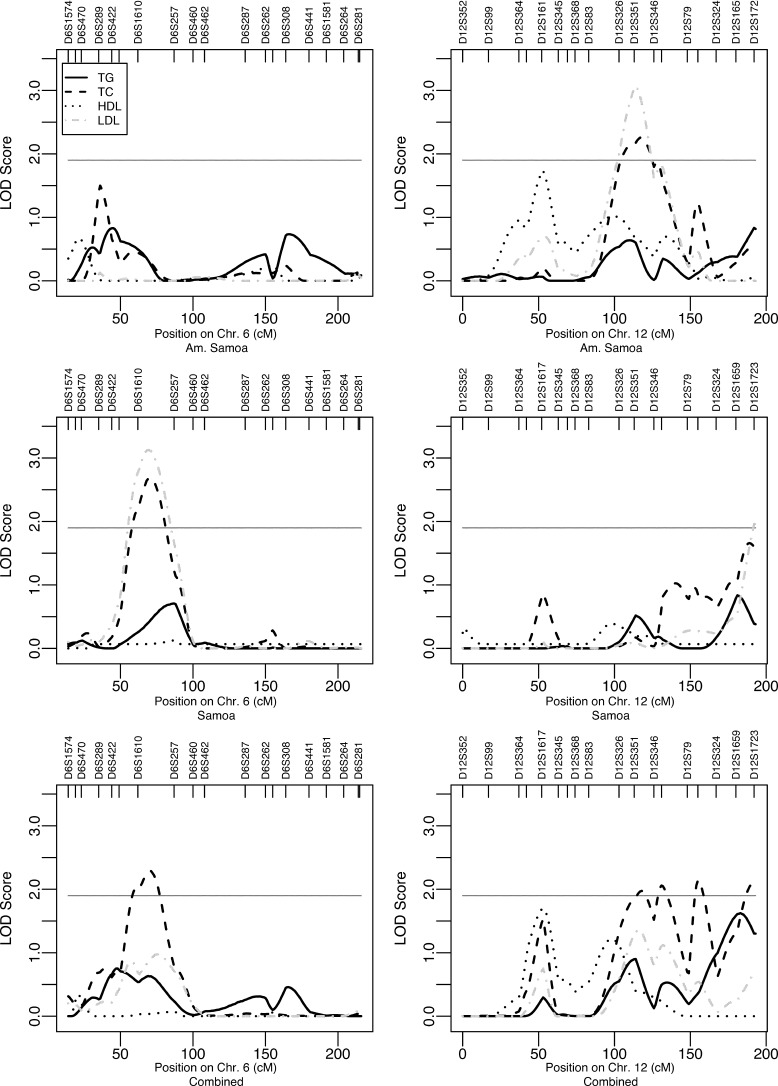
In the study samples from the Samoan islands, the heritability estimates of TC, LDL, HDL, and TG are all significantly different from zero (P < 0.01) and range from 0.18 to 0.71 ( Table 2 ). We detect 11 autosomal regions located on chromosomes 4p-q, 6p, 7q, 9q, 11q, 12q, 13q, 15q, 16p, 18q, and 19p-q with suggestive linkage to serum lipid levels ( Table 3 ; see supplementary Figure IIa, b). For simplified comparability between our study and previous studies in which multiple corrections were not performed, we chose to report both the corrected LOD scores (LODmtc) and the uncorrected LOD scores. Our overall strongest univariate linkage signal was detected on chromosome 6p21-p12 for LDL in the Samoa study sample. We detected a suggestive LOD score of 2.67 when using the initial covariate set. This signal increased to 3.13 when adjusting for BMI in addition to the other covariates ( Fig. 1 , left column). After performing multiple testing correction (mtc) for the two models used for LDL in the Samoa study sample, these LOD scores correspond to LODmtc = 2.37 and 2.83, respectively. In this region, we also detected linkage to TC in the Samoa study sample (LOD = 2.68) and in the combined study samples (LOD = 2.29) (Fig. 1, left column). The second most promising QTL for LDL was detected on chromosome 12q21-q23 (LOD = 3.07) in the American Samoa sample. On the same arm of the chromosome, on 12q24-qtel, a QTL for LDL was detected in the Samoa study sample (LOD = 1.96, LODmtc = 1.66). In addition, the broad region on 12q21-qtel hosts multiple linkage signals (LOD = 1.96–2.26) for TC in American Samoa and in the combined study sample (Fig. 1, right column). The highest linkage signal for TC in the Samoa sample was detected on chromosome 4p14-q12 (LOD = 2.93) and in the American Samoa sample on chromosome 7q35-q36 (LOD = 2.27). Furthermore, suggestive linkage signals were detected for LDL on chromosomes 9q21 (LOD = 2.21, LODmtc = 1.91), 11q23-q24 (LOD = 2.03, LODmtc = 1.73), 13q33 (LOD = 2.25), 16p13 (LOD = 2.00), 18q23-qtel (LOD = 1.91), and 19p13-q12 (LOD = 2.25) and for TC on chromosome 19q13 (LOD = 1.94). The only signal with suggestive linkage for HDL was detected on chromosome 15q14 (LOD = 1.96, LODmtc = 1.66) in the Samoa sample set (Table 3). No suggestive linkage signal was detected for TG. On the X chromosome, on Xq23, we detected a suggestive linkage signal (LOD = 2.30, LODmtc = 2.00) for LDL in the Samoa sample set. No linkage signal of >1.56 was detected on the X chromosome for any of the other traits. None of the QTLs detected with the complete study samples could be detected by any of the larger pedigrees (>40 genotyped individuals) alone when investigated separately with SOLAR.
TABLE 2.
Heritability estimates and overview of the included covariates
| Covariates
d
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Data Set | No. a | h2 (SEM) b | Variance (%) c | A | A2 | S | A*S | A2*S | C | D | P | E | M | B |
| TC | American Samoa | 553 | 0.37 (0.11) | 19 | + | + | + | − | − | + | − | + | − | − | NE/− |
| Samoa | 646 | 0.59 (0.08) | 23 | + | + | + | − | + | − | − | − | − | − | NE/− | |
| Combined | 1,097 | 0.55 (0.07) | 20 | + | + | + | − | + | + | − | + | − | − | NE/− | |
| HDL | American Samoa | 528 | 0.44 (0.11) | 6 | − | + | + | − | − | − | + | − | − | − | NE |
| 516 | 0.47 (0.11) | 9 | − | − | + | − | − | + | + | − | − | − | + | ||
| Samoa | 518 | 0.41 (0.10) | 5 | + | + | + | − | − | + | + | − | − | − | NE | |
| 517 | 0.40 (N.A.) | 15 | − | − | + | − | − | + | + | − | − | − | + | ||
| Combined | 989 | 0.51 (0.07) | 7 | + | + | + | − | − | + | + | − | + | + | NE | |
| 988 | 0.52 (0.07) | 16 | − | − | + | − | − | + | + | − | + | + | + | ||
| LDL | American Samoa | 560 | 0.34 (0.10) | 17 | + | + | − | − | − | − | − | + | − | − | NE/− |
| Samoa | 518 | 0.71 (0.08) | 18 | + | + | − | + | + | − | + | − | − | − | NE | |
| 517 | 0.69 (0.08) | 20 | + | + | + | + | + | − | + | − | − | − | + | ||
| Combined | 1,021 | 0.57 (0.07) | 17 | + | + | + | − | + | − | + | + | − | − | NE/− | |
| TG | American Samoa | 508 | 0.23 (0.11) | 13 | + | + | + | − | − | − | + | − | + | − | NE |
| 528 | 0.18 (0.10) | 16 | + | + | + | − | − | − | + | − | − | − | + | ||
| Samoa | 634 | 0.48 (0.09) | 20 | + | + | + | + | + | − | − | − | − | + | NE | |
| 515 | 0.46 (0.11) | 27 | + | + | + | + | + | − | + | − | − | + | + | ||
| Combined | 1,006 | 0.41 (0.08) | 15 | + | + | + | − | + | − | + | − | + | + | NE | |
| 1,030 | 0.35 (0.07) | 23 | + | + | + | − | + | − | + | − | − | + | + | ||
A, age; A2, age2; S, sex; A*S, age × sex; A2*S, age2 × sex; B, body mass index; C, cigarette smoker; D, drinking alcohol; P, physical activity; E, education; M, material lifestyle index; N.A., SEM could not be computed by SOLAR; NE, covariate not examined; TC, total cholesterol; TG, triglyceride.
Number of included individuals.
h2, heritability estimates with SEM for all traits are significantly different from zero (P < 10−2).
Variance explained by the included covariates.
Minus signs (−) indicate nonsignificant covariates and plus signs (+) indicate significant covariates (P ≤ 0.1) included in the polygenic model.
TABLE 3.
Chromosomal regions with univariate multipoint LOD scores ⩾ 1.9
| Cytogenetic Position | Closest Marker(s) | Trait | Data Set | LOD Score a (Included Covariates) |
|---|---|---|---|---|
| 4p14-q12 | D4S405-D4S1592 | TC | Samoa | 2.93 (A, A2, S, A2*S) |
| 6p21-p12 | D6S1610-D6S257 | TC | Combined | 2.29 (A, A2, S, A2*S, C, P) |
| D6S1610-D6S257 | TC | Samoa | 2.68 (A, A2, S, A2*S) | |
| D6S1610-D6S257 | LDL | Samoa | 2.67 b (A, A2, A*S, A2*S, D) | |
| D6S1610-D6S257 | LDL | Samoa | 3.13 b (A, A2, S, A*S, A2*S, D, B) | |
| 7q35-q36 | D7S661-D7S636 | TC | American Samoa | 2.27 (A, A2, S, C, P) |
| 9q21 | D9S273-D9S175 | LDL | Samoa | 2.15 b (A, A2, A*S, A2*S, D) |
| D9S273-D9S175 | LDL | Samoa | 2.21 b (A, A2, S, A*S, A2*S, D, B) | |
| 11q23-q24 | D11S925-D11S4151 | LDL | Samoa | 1.96 b (A, A2, A*S, A2*S, D) |
| D11S925-D11S4151 | LDL | Samoa | 2.03 b (A, A2, S, A*S, A2*S, D, B) | |
| 12q21.33-q23.1 | D12S351-D12S346 | TC | American Samoa | 2.26 (A, A2, S, C, P) |
| D12S351-D12S346 | LDL | American Samoa | 3.07 (A, A2, P) | |
| D12S351-D12S346 | TC | Combined | 1.96 (A, A2, S, A2*S, C, P) | |
| 12q23.3 | D12S78 | TC | Combined | 2.05 (A, A2, S, A2*S, C, P) |
| 12q24.23 | D12S86 | TC | Combined | 2.14 (A, A2, S, A2*S, C, P) |
| 12q24.33-qtel | D12S1723-tel | TC | Combined | 2.12 (A, A2, S, A2*S, C, P) |
| D12S1723-tel | LDL | Samoa | 1.96 b (A, A2, S, A*S, A2*S, D, B) | |
| 13q33 | D13S173 | LDL | Combined | 2.25 (A, A2, S, A2*S, D, P) |
| 15q14 | D15S1007-D15S1012 | HDL | Samoa | 1.96 b (S, C, D, B) |
| 16p13 | D16S423-D16S404 | LDL | American Samoa | 2.00 (A, A2, P) |
| 18q23-qtel | D18S70-tel | LDL | American Samoa | 1.91 (A, A2, P) |
| 19p13-q12 | D19S226-D19S414 | LDL | American Samoa | 2.25 (A, A2, P) |
| 19q13 | D19S220 | TC | American Samoa | 1.94 (A, A2, S, C, P) |
| Xq23 | DXS8055 | LDL | Samoa | 2.16 b (A, A2, A*S, A2*S, D) |
| DXS8055 | LDL | Samoa | 2.30 b (A, A2, S, A*S, A2*S, D, B) |
Significant covariates (P ≤ 0.1) included in the polygenic model.
The corrected LOD score (LODmtc) can be calculated by subtracting 0.3 from the uncorrected LOD score shown. LOD scores ⩾ 3 are highlighted in boldface.
Fig. 1.
Univariate linkage results [log of the odds (LOD) scores] from chromosome (Chr.) 6 (left column) and chromosome 12 (right column) from the three study samples when all covariates, including body mass index (BMI; see Table 3 for details), were screened for. Suggestive linkage is indicated with the horizontal lines (LOD = 1.9). cM, centimorgan; TC, total cholesterol; TG, triglyceride.
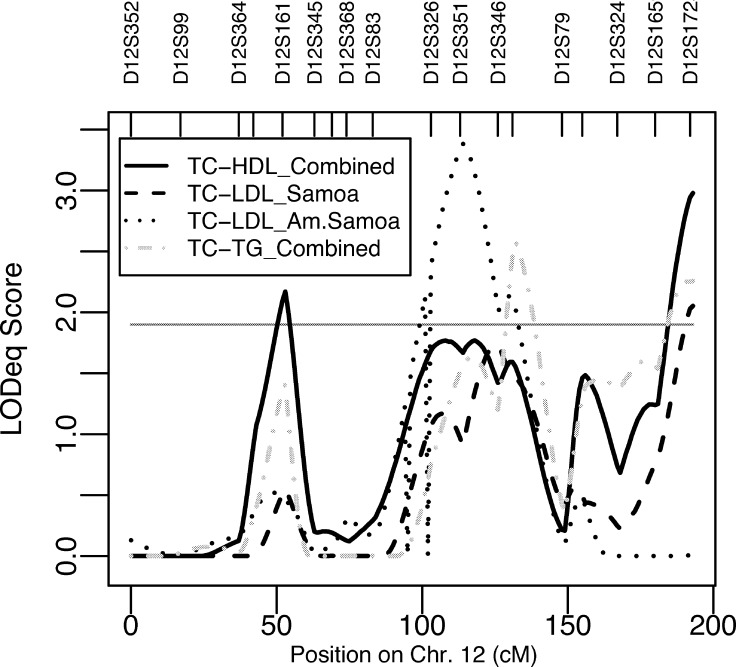
In the region on chromosome 6p21-p12 where we detected a LOD score of 3.13 to LDL in the Samoa study sample, we detected a maximum LODeq score of 2.25 (LODmtc = 1.95) for the bivariate trait TC-LDL in the Samoa sample ( Table 4 ). Furthermore, on chromosome 12q21-q23, where we detected a LOD score of 3.07 to LDL, we detected a genome-wide significant LODeq score of 3.38 to the bivariate trait TC-LDL in the American Samoa sample (Table 4; Fig. 2 ). In this region, we also detected suggestive bivariate linkage (LODeq = 2.05, LODmtc = 1.75) to TC-LDL in the Samoa sample (Table 4; Fig. 2). In addition, we detected suggestive bivariate linkage to TC-LDL on 7q34-q35 (LODeq = 2.25) and on 19p13-q12 (LODeq = 2.14) in American Samoa (Table 4). In the combined sample, we detected suggestive linkage to TC-HDL on chromosome 12p12 (LODeq = 2.17, LODmtc = 1.87) and on 12q24 (LODeq = 3.22, LODmtc = 2.92) as well as to TC-TG in two peaks on chromosome 12q23-12q24 (LODeq = 2.58 and 2.26, LODmtc = 2.28 and 1.96) (Table 4; Fig. 2). P values for tests for compete pleiotropy and coincident linkage are shown in Table 4, and genetic correlation (ranging from 0.15 to 0.96) and environmental correlations (ranging from 0.06 to 0.90) between bivariate traits are shown in Table 5 .
TABLE 4.
Bivariate multipoint LODeq scores ⩾ 1.9 and test for pleiotropy and coincident linkage
| Cytogenetic Position | Closest Marker(s) | Trait | Data Set | LODeq Score (1 Degree of Freedom) | Covariates a | Complete Pleiotropy b | Coincident Linkage b |
|---|---|---|---|---|---|---|---|
| 6p21-p12 | D6S1610-D6S257 | TC-LDL | Samoa | 2.13 c | A, A2, STC, A•SLDL, A2•S, DLDL | 0.500 | 9.8 × 10−4 |
| 6p21-p12 | D6S1610-D6S257 | TC-LDL | Samoa | 2.25 c | A, A2, S, A•SLDL, A2•S, DLDL, BLDL | 0.500 | 8.1 × 10−16 |
| 7q34-q35 | D7S661 | TC-LDL | American Samoa | 2.62 | A, A2, STC, CTC,P | 0.020 | 0.002 |
| 12p12 | D12S1617 | TC-HDL | Combined | 2.17 c | ATC, A2 TC, S, A2•STC, C, PTC, DHDL, EHDL, MHDL, BHDL | ||
| 12q21 | D12S351 | TC-LDL | American Samoa | 3.38 | A, A2, STC, CTC, P | 0.235 | 5.0 × 10−5 |
| 12q21 | D12S346 | TC-LDL | Samoa | 2.05 c | A, A2, S, A•SLDL, A2•S, DLDL, BLDL | 0.500 | 7.3 × 10−4 |
| 12q23 | D12S78 | TC-TG | Combined | 2.58 c | A, A2, S, A2•S, CTC, PTC, DTG, MTG, BTG | 0.028 | 0.625 |
| 12q24 | D12S1723 | TC-TG | Combined | 2.26 c | A, A2, S, A2•S, CTC, PTC, DTG, MTG, BTG | ||
| 12q24 | D12S1723 | TC-HDL | Combined | 3.22 c | A, A2, S, A2•STC, C, PTC, DHDL, EHDL, MHDL | 0.008 | 0.017 |
| 12q24 | D12S1723 | TC-HDL | Combined | 3.00 c | ATC, A2 TC, S, A2•STC, C, PTC, DHDL, EHDL, MHDL, BHDL | N.A. | N.A. |
| 19p13-q12 | D19S226-D19S414 | TC-LDL | American Samoa | 2.14 | A, A2, STC, CTC, P | 0.395 | 0.001 |
Covariates included in the polygenic model. Subscripts indicate trait-specific covariates.
P value for test of complete pleiotropy (ρq constrained to 1 or −1) and no coincident linkage (ρq constrained to 0) for the maximum LODeq per chromosome. N.A. indicates that convergence failure occurred in SOLAR.
The corrected LOD score (LODmtc) can be calculated by subtracting 0.3 from the uncorrected LOD score shown. LODeq ⩾ 3 is indicated in boldface.
Fig. 2.
Bivariate linkage results (LODeq scores) from chromosome 12 when the covariate set including BMI (see Table 4 for details) was used. Suggestive linkage is indicated with the horizontal line (LODeq = 1.9).
TABLE 5.
Genetic (ρg) and environmental (ρe) correlations between trait combinations included in the bivariate analysis
| Trait | Data Set | ρg | SEM | ρe | SEM | Covariates a |
|---|---|---|---|---|---|---|
| TC-LDL | American Samoa | 0.96 b | 0.02 | 0.89 b | 0.02 | A, A2, STC, CTC, P |
| Samoa | 0.96 b | 0.02 | 0.88 b | 0.03 | A, A2, STC, A•SLDL, A2•S, DLDL | |
| Samoa | 0.95 b | 0.02 | 0.90 b | 0.02 | A, A2, S, A•SLDL, A2•S, DLDL, BLDL | |
| TC-HDL | Combined | 0.15 | 0.12 | 0.06 | 0.10 | A, A2, S, A2•STC, C, PTC, DHDL, EHDL, MHDL |
| Combined | 0.15 | 0.11 | 0.06 | 0.10 | ATC, A2 TC, S, A2•STC, C, PTC, DHDL, EHDL, MHDL, BHDL | |
| TC-TG | Combined | 0.17 | 0.14 | 0.35 b | 0.08 | A, A2, S, A2•S, CTC, PTC, DTG, MTG, BTG |
Covariates included in the polygenic model. Subscripts indicate trait-specific covariates.
Correlations are different from zero at P < 4.3 × 10−5.
DISCUSSION
Influence of environmental factors
In this study, we performed a genome-wide linkage investigation to search for loci influencing the human lipid profile in families from the Samoan islands. Since environmental factors exert large effects on variation in lipid-related traits, we attempted to adjust for environmental factors (e.g., cigarette smoking, alcohol consumption, physical activity, education, and MLSI) by including covariates of significant effect in the VC models. To our knowledge, this is the first study in which material lifestyle has been considered a covariate in VC analyses of lipid-related traits. Assessment of material lifestyle at the individual subject level allows for adjustments of the effect on lipids of the general household economic environment, such as diet (8). We observed multiple differences between the study samples regarding which covariates had a significant effect on the studied traits (Table 2). These data suggest that specific environmental factors are of different significance in the two polities. Some of these differences are likely due to the greater impact of economic modernization on American Samoa relative to Samoa (11, 20). However, to account for such differences in economic modernization between and within the polities, we created the MLSI. We reasoned that the MLSI, which is based on 12 material lifestyle components for each individual (see Subjects and Methods), would reflect variation in lifestyle due to variation in financial means. In agreement with previous reports (8 –11), American Samoa on average has a higher material life standard (as measured by the MLSI) than Samoa. Surprisingly, the MLSI did not have a significant effect on any of the lipid traits in the American Samoan sample and only a significant effect on TG in the samples from Samoa. When the two samples were combined, MLSI was included as a significant covariate for HDL and TG but not for LDL and TC. This suggests that the material lifestyle, or factors in strong correlation with the MLSI, such as dietary intake (8), might be of importance for variation in HDL and TG levels but is less likely to have any major effect on variation in LDL and TC levels. In addition to diet, the influence of MLSI on HDL and TG may also be due to other unmeasured socioeconomic and behavioral factors associated with material lifestyle. More detailed research on individual-, household-, and community-level environmental factors influencing blood lipid levels may improve the ability to detect genetic influences. Further support that TC and LDL are influenced by similar environmental factors is indicated by the relatively high environmental correlations (ρe) between the two traits (ρe ⩾ 0.88; Table 5), while correlations for the other trait combinations investigated are fairly low (ρe ⩽ 0.35).
Susceptibility loci
In this study, we report suggestive linkage with LOD scores of >3 for LDL on chromosome 6p21-p12 (LOD = 3.13, LODmtc = 2.83) in Samoa and on chromosome 12q21-q23 (LOD = 3.07) in American Samoa. In addition, we detected QTLs with suggestive linkage for blood lipid traits on chromosomes 4p-q, 6p, 7q, 9q, 11q, 12q, 13q, 15q, 16p, 18q, 19p-q, and Xq23. Furthermore, we detected bivariate genome-wide significant linkage to TC-LDL on chromosome 12q21 (LODeq = 3.38) in American Samoa and bivariate suggestive linkage to TC-LDL, TC-HDL, and/or TC-TG (LODeq ranging from 2.05 to 3.22) on chromosomes 6p, 7q, 12p, 12q, and 19p-q. Coincident linkage was strongly rejected in favor of pleiotropy for TC-LDL in the American Samoan and in the Samoan study samples on chromosomes 6, 12, and 19. On chromosome 7q, where bivariate linkage was detected to TC-LDL in American Samoa, both complete pleiotropy and coincident linkage could be rejected, suggesting that some but not complete pleiotropy occurs in this region. Similarly, on chromosome 12q in the combined study sample, some but not complete pleiotropy may occur for TC-HDL. Complete pleiotropy was strongly rejected in favor of coincident linkage for TC-TG on chromosome 12. Furthermore, the genetic correlation (ρe) between TC and LDL suggests that a common set of genes are involved in the regulation of these two traits (Table 5). The high correlation that was seen for TC and LDL (ρe = 0.96) is likely to result in a fairly similar linkage pattern for the two traits. This was seen, for example, in the Samoan study sample on chromosome 6 and in the American Samoan study sample on chromosome 12. However, despite these encouraging results for common genetic components for TC and LDL, one should keep in mind that TC is a measure of multiple lipoprotein fractions, in which LDL normally constitutes a large proportion. Therefore, it is possible that the genetic contribution to TC in fact reflects its correlation to LDL. Thus, the information gained from genetic studies of TC might be of limited importance.
To our knowledge the region on chromosome 6p21-p12, detected by both univariate and bivariate analyses, has not been reported previously as linked to blood lipid traits in humans. However, the chromosomal region on 6p12-6q12, which directly flanks this QTL, has previously been linked to HDL (42). The ∼34 centimorgan region spanning the 1.5 univariate LOD drop interval is a gene-rich region that demands further fine-mapping to define potential candidate genes. Interestingly, all regions but 18q and Xq23 with suggestive univariate linkage to LDL and/or TC are human ortholog regions to QTLs for plasma non-HDL (i.e., LDL and VLDL) previously reported in mice (43). Furthermore, the regions where the QTLs on chromosome 4 (44), 7q (45 –48), 9q (47), 11q (46, 49, 50), 12q (51), 15q (48, 52, 53), and 19q (44, 51) are located have been suggested to host QTLs for blood lipid traits in humans. The region centromeric of the QTL detected on 18q has been suggested as a susceptibility region for LDL in humans (54), and recently, additional support for QTLs to TC were reported on chromosomes 7q32-q36 and 19q13 in a meta-analysis including nine data sets ascertained for type 2 diabetes mellitus (55). Recently, genome-wide association studies have reported significant association between LDL and multiple single-nucleotide polymorphisms on chromosome 19p13-q13 (56 –59), which overlaps the region where we detected linkage to LDL and to TC-LDL. Within the region on chromosome 12q24 where we detected linkage to TC-HDL, one of the recent genome-wide association studies reported significant association to HDL (59). This same study also reported association for LDL to a single-nucleotide polymorphism located on 6p21, where we detected linkage to LDL, TC, and TC-LDL (59).
Overlap across the three study samples
None of the linkage signals detected in the present study were simultaneously detected in American Samoa, in Samoa, and in the combined study sample. However, a few regions were detected in two of the three analyses: the regions on 6p21-p12 and 12q24-tel were detected in Samoa as well as in the combined study sample, and the region on 12q21-q23 was detected in American Samoa and in the combined study sample. These peaks detected in a broad region of the q arm of chromosome 12 are partly but not fully overlapping, which might suggest that chromosome 12q may host at least one and possibly multiple susceptible loci for the lipid traits in the population from the Samoan islands. To further investigate the 12q region, additional mapping is required.
Even though we adjusted the lipid-related traits for multiple carefully measured environmental factors, we still observed multiple chromosomal regions of different significance in the three study samples. Since the two Samoan polities were shown to have a common evolutionary background with no differences in linkage disequilibrium structure (7, 18, 19), this lack of overlap between the study samples suggests potential gene-environment interaction and the influence of environmental factors not accounted for in the present study. Furthermore, the low degree of overlap between the linkage signals in the study samples might be explained by the relatively larger pedigree structures in the American Samoan sample compared with the Samoan sample, which could result in differences in power to detect linkage (60).
Conclusion
In conclusion, we have found genome-wide significant support for bivariate linkage to TC-LDL in American Samoa and suggestive support for bivariate linkage in Samoa as well as suggestive univariate linkage to TC and/or LDL in all three study samples on chromosome 12q. Furthermore, on chromosome 6p21-p12, we detected suggestive support for linkage to lipid traits in Samoa and in the combined study samples. Chromosomes 6p and 12q may harbor promising susceptibility loci influencing the lipid traits; however, the low degree of overlap between the three study samples strongly encourages further studies of the lipid-related traits in which additional attention will be given to the selection and inclusion of environmental factors.
Supplementary Material
Acknowledgments
The authors are very thankful to the leaders of the Department of Health, American Samoa Government, and the Ministry of Health, Government of Samoa, to local political officials, and to the study participants for their contributions to this research.
Published, JLR Papers in Press, July 1, 2008.
Footnotes
Our work is supported by National Institutes of Health Grant R01 DK-59642 (S.T.M.). K.Å. is supported by the Swedish Research Council.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and one table.
References
- 1. Mathers C. D., and D. Loncar. 2006. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3 e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hawkins M. A. 2004. Markers of increased cardiovascular risk: are we measuring the most appropriate parameters? Obes. Res. 12 (Suppl. 2): 107–114. [DOI] [PubMed] [Google Scholar]
- 3. Kannel W. B., T. R. Dawber, A. Kagan, N. Revotskie, and J. Stokes 3rd. 1961. Factors of risk in the development of coronary heart disease:–six year follow-up experience. The Framingham Study. Ann. Intern. Med. 55 33–50. [DOI] [PubMed] [Google Scholar]
- 4. Blair S. N., J. B. Kampert, H. W. Kohl 3rd, C. E. Barlow, C. A. Macera, R. S. Paffenbarger, Jr., and L. W. Gibbons. 1996. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. J. Am. Med. Assoc. 276 205–210. [PubMed] [Google Scholar]
- 5. Dai F., E. D. Keighley, G. Sun, S. R. Indugula, S. T. Roberts, K. Aberg, D. Smelser, J. Tuitele, L. Jin, R. Deka, et al. 2007. Genome-wide scan for adiposity-related phenotypes in adults from American Samoa. Int. J. Obes. (Lond). 31 1832–1842. [DOI] [PubMed] [Google Scholar]
- 6.Dai, F., G. Sun, K. Aberg, E. D. Keighley, S. R. Indugula, S. T. Roberts, D. Smelser, S. Viali, L. Jin, R. Deka, et al. 2008. A whole genome linkage scan identifies multiple chromosomal regions influencing adiposity-related traits among Samoans. Ann. Hum. Genet. Epub ahead of print. 10.1111/j.1469-1809.2008.00462. [DOI] [PMC free article] [PubMed]
- 7. Tsai H. J., G. Sun, D. Smelser, S. Viali, J. Tufa, L. Jin, D. E. Weeks, S. T. McGarvey, and R. Deka. 2004. Distribution of genome-wide linkage disequilibrium based on microsatellite loci in the Samoan population. Hum. Genomics. 1 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galanis D. J., S. T. McGarvey, C. Quested, B. Sio, and S. A. Afele-Fa'amuli. 1999. Dietary intake of modernizing Samoans: implications for risk of cardiovascular disease. J. Am. Diet. Assoc. 99 184–190. [DOI] [PubMed] [Google Scholar]
- 9. Keighley E. D., S. T. McGarvey, P. Turituri, and S. Viali. 2006. Farming and adiposity in Samoan adults. Am. J. Hum. Biol. 18 112–122. [DOI] [PubMed] [Google Scholar]
- 10. McGarvey S. T. 1991. Obesity in Samoans and a perspective on its etiology in Polynesians. Am. J. Clin. Nutr. 53 (Suppl.): 1586–1594. [DOI] [PubMed] [Google Scholar]
- 11. McGarvey S. T. 2001. Cardiovascular disease (CVD) risk factors in Samoa and American Samoa, 1990–95. Pac. Health Dialog. 8 157–162. [PubMed] [Google Scholar]
- 12. Heath S. C. 1997. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am. J. Hum. Genet. 61 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Almasy L., and J. Blangero. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amos C. I. 1994. Robust variance-components approach for assessing genetic linkage in pedigrees. Am. J. Hum. Genet. 54 535–543. [PMC free article] [PubMed] [Google Scholar]
- 15. Garg A., and V. Simha. 2007. Update on dyslipidemia. J. Clin. Endocrinol. Metab. 92 1581–1589. [DOI] [PubMed] [Google Scholar]
- 16.Census of Population and Housing, American Samoa 2000. U. S. Department of Commerce, Washington, DC, 2004.
- 17.Census of Population and Housing 2001. Government Printing House, Apia, Samoa, 2003.
- 18. Deka R., S. T. McGarvey, R. E. Ferrell, M. I. Kamboh, L. M. Yu, C. E. Aston, L. Jin, and R. Chakraborty. 1994. Genetic characterization of American and Western Samoans. Hum. Biol. 66 805–822. [PubMed] [Google Scholar]
- 19. Tsai H. J., G. Sun, D. E. Weeks, R. Kaushal, M. Wolujewicz, S. T. McGarvey, J. Tufa, S. Viali, and R. Deka. 2001. Type 2 diabetes and three calpain-10 gene polymorphisms in Samoans: no evidence of association. Am. J. Hum. Genet. 69 1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keighley, E. D., S. T. McGarvey, C. Quested, C. McCuddin, S. Viali, and U. A. Maga. 2007. Nutrition and health in modernizing Samoans: temporal trends and adaptive perspectives. In Health Changes in the Asia-Pacific Region: Biocultural and Epidemiological Approaches. R. Ohtsuka, S. J. Ulijaszek, editors. Cambridge University Press, Cambridge, UK. 149–191.
- 21. Allain C. C., L. S. Poon, C. S. Chan, W. Richmond, and P. C. Fu. 1974. Enzymatic determination of total serum cholesterol. Clin. Chem. 20 470–475. [PubMed] [Google Scholar]
- 22. Gidez L. I., G. J. Miller, M. Burstein, S. Slagle, and H. A. Eder. 1982. Separation and quantitation of subclasses of human plasma high density lipoproteins by a simple precipitation procedure. J. Lipid Res. 23 1206–1223. [PubMed] [Google Scholar]
- 23. Friedewald W. T., R. I. Levy, and D. S. Fredrickson. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18 499–502. [PubMed] [Google Scholar]
- 24. Fernandez J. R., C. Etzel, T. M. Beasley, S. Shete, C. I. Amos, and D. B. Allison. 2002. Improving the power of sib pair quantitative trait loci detection by phenotype winsorization. Hum. Hered. 53 59–67. [DOI] [PubMed] [Google Scholar]
- 25. Shete S., T. M. Beasley, C. J. Etzel, J. R. Fernandez, J. Chen, D. B. Allison, and C. I. Amos. 2004. Effect of winsorization on power and type 1 error of variance components and related methods of QTL detection. Behav. Genet. 34 153–159. [DOI] [PubMed] [Google Scholar]
- 26. Box G. E. P., and D. R. Cox. 1964. An analysis of transformations (with discussion). J. R. Stat. Soc. [Ser. A]. 26 211–252. [Google Scholar]
- 27. Allison D. B., M. C. Neale, R. Zannolli, N. J. Schork, C. I. Amos, and J. Blangero. 1999. Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci-mapping procedure. Am. J. Hum. Genet. 65 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wigginton J. E., and G. R. Abecasis. 2005. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 21 3445–3447. [DOI] [PubMed] [Google Scholar]
- 29. Boehnke M., and N. J. Cox. 1997. Accurate inference of relationships in sib-pair linkage studies. Am. J. Hum. Genet. 61 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Epstein M. P., W. L. Duren, and M. Boehnke. 2000. Improved inference of relationship for pairs of individuals. Am. J. Hum. Genet. 67 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McPeek M. S., and L. Sun. 2000. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am. J. Hum. Genet. 66 1076–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun L., K. Wilder, and M. S. McPeek. 2002. Enhanced pedigree error detection. Hum. Hered. 54 99–110. [DOI] [PubMed] [Google Scholar]
- 33. Mukhopadhyay N., L. Almasy, M. Schroeder, W. P. Mulvihill, and D. E. Weeks. 2005. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 21 2556–2557. [DOI] [PubMed] [Google Scholar]
- 34. O'Connell J. R., and D. E. Weeks. 1998. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kong X., K. Murphy, T. Raj, C. He, P. S. White, and T. C. Matise. 2004. A combined linkage-physical map of the human genome. Am. J. Hum. Genet. 75 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lander E., and L. Kruglyak. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11 241–247. [DOI] [PubMed] [Google Scholar]
- 37. Risch N. 1991. A note on multiple testing procedures in linkage analysis. Am. J. Hum. Genet. 48 1058–1064. [PMC free article] [PubMed] [Google Scholar]
- 38. Lange K., R. Cantor, S. Horvath, M. Perola, C. Sabatti, J. Sinsheimer, and E. Sobel. 2001. Mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am. J. Hum. Genet. 69 (Suppl.): A1886. [Google Scholar]
- 39. Almasy L., T. D. Dyer, and J. Blangero. 1997. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet. Epidemiol. 14 953–958. [DOI] [PubMed] [Google Scholar]
- 40. Comuzzie A. G., M. C. Mahaney, L. Almasy, T. D. Dyer, and J. Blangero. 1997. Exploiting pleiotropy to map genes for oligogenic phenotypes using extended pedigree data. Genet. Epidemiol. 14 975–980. [DOI] [PubMed] [Google Scholar]
- 41. Swinburn B. A., S. J. Ley, H. E. Carmichael, and L. D. Plank. 1999. Body size and composition in Polynesians. Int. J. Obes. Relat. Metab. Disord. 23 1178–1183. [DOI] [PubMed] [Google Scholar]
- 42. Canizales-Quinteros S., C. A. Aguilar-Salinas, E. Reyes-Rodriguez, L. Riba, M. Rodriguez-Torres, S. Ramirez-Jimenez, A. Huertas-Vazquez, V. Fragoso-Ontiveros, A. Zentella-Dehesa, J. L. Ventura-Gallegos, et al. 2003. Locus on chromosome 6p linked to elevated HDL cholesterol serum levels and to protection against premature atherosclerosis in a kindred with familial hypercholesterolemia. Circ. Res. 92 569–576. [DOI] [PubMed] [Google Scholar]
- 43. Wang X., and B. Paigen. 2005. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr. Opin. Lipidol. 16 127–137. [DOI] [PubMed] [Google Scholar]
- 44. North K. E., H. H. Goring, S. A. Cole, V. P. Diego, L. Almasy, S. Laston, T. Cantu, B. V. Howard, E. T. Lee, L. G. Best, et al. 2006. Linkage analysis of LDL cholesterol in American Indian populations: the Strong Heart Family Study. J. Lipid Res. 47 59–66. [DOI] [PubMed] [Google Scholar]
- 45. Lin J. P. 2003. Genome-wide scan on plasma triglyceride and high density lipoprotein cholesterol levels, accounting for the effects of correlated quantitative phenotypes. BMC Genet. 4 (Suppl. 1): 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Y., D. F. Wyszynski, D. M. Waterworth, S. D. Wilton, P. J. Barter, Y. A. Kesaniemi, R. W. Mahley, R. McPherson, G. Waeber, T. P. Bersot, et al. 2005. Multiple QTLs influencing triglyceride and HDL and total cholesterol levels identified in families with atherogenic dyslipidemia. J. Lipid Res. 46 2202–2213. [DOI] [PubMed] [Google Scholar]
- 47. Li W. D., C. Dong, D. Li, C. Garrigan, and R. A. Price. 2005. A genome scan for serum triglyceride in obese nuclear families. J. Lipid Res. 46 432–438. [DOI] [PubMed] [Google Scholar]
- 48. Sonnenberg G. E., G. R. Krakower, L. J. Martin, M. Olivier, A. E. Kwitek, A. G. Comuzzie, J. Blangero, and A. H. Kissebah. 2004. Genetic determinants of obesity-related lipid traits. J. Lipid Res. 45 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rotter J. I., X. Bu, R. M. Cantor, C. H. Warden, J. Brown, R. J. Gray, P. J. Blanche, R. M. Krauss, and A. J. Lusis. 1996. Multilocus genetic determinants of LDL particle size in coronary artery disease families. Am. J. Hum. Genet. 58 585–594. [PMC free article] [PubMed] [Google Scholar]
- 50. Allayee H., B. E. Aouizerat, R. M. Cantor, G. M. Dallinga-Thie, R. M. Krauss, C. D. Lanning, J. I. Rotter, A. J. Lusis, and T. W. de Bruin. 1998. Families with familial combined hyperlipidemia and families enriched for coronary artery disease share genetic determinants for the atherogenic lipoprotein phenotype. Am. J. Hum. Genet. 63 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heijmans B. T., M. Beekman, H. Putter, N. Lakenberg, H. J. van der Wijk, J. B. Whitfield, D. Posthuma, N. L. Pedersen, N. G. Martin, D. I. Boomsma, et al. 2005. Meta-analysis of four new genome scans for lipid parameters and analysis of positional candidates in positive linkage regions. Eur. J. Hum. Genet. 13 1143–1153. [DOI] [PubMed] [Google Scholar]
- 52. Duggirala R., J. Blangero, L. Almasy, T. D. Dyer, K. L. Williams, R. J. Leach, P. O'Connell, and M. P. Stern. 2000. A major susceptibility locus influencing plasma triglyceride concentrations is located on chromosome 15q in Mexican Americans. Am. J. Hum. Genet. 66 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen W., S. Li, S. R. Srinivasan, E. Boerwinkle, and G. S. Berenson. 2007. A genome scan for loci influencing levels and trends of lipoprotein lipid-related traits since childhood: the Bogalusa Heart Study. Atherosclerosis. 190 248–255. [DOI] [PubMed] [Google Scholar]
- 54. Bosse Y., L. Perusse, and M. C. Vohl. 2004. Genetics of LDL particle heterogeneity: from genetic epidemiology to DNA-based variations. J. Lipid Res. 45 1008–1026. [DOI] [PubMed] [Google Scholar]
- 55. Malhotra A., S. C. Elbein, M. C. Ng, R. Duggirala, R. Arya, G. Imperatore, A. Adeyemo, T. I. Pollin, W. C. Hsueh, J. C. Chan, et al. 2007. Meta-analysis of genome-wide linkage studies of quantitative lipid traits in families ascertained for type 2 diabetes. Diabetes. 56 890–896. [DOI] [PubMed] [Google Scholar]
- 56. Kathiresan S., O. Melander, C. Guiducci, A. Surti, N. P. Burtt, M. J. Rieder, G. M. Cooper, C. Roos, B. F. Voight, A. S. Havulinna, et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sandhu M. S., D. M. Waterworth, S. L. Debenham, E. Wheeler, K. Papadakis, J. H. Zhao, K. Song, X. Yuan, T. Johnson, S. Ashford, et al. 2008. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 371 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wallace C., S. J. Newhouse, P. Braund, F. Zhang, M. Tobin, M. Falchi, K. Ahmadi, R. J. Dobson, A. C. Marcano, C. Hajat, et al. 2008. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 82 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Willer C. J., S. Sanna, A. U. Jackson, A. Scuteri, L. L. Bonnycastle, R. Clarke, S. C. Heath, N. J. Timpson, S. S. Najjar, H. M. Stringham, et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dolan C. V., D. I. Boomsma, and M. C. Neale. 1999. A note on the power provided by sibships of sizes 2, 3, and 4 in genetic covariance modeling of a codominant QTL. Behav. Genet. 29 163–170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.