Abstract
Chronic hepatitis C virus (HCV) infection remains a major global health burden while current interferon-based therapy is suboptimal. Efforts to develop more effective antiviral agents mainly focus on two viral targets: NS3-4A protease and NS5B polymerase. However, resistant mutants against these viral specific inhibitors emerge quickly both in vitro and in patients, particularly in the case of monotherapy. An alternative and complementary strategy is to target host factors such as cyclophilins that are also essential for viral replication. Future HCV therapies will most likely be combinations of multiple drugs of different mechanisms to maximize antiviral activity and to suppress the emergence of resistance. Here, the effects of combining a host cyclophilin inhibitor NIM811 with other viral specific inhibitors were investigated in vitro using HCV replicon. All of the combinations led to more pronounced antiviral effects than any single agent, with no significant increase of cytotoxicity. Moreover, the combination of NIM811 with a nucleoside (NM107) or a non-nucleoside (thiophene-2-carboxylic acid) polymerase inhibitor was synergistic, while the combination with a protease inhibitor (BILN2061) was additive. Resistant clones were selected in vitro with these inhibitors. Interestingly, it was much more difficult to develop resistance against NIM811 than viral specific inhibitors. No cross-resistance was observed among these inhibitors. Most notably, NIM811 was highly effective in blocking the emergence of resistance when used in combination with viral protease or polymerase inhibitors. Taken together, these results illustrate the significant advantages of combining inhibitors targeting both viral and host factors as key components of future HCV therapies.
Hepatitis C virus (HCV) infection presents a significant global health challenge with approximately 170 million people or 3% of the world population chronically infected and an additional 3 to 4 million more people infected each year (according to World Health Organization estimates). Although only 25% of new infections are symptomatic, 60 to 80% of patients develop chronic liver disease, of whom an estimated 20% progress to cirrhosis with a 1 to 4% annual risk of developing hepatocellular carcinoma (19). Overall, HCV is responsible for 50 to 76% of all liver cancer cases and two-thirds of all liver transplants in developed countries. Ultimately, 5 to 7% of infected patients will die from the consequences of HCV infection (according to World Health Organization estimates).
The current standard therapy for HCV infection is pegylated alpha interferon (IFN-α) in combination with ribavirin. However, fewer than 50% of patients with genotype 1 virus, the predominant HCV genotype in developed countries, are successfully treated with IFN-based therapies. Moreover, both IFN and ribavirin induce significant adverse effects, including flu-like symptoms (fever and fatigue), hematologic complications (leukopenia, thrombocytopenia), and neuropsychiatric issues (depression, insomnia) associated with IFN and significant hemolytic anemia associated with ribavirin. Also, ribavirin is teratogenic and cannot be given to pregnant women. Therefore, the majority of HCV patients are not being treated with the current standard of care. More effective and better tolerated therapies are greatly needed.
HCV is a 9.6-kb positive-sense, single-stranded RNA virus. It encodes a large single open reading frame corresponding to a polyprotein precursor of about 3,000 amino acids, which is proteolytically processed by cellular signal peptidases and HCV-encoded proteases into at least 10 individual proteins, in the order of C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B. Drug discovery efforts for new antivirals have been mainly focusing on two viral proteins, the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase, both of which have enzymatic activities essential for viral replication. However, such approaches may not be sufficient given the high replication rate and high mutation rate of the virus, which can frequently produce resistant mutations in viral genomes, thereby compromising the effectiveness of viral specific inhibitors. An alternative and complementary strategy is to target host factors that are also required for viral replication. Cyclophilins, a family of cellular peptidyl-prolyl isomerases required for HCV replication, represent such an opportunity (6, 16, 25). Previously, we demonstrated that NIM811, a cyclosporine derivative that binds to cyclophilins with high affinity but lacks calcineurin-mediated immunosuppressive activity, has potent anti-HCV activities in vitro (14). This compound is currently in clinical development for hepatitis C treatment. Another nonimmunosuppressive cyclophilin inhibitor, DEBIO-025, also showed antiviral activity in vitro (18) and achieved proof-of-concept efficacy in HCV patients (5).
An often-hypothesized advantage of targeting host factors is that such inhibitors may be less prone to select for resistant mutations in the viral genome and may make for effective combinations with specific inhibitors of viral proteins. HCV has a low-fidelity polymerase that lacks proofreading function. As a result, there is a huge population of viral quasispecies preexisting in every infected patient, and mutants that confer resistance to antiviral agents have a growth advantage and can be rapidly selected and accumulate during antiviral treatment. The use of multiple antiviral agents in combination may help to suppress the emergence of resistant virus in two ways. First, combination therapies can result in a greater decrease in the viral load, thereby limiting the frequency with which mutations (that have a set probability of occurring) arise in the viral population. Using antiviral agents in combination also creates a higher genetic barrier to the development of resistance in that resistant viruses to a combination therapy likely require occurrence of multiple mutations. This is especially true when the combined antiviral agents have distinct mechanisms of action and thus different resistance profiles. In general, the suppression of resistance by combination therapy is of key importance for maintaining the utility of current effective antiviral agents for future generations. The additive or synergistic antiviral effects between antiviral agents may also permit a reduction in the dose or dosing frequency of individual agents, thereby minimizing potential toxicity and adverse effects associated with high doses of single agents. The success of combination therapy is best exemplified by the treatment of human immunodeficiency virus (HIV) infection, where cocktails of multiple drugs including nucleoside and non-nucleoside reverse transcriptase inhibitors and protease inhibitors are necessary to maintain the suppression of viral replication and emergence of resistance.
Considering the significant advantages of combination therapy over monotherapy, it is most likely that future HCV therapy will be a combination of multiple drugs of different mechanisms, similar to that of HIV. Since it is not feasible to study all of the possible combinations in the clinical setting given the number of investigational drugs being developed for HCV, it is of great value to examine the effect of potential drug-drug combinations in vitro. In the present study, we investigated the combinations of a host factor (cyclophilin) inhibitor, NIM811, with several other compounds representing the three main classes of virus specific inhibitors: BILN2061 (ciluprevir), the first NS3-4A protease inhibitor that showed proof-of-concept efficacy in HCV patients (8); a non-nucleoside NS5B polymerase inhibitor thiophene-2-carboxylic acid (2); and NM107, the active moiety of NM283 (valopicitabine), the first nucleoside NS5B inhibitor that showed clinical efficacy (20). All of the combinations examined led to substantially more potent antiviral effects than any single agent with no significant increase in cytotoxicity. Interestingly, the combinations of NIM811 with the nucleoside or non-nucleoside polymerase inhibitor were synergistic, while the combination with the protease inhibitor appeared to be additive. Resistant clones were obtained for all of the inhibitors, although it proved much more difficult to develop resistance against NIM811. Importantly, there was no cross-resistance observed among these inhibitors that act by different mechanisms. The frequency of resistance against NS3-4A protease or NS5B polymerase inhibitors was dramatically reduced when used in combination with NIM811. These results clearly highlight the potential value of antiviral agents targeting host factors such as cyclophilins, particularly in the context of combination therapies.
MATERIALS AND METHODS
Compounds.
NIM811, BILN2061, POL-1, and NM107 were prepared at Novartis. The compounds were stored at −20°C as 20 mM dimethyl sulfoxide (DMSO) stock solutions until being used in the assay. IFN-α was purchased from Calbiochem and stored at −80°C.
Cells.
Huh-luc/neo-ET, a subgenomic genotype 1b (Con 1) HCV replicon cell line containing a luciferase reporter gene, was kindly provided by Ralf Bartenschlager (11). The cells were cultured in Dulbecco modified Eagle medium, supplemented with 2 mM l-glutamine, 1× nonessential amino acids, 10% heat-inactivated fetal bovine serum (ΔFBS), and 0.25 mg of G418 (Invitrogen, Carlsbad, CA)/ml. Another subgenomic genotype 1b (Con 1) HCV replicon cell line, clone A, was provided by Charles Rice and Apath LLC (St. Louis, MO) (1) and was maintained in the same medium above except for 1 mg of G418/ml. G418 was included in the culture medium to provide the selection pressure for maintaining the level of HCV replicon in the cells since both HCV replicons encode neomycin phosphotransferase, which degrades G418.
Luciferase-based HCV replicon assay.
The antiviral activity and cytotoxicity of compounds were determined by using the HCV replicon cell line (Huh-Luc/neo-ET) that contains a luciferase reporter gene. Since the expression of the luciferase reporter is under the control of HCV RNA replication and the turnover of luciferase protein is rapid, the luciferase activity is representative of the amount of HCV RNA present in the cells. Prior to compound treatment, 5,000 replicon cells were seeded in each well of 96-well tissue culture plates and were allowed to attach in complete culture medium without G418 overnight. On the next day, the culture medium was replaced with medium containing serially diluted compounds in the presence of 10% ΔFBS and 0.5% DMSO. After 48 h of compound treatment, the remaining luciferase activity in the cells was determined by using BriteLite reagent (Perkin-Elmer, Wellesley, MA) with an LMaxII plate reader (Molecular Probes/Invitrogen). The percentage of inhibition was calculated as: % inhibition = 1 − (average of compound-treated cells)/(average of untreated control cells). To evaluate the potential cytotoxicity of compounds, the viability of the replicon cells after 48 h of compound treatment was determined by using a tetrazolium compound-based assay (CellTiter 96 AQueous One Solution cell proliferation assay; Promega, Madison, WI). The percentage of cytotoxicity was calculated as: % cytotoxicity = 1 − (average of compound-treated cells)/(average of untreated control cells).
Synergy analysis.
To determine whether the antiviral effect for the combination of two compounds was synergistic, additive, or antagonistic, a mathematical model based on the Bliss Independence theory, MacSynergy (21), was used to analyze the data obtained from the 48-h luciferase-based HCV replicon assay. In this model, a theoretical additive effect for any given concentrations of two compounds is calculated by using the equation: Z = X + Y(1 − X), where X and Y represent the inhibition produced by either compound alone, respectively, and Z represents the effect produced by the combination of two compounds if they were simply additive. For synergy analysis, the theoretical additive effects for various concentrations of the two compounds were compared to (subtracted from) the actual experimental effects and were plotted as a three-dimensional differential surface, which would appear as a horizontal plane at 0% if the combination were additive. Any peak above this plane (positive values) would indicate synergy, whereas any depression below it (negative values) would indicate antagonism. The 95% confidence intervals for the experimental dose-response values were analyzed to determine statistically significant effects.
Nine-day HCV RNA reduction assay.
HCV replicon cells (clone A) were seeded at a low density of 500 cells per well in 96-well plates such that the cells would not become overconfluent after nine continuous days in culture. The cells were treated with compounds serially diluted in Dulbecco modified Eagle medium containing 10% ΔFBS and 0.2% DMSO but no G418. The culture medium was replaced every 3 days with fresh medium containing compounds or 0.2% DMSO control. After the cells were treated for 3, 6, or 9 days, the total RNA was extracted using an RNeasy 96 kit (Qiagen, Valencia, CA). The level of HCV RNA was measured by a real-time quantitative reverse transcription-PCR (qRT-PCR) assay (TaqMan; Applied Biosystems, Foster City, CA) using HCV-specific primers (5′-TCT TCA CGC AGA AAG CGT CTA-3′ and 5′-CTG GCA ATT CCG GTG TAC T-3′) and probe (5′-6-FAM-TCC TGG AGG CTG CAC GAC ACT CAT A-TAMRA-3′). The absolute copy numbers of HCV RNA were determined by using a standard curve that was established with known quantities of in vitro-transcribed RNA. The level of HCV RNA was normalized for each sample against the amount of total RNA extracted, which was determined by using a Quant-iT RNA assay kit (Molecular Probes/Invitrogen). Each data point represents the average of five replicates in cell culture. The HCV RNA reduction after each period of treatment was calculated by comparing the remaining level of HCV RNA in compound-treated cells to that of control cells treated with 0.2% DMSO for the same duration.
Generation of resistant replicon cell lines.
Resistant HCV replicon clones against the NS3-4A protease inhibitor BILN2061, the non-nucleoside NS5B polymerase inhibitor POL-1, or the nucleoside NS5B polymerase inhibitor NM107 were selected by continuously culturing replicon cells (clone A) in the presence of 1 μM BILN2061, 1 μM POL-1, or 20 μM NM107, respectively, with 1 mg of G418/ml and 0.2% DMSO (to dissolve compounds) over a period of 1 to 2 months. It was much more difficult to develop resistance against NIM811, which took over 6 months, during which cells were cultured with stepwise increasing concentration of cyclosporine or NIM811. The final NIM811-resistant clone was selected and maintained in the presence of 5 μM NIM811 with 0.25 mg of G418/ml and 0.2% DMSO. No single colonies were picked in these selection processes. Cells were pooled together for each compound treatment every time during the passage. All resistant cell lines were passaged at least three times prior to compound susceptibility testing. To determine the fold change in 50% effective concentration (EC50) for each compound, a separate control (wild-type) replicon cell line was generated by continuous culture of the clone A cells in the presence of 0.2% DMSO for the same length of time as required to establish each of the resistant cell lines. All cultures were split by 1:4 to 1:8 ratios every 3 to 4 days or when the cell monolayer reached 90 to 95% confluence. For sequence analysis, total RNA was isolated from the selected resistant and control wild-type clone A cell lines by using an RNeasy minikit (Qiagen), and the HCV replicon RNA was amplified by using a High Fidelity Two-Step RT-PCR & Go kit (QBiogene).
qRT-PCR-based HCV replicon assay.
The antiviral activities of compounds against the resistant and control replicon cell lines were determined by a qRT-PCR-based assay. Briefly, 10,000 cells were seeded in each well of 96-well tissue culture plates and were allowed to attach in complete culture medium without G418 overnight. On the next day, the culture medium was replaced with medium containing serially diluted compounds in the presence of 2% ΔFBS and 0.5% DMSO, without G418. After a 48-h compound treatment, total RNA was extracted, and the level of HCV RNA was measured by real-time qRT-PCR as described above. Each data point represents the average of six replicates in cell culture. The magnitude of HCV RNA reduction for each treatment was calculated as the percentage of inhibition relative to DMSO only-treated control, where % inhibition = 1 − (average of compound-treated cells)/(average of control cells). To monitor cytotoxic effect of the compounds, the viability of the replicon cells following 48 h of compound treatment was determined by using a tetrazolium compound-based assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega). The percentage of cytotoxicity was calculated as: % cytotoxicity = 1 − (average of compound-treated cells)/(average of control cells).
Comparison of resistance frequency.
HCV replicon cells (clone A) were seeded at a low density (10% confluence) in six-well plates in complete culture medium containing 10% ΔFBS and 1 mg of G418/ml and allowed to attach overnight. The culture medium was then replaced with medium containing various concentrations of compounds alone or in combination in the presence of 1 mg of G418/ml, 0.2% DMSO, and 10% ΔFBS. The medium was changed every 3 days with fresh compounds or 0.2% DMSO (control). After 8 to 10 days in culture, the cells reached confluence and were split 1:10 once to fresh six-well plates. The cells were continuously cultured for an additional 3 weeks with medium refreshed every 3 days but without being split again, until macroscopic colonies of resistant cells formed. To enhance visualization of colonies, cells were washed with phosphate-buffered saline and fixed with 70% ethanol and 1% crystal violet for 30 min at room temperature. The fixative was then aspirated, and the cells were washed with water until the residual crystal violet was completely removed. Images of each well were captured by using the ChemiDocXRS system (Bio-Rad), and colonies were counted by using the Quantity One v4.4 software (Bio-Rad). For sequence analysis, the colonies were pooled together from each well of the plate, total RNA was isolated by using the RNeasy minikit, and HCV RNA was amplified by using the High Fidelity Two-Step RT-PCR & Go kit.
RESULTS
Combination of NIM811 with HCV protease or polymerase inhibitors resulted in enhanced antiviral activity without significant increase of cytotoxicity.
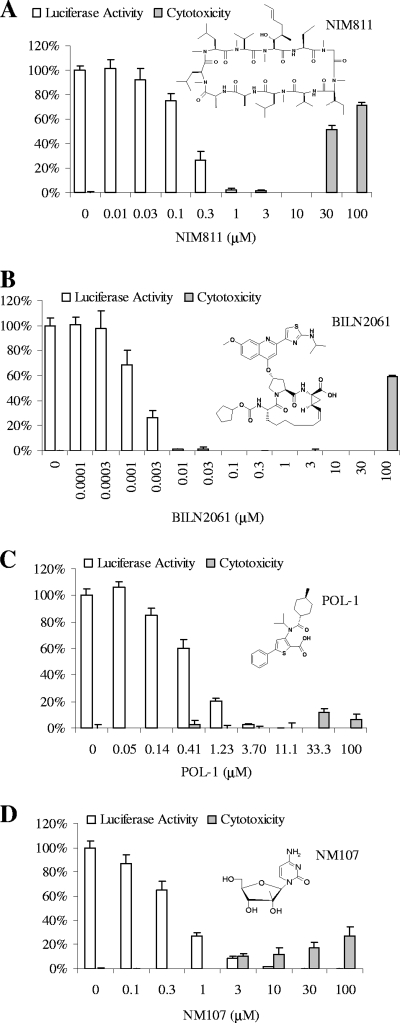
The combination of NIM811 with an HCV NS3-4A protease inhibitor BILN2061 (8), with a non-nucleoside NS5B polymerase inhibitor POL-1 (2), or with a nucleoside NS5B polymerase inhibitor NM107 (19) was investigated in the luciferase-based HCV replicon assay. First, the anti-HCV activity and cytotoxicity of each compound alone were determined in the assay. As shown in Fig. 1A, treatment of replicon cells with NIM811 for 48 h resulted in a concentration-dependent inhibition of HCV replication, as indicated by the reduction of luciferase reporter activity. The average EC50 of NIM811 from three independent experiments was determined to be 0.18 μM. No significant cytotoxicity was observed with NIM811 at concentrations below 30 μM, indicating the inhibition of HCV replicon was specific. Similar results were obtained for BILN2061 (Fig. 1B, EC50 = 2.2 nM, 50% cytotoxic concentration [CC50] > 30 μM), POL-1 (Fig. 1C, EC50 = 0.52 μM, CC50 > 100 μM), and NM107 (Fig. 1D, EC50 = 0.48 μM, CC50 > 100 μM).
FIG. 1.
Chemical structures of various HCV inhibitors and their activities in the 48-h replicon assay as single treatment. HCV replicon cells (Huh-luc/neo-ET) were incubated with various concentrations of NIM811 (A), an NS3-4A protease inhibitor BILN2061 (B), a non-nucleoside NS5B polymerase inhibitor POL-1 (C), or a nucleoside HCV NS5B polymerase inhibitor NM107 (D) for 48 h. To determine antiviral activity (white bars), the remaining luciferase activities in compound treated cells were measured and normalized against the luciferase activities for untreated controls. To monitor the cytotoxic effect (□), the viability of the replicon cells following compound treatment was determined by using a tetrazolium compound-based assay and was compared to that of untreated control cells. Each data point represents the average of four replicates in cell culture. EC50 and CC50 values were determined by four-parameter curve fitting and represent the average values from at least three independent assays.
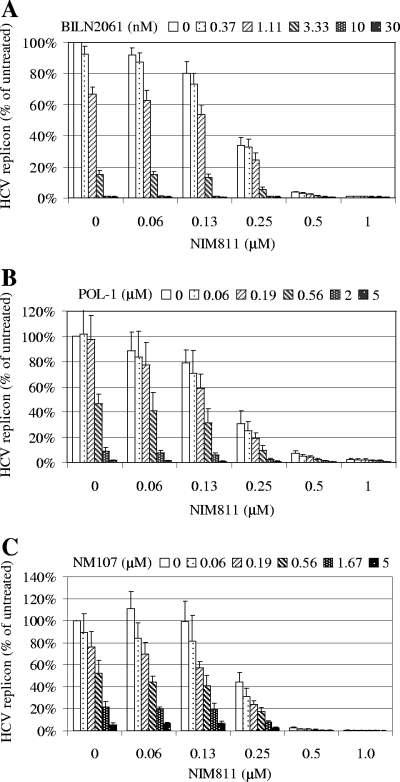
To evaluate the effect of various combinations, HCV replicon cells (Huh-Luc/neo-ET) were treated with five different concentrations of NIM811 in the absence or presence of five different concentrations of BILN2061, POL-1, or NM107 for 48 h. As shown in Fig. 2, there was a concentration-dependent inhibition of HCV replicon replication with these compounds either alone or in combination. Importantly, the combination of two compounds (NIM811 plus BILN2061, NIM811 plus POL-1, or NIM811 plus NM107) at various concentrations always resulted in a greater inhibition than either compound alone at the same concentration. Furthermore, no significant increase in cytotoxicity was observed for any of the combinations (not shown).
FIG. 2.
Combination of NIM811 with various HCV protease or polymerase inhibitors in the 48-h HCV replicon assay. HCV replicon cells (Huh-luc/neo-ET) were treated with various concentrations of NIM811 in combination with various concentrations of BILN2061 (A), POL-1 (B), or NM107 (C) for 48 h. Antiviral activities were determined by measuring the reduction of luciferase activities in the cells. The average percentages of inhibition for each compound treatment compared to untreated control are shown. Each data point represents the average of eight replicates in cell culture.
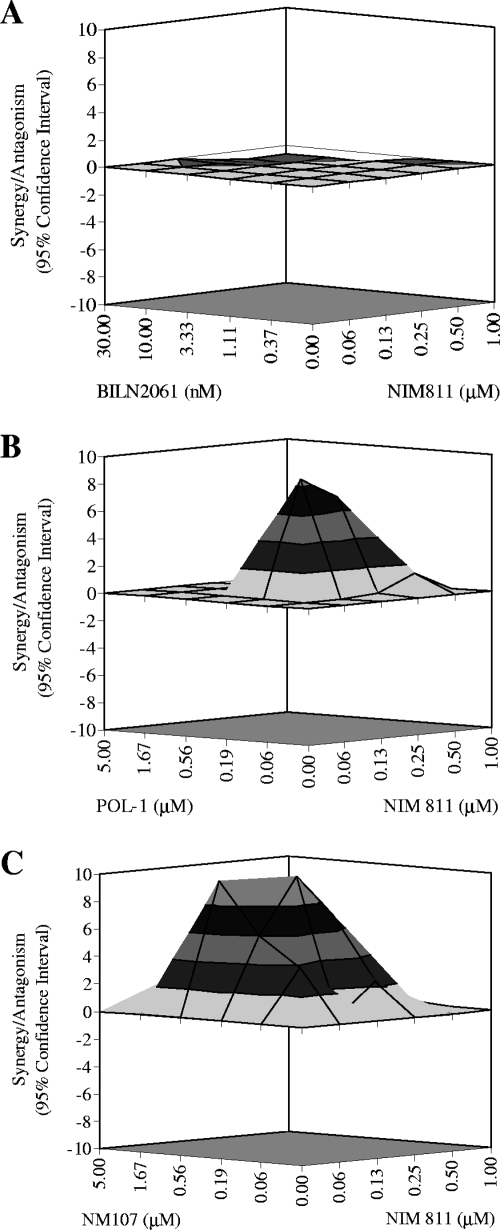
To determine whether the enhanced antiviral effects observed for NIM811 with inhibitors of viral protease or polymerase were synergistic, additive, or antagonistic, the data shown in Fig. 2 were further analyzed by using a mathematic model, MacSynergy (21), as described in Materials and Methods. In this model, the effect of combination was characterized by comparing the actual experimental results to theoretical additive effects, which were calculated from the dose-response curves for single compound treatments based on Bliss Independence theory. When presented as a three-dimensional differential surface plot, synergy would be observed as a convex surface, antagonism as a concave surface, and additive effect as a planar surface. Only statistically significant effects based on the 95% confidence interval were considered at any given concentrations of the two compounds. As shown in Fig. 3A, the combination of NIM811 with the HCV protease inhibitor BILN2061 resulted in antiviral effects that were not significantly different from the theoretical additive effects at the various concentrations of the two compounds tested, i.e., the effect of this combination was mainly additive. In contrast, as shown in Fig. 3B and C, the combinations of NIM811 with either a non-nucleoside (POL-1) or a nucleoside (NM107) inhibitor of HCV polymerase produced antiviral effects that were significantly stronger than theoretical additive effects, suggesting that these combinations were synergistic. The synergism observed between NIM811 and HCV polymerase inhibitors was reproducible in multiple independent experiments and was also confirmed with several other nucleoside and non-nucleosides polymerase inhibitors (not shown).
FIG. 3.
Synergy analysis for the combination of NIM811 with HCV protease or polymerase inhibitors. The results of combinations shown in Fig. 2 were analyzed in a mathematical model, MacSynergy, as described in Materials and Methods. The three-dimensional response surface plot shown represents the differences between the actual experimental effects and the theoretical additive effects at various concentrations of the two compounds. Only statistically significant (95% confidence interval) differences between the two were considered at any given concentration. NIM811 in combination with the NS3-4A protease inhibitor BILN2061 produced an antiviral effect that was comparable to the theoretical additive effect (A), whereas NIM811 in combination with the non-nucleoside NS5B polymerase inhibitor POL-1 (B) or the nucleoside NS5B polymerase inhibitor NM107 (C) produced synergistic antiviral effects that were greater than the theoretical additive effects.
Combination of NIM811 with HCV protease or polymerase inhibitors facilitated multilog viral RNA reduction in the replicon cells after prolonged treatment.
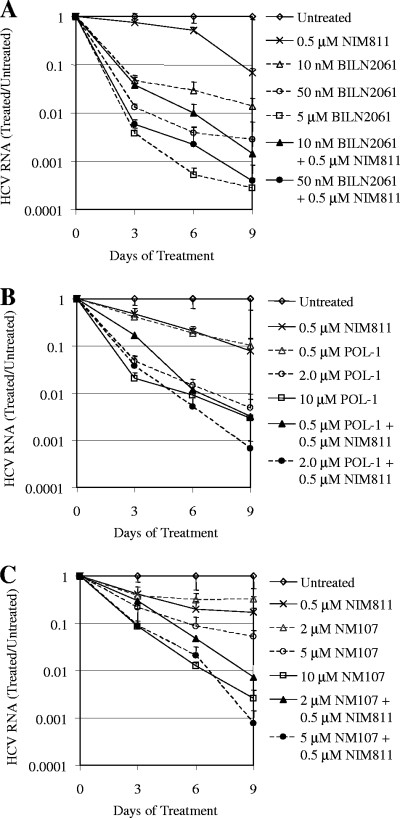
Currently, a successful HCV therapy requires treating patients for 6 to 12 months, during which time the viral load is reduced to an undetectable level. Therefore, in addition to the standard 48-h assay measuring only 50 to 90% of the inhibitory activity of compounds, it is highly relevant to evaluate antiviral agents for their abilities to induce multilog viral RNA reduction after prolonged treatment in vitro. HCV replicon cells (clone A) were treated with various concentrations of the viral protease or polymerase inhibitors alone or in combination with 0.5 μM NIM811 for 3, 6, and 9 consecutive days. At the end of each treatment, total RNA was extracted from cells, and the quantity of HCV RNA was determined by qRT-PCR and normalized against the amount of total RNA extracted. The level of remaining HCV RNA with each compound treatment was then compared to that of untreated control cells from the same time point to determine the log reduction of HCV RNA over time. As shown in Fig. 4, there was a concentration- and time-dependent reduction of HCV RNA with all of the different compound treatments. Importantly, the combinations of NIM811 with protease or polymerase inhibitors resulted in a greater reduction of HCV RNA compared to any single agent alone.
FIG. 4.
Combination of NIM811 with HCV protease or polymerase inhibitors facilitated multilog HCV RNA reduction after prolonged treatment. HCV replicon cells (clone A) were treated with various concentrations of BILN2061 (A), POL-1 (B), or NM107 (C) alone or in combination with NIM811 for 3, 6, or 9 days. Culture medium was replaced every 3 days with fresh compounds. At the end of each treatment, the quantity of HCV RNA was determined by qRT-PCR and was normalized against the amount of total RNA extracted from each sample. The level of remaining HCV RNA in compound-treated cells was compared to that of untreated cells at the same time point to calculate the log reduction of HCV RNA. Each data point represents the average of five replicates in cell culture.
In the experiment with the HCV protease inhibitor (Fig. 4A), 10 nM BILN2061 resulted in a 1.9-log reduction in HCV RNA after treating the replicon cells for 9 days, and 0.5 μM NIM811 resulted in a 1.2 log reduction. In contrast, the combination of the two led to a significantly greater (2.8-log) reduction of HCV RNA. Moreover, 50 nM BILN2061 reduced HCV RNA by 2.6 log after 9 days, whereas its combination with 0.5 μM NIM811 resulted in a 3.4 log reduction, which was almost as potent as a 100-fold-higher concentration (5 μM) of BILN2061 alone (3.6 log).
In the experiment with the non-nucleoside HCV NS5B polymerase inhibitor (Fig. 4B), 0.5 μM POL-1 resulted in a 1.0-log reduction in HCV RNA after 9 days, 0.5 μM NIM811 resulted in a 1.1 log reduction, whereas the combination of the two led to a 2.5-log reduction. Moreover, 2 μM POL-1 reduced HCV RNA by 2.3 logs after 9 days. Although a fivefold-higher concentration (10 μM) of POL-1 alone did not induce much further reduction (2.5 log), the combination of 2 μM POL-1 with 0.5 μM NIM811 was much more effective with a 3.2-log viral RNA reduction after 9 days.
In the experiment with the nucleoside polymerase inhibitor (Fig. 4C), 2 μM NM107 resulted in only a 0.5-log reduction in HCV RNA after 9 days, 0.5 μM NIM811 resulted in a 0.8-log reduction in the same experiment, whereas the combination of the two led to a 2.1-log reduction. Furthermore, the combination of 0.5 μM NIM811 and 5 μM NM107 resulted in a 3.1-log HCV RNA reduction, which was much better than either 5 μM or 10 μM NM107 alone (1.3 and 2.6 logs, respectively). These results confirmed that the significant enhancement of antiviral activity with the combinations of NIM811 and HCV protease or polymerase inhibitors can be sustained and would lead to more pronounced antiviral effects over the time.
No cross-resistance was observed between NIM811 and HCV protease or polymerase inhibitors.
One of the key reasons for using antiviral agents in combination is to suppress the emergence of resistance. If two drugs have the same or similar resistance profile, it would limit the utility of using them in combination. Therefore, it is important to characterize the cross-resistance profile of NIM811 with other HCV inhibitors before considering them for combination therapy. To accomplish this, drug-resistant clones were selected in vitro via continuous culture of HCV replicon cells in the presence of these inhibitors. As described in Materials and Methods, G418 was included in the culture medium to provide the selection pressure: the replication of wild-type HCV replicon was blocked in the presence of HCV inhibitor, which resulted in a decreased expression of neomycin phosphotransferase and therefore cell death in the presence of G418. In contrast, cells containing resistant mutant replicons capable of replicating in the presence of inhibitor would maintain the level of neomycin phosphotransferase sufficient to protect the cells from G418-mediated death. While the selections of resistant cell lines for viral protease inhibitor BILN2061 or viral polymerase inhibitors POL-1 or NM107 were relatively rapid and reproducible, a NIM811-resistant cell line was significantly more difficult to generate and required a stepwise-selection with increasing concentrations of compounds over a >6-month period. These resistant clones were then subject to testing for drug susceptibility and sequencing for identification of potential resistant mutations. Specific mutations—D168V in NS3 protease, M423V in NS5B polymerase, and S282T in NS5B polymerase—were identified in BILN2061, POL-1, and NM107 resistant clones, respectively, which are known to be associated with resistance against these compounds (9, 10, 12, 13, 15, 23, 24). Sequence analysis revealed that the NIM811-resistant clone does not contain these three mutations or any resistant mutations that have been previously reported for viral protease or polymerase inhibitors. Further characterization of the mechanism of resistance to NIM811 is in progress. As shown in Table 1, each of these cell lines was highly resistant to the inhibitor used for selecting the clone. Importantly, no significant loss of activity was observed for the NIM811-resistant clone to viral protease or polymerase inhibitors. Reciprocally, the HCV replicon clones resistant to BILN2061, POL-1, or NM107 remained fully susceptible to NIM811.
TABLE 1.
Characterization of cross-resistance between NIM811 and other HCV inhibitors
| HCV replicon cell line | Fold change in EC50
|
|||
|---|---|---|---|---|
| NIM811 | BILN2061 | POL-1 | NM107 | |
| NIM811-resistant clone | 19.7 | 1.0 | 2.0 | 0.8 |
| BILN2061-resistant clone | 0.7 | 484.2 | 0.3 | 0.4 |
| POL-1-resistant clone | 3.7 | 0.4 | 57.0 | 1.0 |
| NM107-resistant clone | 0.6 | 1.9 | 1.1 | 17.7 |
aHCV replicon clones resistant to various inhibitors were selected in vitro. Their susceptibilities to different classes of inhibitors were determined and are reported as the fold change in EC50 compared to those of the wild-type replicon cell line. The EC50s of NIM811, BILN2061, POL-1, and NM107 in the wild-type replicon cells (clone A) were 0.30 ± 0.27 μM, 1.55 ± 0.44 nM, 0.12 ± 0.06 μM, and 1.85 ± 0.64 μM, respectively.
NIM811 suppressed the emergence of resistance against HCV protease or polymerase inhibitors.
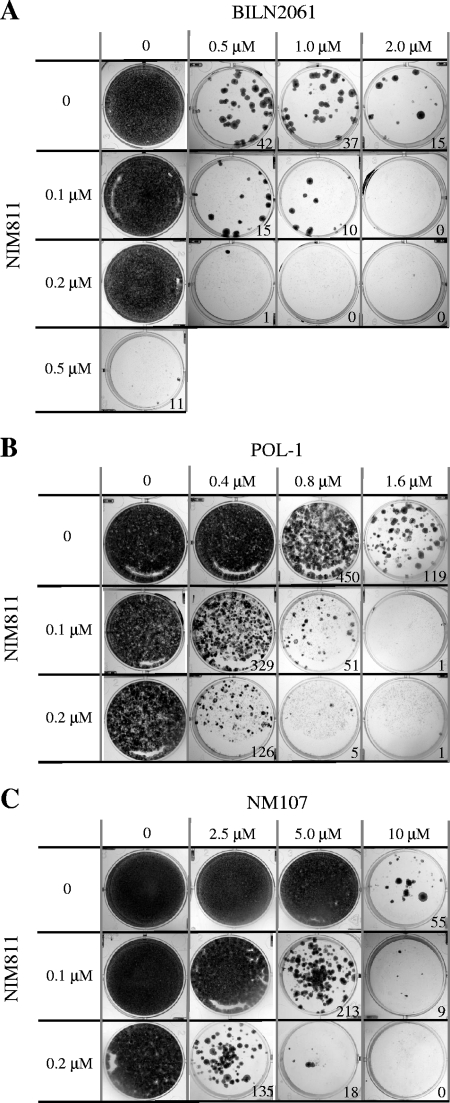
The likelihood or the frequency of resistance development with HCV inhibitors can be assessed in vitro using the replicon cells. HCV replicon cells were seeded at a low density and treated with various concentrations of inhibitors either alone or in combination. As described above, G418 was included to provide the selective pressure on HCV replicon cells such that cells bearing wild-type replicon died while cells bearing resistant replicon grew and formed visible colonies after about 3 weeks of treatment. The number of colonies reflects the frequency of resistance. As shown in Fig. 5, there was an inverse correlation between the number of colonies and the concentration of compounds, i.e., the higher the inhibitor concentration the lower the resistance frequency. Notably, NIM811 was very effective in blocking the emergence of resistance compared to polymerase or protease inhibitors with respect to their relative EC50s. Only 11 colonies (of relatively small size) survived after the treatment with 0.5 μM (2.8× the EC50) NIM811 compared to 42 colonies with 0.5 μM (227× the EC50) BILN2061, 119 colonies with 1.6 μM (3.1× the EC50) POL-1, and 55 colonies with 10 μM (21× the EC50) NM107. It should be noted that the observed effect was not due to the cytotoxicity of the compounds since the concentrations used in the study were much lower than their respective CC50 values and the growth of the cells was not significantly affected by the compounds when G418 was absent (data not shown). Two lower concentrations, 0.1 and 0.2 μM, of NIM811 were selected to investigate the effect of combinations. Although at these low levels NIM811 alone was not able to sufficiently suppress viral replication and sensitize cells to G418-mediated death, it efficiently reduced the frequency of resistant colony formation against viral protease or polymerase inhibitors when used in combination. For example, while treatment with a 0.5 μM concentration of the protease inhibitor BILN2061 resulted in 42 resistant HCV replicon cell colonies, combination with 0.1 μM NIM811 reduced the number of colonies to 15, and combination with 0.2 μM NIM811 further reduced the frequency to just one colony (Fig. 5A). Likewise, although treatment with 0.8 μM concentrations of the non-nucleoside polymerase inhibitor POL-1 resulted in approximately 450 resistant colonies, the addition of 0.1 μM NIM811 reduced this number to 51 colonies, and 0.2 μM NIM811 further reduced it to just five colonies (Fig. 5B). For the nucleoside polymerase inhibitor NM107, the number of resistant colonies was reduced from near confluence with 5.0 μM NM107 alone to 213 colonies with the addition of 0.1 μM NIM811 and only 18 colonies with 0.2 μM NIM811 (Fig. 5C).
FIG. 5.
Combination of NIM811 with HCV protease or polymerase inhibitors suppressed the emergence of resistance. HCV replicon cells (clone A) were seeded at low density and treated with various concentrations of BILN2061 (A), POL-1 (B), or NM107 (C) alone or in combination with NIM811 in the presence of G418 selection as described in Materials and Methods. As soon as macroscopic colonies were visible and G418-sensitive cells died and completely detached from well surfaces, the cells were fixed and stained with crystal violet/ethanol. The numbers of colonies were counted by using Bio-Rad Quantity One v4.4 software and are shown in the graph.
To characterize the resistant colonies produced in these experiments, HCV replicon RNAs were isolated from the cells pooled together and sequenced to confirm the presence of specific mutations in NS3 protease or NS5B polymerase that are known to be associated with resistance against BILN2061, POL-1, or NM107, respectively. As expected, HCV replicon colonies produced via treatments with BILN2061 or BILN2061 plus NIM811 only contained the known resistance-associated mutation D168V in the NS3 coding region (10, 12, 23), those treated with POL-1 or POL-1 plus NIM811 only contained the known resistance-associated mutation M423V in the NS5B coding region (9, 23), whereas those treated with NM107 or NM107 plus NIM811 only contained the known resistance-associated mutations S282T in NS5B (13, 15). No additional mutation was selected with the combination of NIM811 at the two low concentrations used in these experiments. Overall, the extent to which NIM811 reduced the frequency of resistance that emerged against HCV protease or polymerase inhibitors was considerable and reproducible in independent experiments and provides further rationale for the use of NIM811 in combination therapy for HCV.
DISCUSSION
The current standard therapy for chronic hepatitis C is the combination of pegylated IFN-α and ribavirin, which is of limited efficacy and has significant side effects. Extensive efforts have been made to develop more potent and specific antiviral agents. However, given the high replication efficiency and high mutation rate of the virus, it is almost certain that resistant viruses will emerge during long-term treatment with specific antivirals. Therefore, it is anticipated that the future therapy for HCV, like that of HIV, will be a combination of multiple drugs of different mechanisms. NIM811, an HCV inhibitor with a novel mechanism targeting host factor cyclophilins, presents a unique opportunity for diverse combinations. In the present study, the combinations of NIM811 with specific inhibitors of HCV NS3-4A protease or NS5B polymerase were evaluated in vitro for their effects on antiviral activity, as well as toxicity. In the standard 48-h HCV replicon assay, the combinations of NIM811 with any of the other HCV inhibitors produced a greater inhibition of HCV replication than any of the single agents alone. None of the combinations resulted in a significant increase of cytotoxicity. Moreover, the effect of the combinations was determined to be synergistic for NIM811 with a nucleoside or a non-nucleoside viral polymerase inhibitor and additive for NIM811 with a viral protease inhibitor based on mathematical modeling. The synergism observed between NIM811 and viral polymerase inhibitors appears to be generalizable since it was also demonstrated with at least three other nucleoside and non-nucleoside polymerase inhibitors (data not shown). Interestingly, Watashi et al. reported that cyclophilin B binds to NS5B and enhances its RNA-binding activity (26). The effect of combination between a cyclophilin inhibitor and an HCV polymerase inhibitor is consistent with the notion that synergism is often observed as the result of interactions between two related targets or pathways.
The HCV titer in chronically infected patients is typically 106 to 107 copies/ml, and the goal of antiviral therapy is to completely eradicate the virus to an undetectable level, which requires multiple log reduction in viral load following months of treatment. To model the effect in vitro, we treated HCV replicon cells for a prolonged period and then compared the activities of combinations to those of single agents. All of the compound treatments resulted in a concentration- and time-dependent reduction of HCV RNA. There were about 103 copies of HCV RNA in each untreated replicon cell, or 107 copies in each RNA sample extracted from 104 cells and then measured by qRT-PCR. Since the lower limit of quantitation by qRT-PCR was 102 copies per sample, theoretically a 5-log reduction in HCV RNA can be detected in this assay. However, the maximum HCV RNA reduction observed after 9 days appeared to be 3 to 4 logs, which was likely limited by the half-life or degradation of remaining HCV replicon RNA even if its replication was 100% blocked. For the viral protease inhibitor, 5 μM BILN2061 alone resulted in a >3-log HCV RNA reduction after 9 days of treatment. In contrast, a 100-fold-lower concentration (50 nM) of the compound BILN2061 achieved the same effect when used in combination with 0.5 μM NIM811. For the non-nucleoside polymerase inhibitor POL-1, combination with NIM811 was able to overcome the apparent concentration-dependent plateau in antiviral activity observed for POL-1 alone at 2 to 10 μM. For NM107, the combination with NIM811 was also significantly more potent than single agents. These results support the rationale for including NIM811 in combination therapies aimed to maximize long-lasting antiviral response and potentially reduce dosages of individual drugs.
A key advantage to target host factors is that it may present a greater genetic barrier to the emergence of viral escape mutants. It has been demonstrated both in vitro and in patients that drug-resistant mutants can emerge quickly, even with the most potent inhibitors of viral protease or polymerase (7). This has been attributed to two factors: (i) HCV replicates at a high rate in patients, producing an estimate of 1010 to 1012 virions per day (17), and (ii) the RNA-dependent RNA polymerase of the virus lacks proofreading function and has an error rate of about 10−4 mutations per genome per replication cycle. As a result, there is an extremely high degree of heterogeneity of viral population (quasispecies) in each patient. Since virus-specific inhibitors typically bind to a defined pocket of viral protein, mutations in viral genomes that disrupt the binding would reduce the effectiveness of inhibitors and lead to resistance. It is postulated that inhibitors targeting host factors may offer certain advantages in this regard since the inhibitors do not bind directly to the viral targets, which have a diverse genetic background and are easier to mutate. Indeed, we, as well as others, have observed that it appeared to be much more difficult to develop resistance against NIM811 or other cyclophilin inhibitors in vitro than against direct inhibitors of viral protease and polymerase (3, 27). When we compared the frequency of resistance in vitro, a much lower concentration of NIM811 was required to block the emergence of resistance compared to protease or polymerase inhibitors with regard to their relative EC50s. This is likely because multiple cyclophilins are involved in the viral life cycle and multiple mutations need to be acquired to confer resistance against cyclophilin inhibitors (3, 4, 22, 27). In contrast, a single point mutation is typically sufficient to confer high level of resistance against viral protease or polymerase inhibitors. In other words, cyclophilin inhibitors such as NIM811 may create a higher genetic barrier than those of viral specific inhibitors. The mechanism of resistance against NIM811 appears to be different from those of protease or polymerase inhibitors, as suggested by the cross-resistance study. This is important especially when combination therapies are being considered: if treatment with one of the antiviral agents produces virus that is cross-resistant to other agents in the combination, then that combination therapy will be rendered ineffective. Furthermore, treatment of patients with a drug that selects for cross-resistant virus could result in the spread of multiresistant, treatment-intractable viruses in the population. Importantly, the NIM811-resistant HCV replicon cell line was fully susceptible to viral protease and polymerase inhibitors, while HCV replicon cell lines that were resistant to protease or polymerase inhibitors remained fully sensitive to NIM811. Therefore, development of cross-resistance does not appear to be a concern for NIM811.
Given its novel mechanism of action and unique resistance profile, NIM811 presents a new opportunity for combination therapy in blocking the emergence of resistance. This was evaluated in vitro by measuring the frequency of virus-resistant colonies in the presence of inhibitors. NIM811 substantially reduced the number of resistant colonies that emerged with viral protease or polymerase inhibitors when used in combination. Notably, NIM811 appeared to be even more effective in blocking the emergence of resistance since relatively low levels (0.1 to 0.2 μM) of NIM811 was needed in this experiment compared to those required to significantly reduce HCV RNA (0.5 to 1 μM). One of the potential challenges for host factor inhibitors is to identify the therapeutic doses that do not affect the normal functions of host targets and therefore do not lead to significant side effects. These results suggest an interesting strategy and a potential development path of using low doses of certain inhibitors, even in the absence of significant antiviral effect on their own, to combine with other inhibitors with the sole purpose of suppressing the rate of resistance. Whether and how this can be studied in the clinical setting may require innovative study design and regulatory approval, but it should be worthwhile to explore since resistance will likely become one of the most important factors for the success of future specifically targeted antiviral therapies.
We have demonstrated here that NIM811, an inhibitor targeting host factor cyclophilins, led to synergistic antiviral effects when used in combination with HCV polymerase inhibitors and additive effects when used in combination with viral protease inhibitor. Resistance to NIM811 appeared to be difficult to develop in vitro and remained fully susceptible to viral specific inhibitors. Furthermore, NIM811 is highly effective in suppressing the emergence of resistance when used in combination with viral protease or polymerase inhibitors. These data support the two-pronged antiviral strategy of targeting both host and viral factors. Further investigation of NIM811 in combination with other HCV inhibitors in chronic hepatitis C patients is warranted.
Acknowledgments
We thank Thomas G. Evans, Michael P. Cooreman, Meimei Huang, Roger Fujimoto, Prakash Raman, and Pascal Rigollier for their support of the study. We also thank Brigitte Wiedmann, Jennifer Philips, and Thomas G. Evans for critically reviewing the manuscript.
Footnotes
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Chan, L., O. Pereira, T. J. Reddy, S. K. Das, C. Poisson, M. Courchesne, M. Proulx, A. Siddiqui, C. G. Yannopoulos, N. Nguyen-Ba, C. Roy, D. Nasturica, C. Moinet, R. Bethell, M. Hamel, L. L'Heureux, M. David, O. Nicolas, P. Courtemanche-Asselin, S. Brunette, D. Bilimoria, and J. Bédard. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2: tertiary amides. Bioorg. Med. Chem. Lett. 14:797-800. [DOI] [PubMed] [Google Scholar]
- 3.Coelmont, L., J. Paeshuyse, S. Kaptein, I. Vliegen, A. Kaul, E. De Clercq, B. Rosenwirth, and R. Crabbe. 2007. The cyclophilin inhibitor DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro and has a unique resistance profile, abstr. O-61. 14th International Symposium on Hepatitis C Virus and Related Viruses, Glasgow, Scotland.
- 4.Fernandes, F., D. S. Poole, S. Hoover, R. Middleton, A. C. Andrei, J. Gerstner, and R. Striker. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 46:1026-1033. [DOI] [PubMed] [Google Scholar]
- 5.Flisiak, R., A. Horban, P. Gallay, M. Bobardt, S. Selvarajah, A. Wiercinska-Drapalo, E. Siwak, I. Cielniak, J. Higersberger, J. Kierkus, C. Aeschlimann, P. Grosgurin, V. Nicolas-Métral, J. M. Dumont, H. Porchet, R. Crabbé, and P. Scalfaro. 2008. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology 47:817-826. [DOI] [PubMed] [Google Scholar]
- 6.Inoue, K., K. Sekiyama, M. Yamada, T. Watanabe, H. Yasuda, and M. Yoshiba. 2003. Combined interferon alpha2b and cyclosporin A in the treatment of chronic hepatitis C: controlled trial. J. Gastroenterol. 38:567-572. [DOI] [PubMed] [Google Scholar]
- 7.Koev, G., and W. Kati. 2008. The emerging field of HCV drug resistance. Expert Opin. Investig. Drugs 17:303-319. [DOI] [PubMed] [Google Scholar]
- 8.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bös, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagacé, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinàs-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 9.Le Pogam, S., K. Hyunsoon, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W.-R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Nájera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 11.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 12.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, S., J. E. Boerner, C. TiongYip, B. Weidmann, N. S. Ryder, M. P. Cooreman, and K. Lin. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 50:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporine. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 18.Paeshuyse, J., A. Kaul, E. De Clercq, B. Rosenwirth, J. M. Dumont, P. Scalfaro, R. Bartenschlager, and J. Neyts. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43:761-770. [DOI] [PubMed] [Google Scholar]
- 19.Pawlotsky, J.-M. 2006. Therapy of hepatitis C: from empiricism to eradication. Hepatology 43:S207-S220. [DOI] [PubMed] [Google Scholar]
- 20.Pierra, C., A. Amador, S. Benzaria, E. Cretton-Scott, M. D'Amours, J. Mao, S. Mathieu, A. Moussa, E. G. Bridges, D. N. Standring, J. P. Sommadossi, R. Storer, and G. Gosselin. 2006. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J. Med. Chem. 49:6614-6620. [DOI] [PubMed] [Google Scholar]
- 21.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 22.Robida, J. M., H. B. Nelson, Z. Liu, and H. Tang. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomei, L., S. Altamura, G. Paonessa, R. De Francesco, and G. Migliaccio. 2005. HCV antiviral resistance: the impact of in vitro studies on the development of antiviral agents targeting the viral NS5B polymerase. Antivir. Chem. Chemother. 16:225-245. [DOI] [PubMed] [Google Scholar]
- 24.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 26.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 27.Wiedmann, B., X. Puyang, K. Brown, A. Gaither, J. Baryza, J. Boerner, D. Poulin, S. Ma, X. Shen, J. Tao, P. Devay, T. Compton, and K. Lin. 2007. Roles of cyclophilins in HCV replication and mode of action of the cyclophilin inhibitor NIM811, abstr. P-234. 14th International Symposium on Hepatitis C Virus and Related Viruses, Glasgow, Scotland.