Abstract
To identify mutations associated with the virological response (VR) to a tipranavir-ritonavir (TPV/r)-based regimen, 143 patients previously treated with protease inhibitor (PI) were studied. VR was defined by a decrease of at least 1 log10 in, or undetectable, human immunodeficiency virus (HIV) RNA at month 3. The effect of each mutation in the protease, considering all variants at a residue as a single variable, on the VR to TPV/r was investigated. Mutations at six residues were associated with a lower VR (E35D/G/K/N, M36I/L/V, Q58E, Q61D/E/G/H/N/R, H69I/K/N/Q/R/Y, and L89I/M/R/T/V), and one mutation was associated with a higher VR (F53L/W/Y). The genotypic score M36I/L/V − F53L/W/Y + Q58E + H69I/K/N/Q/R/Y + L89I/M/R/T/V was selected as providing a strong association with VR. For the seven patients with a genotypic score of −1 (viruses with only mutation at codon 53), the percentage of responders was 100% and the percentages were 79%, 56%, 33%, 21%, and 0% for those with scores of 0, 1, 2, 3, and 4, respectively. The percentage of patients showing a response to TPV/r was lower for patients infected with non-clade B viruses (n = 16, all non-B subtypes considered together) than for those infected with clade B viruses (n = 127) (25% and 59%, respectively; P = 0.015). Most mutations associated with VR to TPV/r had not previously been associated with PI resistance. This is consistent with phenotypic analysis showing that TPV has a unique resistance profile. Mutations at five positions (35, 36, 61, 69, and 89) were observed significantly more frequently in patients infected with a non-B subtype than in those infected with the B subtype, probably explaining the lower VR observed in these patients.
Tipranavir (TPV) is a recently approved nonpeptidic protease inhibitor (PI) with antiviral activity against multi-PI-resistant clinical human immunodeficiency virus type 1 (HIV-1) isolates. Its average 50% effective concentration for these isolates is 240 nmol/liter (range, 50 to 380 nmol/liter) (9, 13, 14). TPV-resistant viruses were selected in vitro in a previous study from serial passages of wild-type HIV-1 NL4-3 in the presence of increasing concentrations of TPV in cell culture. HIV-1 variants with 70-fold-decreased susceptibility to TPV were selected after 9 months in passage. Ten mutations were identified, arising in the following order: L33F, I84V, K45I, I13V, V32I, V82L, M36I, A71V, L10F, and I54V (4).
The efficacy of ritonavir-boosted tipranavir (TPV/r) in HIV-infected patients was examined in two phase III trials. These patients were highly treatment experienced and displayed stronger virological and immunological responses to TPV/r than to other ritonavir-boosted PIs (2, 7). Previous analyses of phase II and III clinical trials with TPV/r in PI-experienced patients were conducted to determine the association of protease mutations with reduced susceptibility and virological response (VR) to TPV (1a). A TPV mutation score was generated from these analyses, incorporating a set of 16 protease amino acid positions and 21 mutations (10V, 13V, 20M/R/V, 33F, 35G, 36I, 43T, 46L, 47V, 54A/M/V, 58E, 69K, 74P, 82L/T, 83D, and 84V). HIV-1 isolates with a greater number of these TPV resistance-associated mutations had reduced phenotypic susceptibility and VR to TPV. Parkin et al. proposed revisions to the TPV mutation score based on the analysis of 1,411 clinical samples from the Monogram database. They added new mutations and weighted other specific mutations that were associated with lower-than-expected (10I, 11L, 32I, 36L, 46I, 47V, 54A, 55R, 60E, 71L, 73T, 82T, 84V, 89V, and 90 M) or higher-than-expected (10F/V, 13V, 20R, 24I, 30N, 36I, 46L, 50L/V, 54L, 76V, 82I, and 88D) susceptibility to TPV (12a). The U.S. Food and Drug Administration determined in Boehringer Ingelheim studies that at least five of eight mutations present at baseline (I13, V32, M36, I47, Q58, D60, V82, and I84) were associated with a poorer VR rate at week 24 and that the most common substitutions emerging in patients with virological failure were L33V/I/F, V82T, and I84V (12). These studies showed that resistance to TPV is complex, involving mutations that have not previously been associated with resistance to other PIs.
The aim of this study was to determine, using simple and previously described methods, mutations associated with VR to TPV/r in a population of PI-experienced patients.
(This work was presented at the 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, February 2007 [11a], and at the XVI International HIV Drug Resistance Workshop, Barbados, Barbados, 12 to 16 June 2007 [11b].)
MATERIALS AND METHODS
Patients and antiretroviral regimens.
One hundred forty-three PI-experienced patients were recruited to the study. All patients were treated with ritonavir (200 mg twice a day [b.i.d.]) plus TPV (500 mg b.i.d.) with a background regimen comprising nucleoside reverse transcriptase (RT) inhibitors (NRTI) and/or nonnucleoside RT inhibitors (NNRTI) and/or enfuvirtide (ENF). TPV and ritonavir were the only PIs used in the antiretroviral combinations. Sociodemographic data, clinical data, and treatment histories were collected for all patients recruited. Inclusion criteria and all data were checked by the study monitor. The main characteristics of the study population are shown in Table 1. At baseline, the median numbers (interquartile ranges [IQR]) of major and minor PI resistance mutations using the International AIDS Society-USA (IAS-USA) panel list were 3 (0 to 7) and 9 (1 to 14), respectively (8). Participating laboratories belonged to the Agence Nationale de Recherches sur le SIDA (ANRS) AC11 network and participated in the ANRS quality control assessment of HIV-1 drug resistance sequencing (3).
TABLE 1.
Main baseline characteristics of the study population (n = 143)
| Parameter | Value |
|---|---|
| % Male | 85 |
| % Infected with subtype B | 89 |
| Median plasma HIV-1 RNA (log10 copies/ml) | |
| (IQR) | 4.7 (3.9-5.2) |
| Median CD4 cell count/mm3 (IQR) | 115 (25-224) |
| Previous antiretroviral treatment | |
| Median no. of antiretroviral drugs (IQR) | 11 (9-12) |
| Median no. of NRTI (IQR) | 6 (5-7) |
| Median no. of PIs (IQR) | 4 (3-5) |
| % of patients treated with NNRTI | 80 |
| % of patients treated with ENF | 28 |
| % of patients with TPV/r (500/200 mg b.i.d.) cotreatment | |
| NRTI | 97 |
| NRTI + NNRTI | 15 |
| NRTI + ENF | 55 |
Genotypic resistance testing.
Sequences of the protease and RT genes were determined at baseline in each laboratory using the ANRS consensus technique (http://www.hivfrenchresistance.org), the Bayer TrueGene kit, the Abbott ViroSeq kit, or an in-house method. All protease and RT gene mutations were identified from the International AIDS Society-USA resistance testing panel (September 2006) (8). The lower limit of quantification (LOQ) was 200 copies/ml or 50 copies/ml according to the virological center.
TPV plasma concentration measurements.
TPV plasma concentrations were measured by a specific and validated high-performance liquid chromatographic assay coupled to UV detection at 240 nm as previously described (12b). All patients had detectable TPV plasma concentrations (>100 ng/ml) and were retained for the determination of the resistance score.
Statistical methods.
The end point for the analysis was the percentage of responders at month 3. VR was defined by a decrease of at least 1 log10 in HIV RNA from baseline or HIV RNA less than the LOQ at month 3. These criteria were also used in the RESIST (Randomized Evaluation of Strategic Intervention in Multidrug Resistant Patients with Tipranavir) studies and the FDA TPV resistance analysis (7, 12). Potential associations between each protease mutation (codons 1 to 99) and VR were determined using Fisher's exact test. Multiple mutations arising at a given position were grouped together as a single variable. Mutations present in at least 10% of patients giving a P value lower than 0.10 in the above-mentioned univariate analysis were retained. They were then analyzed using the removing procedure with a nonparametric test to select the combination of mutations most strongly associated with VR (5, 10, 11, 15). The Cochran-Armitage (CA) test was used. The removing procedure begins with all mutations from the univariate analysis (k) retained. The first step is to calculate the P value with the CA test to give a score incorporating these initial k mutations. All combinations of k − 1 mutations are investigated one by one; a combination is retained if it yields a lower P value with the CA test than the P value obtained with the k mutations. In the second step, mutations are again removed one by one to compare the combinations of k − 2 mutations; a combination yielding a P value lower than the P value obtained with k − 1 mutations is again retained, and so on. The procedure ends when removing a mutation does not provide a lower P value than the previous one. The removing procedure was selected because scores obtained using this technique tend to be more strongly associated with the VR than those obtained using the adding procedure (7).
A series of univariate logistic regression was fitted to the data to retain, in the multivariate analysis, variables associated with the VR (P < 0.10). The final multivariate model was selected using a stepwise procedure. We used mutations present in the RT gene and the ANRS algorithm (http://www.hivfrenchresistance.org) to determine whether patients receiving a particular NRTI or NNRTI had resistant or susceptible virus strains.
The statistical program used for analyses was SAS (version 9.0).
RESULTS
Effect of PI resistance mutations on VR.
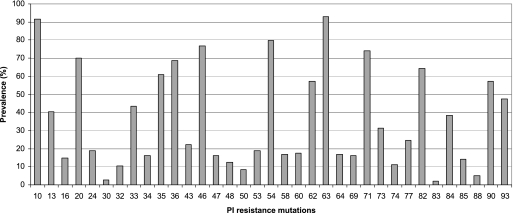
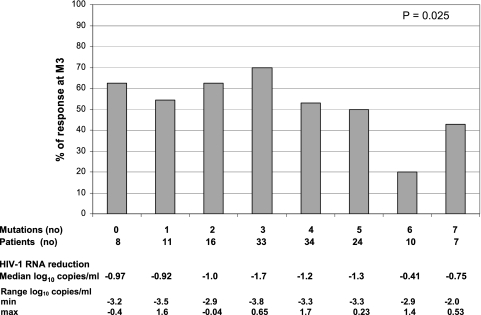
Overall, 79 (55%) patients receiving a TPV/r-containing regimen displayed a VR at month 3. Four patients had a viral load (VL) less than the LOQ, and 32 had a decrease in VL of at least 1 log at month 3. Forty-three patients met both criteria defining a VR. The median decreases (IQR) in plasma HIV RNA were −1.16 log copies/ml (−3.82 to 1.16 log copies/ml) between baseline and month 3 and −0.83 log copies/ml (−4.00 to 0.58 log copies/ml) between baseline and month 6. The number of IAS-USA panel mutations (major only or minor and major) was not associated with the VR at month 3 (P = 0.82 and P = 0.20, respectively). The prevalence of IAS-USA panel PI resistance mutations among the viruses studied is shown in Fig. 1. The TPV mutation score (10V, 13V, 20 M/R/V, 33F, 35G, 36I, 43T, 46L, 47V, 54A/M/V, 58E, 69K, 74P, 82L/T, 83D, and 84V), developed from the Boehringer Ingelheim studies, was associated with VR at month 3; however, no clear trend in the distribution of the VR was observed (P = 0.025) (Fig. 2) (1a). A weighted TPV score, based on 15 mutations (10V, 24I, 36I, 43T, 46L, 47V, 50L/V, 54A/M/V, 54L, 58E, 74P, 76V, 82L/T, 83D, 84V), was recently generated (14a). Among patients found susceptible (score ≤ 3), 66% were responders; among those who were partially susceptible (3 < score ≤ 10), 60% were responders; and 20% of resistant patients (score > 10) were responders (P = 0.0002). Finally, a modified version of the score developed by Boehringer Ingelheim was proposed, based on 22 mutations (I47V, I54A, V82T, I84V, L10V, I13V, K20R, M46L, V11L, V32I, A71L, G73T, L89V, L10I, M46I, L90M, L24I, D30N, I50L/V, I54L, L76V, and V82I) (12a). Some mutations increased the resistance score (by 1, 2, or 0.5), whereas other mutations decreased the resistance score (reducing the score by 1 or 0.5). In our sample, 17/25 (68%) patients with a score ≤2, 45/73 (62%) patients with a score >2 and ≤5, 17/41 (42%) patients with a score >5 and ≤8, and 1/5 (20%) of patients with a score >8 were responders (P = 0.003).
FIG. 1.
Values for the prevalence of IAS-USA PI resistance mutations among the 143 isolates.
FIG. 2.
Percentages of patient responders at month 3 (M3) as a function of the number of mutations for the TPV mutation score developed by Boehringer Ingelheim (10V, 13V, 20 M/R/V, 33F, 35G, 36I, 43T, 46L, 47V, 54A/M/V, 58E, 69K, 74P, 82L/T, 83D, and 84V).
In univariate analysis, mutations at six positions (all variants of each position considered as a single variable) were associated with a lower VR to TPV/r (P < 0.1) (E35D/G/K/N, M36I/L/V, Q58E, Q61D/E/G/H/N/R, H69I/K/N/Q/R/Y, and L89I/M/R/T/VP) and one mutation was associated with a higher VR (F53Y/L/W). In addition, for positions displaying one predominant variant (e.g., E35D, M36I, F53L, H69K, and L89M), independent statistical analysis has been done to determine whether the results are driven by these amino acid variants only and whether the inclusion of the others is actually not supported by the data. The P values of the corresponding Fisher exact tests were as follows: E35D, 0.13, versus E35D/G/K/N, 0.006; M36I, 0.04, versus M36I/L/V, 0.004; F53L, 0.067, versus F53L/W/Y, 0.033; H69K, 1.22 × 10−4, versus H69I/K/N/Q/R/Y, 7.22 × 10−6; L89M, 0.77, versus L89I/M/R/T/V, 0.03. The score obtained using such an analysis with the preponderant variant provided a greater P value than the P value generated with all variants at a given position considered as a single variable (P = 1.2 × 10−6 versus 1.5 × 10−7).
Table 2 shows univariate analysis of the VR as a function of the presence of mutated or wild-type codons at specific sites of the protease gene. Mutations at codons 1 to 9, 18, 21, 22, 23, 25 to 31, 38 to 40, 42, 44, 45, 49 to 52, 56, 59, 65, 67, 68, 70, 75, 78 to 81, 83, 86 to 88, 91, 92, and 94 to 99 in the protease gene could not be evaluated because their prevalence was <10%.
TABLE 2.
Univariate analysis of VR as a function of the presence of mutated or wild-type codons at specific sites of the protease gene
| Position | Amino acid | No. of patients | % Responders | P |
|---|---|---|---|---|
| 35 | E (wild-type) | 56 | 70 | 0.0061 |
| D | 72 | 46 | ||
| G | 2 | |||
| K | 2 | |||
| N | 11 | |||
| 36 | M (wild-type) | 45 | 73 | 0.0037 |
| I | 87 | 47 | ||
| L | 7 | |||
| V | 4 | |||
| 53 | F (wild-type) | 116 | 51 | 0.033 |
| L | 23 | 74 | ||
| W | 1 | |||
| Y | 3 | |||
| 58 | Q (wild-type) | 119 | 59 | 0.072 |
| E | 24 | 38 | ||
| 61 | Q (wild-type) | 124 | 58 | 0.088 |
| D | 2 | 37 | ||
| E | 4 | |||
| G | 1 | |||
| H | 4 | |||
| N | 7 | |||
| R | 1 | |||
| 69 | H (wild-type) | 120 | 63 | 7.22 × 10−5 |
| I | 1 | 17 | ||
| K | 14 | |||
| N | 1 | |||
| Q | 1 | |||
| R | 4 | |||
| Y | 2 | |||
| 89 | L (wild-type) | 107 | 61 | 0.03 |
| I | 7 | 39 | ||
| M | 12 | |||
| R | 1 | |||
| T | 1 | |||
| V | 15 |
Boosted TPV/r genotypic score. (i) Removing procedure.
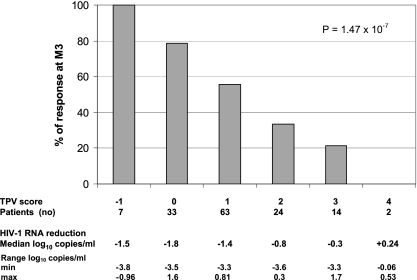
Among the mutations selected by the univariate analysis, the removing procedure did not retain mutations at codons 35 and 61, and this led to selection of the genotypic score 36I/L/V − 53Y/L/W + 58E + 69I/K/N/Q/R/Y + 89I/M/R/T/V as providing the strongest association with VR (P = 1.47 × 10−7). Figure 3 shows the percentage of responders at month 3 as a function of genotypic score. For the seven patients with a genotypic score of −1 (viruses with mutation only at codon 53), the percentage of responders was 100%, and percentages were 79% (n = 33), 56% (n = 63), 33% (n = 24), 21% (n = 14), and 0% (n = 2) for those with a genotypic scores of 0, 1, 2, 3, and 4, respectively. Values for the prevalence of mutations included in the TPV/r score for patients grouped by genotypic score are shown in Table 3.
FIG. 3.
Percentages of patient responders at month 3 (M3) as a function of the number of mutations for the TPV mutation score (36I/L/V − 53Y/L/W + 58E + 69I/K/N/Q/R/Y + 89I/M/R/T/V).
TABLE 3.
Prevalence of mutations included in the TPV/r score found in patients grouped by genotypic score
| Position | Prevalence (%) for TPV/r scorea of:
|
|||||
|---|---|---|---|---|---|---|
| −1 (7) | 0 (33) | 1 (63) | 2 (24) | 3 (14) | 4 (2) | |
| 36 | 0 | 36 | 73 | 100 | 100 | 100 |
| 53 | 100 | 36 | 7.9 | 8.3 | 7.1 | 0 |
| 58 | 0 | 0 | 9.5 | 45.8 | 35.7 | 100 |
| 69 | 0 | 0 | 7.9 | 20.8 | 78.6 | 100 |
| 89 | 0 | 0 | 17.5 | 41.7 | 92.9 | 100 |
Numbers of patients are in parentheses.
According to the IAS-USA panel list, the median numbers of major PI mutations in patients with a TPV/r genotypic scores of −1, 0, 1, 2, 3, and 4 were 4, 3, 3, 3, 3 and 2, respectively. Based on these TPV/r genotypic scores, the prevalence values for strains classified as resistant (a score of at least +3) and sensitive (a score of −1 to +2) to TPV/r were 26% and 74%, respectively, in our database of treated patients experiencing virological failure (473 virological failures in 2006).
(ii) Multivariate analysis.
The following variables provided a P value <0.10 in a univariate logistic model and were potentially included in the selection of the final multivariate model: previous ENF treatment regimen, HIV-1 subtype B versus non-B, susceptibility to efavirenz in patients receiving efavirenz in the background regimen, TPV score (score < 2 or ≥ 2). In the final model, the following were retained as independently associated with the VR: previous ENF treatment (n = 38, odds ratio [OR] = 4.18, P = 0.0012) associated with a poorer VR and susceptibility to efavirenz in patients receiving efavirenz in the background regimen (n = 13, OR = 0.11, P = 0.015) associated with higher VR and TPV genotypic score (score < or ≥ 2) (OR = 7.5, P < 0.001).
Effect of HIV-1 subtype on VR to TPV/r at month 3.
Sixteen of 146 patients were infected with a non-B HIV-1 subtype (1 with A, 1 with D, 2 with F, 3 with G, 1 with H, 2 with J, 4 with CRF02_AG, 1 with B/F, and 1 with CRF09) in this study. Viral subtype (B versus non-B) was associated with VR at month 3. The VR at month 3 of patients infected with a non-B HIV-1 subtype was lower than that of patients infected with HIV-1 subtype B (25% versus 59%, P = 0.015).
Among mutations previously identified as being associated with VR at month 3 in univariate analysis, mutations at five positions (35, 36, 61, 69, and 89) were significantly more frequent in non-B subtypes than in subtype B (Table 4).
TABLE 4.
Prevalence of mutations associated with VR at month 3 in univariate analysis as a function to HIV-1 subtype
| Position | Amino acid | No. (%) of patients with infections of subtype:
|
P | |
|---|---|---|---|---|
| B | Non-B | |||
| 35 | E (wild type) | 54 (43) | 2 (13) | 0.028 |
| D/G/K/N | 73 (57) | 14 (88) | ||
| 36 | M (wild type) | 45 (35) | 0 (0) | 0.003 |
| I/L/V | 82 (65) | 16 (100) | ||
| 53 | F (wild type) | 103 (81) | 13 (81) | 1 |
| L/W/Y | 24 (19) | 3 (19) | ||
| 58 | Q (wild type) | 107 (84) | 12 (75) | 0.47 |
| E | 20 (16) | 4 (25) | ||
| 61 | Q (wild type) | 117 (92) | 7 (44) | 1.20 × 10−5 |
| D/E/G/H/N/R | 10 (8) | 9 (56) | ||
| 69a | H (wild type) | 117 (92) | 3 (19) | 5.35 × 10−5 |
| I/K/N/Q/R/Y | 10 (8) | 13 (81) | ||
| 89 | L (wild type) | 104 (82) | 3 (19) | 7.8 × 10−7 |
| I/M/R/T/V | 23 (18) | 13 (81) | ||
Viruses with a mutation at position 69 also have a mutation at position 89.
Pharmacological measurements.
The median concentration of TPV in plasma at month 3 was 27,048 ng/ml. TPV concentration at month 3 was not associated with the VR at month 3.
DISCUSSION
The development of genotypic resistance scores is based on the assessment of the effect of genotypic profiles at baseline on the subsequent VR. This provides objective information for guidance on drug selection, in particular for treatment-experienced patients. In this study, we determined a genotypic resistance score for TPV/r in PI-experienced patients using a stepwise methodology described previously (5, 10, 11, 15).
We observed VR—defined as a decrease of at least 1 log10 of HIV RNA from baseline or an HIV RNA less than the LOQ at month 3—in 55% of our patients. Changes at five codons were found to constitute a genotypic resistance score significantly associated with VR to TPV/r: 36I/L/V − 53Y/L/W + 58E + 69I/K/N/Q/R/Y + 89I/M/R/T/V. For the seven patients with a genotypic score of −1 (viruses with only mutation at codon 53), the percentage of responders was 100%, and percentages were 79% (n = 33), 56% (n = 63), 33% (n = 24), 21% (n = 14), and 0% (n = 2) for those with a genotypic scores of 0, 1, 2, 3, and 4, respectively. Based on these results, the French HIV resistance guidelines recommend considering viruses in patients with a score of +3 as resistant to TPV/r and viruses with a score of +2 as possibly resistant. Although the score previously developed by Boehringer Ingelheim was predictive of VR in this data set, it was not possible to determine a clinical cutoff using this score. It is not surprising that a score derived from another data set predicts the VR less accurately than a score derived from this data set. Indeed, this may be due to using different patient populations, resistance methodologies, viruses, and statistical approaches.
Some of the five mutations identified in the TPV genotypic resistance score (at positions 36, 58, and 69) were previously identified in the Boehringer Ingelheim TPV mutation score (1a). Although the 89M/V mutation belonged to the list of mutations assessed in stepwise multiple regression analyses of phase III, it was not retained in this final score (1a). In our study, F53Y/L/W was associated with a higher VR at month 3. Interestingly, the F53L mutation was previously associated with increased susceptibility to TPV in a study by Virco (1). None of the five mutations identified by the TPV genotypic resistance score are considered major IAS-USA panel mutations, and most have not previously been associated with resistance to other PIs. These findings could explain the activity of TPV against viruses harboring such classical PI mutations (i.e., patients with a genotypic score of −1 had a median of four major PI mutations). This is consistent with another analysis of the RESIST and 1182.51 studies showing that isolates with PI resistance mutations at 24I, 48V, 50V, 54S, 54T, and 82A retained susceptibility to TPV, whereas isolates with 84V, 89V, 54M, 47V, and 74P mutations were cross-resistant to TPV and other approved PIs (14b). Thus, the use of the TPV genotypic resistance is likely to be restricted to populations with similar PI mutation profiles, since mutations associated with this new score are almost always present in combination with these more classical PI resistance mutations. Indeed, their effects in different backgrounds or in combination with other mutations may be quite different.
Interestingly, although the result is based on a small number of patients, the VR to TPV/r seemed to be lower in patients infected with non-clade B viruses than in patients infected with clade B viruses. The low numbers of samples for any given subtype is a potential limitation. However, this result seems to be related to the fact that, among the mutations associated with VR to TPV at month 3 in univariate analysis, five positions (35, 36, 61, 69, and 89) were significantly more frequent in non-B subtype viruses. Subtype, however, was not retained in the final multivariate model, and in vitro studies have shown that TPV displays similar antiviral activities against non-B and B subtype isolates (1b). This discrepancy should be investigated by further clinical studies exploring the VR to TPV/r in a larger number of patients infected with non-B subtype strains. It also highlights the need to develop scoring algorithms targeted more specifically to prevalent non-B subtypes.
In the multivariate analysis, previous treatment with ENF was associated with a poorer VR whereas susceptibility to efavirenz in patients receiving efavirenz in the background regimen and a TPV genotypic score of <2 were associated with a better VR. The fact that previous treatment with ENF was associated with a poorer VR could be explained: these patients were infected with viruses more resistant to all currently available antiretrovirals than those who did not received ENF in the past. In addition, 38 of 79 (48%) patients who use ENF in combination with TPV/r had previously used it, thus recycling it.
One limitation of this study is that all mutations were considered as having equal importance. We did not weight the different mutations in the score, because there is no standardized weighting method currently available, despite a number of different approaches having been described (6, 14a). Another potential limitation is that multiple amino acids at a given protease position were grouped together as a single variable. However, independent statistical analysis for positions with a predominant single variant did not support the possibility that the results were dependent on a single amino acid. Indeed, for each mutation tested, the P value of the corresponding Fisher exact test was always higher (sometimes above 5%) than the P value obtained from the regrouping of all variants. In addition, the score obtained from analysis of the single dominant variant provided a P value greater than the P value obtained with the score for all variants considered as a single variable (P = 1.2 × 10−6 versus 1.5 × 10−7).
This work identified mutations not previously associated with a strong effect on PI resistance, suggesting that TPV has a unique resistance profile. This is consistent with previous phenotypic analysis. This mutation profile may partly explain TPV's activity against viruses harboring classical PI resistance mutations. This TPV/r resistance score is based on a relatively large number of patients with PI resistance mutations at baseline and could provide a useful tool for the prediction of VR to TPV/r in PI-experienced patients. However, it requires cross-validation in a different population study.
Acknowledgments
The Tipranavir Agence Nationale de Recherches sur le SIDA study group consisted of the following: Avicenne, C. Alloui; Bordeaux, B. Masquelier; Creteil, M. Bouvier; Fort de France, G. Dos Santos; Grenoble, A. Signori-Schmuck; Montpellier, B. Montes; Paris-Bichat Claude Bernard, D. Descamps and F. Brun-Vézinet; Paris-HEGP, A. Si Mohamed and C. Charpentier; Paris-Necker, M. L. Chaix; Paris-Pitié-Salpêtrière, A. Derache, A. G. Marcelin, V. Calvez, and P. Flandre; Paris-Tenon, C. Amiel and V. Schneider; Rennes, A. Ruffault and A. Maillard; Toulouse, J. Izopet; Tours, F. Barin.
Personnel at the clinical centers were as follows: Avicenne, O. Bouchaud; Bordeaux, P. Morlat and D. Neau; Créteil, Y. Lévy and S. Dominguez; Fort de France, A. Cabié; Grenoble, P. Leclercq; Montpellier, J. Reynes; Paris-Bichat Claude Bernard, P. Yéni; Paris-HEGP, L. Weiss; Paris-Necker, J. P. Viard; Paris-Pitié-Salpêtrière, C. Katlama; Paris-Tenon, G. Pialoux; Rennes, C. Michelet; Toulouse, B. Marchou; Tours, J. M. Besnier.
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Bacheler, L. T., H. Vermeiren, B. Winters, H. Van Der Borght, and D. Mayers. 2006. Abstr. 4th Eur. HIV Drug Resist. Workshop, abstr. 40.
- 1a.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.Bonneau, P. R., Y. Lie, and R. Bethell. 2007. Abstr. 11th Eur. AIDS Conf., abstr. P3.3/02.
- 2.Cahn, P., J. Villacian, A. Lazzarin, C. Katlama, B. Grinsztejn, K. Arasteh, P. Lopez, N. Clumeck, J. Gerstoft, N. Stavrianeas, S. Moreno, F. Antunes, D. Neubacher, and D. Mayers. 2006. Ritonavir-boosted tipranavir demonstrates superior efficacy to ritonavir-boosted protease inhibitors in treatment-experienced HIV-infected patients: 24-week results of the RESIST-2 trial. Clin. Infect. Dis. 43:1347-1356. [DOI] [PubMed] [Google Scholar]
- 3.Descamps, D., C. Delaugerre, B. Masquelier, A. Ruffault, A. G. Marcelin, J. Izopet, M. L. Chaix, V. Calvez, F. Brun-Vezinet, and D. Costagliola. 2006. Repeated HIV-1 resistance genotyping external quality assessments improve virology laboratory performance. J. Med. Virol. 78:153-160. [DOI] [PubMed] [Google Scholar]
- 4.Doyon, L., S. Tremblay, L. Bourgon, E. Wardrop, and M. G. Cordingley. 2005. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antivir. Res. 68:27-35. [DOI] [PubMed] [Google Scholar]
- 5.Flandre, P., A. G. Marcelin, J. Pavie, N. Shmidely, M. Wirden, O. Lada, M. C. Bernard, J. M. Molina, and V. Calvez. 2005. Comparison of tests and procedures to build clinically relevant genotypic scores: application to the Jaguar study. Antivir. Ther. 10:479-487. [PubMed] [Google Scholar]
- 6.Flandre, P., A. G. Marcelin, V. Soriano, S. Yerly, C. Katlama, and V. Calvez. 2006. Abstr. XV Int. HIV Drug Resist. Workshop, abstr. 67.
- 7.Hicks, C. B., P. Cahn, D. A. Cooper, S. L. Walmsley, C. Katlama, B. Clotet, A. Lazzarin, M. A. Johnson, D. Neubacher, D. Mayers, and H. Valdez. 2006. Durable efficacy of tipranavir-ritonavir in combination with an optimised background regimen of antiretroviral drugs for treatment-experienced HIV-1-infected patients at 48 weeks in the Randomized Evaluation of Strategic Intervention in multi-drug reSistant patients with Tipranavir (RESIST) studies: an analysis of combined data from two randomised open-label trials. Lancet 368:466-475. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 9.Larder, B. A., K. Hertogs, S. Bloor, C. H. van den Eynde, W. DeCian, Y. Wang, W. W. Freimuth, and G. Tarpley. 2000. Tipranavir inhibits broadly protease inhibitor-resistant HIV-1 clinical samples. AIDS 14:1943-1948. [DOI] [PubMed] [Google Scholar]
- 10.Marcelin, A. G., P. Flandre, C. de Mendoza, B. Roquebert, G. Peytavin, L. Valer, M. Wirden, S. Abbas, C. Katlama, V. Soriano, and V. Calvez. 2007. Clinical validation of saquinavir/ritonavir genotypic resistance score in protease-inhibitor-experienced patients. Antivir. Ther. 12:247-252. [PubMed] [Google Scholar]
- 11.Marcelin, A. G., P. Flandre, J. Pavie, N. Schmidely, M. Wirden, O. Lada, D. Chiche, J. M. Molina, and V. Calvez. 2005. Clinically relevant genotype interpretation of resistance to didanosine. Antimicrob. Agents Chemother. 49:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Marcelin, A. G., B. Masquelier, D. Descamps, J. Izopet, C. Charpentier, C. Alloui, M. Bouvier-Alias, A. Signori-Schmuck, B. Montes, M.-L. Chaix, C. Amiel, G. Dos Santos, A. Ruffault, F. Barin, G. Peytavin, M. Lavignon, P. Flandre, and V. Calvez. 2007. Abstr. 14th Conf. Retrovir. Opportunistic Infect., abstr. 612.
- 11b.Marcelin, A. G., B. Masquelier, D. Descamps, J. Izopet, C. Charpentier, C. Alloui, M. Bouvier-Alias, A. Signori-Schmuck, B. Montes, M.-L. Chaix, C. Amiel, G. Dos Santos, A. Ruffault, F. Barin, G. Peytavin, M. Lavignon, P. Flandre, and V. Calvez. 2007. Abstr. XVI Int. HIV Drug Resist. Workshop, abstr. 74.
- 12.Naeger, L. K., and K. A. Struble. 2007. Food and Drug Administration analysis of tipranavir clinical resistance in HIV-1-infected treatment-experienced patients. AIDS 21:179-185. [DOI] [PubMed] [Google Scholar]
- 12a.Parkin, N. T., and C. Chappey. 2006. Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 637.
- 12b.Peytavin, G., A. G. Marcelin, A. Rouault, M. Bonmarchand, H. Ait-Mohand, B. Cassard, A. Simon, L. Schneider, D. Costagliola, V. Calvez, and C. Katlama. 2006. Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 591.
- 13.Poppe, S. M., D. E. Slade, K. T. Chong, R. R. Hinshaw, P. J. Pagano, M. Markowitz, D. D. Ho, H. Mo, R. R. Gorman III, T. J. Dueweke, S. Thaisrivongs, and W. G. Tarpley. 1997. Antiviral activity of the dihydropyrone PNU-140690, a new nonpeptidic human immunodeficiency virus protease inhibitor. Antimicrob. Agents Chemother. 41:1058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusconi, S., S. La Seta Catamancio, P. Citterio, S. Kurtagic, M. Violin, C. Balotta, M. Moroni, M. Galli, and A. d'Arminio-Monforte. 2000. Susceptibility to PNU-140690 (tipranavir) of human immunodeficiency virus type 1 isolates derived from patients with multidrug resistance to other protease inhibitors. Antimicrob. Agents Chemother. 44:1328-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Scherer, J. R., C. A. Boucher, J. D. Baxter, J. M. Schapiro, V. Kohlbrenner, and D. B. Hall. 2007. Abstr. 11th Eur. HIV Drug Resist. Workshop, abstr. P3.4/07.
- 14b.Scherer, J. R., V. Kohlbrenner, D. B. Hall, J. D. Baxter, J. M. Schapiro, and C. A. Boucher. 2007. Abstr. 5th Eur. HIV Drug Resist. Workshop, abstr. 100.
- 15.Vora, S., A. G. Marcelin, H. F. Gunthard, P. Flandre, H. H. Hirsch, B. Masquelier, A. Zinkernagel, G. Peytavin, V. Calvez, L. Perrin, and S. Yerly. 2006. Clinical validation of atazanavir/ritonavir genotypic resistance score in protease inhibitor-experienced patients. AIDS 20:35-40. [DOI] [PubMed] [Google Scholar]