Abstract
NXL101 is one of a new class of quinoline antibacterial DNA gyrase and topoisomerase IV inhibitors showing potent activity against gram-positive bacteria, including methicillin- and fluoroquinolone-resistant strains. NXL101 inhibited topoisomerase IV more effectively than gyrase from Escherichia coli, whereas the converse is true of enzymes from Staphylococcus aureus. This apparent target preference is opposite to that which is associated with most fluoroquinolone antibiotics. In vitro isolation of S. aureus mutants resistant to NXL101 followed by cloning and sequencing of the genes encoding gyrase and topoisomerase IV led to the identification of several different point mutations within, or close to, the quinolone resistance-determining region (QRDR) of GyrA. However, the mutations were not those that are most frequently associated with decreased sensitivity to quinolones. A fluoroquinolone-resistant mutant variant of gyrase generated in vitro was highly resistant to inhibition by the fluoroquinolones ciprofloxacin and moxifloxacin but remained fully susceptible to inhibition by NXL101. Two mutant gyrases constructed in vitro, with mutations in gyrA engineered according to those most frequently found in S. aureus strains resistant to NXL101, were insensitive to inhibition by NXL101 and had a diminished sensitivity to ciprofloxacin and moxifloxacin. Certain combinations of mutations giving rise to NXL101 resistance and those giving rise to fluoroquinolone resistance may be mutually exclusive.
Most characterized bacteria possess two distinct type II topoisomerases, DNA gyrase and topoisomerase IV, that catalyze processes essential for DNA replication and cell viability. Topoisomerase IV is responsible for the decatenation of daughter chromosomes formed during cell division and also unlinks catenanes generated by site-specific recombination. Gyrase is uniquely responsible for the introduction of negative supercoils into DNA that are required for initiation of DNA replication and also removes positive supercoils to allow fork progression (3, 4, 6, 7, 12). Changes in the various topologies of DNA that are brought about by the catalytic activity of type II topoisomerases are thought to be mediated by the strand passage activity of the enzymes. Despite the variety of reactions catalyzed by the various type II enzymes (DNA relaxation and supercoiling, catenation and decatenation, and knotting and unknotting), the reactions are effected according to a broadly similar mechanism and by enzymes that share common structural features. Gyrase is an A2B2 heterotetramer encoded by the gyrA and gyrB genes. Similarly, topoisomerase IV is an A2B2 enzyme encoded by parC and parE (grlA and grlB in S. aureus); the GyrA and GyrB proteins share a high degree of amino acid sequence identity with ParC and ParE, respectively, within an organism (12).
For several decades, these enzymes have been successfully exploited as the molecular targets of the quinolone and fluoroquinolone antibiotics (31). The fluoroquinolones bind to the topoisomerase-DNA complex to form a tripartite complex which usually, if not always, stabilizes the covalent attachment of cleaved DNA to the GyrA or ParC subunit (the so-called “cleaved complex”) that is a natural transitory intermediate in the catalytic cycle of topoisomerases (3). It is widely believed that it is the double-strand DNA breaks stabilized in this way which ultimately lead to lethal DNA damage. The single most common cause of resistance to fluoroquinolones among clinical isolates is the appearance of mutations in the genes encoding the subunits of gyrase and topoisomerase IV. Mutations in gyrA are those the most frequently found in gram-negative bacteria, and mutations in parC/grlA are generally the most common in gram-positive bacteria (16). Incremental increases in resistance occur in a stepwise fashion as a mutation of the primary target that lowers affinity for the antibiotic will generally produce a bacterium whose tolerance of the antibiotic is dependent upon the susceptibility of the secondary target. Subsequent mutations can of course occur and accumulate with the passage of successive generations to produce n step mutations which demonstrate progressively lower susceptibility.
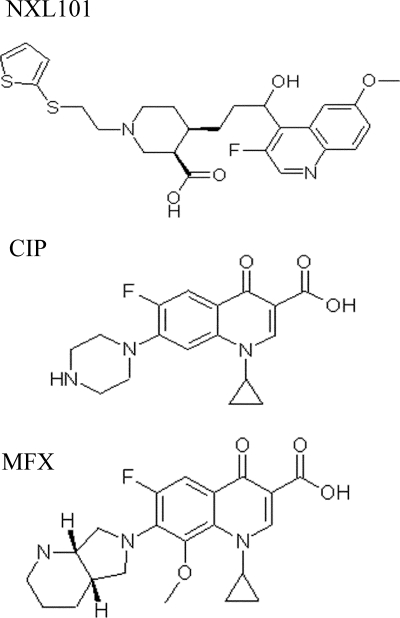
Improvement in activity against gram-positive bacteria would presumably come from the development of inhibitors which are highly active against both gyrase and topoisomerase IV. The selection and development of fluoroquinolones during recent years have in part been driven by such a goal. Some experimental compounds and some of the most recently developed antibiotics such as moxifloxacin and gemifloxacin appear to have a preference for gyrase over topoisomerase IV in Streptococcus pneumoniae (1, 11, 14, 29), although not in Staphylococcus aureus (18, 19). NXL101 {(3R,4R)-4-[(S)-3-(3-fluoro-6-methoxy-quinolin-4-yl)-3-hydroxy-propyl]-1-[2-(thiophen-2-ylsulfanyl)-ethyl]-piperidine-3-carboxylic acid) is a member of a novel chemical class of quinoline antibacterial agents (Fig. 1). In vitro, NXL101 is highly active against a range of refractory multiresistant gram-positive pathogens involved in nosocomial and community-acquired infections, including fluoroquinolone-resistant strains (23; M. Borgonovi, C. Delachaume, A.-M. Girard, J. Lowther, and J. Hodgson, presented at the 16th European Congress of Clinical Microbiology and Infectious Diseases, Nice, France, 1 to 4 April 2006 [poster P-1567]). The compound has also demonstrated potent in vivo activity against multiresistant S. aureus in murine septicemia and thigh muscle infections (24) and against fluoroquinolone-resistant S. pneumoniae in a murine pneumonia model (22) and has been shown to achieve homogeneous and potent bactericidal concentrations in human volunteer plasma (34).
FIG. 1.
Molecular structure of NXL101 {(3R,4R)-4-[(S)-3-(3-fluoro-6-methoxy-quinolin-4-yl)-3-hydroxy-propyl]-1-[2-(thiophen-2-ylsulfanyl)-ethyl]-piperidine-3-carboxylic acid)}, ciprofloxacin (CIP), and moxifloxacin (MFX).
The aim of this study was to evaluate the activity of NXL101 against purified S. aureus and E. coli gyrase and topoisomerase IV enzymes, including mutant variants with reduced sensitivity to fluoroquinolones or to NXL101. The levels of inhibition of topoisomerase activities were evaluated in vitro and compared with those of the fluoroquinolone antibiotics ciprofloxacin and moxifloxacin. The potential for certain mutations conferring reduced sensitivity to either fluoroquinolones or to NXL101 to coexist within the same GyrA protein was also investigated.
MATERIALS AND METHODS
Materials were purchased from the following suppliers: ciprofloxacin HCl salt, USP, Rockville, MD; moxifloxacin, Bayer AG; NXL101, Novexel; kanamycin, Sigma; and levofloxacin, Aventis. E. coli DNA gyrase and topoisomerase IV enzymes, pBR322 plasmid DNA (relaxed, supercoiled, and linear), kinetoplast DNA (kDNA), and assay buffers were obtained from John Innes Entreprises. S. aureus strains 8325, COL, MSSA476, MRSA252, USA400, MW2, N315, and Mu50 were obtained from NARSA (Network on Antibiotic Resistant Staphylococcus aureus).
Production of WT and mutant GyrA proteins. (i) S. aureus gyrase A production.
Plasmid pSAGA3.1 (33) contains coding sequences for a thioredoxin-GyrA fusion protein with an N-terminal six-His (His6) tag and a cleavage site for tobacco etch virus (TEV) protease to release native GyrA subunit. An overnight culture of E. coli TOP10 with pSAGA3.1 plasmid in LB broth containing ampicillin (100 μg.ml−1) was used to inoculate 1 liter of fresh antibiotic containing broth at an optical density at 600 nm (OD600) of 0.2. The cells were grown at 37°C with vigorous shaking to mid-logarithmic phase (OD600 of 0.8) and induced by the addition of arabinose to a final concentration of 0.2% with further incubation for 3 h at 25°C. The cell pellet was harvested by centrifugation at 4°C, frozen, and stored at −70°C until protein isolation.
(ii) S. aureus gyrase B production.
Plasmid pTrcHis-SAGyrB contains the GyrB coding sequence fused to a His6 tag at the N terminus (20). An overnight culture of E. coli BL21(DE3)with pTrcHis-SAGyrB plasmid in LB broth containing ampicillin (100 μg ml−1) was used to inoculate 1 liter of fresh antibiotic-containing broth at an OD600 of 0.2. The cells were grown at 37°C with vigorous shaking to mid-logarithmic phase (OD600 of 0.8) and induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM with further incubation for 4 h at 37°C.
The cell pellet was harvested by centrifugation at 4°C, frozen, and stored at −70°C until protein isolation.
(iii) Mutant S. aureus gyrase.
Mutations were introduced in the wild-type (WT) S. aureus gyrA gene by a single PCR with one pair of complementary primers containing the mutation of interest, using the QuickChange site-directed mutagenesis system (Stratagene) according to the manufacturers' recommendations. A 50-μl reaction mixture contained 10 ng of pSAGA3.1 DNA template, 10 pmol of each of the primers, 200 μM of each deoxynucleoside triphosphate (dNTP), and 2.5 U Pfu DNA polymerase. The temperature cycles were as follows: incubation at 95°C for 30 s, followed by 30 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 10 min and a final extension at 68°C for 7 min. The following primers were used: SAGYRA-M121K-F (5′-GGCGCAGCAGCAAAGCGTTATACTGAAGCG-3′), SAGYRA-M121K-R (5′-CGCTTCAGTATAACGCTTTGCTGCTGCGCC-3′), SAGYRA-S84L+E88K-F (5′-CCCTCATGGTGACTTATCTATTTATAAAGCAATGGTACGTATGGC-3′), SAGYRA-S84L+E88K-R (5′-GCCATACGTACCATTGCTTTATAAATAGATAAGTCACCATGAGGG-3′), SAGYRA-D83N-F (5′-GGGTAAATATCACCCTCATGGTAACTCATCTATTTATGAAGC-3′), and SAGYRA-D83N-R (5′-GCTTCATAAATAGATGAGTTACCATGAGGGTGATATTTACCC-3′).
The entire S. aureus gyrA DNA sequences were verified by DNA sequencing, pSAGA3.1 plasmids containing the mutations of interest were transformed in E. coli TOP10 cells, and the overexpression of mutated proteins was induced as for the native S. aureus gyrA.
Production of S. aureus topoisomerase IV and mutants. (i) S. aureus GrlA and -B production.
Two hundred microliters of a culture of S. aureus RN4220 at an OD600 of 1.0 was centrifuged, and the pellet was homogenized in 50 μl TES [N-tris(hydroxymethyl)methyl-2-aminoethane sulfonic acid] buffer including lysostaphin at 40 μg ml−1 and incubated for 1 h at 37°C. PCR was performed on a GeneAmp PCR system 9700 (Applied Biosystems), on 0.5 μl of the digested cell suspension using ProofStar polymerase (Qiagen). Primers for both genes carried an EcoRI (forward primers) or NotI (reverse primers) site. PCR conditions were an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 3 min, followed by a final extension at 72°C for 7 min. The sequences (5′→3′) of the primers used are as follows: Sa grlA-F, GGAGGAGAATTCTAATGAGTGAAATAATTCAAGATTT; Sa grlA-R, ATTGCAGCGGCCGCTTAGCTAATATACATGTCTATT; Sa grlB-F, AAATCAGAATTCCAATGAATAAACAAAATAATTAT; and Sa grlB-R, AATTATGCGGCCGCCTAGATTTCCTCCTCATCA. PCR products were purified using a High Pure PCR product purification kit (Roche). Purified PCR products and pET28c(+) (His tag, N terminal; Novagen) were digested with EcoRI and NotI. Digested fragments were purified following electrophoresis on a 1% agarose-Tris-borate-EDTA gel using the QIAquick gel extraction kit (Qiagen) and eluted in Tris-HCl (pH 8.0). Digested pET28c and genes were ligated using a Ready-To-Go T4 DNA ligase kit (Amersham) to generate vectors for expression of GrlA (pET28-GrlA) and GrlB (pET28-GrlB). Protein production was performed according to published procedures (20) with the following modifications. Induction was performed in E. coli BL21(DE3) cells transformed with the plasmid of interest by addition of IPTG to 1 mM at 30°C for 2 h in LB broth supplemented with kanamycin at 100 μg/ml.
(ii) Mutant GrlA.
The GrlA Ser80Phe mutation was introduced using the QuickChange mutagenesis kit (Stratagene). The sequences (5′→3′) of the primers used are as follows: Sa grlA-S80F-F, CATCCACATGGAGACTTCTCAGTGTACGAAGCAATG; and Sa grlB-S80F-R, CATTGCTTCGTACACTGAGAAGTCTCCATGTGGATG.
Plasmid amplification was performed after transformation of XL1-Blue supercompetent cells (Stratagene), and the sequences of the insert were verified with standard T7 and T7term forward and reverse primers, respectively. Sequencing was performed by MWG-Biotech, (Ebersberg, Germany) on an ABI capillary sequencer. Results were analyzed with Vector NTI 9.0 (Invitrogen).
Purification of proteins.
GyrA was purified from 1-liter cell cultures according to published procedures (33), with modifications as described below. After centrifugation, cell pellets were resuspended in lysis buffer: 20 mM K2HPO4, 2.5 mM dithiothreitol (DTT) buffer supplemented with a protease inhibitor cocktail (Complete EDTA free; Roche). Lysis buffer (65 μl) was used to resuspend the cells corresponding to a 20-ml cell culture at OD600 of 1.0. Three hundred and fifty milligrams of glass beads (Sigma) was added, and the mixture was vortexed for 10 min. After addition of 900 μl lysis buffer, the suspension was centrifuged for 15 min at 30,000 × g. The pellet was discarded and the cell lysate supplemented with 20 mM imidazole and loaded onto a 5-ml equilibrated HisTrap FF crude column (GE Healthcare) at 0.15 ml min−1. After washing (buffer containing 20 mM sodium phosphate, 500 mM NaCl, and 20 mM imidazole [pH 7.4]), proteins were eluted with buffer containing 20 mM sodium phosphate, 500 mM NaCl, and 300 mM imidazole (pH 7.4). The eluate (20 ml) was concentrated to ∼4 ml using 30,000 NMWL Amicon Ultra-4 centrifugal filter devices (Millipore). Exchange of the buffer for a buffer containing 50 mM Tris-HCl, 100 mM KCl, 2 mM DTT (pH 7.5) (topoisomerase storage buffer) was done by three serial concentration cycles using the same device. AcTEV protease (1,200 U) (Invitrogen) was added to the concentrate, and this mixture was incubated overnight at 4°C. The mixture was then diluted threefold with topoisomerase storage buffer and loaded on the HisTrap FF crude column to adsorb noncleaved fusion protein, the thioredoxin tag and the AcTEV protease. The flow-through fraction was concentrated, and the buffer was changed to a 2× concentrated topoisomerase storage buffer (100 mM Tris-HCl, 200 mM KCl, 4 mM DTT [pH 7.5]). Protein concentration was measured by the Bradford method (Bio-Rad protein assay). The concentrated gyrase A sample was diluted twofold with glycerol, and aliquots were flash frozen in liquid nitrogen and stored at −80°C. The same protocol as that described above was used for purification of GyrB, GrlA, and GrlB, except the AcTEV cleavage step was omitted. Protein purity was estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on Tris-Glycine 8 to 16% acrylamide gels under reducing conditions (Novex; Invitrogen) and Coomassie blue staining, scanning, and band quantification using ImageJ (http://rsb.info.nih.gov/ij/) and Protein 230 Chips on an Agilent 2100 Bioanalyzer. DNA gyrases and topoisomerase IV (WT or mutant) were reconstituted by incubation of equimolar amounts of GyrA and GyrB subunits or GrlA and GrlB subunits, respectively, for 30 min at room temperature. The activities of the holoenzymes were stable for at least 1 month when stored at −80°C.
Generation and sequencing of eQRDR DNA.
Generation and sequencing of extended QRDR (eQRDR) DNA were performed as follows. For cell wall hydrolysis, S. aureus strains were grown overnight at 37°C in LB medium under shaking. Two hundred microliters of a culture of S. aureus at an OD600 of 1.0 was centrifuged for 10 min at 3,000 × g at 4°C. The pellet was mixed with 50 μl of TES buffer plus 40 μg of lysostaphin and incubated 1 h at 37°C.
PCR was performed on 0.125 to 0.5 μl of the cell-wall-hydrolyzed suspension using the Expand high-fidelity polymerase (Roche). The PCR conditions were an initial denaturation at 92°C for 2 min, followed by 40 cycles of denaturation at 92°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min. The amplicon sizes were 770 bp (gyrA), 730 bp (gyrB), 787 bp (grlA), and 717 bp (grlB). PCR products were purified on columns using the High Pure PCR purification kit (Roche) and quantified on E-Gel (Invitrogen) with SmartLadder as a reference.
The sequences (5′→3′) of the primers used are as follows: Sa gyrA QRDR-F, CGCTGTGAACTGAACTTTTGAAGGAG; Sa gyrA QRDR-R, GAGAACGCATTTGAATTGAACCACG; Sa gyrB QRDR-F, CAAACATGGTGATCCTCAATTCGAAG; Sa gyrB QRDR-R, TCGTGCAATAGACCATTTTGGTGTTG; Sa grlA QRDR-F, GACAAAGTACAACCTAGACGTGAATGG; Sa grlA QRDR-R, ATACCACCAGTTGGAAAATCAGGAC; Sa grlB QRDR-F, ACAGCTGTTGTGTCTGTTCGTATTCC; and Sa grlB QRDR-R, CTCTTCGTCTGTCCAAGCGTATTC.
Sequencing was performed by MWG-Biotech, and the results were analyzed with Vector NTI 9.0. Sequences covered at least amino acids 1 to 220 for GyrA, 370 to 550 for GyrB, 1 to 200 for GrlA, and 350 to 530 for GrlB.
Topoisomerase assays.
Assays of E. coli enzymes were performed using materials from John Innes Enterprises according to the supplier's protocols and procedures described below. Assays of S. aureus enzymes were performed under similar conditions with slight modifications described below.
All reaction mixtures (25 μl) were incubated at 37°C for 1 h. Supercoiling, decatenation, and relaxation reactions were stopped by addition of 6 μl of blue/orange loading dye (Promega) containing 1% SDS. Reactions were analyzed by separation of DNA topomers on agarose gels, ethidium bromide staining, and quantitation with ImageQuant (GE Healthcare). IC50s were calculated with GraFit (Erythacus) software. IC50s (mean of at least two independent determinations) correspond to the concentration of drug required to inhibit 50% of the reaction.
(i) DNA supercoiling.
E. coli gyrase (5 nM) or S. aureus gyrase (2 nM) was incubated with 0.3 μg of relaxed pBR322 DNA in a 25-μl reaction mixture at 37°C for 1 h under the following conditions: 35 mM Tris HCl (pH 7.5), 1 mM ATP, 24 mM KCl, 4 mM MgCl2, 2 mM DTT, 1.8 mM spermidine, 6.5% (wt/vol) glycerol, and 0.1 mg ml−1 bovine serum albumin. Assays of S. aureus gyrase contained 500 mM potassium glutamate. Each reaction was analyzed by electrophoresis through a 0.8% agarose gel (run at 65 V for 3 h). IC50s corresponded to a 50% reduction of the supercoiled DNA.
(ii) Decatenation activity of topoisomerase IV.
The conversion of kDNA to the monomer was performed using 0.2 nM E. coli topoisomerase IV or 15 nM S. aureus topoisomerase IV. Topoisomerase IV enzymes were incubated with 0.2 μg of kDNA in a 25-μl reaction at 37°C for 1 h with 50 mM HEPES.KOH (pH 7.6), 10 mM magnesium acetate, 100 mM potassium glutamate, 10 mM DTT, 1 mM ATP, and 50 μg ml−1 bovine serum albumin. Assays of S. aureus topoisomerase IV contained 250 mM potassium glutamate (20). Each reaction was analyzed by electrophoresis through a 1% agarose gel (run at 75 V for 3 h). IC50s corresponded to a 50% reduction of the decatenated DNA.
(iii) Relaxation activity of topoisomerases.
DNA gyrase relaxation activity was carried out as for the supercoiling assay except that ATP was omitted. Topoisomerase IV relaxation activity was carried out as for decatenation assay with 1 mM ATP. In all cases, reaction mixtures contained 0.3 μg of supercoiled pBR322 DNA as substrate, 100 mM potassium glutamate and appropriate amount of enzymes to fully relax the supercoiled DNA in the absence of inhibitor: 2 nM for E. coli topoisomerase IV, 30 nM for S. aureus topoisomerase IV, and 60 nM for both gyrases. Each reaction was analyzed by electrophoresis through a 0.8% agarose gel (run at 65 V for 3 h). IC50s were evaluated by quantification of the supercoiled DNA.
(iv) Topoisomerase-mediated DNA cleavage.
Reactions were performed under the same conditions as the relaxation assay with the following enzyme concentrations: 20 nM for E. coli topoisomerase IV, 30 nM for S. aureus topoisomerase IV, 60 nM E. coli gyrase, 60 nM S. aureus WT gyrase, 600 nM S. aureus mutant gyrases. After a 1-h reaction, 0.2% SDS and 0.1 mg ml−1 proteinase K were added before a further incubation at 37°C for 30 min. CC50 corresponds to the concentration of drug causing half-maximum production of the linear pBR322 DNA. After addition of 6 μl of blue/orange loading dye, each reaction was analyzed by electrophoresis through 0.8% agarose gel (run at 65 V for 3 h).
Serial passage of NXL101-resistant S. aureus in the presence of fluoroquinolones.
MICs were determined using the NCCLS method (27) for antimicrobial susceptibility testing with Mueller-Hinton (MH) broth. After 18 h of incubation at 37°C, the MIC was recorded and cells were recovered from the tube containing the highest concentration of antibiotic that allowed full growth compared to the control tube. These cells were diluted to inoculate a new set of tubes at 5.105 CFU ml−1. This procedure was repeated up to 10 times. Resistance stability was determined by 24-h serial passages in antibiotic-free media.
Single-step mutant generation and analysis.
Seven S. aureus strains for which the complete genome is known (8325, COL, MSSA476, MRSA252, USA400, MW2, N315, and Mu50) and 43 S. aureus clinical strains were used for the test of acquisition of resistance. The strains exhibited the phenotypes methicillin susceptible or resistant and fluoroquinolone susceptible or resistant.
Continuous gradients of NXL101 were established in petri dishes by placement of six strips of sterile filter paper (3 mm) loaded with 0.24, 0.97, 3.9, 15.6, 62.5, and 250 mg liter−1 NXL101 at regular intervals on the surface of MH agar and allowed to diffuse at 4°C for 24 h. The inoculation was accomplished from a streak of solid agar medium in the direction of the agar gradient. Six different inocula (105 to 1011 CFU ml−1) for each strain were used. The petri dishes were incubated at 37°C for 24 h. NXL101-resistant clones were collected, and MICs were determined.
RESULTS
Enzyme inhibition assays.
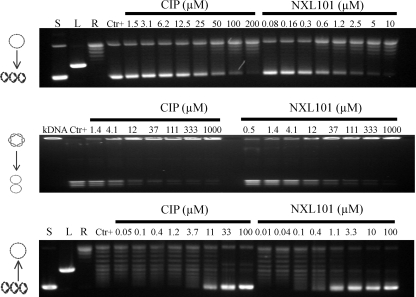
Figure 2 shows typical gels generated for the assessment of inhibition of supercoiling, decatenation, and relaxation of DNA by NXL101 and comparators. Table 1 shows the average IC50 values, calculated from these and similar gels, of NXL101, ciprofloxacin, and moxifloxacin obtained for inhibition of such activities. When considering the effect on S. aureus gyrase reactions, it is apparent that NXL101 is a considerably more potent inhibitor of both supercoiling and relaxation than either moxifloxacin or ciprofloxacin. When considering E. coli gyrase, however, the opposite is true; the fluoroquinolones are considerably more potent inhibitors of both supercoiling and relaxation reactions. The effects against topoisomerase IV enzymes exhibit no such clear trend. NXL101 activity against S. aureus topoisomerase IV relaxation is similar to those of the fluoroquinolones, whereas its activity against S. aureus topoisomerase IV decatenation is notably poorer. Conversely, against E. coli topoisomerase IV decatenation NXL101 activity is rather similar to that of the fluoroquinolones, whereas its activity against topoisomerase IV-catalyzed DNA relaxation is notably better.
FIG. 2.
Typical gels generated for the assessment of inhibition of supercoiling (top), decatenation (middle), and relaxation (bottom) of DNA by NXL101 and comparators. S, L, and R, supercoiled, linear, and relaxed DNA gel markers, respectively. CIP, ciprofloxacin.
TABLE 1.
IC50 values derived from scans of gels as exemplified in Fig. 2
| Antibiotic | IC50 (μM) for enzymea:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Supercoiling
|
Decatenation
|
Relaxation
|
||||||
| Gyrase
|
Topoisomerase IV
|
Gyrase
|
Topoisomerase IV
|
|||||
| E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | E. coli | S. aureus | |
| Ciprofloxacin | 0.2 | 27 | 2.1 | 6.1 | 0.2 | 7.7 | 3.5 | 0.2 |
| Moxifloxacin | 1.1 | 17 | 2.0 | 3.8 | 0.8 | 3.3 | 2.8 | 1.0 |
| NXL101 | 11 | 1.0 | 2.2 | 18 | 23 | 0.4 | 0.2 | 0.3 |
The IC50 is the concentration of compound required to inhibit 50% of the reaction. Each value is the average of at least two assays.
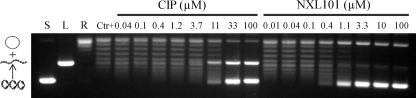
These intriguing data led to experiments to further clarify the inhibitory activity of NXL101 by assessing the propensity to stabilize the cleaved complex in the way that fluoroquinolones are thought to. Figure 3 shows a typical gel generated for assessment of the stabilization of the cleaved complex by NXL101 and its comparators. Although evidence of cleaved-complex formation could readily be generated by low concentrations of both moxifloxacin and ciprofloxacin against gyrase and topoisomerase IV from both S. aureus and E. coli, no such evidence was seen for NXL101 at concentrations up to 200 μΜ against either of the S. aureus enzymes or against gyrase from E. coli (Table 2). However, against E. coli topoisomerase IV, NXL101 was able to stabilize the cleaved complex at submicromolar concentrations 10-fold lower than those required of the fluoroquinolones.
FIG. 3.
Typical gel generated for assessment of the stabilization of the cleaved complex by NXL101 and its comparators. S, L, and R, supercoiled, linear, and relaxed DNA gel markers, respectively.
TABLE 2.
CC50 values derived from scans of gels as exemplified in Fig. 3
| Antibiotic | Cleaved-complex CC50 (μM) for enzymea:
|
|||
|---|---|---|---|---|
| Gyrase
|
Topoisomerase IV
|
|||
| E. coli | S. aureus | E. coli | S. aureus | |
| Ciprofloxacin | 0.2 | 11 | 0.6 | 0.5 |
| Moxifloxacin | 0.6 | 2.5 | 0.5 | 0.8 |
| NXL101 | >200 | >200 | 0.05 | >200 |
CC50 is the concentration of compound required to inhibit production of linear pBR322 by 50%. Each value is the average of at least two assays.
Resistance mutations.
Various strains of S. aureus were either subjected to serial passage in media of progressively increasing concentrations of antibiotic or were selected by plating onto antibiotic-containing agar. The DNA coding sequence for the entirety of each of the four topoisomerase subunits (gyrA, gyrB, grlA, and grlB) from all three strains of both WT and NXL101-resistant status was determined. In the three strains tested, all mutations appeared uniquely within the gyrA coding sequence, and none appeared within gyrB or in topoisomerase IV subunits. Table 3 shows the mutations and the NXL101 MICs for the host strains.
TABLE 3.
Amino acid changes resulting from mutations in gyrA coding sequence and the associated MICs of NXL101 and ciprofloxacin of the host S. aureus strain
| WT or mutant strain | MIC (mg liter−1)
|
|
|---|---|---|
| NXL101 | Ciprofloxacin | |
| Robine | ||
| WT | 0.12 | 0.5 |
| His81Asn mutant | 16 | 0.5 |
| Goodtime | ||
| WT | 0.12 | 4 |
| Arg92Cys mutant | 4 | 4 |
| Bontempsa | ||
| WT | 1 | 4 |
| Met121Lys mutant | 64-128 | 4 |
| Asp83Gly mutant | 16-32 | 4 |
The two mutants and corresponding MICs for the Bontemps strain represent two separate experiments.
The MICs reported were maintained by the bacteria after several days of culture in antibiotic-free media indicating stable mutation as opposed to an adaptive response. Although mutations in other parts of the genome cannot be excluded from involvement in production of the resistant genotype, it is clear that no amino acid changes in topoisomerase IV or in the gyrase B subunit contribute to the elevated MICs. In addition, the resistant phenotype was not reversed by addition of the efflux pump inhibitor reserpine at 20 mg liter−1 (data not shown). It should be noted that in the original S. aureus Bontemps isolate used in these experiments, amino acid position 80 of GrlA is phenylalanine and not serine, as is normally the case. This amino acid is the equivalent of Ser84 in GyrA, and a mutation from serine is often associated with fluoroquinolone resistance.
Cross-resistance.
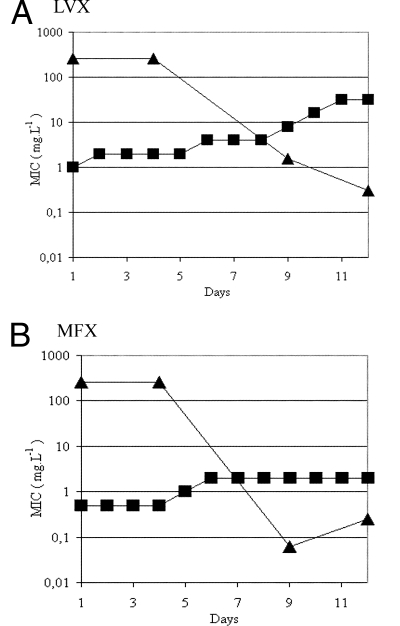
The similarities of NXL101 to fluoroquinolones in mechanism of action, combined with the observation that the Bontemps mutant strain highly resistant to NXL101 (Met121Lys; MIC, 128 mg liter−1) remained susceptible to fluoroquinolones, suggest experiments to determine whether mutations to NXL101 resistance can coexist with those effecting fluoroquinolone resistance. In order to investigate this possibility, the S. aureus strain Bontemps:M121K was subjected to serial passage with increasing concentrations of moxifloxacin or levofloxacin for several days in an attempt to generate mutants resistant to these fluoroquinolones. MICs of both fluoroquinolones were determined on each day (Fig. 4). Samples of cell culture were taken at the start of the experiment, day 1, and on representative days on which increases in the MICs for the fluoroquinolones were seen (days 4, 9, and 12 [Fig. 4]) and plated for single colonies. Ten individual colonies were picked from each plate, and MICs of NXL101 were determined. MICs of bacteria from colonies of each individual plate were in each case identical to within 1 dilution factor. It is apparent in Fig. 4 that as fluoroquinolone resistance increases, so NXL101 resistance decreases. At least two individual colonies (four from the day 1 sample common to both experiments depicted in Fig. 4) from cultures obtained on days 4, 9, and 12 from both the moxifloxacin and levofloxacin experiments were selected, and the coding DNA corresponding to eQRDRs of gyrase and topoisomerase IV were cloned and sequenced. In all cases in which fluoroquinolone resistance increased and NXL101 resistance decreased, the gyrase A serine at position 84 had mutated to leucine and lysine at position 121 had in all cases reverted to methionine (15 separate clones). After a subsequent 5-day passage in antibiotic-free media, DNA sequences remained unchanged compared with those at day 12. These data indicate that either the Ser84Leu mutation and the Met121Lys mutation cannot coexist in the same gyrase A subunit to yield an enzyme with activity sufficient to sustain cell viability or that a bacterium with such a combination suffers a severe fitness cost and is counterselected during cell proliferation.
FIG. 4.
MICs (mg liter−1) of levofloxacin (LVX) (A) and moxifloxacin (MFX) (B) against the S. aureus Bontemps mutant strain possessing, at day 1, a substitution of lysine for methionine at gyrase A primary structure position 121. Squares, fluoroquinolone; triangles, NXL101.
Single-step mutants.
The NXL101-resistant mutants described above were generated in order to explore the nature of the changes to primary structure that result in resistant proteins, the extent of resistance that can be produced, and the relationship between NXL101 resistance and fluoroquinolone resistance. The experiments do not necessarily give an indication of the type of mutations that might arise spontaneously in a clinical setting. An experiment was performed with 50 S. aureus strains in order to isolate and characterize mutants spontaneously resistant to NXL101. Eight strains with various degrees of prior characterization were selected. The Bontemps strain was selected in order to extend the studies described above, and seven strains for which the genome sequence has been determined, and for which the antibiotic sensitivity profile is known, were selected (8325, COL, MSSA476, MRSA252, USA400, MW2, N315, and Mu50). A further 42 clinical isolates were selected which collectively displayed a wide range of quinolone and fluoroquinolone sensitivities. Mutations were selected by plating cultures onto antibiotic gradient plates, and cultures displaying a MIC of ≥2 mg liter−1 were considered to be NXL101 resistant. Resistant colonies were selected and subjected to MIC testing, as were their corresponding WT progenitors, against NXL101 and the quinolones moxifloxacin, sparfloxacin, levofloxacin, ciprofloxacin, and norfloxacin. In total, 24 of the 50 strains yielded resistant strains by this criterion, including all 8 strains of the characterized set. The eQRDRs of gyrA, gyrB, grlA and grlB from all 24 mutant strains and all 24 corresponding progenitor strains were sequenced.
The 10 progenitor strains which were fluoroquinolone resistant all possessed mutations at Ser80 of GrlA, of which 8 strains also possessed mutations in Ser84 of GyrA (Table 4). These are the point mutation sites most frequently associated with the appearance of fluoroquinolone insensitivity in topoisomerase IV and gyrase (30). It is clear that extant resistance to fluoroquinolones mediated by mutations in gyrA did not prevent the appearance of NXL101 resistance mediated by mutations in the same structural gene. Considering the mutations associated with NXL101 resistance, with only one exception (strain N315, GyrB mutation of Asp437Val) all gyrA eQRDR sequences contained a mutation. There were no mutations identified in the topoisomerase IV subunits, implying that gyrase is the primary target of NXL101 in S. aureus. Of the 23 mutant GyrA proteins, 16 were Asp83Asn, 3 were Asp83Gly, 2 were Met121Lys, 1 was Ala94Pro, and 1 was Val45Ala. The GyrA mutation V45A and GyrB mutation D437V were associated with MICs that increased only to ∼2 mg liter−1, the Ala94Pro mutation was associated with those that increased to 16 to 32 mg liter−1, and the remainder were associated with generally more substantial increases of up to 64 mg liter −1. It is noteworthy that neither of the two occurrences of mutation of Met121 occurred in strains resistant to fluoroquinolones. The MICs of none of these mutants were reduced by coincubation with reserpine at 20 mg liter−1.
TABLE 4.
Amino acid changes resulting from mutations in gyrA and grlA eQRDR coding sequence and the associated MICs of NXL101 and various fluoroquinolones against various S. aureus strains before and after selection for NXL101 resistance
| WT or mutant straina | MIC (mg liter−1)b
|
Mutation in QRDR
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Fluoroquinolone resistance (WT strains)
|
NXL101 resistance (GyrA [mutant strains])c | ||||||||
| NXL101 | NOR | CIP | LVX | SPX | MFX | GyrA | GrlA | ||
| Mu50 | |||||||||
| WT | 0.25 | 128 | 32 | 16 | 32 | 8 | S84L | S80F | |
| Mutant | 64 | 128 | 32 | 32 | 64 | 16 | D83N | ||
| N315 | |||||||||
| WT | 0.25 | 8 | 2 | 0.5 | 0.125 | 0.125 | |||
| Mutant | 2 | 2 | 0.5 | 0.25 | 0.125 | 0.125 | —d | ||
| MRSA252 | |||||||||
| WT | 0.125 | 128 | 128 | 16 | 16 | 4 | S84L | S80F | |
| Mutant | 2 | 128 | 128 | 16 | 8 | 4 | V45A | ||
| MSSA476 | |||||||||
| WT | 0.5 | 2 | 1 | 0.5 | 0.125 | 0.125 | |||
| Mutant | 64 | 1 | 0.5 | 0.25 | 0.125 | 0.125 | D83N | ||
| NCTC8325 | |||||||||
| WT | 0.25 | 0.5 | 0.5 | 0.25 | 0.25 | 0.25 | |||
| Mutant | 64 | 0.25 | 0.25 | 0.125 | 0.125 | 0.125 | M121K | ||
| COL | |||||||||
| WT | 0.25 | 2 | 0.5 | 0.5 | 0.25 | 0.125 | |||
| Mutant | 32 | 1 | 0.5 | 0.25 | 0.25 | 0.25 | D83N | ||
| MW2 | |||||||||
| WT | 0.25 | 2 | 1 | 0.25 | 0.125 | 0.125 | |||
| Mutant | 64 | 1 | 1 | 0.25 | 0.125 | 0.125 | M121K | ||
| Bontemps | |||||||||
| WT | 2 | 32 | 4 | 1 | 0.25 | 0.25 | S80F | ||
| Mutant | 32 | 32 | 4 | 2 | 0.5 | 1 | D83G S84T R92C | ||
| 1 | |||||||||
| WT | 0.25 | 32 | 16 | 1 | 0.5 | 0.25 | I45M S80F | ||
| Mutant | 16 | 32 | 8 | 2 | 0.5 | 0.5 | D83G | ||
| 2 | |||||||||
| WT | 0.5 | 4 | 1 | 0.5 | 0.25 | 0.125 | |||
| Mutant | 64 | 4 | 1 | 0.5 | 0.25 | 0.25 | D83N | ||
| 3 | |||||||||
| WT | 0.125 | 2 | 0.5 | 0.25 | 0.125 | 0.125 | |||
| Mutant | 64 | 2 | 1 | 0.25 | 0.25 | 0.125 | D83N | ||
| 4 | |||||||||
| WT | 0.125 | 128 | 64 | 32 | 32 | 8 | S84V | S80F | |
| Mutant | 32 | 128 | 128 | 128 | 128 | 16 | D83N | ||
| 5 | |||||||||
| WT | 0.125 | 2 | 0.5 | 0.25 | 0.125 | 0.125 | |||
| Mutant | 64 | 1 | 1 | 0.125 | 0.125 | 0.125 | D83N | ||
| 6 | |||||||||
| WT | 0.25 | 2 | 0.5 | 0.25 | 0.125 | 0.125 | |||
| Mutant | 32 | 1 | 0.5 | 0.125 | 0.125 | 0.125 | D83N | ||
| 7 | |||||||||
| WT | 0.25 | 64 | 16 | 8 | 16 | 2 | S84L | S80Y | |
| Mutant | 32 | 16 | 16 | 2 | 4 | 2 | D83N | ||
| 8 | |||||||||
| WT | 0.25 | 128 | 32 | 16 | 32 | 4 | M18I S84 L | S80Y | |
| Mutant | 16 | 32 | 32 | 4 | 8 | 2 | D83N | ||
| 9 | |||||||||
| WT | 0.25 | 64 | 32 | 8 | 8 | 2 | S84L | S80Y | |
| Mutant | 32 | 32 | 32 | 4 | 4 | 2 | D83N | ||
| 10 | |||||||||
| WT | 0.125 | 128 | 128 | 32 | 32 | 4 | S84V | S80F | |
| Mutant | 16 | 128 | 128 | 16 | 16 | 8 | D83N | ||
| 11 | |||||||||
| WT | 0.25 | 2 | 1 | 0.5 | 0.25 | 0.125 | |||
| Mutant | 16 | 2 | 2 | 1 | 1 | 1 | D83N | ||
| 12 | |||||||||
| WT | 0.125 | 2 | 0.5 | 0.25 | 0.125 | 0.125 | |||
| Mutant | 16 | 1 | 1 | 0.125 | 0.125 | 0.125 | D83G | ||
| 13 | |||||||||
| WT | 0.125 | 16 | 8 | 1 | 1 | 0.5 | I45M S80F | ||
| Mutant | 16 | 16 | 16 | 1 | 1 | 1 | D83N | ||
| 14 | |||||||||
| WT | 0.125 | 128 | 128 | 32 | 32 | 16 | S84V | S80F | |
| Mutant | 16 | 128 | 128 | 16 | 16 | 16 | D83N | ||
| 15 | |||||||||
| WT | 0.5 | 1 | 0.25 | 0.25 | 0.125 | 0.125 | |||
| Mutant | 16 | 0.5 | 0.13 | 0.125 | 0.125 | 0.125 | A94P | ||
| 16 | |||||||||
| WT | 0.25 | 2 | 0.5 | 0.5 | 0.125 | 0.125 | |||
| Mutant | 64 | 2 | 0.5 | 0.5 | 0.25 | 0.125 | D83N | ||
Mutant strains were isolated by selection on plates containing NXL101.
NOR, norfloxacin; CIP, ciprofloxacin; LVX, levofloxacin; SPX, sparfloxacin; MFX, moxifloxacin.
There were no NXL101 resistance GrlA mutations.
-, a mutation of Asp437 to Val was seen in the GyrB subunit of the NXL101-resistant variant of S. aureus N315.
Inhibition of mutant enzymes.
Three mutant forms of S. aureus GyrA were constructed by site-directed mutagenesis of the gyrA gene. A “classical” fluoroquinolone-insensitive GyrA double mutant (Ser84Leu Glu88Lys) was constructed, and in addition two putatively NXL101-resistant mutants (Asp83Asn and Met121Lys) were constructed. The mutant forms were overexpressed in E. coli, purified, and combined with purified WT GyrB subunit to produce the three mutant DNA gyrases. Each gyrase was tested for inhibition of supercoiling and of cleaved-complex stabilization by moxifloxacin, ciprofloxacin, and NXL101 (Table 5). The IC50 values of both ciprofloxacin and moxifloxacin for inhibition of supercoiling activity of the fluoroquinolone-resistant variant (Ser84Leu Glu88Lys) were increased substantially from 27 μΜ and 17 μΜ, respectively, to over 200 μΜ, whereas sensitivity to NXL101 was unchanged compared to that of WT gyrase. The sensitivity of the two Asp83Asn and Met121Lys mutants to NXL101 was diminished by over 200-fold in each case. IC50 values of both ciprofloxacin and moxifloxacin were increased approximately fourfold against the Met121Lys variant and approximately threefold for moxifloxacin against the Asp83Asn variant. A more substantial decrease in sensitivity of the Asp83Asn variant was seen with ciprofloxacin, producing an IC50 value in excess of 200 μΜ. Data derived from measurement of cleaved-complex stabilization were consistent with this pattern (Table 5). Note that the inability of NXL101 to stabilize the cleaved complex with WT gyrase extends to all three mutant forms.
TABLE 5.
IC50 and CC50 values of moxifloxacin, ciprofloxacin, and NXL101 against the gyrase supercoiling reaction and against cleaved-complex stabilization of WT and mutant S. aureus DNA gyrases
| Antibiotic | IC50 or CC50 (μM) against gyrasea:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WT
|
Mutant
|
|||||||
| S84L E88K
|
M121K
|
D83N
|
||||||
| IC50 | CC50 | IC50 | CC50 | IC50 | CC50 | IC50 | CC50 | |
| Ciprofloxacin | 27 | 11.2 | >200 | >200 | 94 | 24 | >200 | 50 |
| Moxifloxacin | 17 | 2.5 | >200 | >200 | 58 | 5 | 44 | 19 |
| NXL101 | 1.0 | >200 | 0.8 | >200 | >200 | >200 | >200 | >200 |
Shown are IC50 and CC50 values of moxifloxacin, ciprofloxacin, and NXL101 against the gyrase supercoiling reaction (IC50) and against cleaved-complex stabilization (CC50) of WT and mutant forms of S. aureus DNA gyrase.
Several strains possessing a common mutation in the GrlA (ParC) subunit of topoisomerase IV that is associated with fluoroquinolone resistance remain susceptible to NXL101 (Table 4). Although the data presented here strongly implicate gyrase as the principal target of NXL101, it was considered of interest to characterize the effect of this common mutation on NXL101. A mutant GrlA protein, Ser80Phe, was therefore constructed by site-directed mutagenesis, overexpressed, and purified. Decatenation assays were performed on both WT and mutant reconstituted topoisomerase IV enzymes, and inhibition with ciprofloxacin, moxifloxacin, and NXL101 was measured. The IC50 values of decatenation inhibition for moxifloxacin and ciprofloxacin increased ∼50-fold and ∼70-fold, respectively, to around 450 μΜ and 200 μΜ. Although the decrease in potency of inhibition by NXL101 was diminished by the proportionally smaller amount of ∼10-fold, the IC50 value was increased from 20 μΜ to approximately 200 μΜ. Therefore, although NXL101 retains full activity against the commonly encountered Ser84 mutations in GyrA that give rise to fluoroquinolone resistance, its activity against topoisomerase IV mutated at the equivalent position appears to be significantly diminished. This effect is consistent with the proposal that the impact of NXL101 on topoisomerase IV activity in situ in S. aureus is limited as compared with that of the fluoroquinolones.
DISCUSSION
The data reported herein demonstrate that NXL101, a novel antibiotic with excellent activity against gram-positive bacteria, has potent in vitro activity against purified type II topoisomerases. Although NXL101 is active against topoisomerases from E. coli in vitro, there is no significant antibacterial activity except against genetically uncharacterized laboratory hypersusceptible strains (data not shown). A striking characteristic of NXL101 relating to inhibition of the S. aureus topoisomerases is the markedly improved activity against gyrase supercoiling and relaxation reactions compared with those of the fluoroquinolones. This is in stark contrast to the situation with purified E. coli gyrase, where NXL101 activity was significantly poorer than those of ciprofloxacin and moxifloxacin.
Against S. aureus topoisomerase IV-catalyzed relaxation activity, NXL101 has an effect equivalent to (ciprofloxacin) or better than (moxifloxacin) those of the fluoroquinolones, but perhaps surprisingly in light of these data, the activity against decatenation of kinetoplast DNA by S. aureus topoisomerase IV is notably poorer than those of the fluoroquinolones. A similar observation, though quantitatively less extreme, can be made regarding E. coli topoisomerase IV. With respect to relative gyrase and topoisomerase IV activity, NXL101 has aspects of a fluoroquinolone antibiotic profile against gram-positive bacteria that has been pursued for many years, as the limited utility of fluoroquinolones against such bacteria has long been considered to be due to the relatively poor activity against gyrase (32). However, due to its apparently limited activity against topoisomerase IV in situ (see below), NXL101 is a less than optimal antibiotic when considering the proposed advantageous qualities of equal activity against both enzymes.
Perhaps the most marked difference between NXL101 and fluoroquinolones relates to the effect on cleaved-complex stabilization. As the effects of fluoroquinolones and NXL101 on the reactions described above are qualitatively, and sometimes quantitatively, very similar, it is notable that, with the exception of E. coli topoisomerase IV, no evidence for the stabilization of the cleaved complex by NXL101 is apparent under the experimental conditions employed here, even up to 200 μΜ. In the case of ciprofloxacin and moxifloxacin, cleaved-complex formation is apparent with a CC50 value similar to, or lower than, the IC50 for all other measured reactions. The exception of E. coli topoisomerase IV is remarkable in terms of magnitude as the CC50 value of NXL101 is 50 nM, being at least 10 times more potent in stabilizing the cleaved complex than ciprofloxacin or moxifloxacin against any of the four enzymes. The reasons for failure to stabilize the cleaved complex in three cases, and for the pronounced ability to do so in the case of E. coli topoisomerase IV, are not immediately obvious. Although the absence of complex formation with fluoroquinolones acting on S. aureus gyrase has been reported (5), it may be a unique observation as it is not the usual case (2), nor is it the case with enzymes from S. pneumoniae (36). The inability of NXL101 to stabilize the cleaved complex in vitro may be an artifact of the in vitro assay conditions that may vary with physical conditions (pH, ionic strength, etc.) or may indicate genuine differences in mechanisms. In the case of gyrases and S. aureus topoisomerase IV, NXL101 is clearly an effective inhibitor of enzyme function in vitro, as are the fluoroquinolones. It has been proposed that bacterial killing is the result of the (reversible) formation of cleaved complexes followed by chromosome fragmentation that may, or may not, depend on protein synthesis, depending upon the fluoroquinolone in question (10, 25). If the in vitro results reflect the effect of NXL101 on gyrase in situ, then NXL101 must differ in this respect as NXL101 is rapidly cidal against S. aureus, killing 99.9% in ∼6 h (Borgonovi et al., presented at the 16th European Congress of Clinical Microbiology and Infectious Diseases, Nice, France, 1 to 4 April 2006 [poster P-1567]), without any evidence for cleaved-complex formation. The stabilization of the cleaved complex mediated by fluoroquinolones occurs due to the trapping of a covalent adduct of DNA to the enzyme via the active-site tyrosine residue (17). If NXL101 were to bind in such a way that generation of the tyrosine phenolate anion is prevented, or in such a way that an attack by this species on the phosphodiester bond of the DNA is prevented, then the enzymes would be effectively inhibited without formation of the cleaved complex.
Whatever the differences in the molecular mechanism of NXL101 might be, the most prominent characteristic that differentiates NXL101 from the fluoroquinolones is the capacity to inhibit mutant forms of gyrase that are resistant to inhibition by fluoroquinolones.
The most frequently found mutation in the GyrA subunit of gyrase is a change of Ser84, often accompanied by an additional mutation of Glu88. NXL101 retains antibacterial activity against fluoroquinolone-resistant S. aureus and S. pneumoniae (23; M. Borgonovi et al., presented at the 16th European Congress of Clinical Microbiology and Infectious Diseases, Nice, France, 1 to 4 April 2006 [poster P-1567]) (Table 4), and so it would be unsurprising if NXL101 were to retain activity against purified fluoroquinolone-resistant gyrase in vitro. Full activity against DNA supercoiling catalyzed by a double-mutant fluoroquinolone-resistant gyrase was indeed retained and is consistent with antibacterial tests which indicate MICs as low as 0.125 mg liter−1 even in cases where MICs of ciprofloxacin and moxifloxacin are as high as 128 mg liter−1 and 16 mg liter−1, respectively (Table 4).
Although NXL101 is fully active against all of the fluoroquinolone-resistant S. aureus strains tested, it is possible to isolate mutants resistant to NXL101, and seemingly only a single base change to the gyrA gene is required in order to produce this phenotype. Three infrequently occurring mutants were characterized that afford MIC increases of 16-fold (Val45Ala), 32-fold (Arg92Cys and Ala94Pro), and 128-fold (His81Asn). Although Val45 (position 44 in E. coli GyrA) is outside of the QRDR, a mutation of E. coli GyrA (Ala51Val) that is also outside of the QRDR and which confers a small increase in MIC for ciprofloxacin and gatifloxacin has been noted (13), although the equivalent amino acid in S. aureus is a glycine. The mutation Val45Ala occurred in a fluoroquinolone-resistant strain and did not further increase MICs of fluoroquinolones (Table 4). The remaining infrequent mutants were all within the QRDR. Arg92 and Ala94 are equivalent to Arg91 and Ala 93 in E. coli GyrA, which are found at the extreme C-terminal end of helix 4 (the helix containing Ser83 and Asp87) in the crystal structure of the 59-kDa N-terminal breakage-reunion domain (26). These mutations are also without effect on quinolone sensitivities. The His81Asn mutation is the most significant of the infrequent mutants as an increase in MIC for NXL101 from 0.125 to 16 mg liter−1 was observed, although no increase in MIC of ciprofloxacin was apparent (Table 3). The equivalent residue in E. coli GyrA (His80) has been implicated in the breakage-reunion reaction (15). In vitro assays with mutant enzyme indicated that although neither DNA binding affinity nor relaxation rate was reduced, the DNA supercoiling rate was reduced to 6% of that of the WT. However, in S. aureus at least, the mutation does not preclude cell viability. The most frequent spontaneous mutations were those which substituted either glycine or asparagine for aspartate at position 83 in GyrA. The mutant protein seems to confer on the bacterium a significant resistance to NXL101, as does the second most common change, the substitution of lysine for methionine at position 121 of GyrA. Neither mutation causes any perceptible change to antimicrobial fluoroquinolone sensitivities. A mutation of Asp83 to Ala in S. aureus GyrA has been previously documented and linked to sparfloxacin resistance, but not to ciprofloxacin resistance (28), but the sparfloxacin resistance may be linked to a small-colony phenotype transition indirectly associated with the Asp83Ala mutant. The data in Table 4 indicate there is no such reduced susceptibility to sparfloxacin in the case of mutation of Asp83 to either glycine or asparagine. A similar mutation in GyrA from E. coli (Asp82 in E. coli) has been reported (35) in the context of fluoroquinolone resistance, but the E. coli host remained susceptible to nalidixic acid and the MIC for ciprofloxacin rose only to 1 mg liter−1. As the 59-kDa N-terminal E. coli GyrA fragment shares 61% sequence identity and 73% similarity with the S. aureus ortholog, it is reasonable to predict a similar three-dimensional structure. All of the amino acids that were found to mutate during this study are conserved between the two proteins. Figure 5 shows the DNA-binding “gate” region of the E. coli GyrA subunit (26), with the mutated residues and active-site tyrosine displayed. Referring to the S. aureus GyrA numbering system, Asp83 can be seen protruding from the opposite side of helix 4 from Ser84 and Glu88 and the Arg92 and Ala94 residues at the opposite end of helix 4 relative to Asp83. Interestingly, Met121 is spatially very close to Asp83, with the methionyl ɛ carbon at 3.4 Å from the δ2 oxygen of the Asp83 carboxyl group (Fig. 5). A substitution of lysine for Met121 would involve replacement of a long unbranched side chain for another, but extended by one methyl group and, importantly, with introduction of a positive charge at the ζ nitrogen probably even closer to the δ2 oxygen of the Asp83 carboxyl group. This would certainly have a significant effect on the properties of Asp83 by altering the pKa of the carboxylate and/or by causing loss of conformational freedom. Thus, there is a distinct possibility that the resistance to inhibition by NXL101 may not be due to loss of function of Met121 but to loss of function of Asp83 mediated by the introduction of Lys121. It is noteworthy that mutations of methionine mediated by a single base change can in principle result in Ile, Thr, Arg, Leu, and Val as alternatives to Lys, but such substitutions for Met121 did not occur in the three independent S. aureus mutant strains.
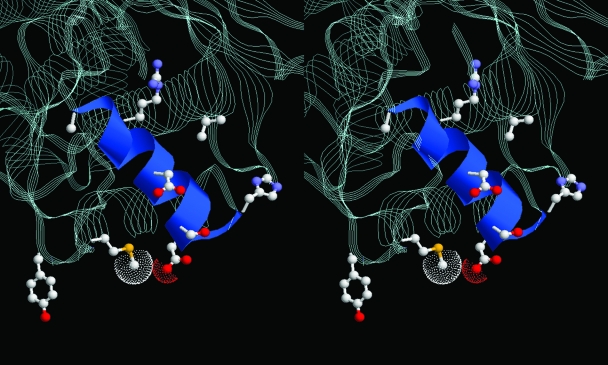
FIG. 5.
Stereo representation of the gate region of E. coli DNA gyrase breakage-reunion domain (RCSB Protein Data Bank accession no. 1AB4) with helix 4 shown in ribbon format. The catalytic tyrosine and the residues that are mutated to effect NXL101 or fluoroquinolone resistance are rendered in ball-and-stick mode. In S. aureus numbering, these are Val45, His80, Asp83, Arg92, Ala94, and Met121 (NXL101 resistance) and Ser84 and Asp/Glu88 (fluoroquinolone resistance). The terminal atoms of the Met121 and Asp83 side chains show van der Waals spheres.
The mechanisms of action of the fluoroquinolones are still far from completely understood even decades after their introduction into clinical practice, not least because of the absence of structural information on the tripartite complex, or on the covalent complex, although structures of several GyrA and ParC subunits, or parts thereof, are available (8, 9, 21, 26). Despite the lack of structural information, some knowledge concerning protein function has been gleaned from studies on interactions with quinolones of diverse structures. It is known that helix 4 of GyrA carries the Ser84 and Glu88 residues important for DNA binding and for fluoroquinolone resistance (26, 32). The principal point mutation in GyrA resulting in NXL101 resistance, Asp83, is located at the extreme end of helix 4, and Met121, the residue whose mutation to lysine results in such a large increase in MIC for NXL101, is located very close to Asp83 in the three-dimensional structure and two residues before the catalytic tyrosine in primary structure. The three-dimensional structure of the breakage-reunion domain of DNA gyrase (26) confirms a close spatial proximity of the NXL101 resistance residues with those associated with fluoroquinolone resistance and in particular emphasizes the prominence of helix 4 in considerations of mechanisms of action of, and resistance to, antibacterial drugs which inhibit DNA gyrase activity, including nonquinolones.
Acknowledgments
We thank David Hooper for supplying plasmids pSAGA3.1 and pTrcHis-SAGyrB.
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Alovero, F. L., X.-S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, V. E., R. P. Zaniewski, F. S. Kaczmarek, T. D. Gootz, and N. Osheroff. 2000. Action of quinolones against Staphylococcus aureus topoisomerase IV: basis for DNA cleavage enhancement. Biochemistry 39:2726-2732. [DOI] [PubMed] [Google Scholar]
- 3.Bates, A. D., and A. Maxwell. 2005. DNA topology. Oxford University Press, Oxford, United Kingdom.
- 4.Bates, A. D., and A. Maxwell. 2007. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry 46:7929-7941. [DOI] [PubMed] [Google Scholar]
- 5.Blanche, F., B. Cameron, F. X. Bernard, L. Maton, B. Manse, L. Ferrero, N. Ratet, C. Lecoq, A. Goniot, D. Bisch, and J. Crouzet. 1996. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob. Agents Chemother. 40:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 7.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 8.Corbett, K. D., A. J. Schoeffler, N. D. Thomsen, and J. M. Berger. 2005. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351:545-561. [DOI] [PubMed] [Google Scholar]
- 9.Corbett, K. D., R. K. Shultzaberger, and J. M. Berger. 2004. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl. Acad. Sci. USA 101:7293-7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica, K., M. Malik, R. J. Kerns, and X. Zhao. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, L. M., K. A. Gould, X.-S. Pan, S. Patel, and V. J. Heaton. 2003. Analysis of dual active fluoroquinolones in Streptococcus pneumoniae. J. Antimicrob. Chemother. 52:312-313. [DOI] [PubMed] [Google Scholar]
- 12.Forterre, P., S. Gribaldo, D. Gadelle, and M. C. Serre. 2007. Origin and evolution of DNA topoisomerases. Biochimie 89:427-446. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, S. M., T. Lu, and K. Drlica. 2001. Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother. 45:2378-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heaton, V. J., J. E. Ambler, and L. M. Fisher. 2000. Potent antipneumococcal activity of gemifloxacin is associated with dual targeting of gyrase and topoisomerase IV, an in vivo target preference for gyrase, and enhanced stabilization of cleavable complexes in vitro. Antimicrob. Agents. Chemother. 44:3112-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hockings, S. C., and A. Maxwell. 2002. Identification of four GyrA residues involved in the DNA breakage-reunion reaction of DNA gyrase. J. Mol. Biol. 318:351-359. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, D. C. 2001. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 32(Suppl. 1):S9-S15. [DOI] [PubMed] [Google Scholar]
- 17.Horowitz, D. S., and J. C. Wang. 1987. Mapping the active site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 262:5339-5344. [PubMed] [Google Scholar]
- 18.Ince, D., X. Zhang, and D. C. Hooper. 2003. Activity of and resistance to moxifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2003. Topoisomerase targeting with and resistance to gemifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:274-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2002. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of garenoxacin (BMS-284756, T-3811ME), a new desfluoroquinolone. Antimicrob. Agents Chemother. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laponogov, I., D. A. Veselkov, M. K. Sohi, X.-S. Pan, A. Achari, C. Yang, J. D. Ferrara, L. M. Fisher, and M. R. Sanderson. 2007. Breakage-reunion domain of Streptococcus pneumoniae topoisomerase IV: crystal structure of a gram-positive quinolone target. PLoS ONE 2:e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levasseur, P., A.-M. Girard, and J. Lowther. 2007. Efficacy of oral NXL101, a novel topoisomerase inhibitor, against fluoroquinolone-susceptible (FQS) and -resistant (FQR) Streptococcus pneumoniae (Sp) strains in a mouse pneumonia model, abstr. F1-2125, p. 260. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother.
- 23.Levasseur, P., C. Delachaume, J. Lowther, and J. Hodgson. 2005. Minimum inhibitory concentrations (MIC) and mutation prevention concentrations (MPC) of NXL101, a novel topoisomerase IV inhibitor, against Staphylococcus aureus including multi-resistant strains, abstr. F-507, p. 184. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother.
- 24.Lowther, J., P. Levasseur, A.-M. Girard, C. Delachaume, M. Borgonovi, and J. Hodgson. 2006. Efficacy of NXL101, a novel topoisomerase inhibitor, against multi-resistant Staphylococcus aureus in murine septicaemia and thigh muscle infections, abstr. F1-1999, p. 239. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother.
- 25.Malik, M., X. Zhao, and K. Drlica. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61:810-825. [DOI] [PubMed] [Google Scholar]
- 26.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 28.Pan, X.-S., P. J. Hamlyn, R. Talens-Visconti, F. L. Alovero, R. H. Manzo, and L. M. Fisher. 2002. Small-colony mutants of Staphylococcus aureus allow selection of gyrase-mediated resistance to dual-target fluoroquinolones. Antimicrob. Agents Chemother. 46:2498-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova, E., J. J. Millichap, G. A. Noskin, and L. R. Peterson. 2000. Intracellular targets of moxifloxacin: a comparison with other fluoroquinolones. J. Antimicrob. Chemother. 45:583-590. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz, F.-J., B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, M. Klootwijk, J. Verhoef, A. Fluit, H.-P. Heinz, K. Kohrer, and M. E. Jones. 1998. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 41:481-484. [DOI] [PubMed] [Google Scholar]
- 31.Sheehan, G., and N. S. Y. Chew. 2003. The history of quinolones, p. 1-10. In A. R. Ronald and D. E. Low (ed.), Fluoroquinolone antibiotics. Birkhäuser Verlag, Basel, Switzerland.
- 32.Strahilevitz, J., A. Robicsek, and D. C. Hooper. 2006. Role of the extended α4 domain of Staphylococcus aureus gyrase A protein in determining low sensitivity to quinolones. Antimicrob. Agents Chemother. 50:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strahilevitz, J., Y. Onodera, and D. C. Hooper. 2006. An improved expression plasmid for affinity purification of Staphylococcus aureus gyrase A subunit. Protein Expr. Purif. 47:10-15. [DOI] [PubMed] [Google Scholar]
- 34.Tarral, A., M. Rangaraju, H. Merdjan, A.-M. Girard, C. Delachaume, and J. Lowther. 2007. Plasma bactericidal activity of NXL101 in healthy volunteers after single intravenous administrations of 50 mg to 700 mg, abstr. A-808, p. 25. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother.
- 35.Truong, Q. C., J.-C. van Nguyen, D. Shlaes, L. Gutmann, and N. J. Moreau. 1997. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob. Agents Chemother. 41:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yague, G., J. E. Morris, X.-S. Pan, K. A. Gould, and L. M. Fisher. 2002. Cleavable-complex formation by wild-type and quinolone-resistant Streptococcus pneumoniae type II topoisomerases mediated by gemifloxacin and other fluoroquinolones. Antimicrob. Agents Chemother. 46:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]