Abstract
Active efflux of antimicrobial agents is one of the most important adapted strategies that bacteria use to defend against antimicrobial factors that are present in their environment. The NorM protein of Neisseria gonorrhoeae and the YdhE protein of Escherichia coli have been proposed to be multidrug efflux pumps that belong to the multidrug and toxic compound extrusion (MATE) family. In order to determine their antimicrobial export capabilities, we cloned, expressed, and purified these two efflux proteins and characterized their functions both in vivo and in vitro. E. coli strains expressing norM or ydhE showed elevated (twofold or greater) resistance to several antimicrobial agents, including fluoroquinolones, ethidium bromide, rhodamine 6G, acriflavine, crystal violet, berberine, doxorubicin, novobiocin, enoxacin, and tetraphenylphosphonium chloride. When they were expressed in E. coli, both transporters reduced the levels of ethidium bromide and norfloxacin accumulation through a mechanism requiring the proton motive force, and direct measurements of efflux confirmed that NorM behaves as an Na+-dependent transporter. The capacities of NorM and YdhE to recognize structurally divergent compounds were confirmed by steady-state fluorescence polarization assays, and the results revealed that these transporters bind to antimicrobials with dissociation constants in the micromolar region.
Neisseria gonorrhoeae is a gram-negative diplococcus which is found only in humans and causes the sexually transmitted disease gonorrhea. Since it is a strictly human pathogen and can colonize male and female genital mucosal surfaces and other sites, it has developed mechanisms to overcome host antimicrobial systems that are important in the innate host defense. One important mechanism that N. gonorrhoeae uses to subvert antimicrobial agents is the expression of multidrug efflux pumps that recognize and actively export a wide variety of toxic (often structurally unrelated) compounds, including antibacterial peptides, long-chain fatty acids, and several clinically useful antibiotics, from the bacterial cell (17, 32, 35, 36).
Recently, four efflux pumps have been identified in N. gonorrhoeae. One such pump is the MtrD inner membrane protein (21) that exists as a component of a tripartite resistance nodulation cell division (RND) efflux system (41). MtrD interacts with a periplasmic membrane fusion protein, MtrC, and an outer membrane channel, MtrE, to mediate the export of hydrophobic antimicrobial agents, including antibiotics, nonionic detergents, certain antibacterial peptides, bile salts, and gonadal steroidal hormones (7, 9, 10, 36). The FarB efflux pump (17), which belongs to the major facilitator (MF) family (8, 22, 31), recognizes antibacterial long-chain fatty acids and exports them out of the cell in conjunction with the FarA membrane fusion protein and the MtrE outer membrane protein channel (17). The MacB transporter was recently described (33), and it belongs to the ATP binding cassette transporter family (13). It is poorly expressed in wild-type gonococci due to a natural mutation in its promoter, but it can recognize and export certain macrolide antibiotics. Finally, N. gonorrhoeae contains the NorM efflux pump (32), which is a member of the multidrug and toxic compound extrusion (MATE) family of efflux transporters (5, 25, 32). The N. gonorrhoeae NorM transporter is homologous to NorM of Vibrio parahaemolyticus (25), YdhE of Escherichia coli (2, 25, 42), VmrA of V. parahaemolyticus (6), and VcrM of Vibrio cholerae (1, 27). As a multidrug efflux pump, the gonococcal NorM appears to recognize a number of cationic toxic compounds, such as ethidium bromide, acriflavine, 2-N-methylellipticinium, and ciprofloxacin (32). The capacity of NorM to export ciprofloxacin was suggested to be of clinical relevance in the development of fluoroquinolone resistance in N. gonorrhoeae, especially when isolates have other mutations that raise the MIC to a level near that seen in treatment failures.
Members of the MATE family of transporters characteristically possess 12 putative transmembrane domains and have been reported in all three kingdoms of life (Eukaryae, Archaeae, and Eubacteriae) (5). Phylogenetic tree analysis suggests that this family possesses three distinct clusters: (i) the bacterial multidrug efflux pumps, including N. gonorrhoeae NorM and E. coli YdhE; (ii) the eukaryotic efflux proteins found in fungi and plants, such as Erc1; and (iii) a branch containing E. coli DinF (5). Among these three clusters, the putative NorM branch exhibits overall identity and similarity of 26 and 47%, respectively.
In order to directly test the capacity of the N. gonorrhoeae NorM pump to recognize putative substrates, including ciprofloxacin, which was, until recently, widely used in the United States to treat gonococcal infections (15), we expressed the gonococcal norM gene in E. coli and investigated its function by using drug susceptibility and transport assays. We also studied the homologous protein YdhE in E. coli and compared its function with that of N. gonorrhoeae NorM. Using a sensitive fluorescence polarization assay (12, 18), we characterized the interactions of these two transporters with various drugs. The results revealed that NorM and YdhE bind to a variety of structurally dissimilar agents in the micromolar range.
MATERIALS AND METHODS
Cloning of norM and ydhE.
The norM open reading frame from the genomic DNA of N. gonorrhoeae strain FA19 was amplified by PCR with primers 5′-GAGGAATAATAAATGCTGCTCGACCTCGACCGC-3′ and 5′-GTTTAAACTCAATGGTGATGGTGATGATGGACGGCCTTGTGTGATTTGACC-3′ to generate a product that would encode a NorM recombinant protein with a six-His tag at the C terminus (32). The corresponding PCR product was extracted from the agarose gel and cloned into pBAD-TOPO, as described by the manufacturer (Invitrogen). To remove the V5 epitope and polyhistidine region of the vector, the plasmid was digested with PmeI and then self-ligated to form the expression vector pBADΩnorM. The recombinant plasmid was transformed into DH5α cells, and transformants were selected on LB agar plates containing 100 μg/ml ampicillin. The presence of the correct norM sequence in the plasmid construct was verified by DNA sequencing.
Similarly, the open reading frame of ydhE from E. coli K-12 genomic DNA was amplified by PCR and TA cloned into pBAD-TOPO with primers 5′-GAGGAATAATAAATGCAGAAGTATATCAGTG-3′ and 5′-GTTTAAACTCAATGGTGATGGTGATGATG GCGGGATGCTCGTTGCAGAATG-3′ to generate a PCR product that would encode a recombinant protein containing a six-His tag at the C terminus. The recombinant plasmid (pBADΩydhE) was transformed into DH5α cells as described above, and the presence of the correct insert in a representative plasmid transformant was verified by DNA sequencing.
Drug susceptibility assays.
The MICs to various antimicrobial agents of E. coli strains (inoculum, 104 CFU/spot) harboring pBADΩnorM, pBADΩydhE, or the pBAD vector were determined by the twofold dilution method with LB agar containing the desired antimicrobial agent (28). Bacterial growth was recorded after 18 to 24 h of incubation at 37°C. Each assay was repeated at least four times to ensure the reproducibility of the results.
Drug accumulation assays.
Assays of norfloxacin and ethidium bromide accumulation were performed as described previously (20, 24, 25). In brief, E. coli AG100AX cells carrying pBADΩnorM, pBADΩydhE, or pBAD were grown in LB broth with 0.01% l-arabinose at 37°C to an optical density at 600 nm (OD600) of 1.0, harvested, washed three times with buffer containing 0.1 M Tris-HCl (pH 7.0), and suspended in the same buffer to an OD600 of 1.0. Norfloxacin was then added to a final concentration of 100 μM. Samples of 1 ml were taken at intervals, centrifuged at 10,000 rpm for 30 s at 4°C, and washed once with the same buffer. After 15 min, carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to a final concentration of 100 μM to disrupt the proton gradient across the membrane. Then, 15 min later, CCCP was removed by centrifugation at 10,000 rpm for 30 s and the cells were resuspended in buffer containing 0.1 M Tris-HCl (pH 7.0) and 0.4% glucose. Each pellet was resuspended in 1 ml of 100 mM glycine-HCl (pH 3.0), shaken vigorously for 1 h at room temperature to release the fluorescent content, and then centrifuged at 15,000 rpm for 10 min at room temperature. The fluorescence of the supernatants was measured at an excitation λ (λex) and an emission λ (λem) of 277 and 448 nm, respectively, by using a Perkin-Elmer LS55 spectrofluorometer equipped with a Hamamatsu R928 photomultiplier. The amount of maximum fluorescence was normalized to 100%.
For ethidium bromide accumulation, the cells were prepared in a manner similar to that described above. To initiate the assay, ethidium bromide was added to the cell suspension at a final concentration of 20 μg/ml. Samples of 1 ml were taken at different time points. After 15 min of incubation, CCCP was added to a final concentration of 100 μM. Then, 15 min later, CCCP was removed by centrifugation at 10,000 rpm for 30 s and the cells were resuspended in buffer containing 0.1 M Tris-HCl (pH 7.0) and 0.4% glucose. The fluorescence of the samples was measured at a λex and a λem of 500 and 580 nm, respectively (3). The amount of maximum fluorescence was normalized to 100%.
Accumulation of ethidium bromide in the presence of Na+.
E. coli AG100AX cells containing pBADΩnorM or pBAD were grown in LB broth with 0.01% l-arabinose at 37°C to an OD600 of 1.0, harvested by centrifugation, washed three times with buffer containing 0.1 M Tris-HCl (pH 7.0), and suspended in the same buffer to an OD600 of 1.0. Ethidium bromide was added to the cell suspension at a final concentration of 20 μg/ml, followed by the addition of 100 mM NaCl or KCl. At each time point, a 1-ml sample was removed, centrifuged at 10,000 rpm for 30 s at 4°C, and washed once with the same buffer. The fluorescence signal was measured at a λex and a λem of 500 and 580 nm, respectively.
Efflux of norfloxacin in the presence of Na+.
For the determination of norfloxacin efflux, cells were grown in LB broth with 0.01% l-arabinose at 37°C to an OD600 of 1.0, harvested, and washed twice with buffer containing 0.1 M Tris-HCl (pH 7.0). To load the cells with norfloxacin, the bacteria were incubated in the same buffer supplemented with 100 μM norfloxacin and 20 μM CCCP at 37°C for 30 min, as described previously (30). The cells were then pelleted, washed twice, and resuspended in the same buffer to an OD600 of 2.0. Samples of 1 ml were taken at intervals, centrifuged at 10,000 rpm for 30 s at 4°C, and washed once with the same buffer. After 10 min, NaCl or KCl was added to a final concentration of 100 mM. Fluorescence measurement of norfloxacin was performed by a procedure similar to that described above.
Purification of transporters.
The N. gonorrhoeae NorM protein containing a six-His tag at the C terminus was overproduced in E. coli TOP10 cells (Invitrogen) possessing pBADΩnorM. The cells were grown in 12 liters of LB medium with 100 μg/ml ampicillin at 37°C. When the OD600 reached 0.5, the culture was treated with 0.2% l-arabinose to induce norM expression and cells were harvested within 3 h. The bacteria collected were resuspended, as described previously (43), in low-salt buffer containing 100 mM sodium phosphate (pH 7.2), 10% glycerol, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and were then disrupted with a French pressure cell. The membrane fraction was collected and washed twice with a high-salt buffer containing 20 mM sodium phosphate (pH 7.2), 2 M KCl, 10% glycerol, 1 mM EDTA, and 1 mM PMSF and once with 20 mM HEPES-NaOH buffer (pH 7.5) containing 1 mM PMSF. The membrane protein was then solubilized in 1% (wt/vol) n-dodecyl-β-d-maltoside (DDM). Insoluble material was removed by ultracentrifugation at 370,000 × g. The extracted protein was loaded into an Ni2+-affinity column; washed with a buffer containing 20 mM HEPES-NaOH (pH 7.5), 50 mM imidazole, and 0.02% DDM; and eluted with a buffer consisting of 20 mM HEPES-NaOH (pH 7.5), 400 mM imidazole, and 0.02% DDM. The eluted protein was then concentrated to 5 mg/ml and loaded into G-200 sizing columns preincubated with 20 mM HEPES-NaOH (pH 7.5) and 0.02% DDM for further purification. The YdhE protein that contains a six-His tag at the C terminus was overproduced in E. coli TOP10/pBADΩydhE and purified as described above for NorM. The purities (>90%) of the NorM and YdhE proteins were judged by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie brilliant blue staining.
Fluorescence polarization assays.
Fluorescence polarization assays (12, 18) were used to determine the drug binding affinities of NorM and YdhE. The experiments were done with a ligand binding solution containing 20 mM HEPES-NaOH (pH 7.5), 0.02% DDM, and 1 μM ligand (rhodamine 6G, ethidium bromide, proflavine, ciprofloxacin, or norfloxacin). The protein solution, which consisted of NorM or YdhE in 20 mM HEPES-NaOH (pH 7.5), 0.02% DDM, and 1 μM ligand, was titrated into the ligand binding solution until the polarization was unchanged. As this is a steady-state approach, the fluorescence polarization measurement was taken after a 5-min incubation for each corresponding concentration of the protein and drug to ensure that the binding had reached equilibrium. It should be noted that the detergent concentration was kept constant at all times to eliminate the change in polarization generated by the drug-DDM micelle interaction. All measurements were performed at 25°C with a Perkin-Elmer LS55 spectrofluorometer equipped with a Hamamatsu R928 photomultiplier. The excitation wavelengths were 527, 483, 447, 330, and 277 nm for rhodamine 6G, ethidium, proflavine, ciprofloxacin, and norfloxacin, respectively. Fluorescence polarization signals (as the change in polarization) were measured at emission wavelengths of 550, 620, 508, 415, and 448 nm for these ligands, respectively. Each titration point recorded was an average of 15 measurements. Data were analyzed by using the equation P = {(Pbound − Pfree)[protein]/(KD + [protein])} + Pfree, where P is the polarization measured at a given total protein concentration, Pfree is the initial polarization of free ligand, Pbound is the maximum polarization of specifically bound ligand, [protein] is the protein concentration, and KD is the dissociation constant. The titration experiments were repeated three times to obtain the average KD value. Curve fitting was accomplished with the ORIGIN program (version 7.5; OriginLab Corporation, Northampton, MA).
RESULTS AND DISCUSSION
Drug specificity of NorM and YdhE in vivo.
To determine if the N. gonorrhoeae NorM multidrug transporter is functionally expressed in E. coli and to compare its activity to that of a second member (YdhE) of the MATE family of efflux pumps, we transformed E. coli AG100AX, an AcrAB-deficient strain (29), with the empty pBAD vector, pBADΩnorM, or pBADΩydhE. We then tested the susceptibilities of the representative transformants bearing these plasmids to a panel of 24 structurally divergent antimicrobials (Table 1). These antimicrobials were selected on the basis of whether or not they are putative substrates of MATE family exporters in the NorM subclass. It is important to stress that in many instances the expression of drug efflux pumps, including members of the MATE superfamily (5), can result in only modest changes (twofold) in bacterial susceptibility to certain antimicrobials, while more significant changes in susceptibility to other agents can be observed in the same system. Indeed, in our drug testing experiments, we detected a range of changes in the susceptibilities of E. coli cells producing either NorM or YdhE.
TABLE 1.
Drug susceptibility
| Drug | MIC (μg/ml)
|
||
|---|---|---|---|
| AG100AX/pBADΩnorM | AG100AX/pBADΩydhE | AG100AX/pBAD | |
| Ethidium bromide | 15.63 | 15.63 | 7.81 |
| Ciprofloxacin | 0.006 | 0.006 | 0.003 |
| Rhodamine 6G | 12.5 | 12.5 | 3.125 |
| Chloramphenicol | 0.625 | 0.625 | 0.625 |
| TPP | 25 | 25 | 12.5 |
| Norfloxacin | 0.032 | 0.016 | 0.008 |
| Ofloxacin | 0.013 | 0.013 | 0.013 |
| Lomefloxacin | 0.032 | 0.016 | 0.016 |
| Tetracycline | 1.25 | 1.25 | 1.25 |
| Enoxacin | 0.063 | 0.063 | 0.031 |
| Doxorubicin | 6.25 | 6.25 | 1.56 |
| Benzalkonium | 0.625 | 0.312 | 0.156 |
| Novobiocin | 3.125 | 3.125 | 1.56 |
| Nalidixic acid | 0.625 | 0.625 | 0.625 |
| Crystal violet | 2.5 | 2.5 | 0.625 |
| CCCP | 6.25 | 6.25 | 6.25 |
| Rifampin | 6.25 | 6.25 | 6.25 |
| Streptomycin | >50,000 | >50,000 | >50,000 |
| Methyl viologen | 156.3 | 156.3 | 156.3 |
| Vancomycin | 250 | 250 | 250 |
| Minocycline | 0.625 | 0.625 | 0.625 |
| Berberine | 250 | 250 | 31.25 |
| Acriflavine | 7.82 | 7.82 | 3.91 |
| Proflavine | 12.5 | 12.5 | 6.25 |
| Gentamicin | 2.5 | 2.5 | 2.5 |
Among the 24 antimicrobials tested, AG100AX cells expressing the N. gonorrhoeae NorM were less sensitive (two- to eightfold) than AG100AX cells containing pBAD to a subset of antibiotics (norfloxacin, doxorubicin, novobiocin, lomefloxacin, enoxacin, and ciprofloxacin), dyes (rhodamine 6G, ethidium bromide, acriflavine, and crystal violet), and quaternary ammonium compounds (berberine, benzalkonium, and tetraphenylphosphonium chloride [TPP]). The YdhE-producing cells displayed a similar drug susceptibility profile (with the exception of that to lomefloxacin) as that of the NorM-producing E. coli cells, suggesting that these two MATE transporters are functionally similar in vivo.
Like YdhE of E. coli, our drug susceptibility experiments showed that NorM functions as a multidrug efflux pump when it is expressed in E. coli. The MICs to 14 of the 24 antimicrobials (Table 1) were observed to increase by two- to eightfold in NorM-producing AG100AX bacteria, which lack the AcrB efflux pump. Similar differences (data not presented) in antimicrobial susceptibility were obtained in AcrB+ E. coli TOP10 cells that were used to obtain recombinant NorM. In that the MICs of norfloxacin, ciprofloxacin, enoxacin, and lomefloxacin were raised in cells expressing NorM or YdhE, our results suggest that, like other MATE transporters, such as BexA of Bacteroides thetaiotaomicron (23), VcmA of Vibrio cholerae (16), NorM of Vibrio cholerae (37) and AbeM of Acinetobacter baumannii (40), fluoroquinolones might be a common substrate for MATE transporters in the NorM cluster.
NorM and YdhE reduce drug accumulation in bacteria.
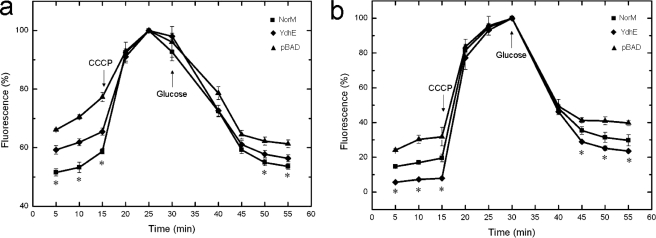
The observed (Table 1) capacity of NorM and YdhE to reduce the susceptibility of E. coli to norfloxacin, ciprofloxacin, and ethidium bromide is consistent with the drug recognition profiles of other members of the MATE efflux transporter family. To confirm the drug susceptibility testing results, we measured the levels of accumulation of norfloxacin and ethidium bromide in AG100AX cells that carried pBADΩnorM, pBADΩydhE, or pBAD. The results showed lower levels of norfloxacin (Fig. 1a) or ethidium bromide (Fig. 1b) accumulation in AG100AX cells producing NorM or YdhE compared to those in control cells harboring the empty pBAD vector.
FIG. 1.
NorM and YdhE reduce the levels of accumulation of norfloxacin and ethidium bromide in E. coli cells. (a) Accumulation of norfloxacin in cells transformed by NorM (AG100AX/pBADΩnorM), YdhE (AG100AX/pBADΩydhE), or an empty vector (AG100AX/pBAD). CCCP was added to the suspensions (first arrow) at a final concentration of 100 μM. After 15 min, glucose was added (second arrow) at a final concentration of 0.4%. (b) Accumulation of ethidium bromide in the same strains. CCCP was added to the suspensions (first arrow) at a final concentration of 100 μM. After 15 min, glucose was added (second arrow) at a final concentration of 0.4%. *, the values for AG100AX/pBADΩnorM and AG100AX/pBADΩydhE cells were significantly different from those for the control (AG100AX/pBAD) (P < 0.05).
Members of the MATE family use energy from the proton motive force (PMF) during export; and loss of the PMF inactivates pump activity, resulting in the enhanced accumulation of substrates, which translates to increased susceptibility to the agents normally recognized by the transporter. Accordingly, we also measured the levels of norfloxacin and ethidium bromide accumulation after the addition of CCCP, a decoupler of the membrane proton gradient. After the addition of CCCP into the assay solution, the levels of accumulation of norfloxacin (Fig. 1a) and ethidium bromide (Fig. 1b) increased drastically in the NorM- and YdhE-expressing cells, with the levels of accumulation being nearly the same in the three different strains within 5 min. It should be noted that the effect of CCCP is reversible simply by removing the CCCP and adding glucose to reenergize the AG100AX cells. The experiments were performed at least three times. Statistical comparisons were assessed by Student's t test, and P values of <0.05 were considered to indicate statistically significant differences (Fig. 1a and b).
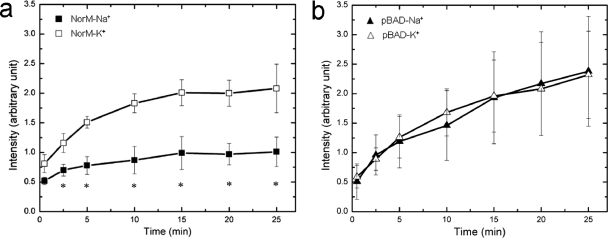
NorM expressed in E. coli behaves as an Na+ ion-dependent transporter.
It has been suggested but not proven that NorM of N. gonorrhoeae is an Na+-drug antiporter (32). Thus, we investigated the levels of accumulation of ethidium bromide in isogenic strains AG100AX/pBADΩnorM and AG100AX/pBAD in the presence of Na+ or K+. In AG100AX/pBADΩnorM cells, a lower level of ethidium bromide accumulation was observed in the presence of 100 mM NaCl than in the presence of 100 mM KCl (Fig. 2a). In contrast, we did not observe a difference in the levels of ethidium bromide accumulation in AG100AX/pBAD cells, regardless of the presence of NaCl or KCl (Fig. 2b). Furthermore, in the presence of NaCl, the level of accumulation of ethidium bromide in the NorM-producing strain was lower than that in AG100AX/pBAD.
FIG. 2.
Sodium ion reduces the level of accumulation of ethidium bromide in cells transformed by NorM. (a) Accumulation of ethidium bromide in AG100AX/pBADΩnorM cells after the addition of 100 mM NaCl or KCl; (b) accumulation of ethidium bromide in AG100AX/pBAD cells (cells carrying the empty vector) after the addition of 100 mM NaCl or KCl. *, the values of the ethidium bromide fluorescence intensity in the presence of 100 mM NaCl were significantly different from those in the presence of 100 mM KCl (P < 0.01).
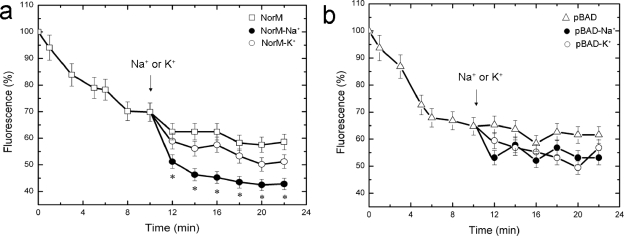
We next measured the efflux of norfloxacin that had accumulated in strain AG100AX/pBADΩnorM or AG100AX/pBAD in the presence of Na+ or K+. Cells were first loaded with norfloxacin; thereafter, 100 mM either NaCl or KCl was added to test the effect on drug efflux. As shown in Fig. 3a, the addition of 100 mM NaCl to AG100AX/pBADΩnorM cells resulted in a greater efflux of the drug from the cells compared to the levels of efflux observed in the presence of KCl or without added salt. This effect was NorM dependent because the addition of either NaCl (100 mM) or KCl (100 mM) to AG100AX/pBAD cells did not affect norfloxacin efflux (Fig. 3b). Taken together with the ethidium bromide accumulation results, our observations support the earlier proposal (32) that the gonococcal NorM protein is an Na+-dependent transporter.
FIG. 3.
Sodium ion enhances norfloxacin efflux via NorM. (a) Norfloxacin efflux from AG100AX/pBADΩnorM cells carrying the norM gene from N. gonorrhoeae; (b) norfloxacin efflux from AG100AX/pBAD cells carrying the empty vector. NaCl or KCl was added to the suspensions (arrow) at a final concentration of 100 mM. *, the values of norfloxacin fluorescence in the presence of 100 mM NaCl were significantly different from those in the presence of 100 mM KCl (P < 0.05).
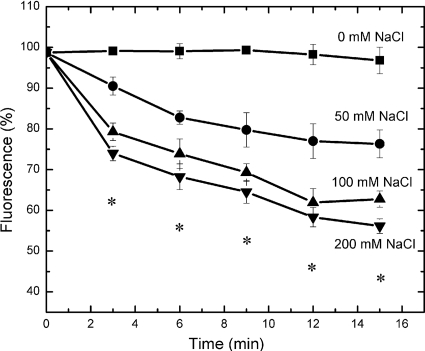
In order to directly test whether efflux of an antimicrobial by NorM is Na+ dependent, we then studied the extrusion of the norfloxacin that had accumulated in AG100AX/pBADΩnorM in the presence of CCCP. Cells were first loaded with norfloxacin. Thereafter, different concentrations of NaCl (50 to 200 mM) were added to test their effects on drug efflux. All these tests were done in the presence of 20 μM CCCP. If NorM requires proton ion as a coupler, then the level of norfloxacin accumulated in the cell should not be reduced under these conditions. However, if the coupling ion is Na+, we should be able to observe norfloxacin efflux in an [Na+]-dependent manner. Figure 4 depicts the effect of Na+ on the extrusion of norfloxacin. The experiment suggests that efflux activity was stimulated by the Na+ ion. The addition of NaCl resulted in a significant change in norfloxacin efflux. A previous study with Pseudomonas aeruginosa PmpM, an H+-dependent transporter in the MATE family, indicated that the Na+ ion has no effect on drug transport (11). Thus, it is highly likely that Na+ is the coupling cation for NorM.
FIG. 4.
Effect of Na+ concentration on norfloxacin extrusion via NorM. Norfloxacin efflux from AG100AX/pBADΩnorM cells carrying the norM gene was measured in the presence of 0 to 200 mM NaCl. *, the values of norfloxacin fluorescence in the presence of 50, 100, or 200 mM NaCl were significantly different from those in the absence of NaCl (P < 0.003), and the values of norfloxacin fluorescence in the presence of 100 or 200 mM NaCl were significantly different from those in the presence of 50 mM NaCl (P < 0.01).
Our drug accumulation and efflux assays confirm the results from an earlier report which suggested that NorM of N. gonorrhoeae is an Na+-drug antiporter (32). First, Na+ was found to enhance the ability of E. coli cells producing NorM to maintain a lower level of accumulation of ethidium bromide compared to the levels of accumulation by cells not producing NorM, even in the presence of Na+ (Fig. 2). Hence, drug export mediated by NorM of N. gonorrhoeae is driven by an Na+ gradient. This export mechanism is also influenced by the integrity of the PMF of the cytoplasmic membrane, since CCCP, a proton conductor, could significantly enhance the level of accumulation of ethidium bromide in NorM-producing bacteria. These accumulation assays, however, provide indirect evidence for efflux. Accordingly, direct support for efflux was obtained in experiments that measured the level of norfloxacin export by NorM-producing E. coli cells. The observed Na+-dependent capacity of NorM to mediate the export of norfloxacin (Fig. 3 and 4) provides direct evidence for this model.
Binding affinities of different drugs in vitro.
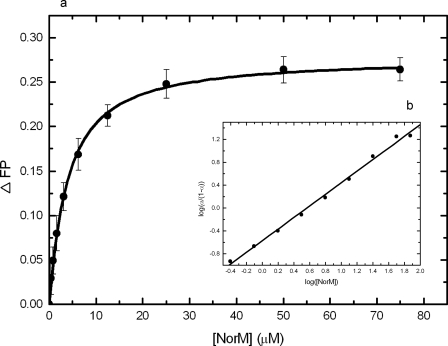
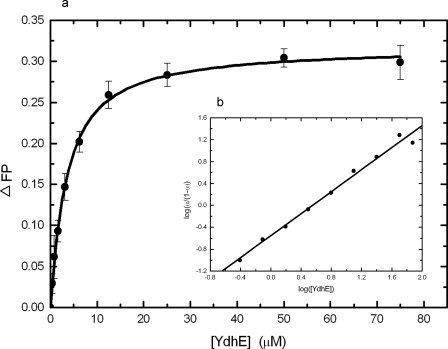
The results of the in vivo antimicrobial susceptibility tests (Table 1) and the accumulation/efflux studies (Fig. 1 to 4) suggested that NorM and YdhE recognize and export structurally diverse compounds. Little is known, however, about the detail of substrate binding by these MATE transporters. In order to directly test the capacities of these proteins to bind to such diverse compounds, we used a fluorescence polarization assay to quantify the binding affinities of several compounds to NorM and YdhE in vitro. This technique has been found to be sensitive and precise enough and allows us to detect for the first time the ligand binding of the E. coli AcrB multidrug efflux pump (an RND transporter) in a detergent environment (39). As an example of the results obtained by this protocol, Fig. 5a illustrates the binding isotherm of NorM in the presence of rhodamine 6G. As presented in Fig. 5a, a simple hyperbolic curve was observed for the binding of rhodamine 6G, and the KD was 3.4 ± 0.2 μM. A Hill plot of the data yielded a Hill coefficient of 1.01 ± 0.03 (Fig. 5b), suggesting a simple drug binding process with a stoichiometry of one NorM monomer per rhodamine 6G ligand. When the data were presented as a Scatchard plot (data not shown), it indicated a similar KD of 3.6 ± 0.1 μM with no sign of heterogeneity in binding sites. In addition, the titration experiments indicated that YdhE binds to rhodamine 6G with a KD of 3.0 ± 0.2 μM (Fig. 6a). This value is comparable to that of NorM, suggesting that these two transporters have similar affinities for this ligand. The Hill plots (Fig. 6b) and the Scatchard plots (data not shown) of the data indicated that the transporter employs a simple binding stoichiometry of a 1:1 monomeric YdhE-to-rhodamine 6G molar ratio.
FIG. 5.
Representative fluorescence polarization of NorM in 0.02% DDM with rhodamine 6G. (a) Binding isotherm of NorM with rhodamine 6G showing a KD of 3.4 ± 0.2 μM in buffer containing 20 mM HEPES-NaOH (pH 7.5) and 0.02% DDM. (b) Hill plot of the data obtained for rhodamine 6G binding to NorM. α, the fraction of bound rhodamine 6G. The plot gives a slope of 1.01 ± 0.03, indicating a simple binding process with no cooperativity. The interception of the plot provides a KD of 3.6 ± 0.1 μM for rhodamine 6G binding.
FIG. 6.
Representative fluorescence polarization of YdhE in 0.02% DDM with rhodamine 6G. (a) Binding isotherm of YdhE with rhodamine 6G showing a KD of 3.0 ± 0.2 μM in buffer containing 20 mM HEPES-NaOH (pH 7.5) and 0.02% DDM. (b) Hill plot of the data obtained for rhodamine 6G binding to YdhE. α, the fraction of bound rhodamine 6G. The plot gives a slope of 1.00 ± 0.04, indicating a simple binding process with no cooperativity. The interception of the plot provides a KD of 3.6 ± 0.2 μM for rhodamine 6G binding.
Fluorescence polarization assays were also employed to elucidate the interaction of a variety of drugs with the NorM and YdhE transporters. The titration experiments indicated that NorM binds to ethidium bromide, proflavine, ciprofloxacin, and norfloxacin with KDs of 12.3 ± 1.3 μM, 33.6 ± 1.9 μM, 121.3 ± 15.7 μM, and 105.6 ± 9.8 μM, respectively (Table 2). These values are in the same range as those of YdhE, which binds to ethidium bromide, proflavine, ciprofloxacin, and norfloxacin with KDs of 9.8 ± 0.9 μM, 22.1 ± 0.9 μM, 90.9 ± 12.4 μM, and 98.4 ± 16.2 μM, respectively (Table 2). This binding was not affected by detergent, since similar KDs for the rhodamine 6G-YdhE complex were obtained in reaction mixtures that included 0.2%, 0.02%, and 0.0075% DDM.
TABLE 2.
KDs and Hill coefficients of NorM and YdhE with five different transported drugs
| Drug | NorM
|
YdhE
|
||
|---|---|---|---|---|
| KD (μM) | Hill coefficient | KD (μM) | Hill coefficient | |
| Rhodamine 6G | 3.4 ± 0.2 | 1.01 ± 0.03 | 3.0 ± 0.2 | 1.00 ± 0.04 |
| Ethidium bromide | 12.3 ± 1.3 | 0.96 ± 0.05 | 9.8 ± 0.9 | 0.97 ± 0.05 |
| Profloxacin | 33.6 ± 1.9 | 0.94 ± 0.03 | 22.1 ± 0.9 | 1.02 ± 0.01 |
| Ciprofloxacin | 121.3 ± 15.7 | 1.08 ± 0.06 | 90.9 ± 12.4 | 1.09 ± 0.06 |
| Norfloxacin | 126.3 ± 11.3 | 1.06 ± 0.03 | 98.4 ± 16.2 | 1.14 ± 0.05 |
To verify the validity of the fluorescence polarization assay, we studied the binding affinity of rhodamine 6G to the purified, detergent-solubilized YdhE efflux pump using equilibrium dialysis. The dialysis data suggest a KD of 3.0 ± 0.1 μM, with a stoichiometry of a 1:1 monomeric YdhE-to-ligand molar ratio, for the binding of YdhE to rhodamine 6G (see Fig. S1 in the supplemental material). This result is in good agreement with that observed by the fluorescence polarization assay, in which the KD of YdhE-rhodamine 6G is 3.0 μM with a 1:1 binding stoichiometry. Apparently, NorM and YdhE bind to these drugs with similar affinities. These values are similar to the KD values for TPP binding in MdfA, an MF transporter, and EmrE, an SMR transporter, as determined by radiolabeled binding assays (19, 26), as well as the most recently determined KDs for AcrB, an RND transporter, with four different ligands, as determined by fluorescence polarization assays (39).
The results from the fluorescence polarization and equilibrium dialysis assays suggest that NorM and YdhE use a binding stoichiometry of a 1:1 monomeric transporter-to-ligand ratio. Thus, it is very likely that these transporters employ a simple drug binding process with no cooperativity. There is, however, a possibility that drug molecules may bind to more than one site in the proteins with similar affinities that may not be able to be distinguished by these techniques. Further experiments by different approaches may need to confirm this drug binding stoichiometry.
pH dependence of drug binding to NorM.
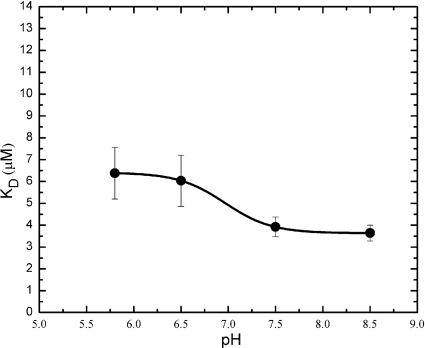
We performed fluorescence polarization experiments to determine the binding of rhodamine 6G to NorM at different pH values. Figure 7 illustrates the decreases in the KD values for rhodamine 6G as the pH increases from 5.8 to 8.5. Within this pH range, the KD value decreases by about 2.6 μM. This change is quite modest compared with the effect of protons on the PMF-dependent efflux pumps AcrB (39) and EmrE (26) in E. coli. The binding affinities for drugs in these two transporters were strikingly dependent on the pH. In the case of AcrB, within a pH range from 5.5 to 8.4, the fluorescence polarization assay results suggest that the change in KD for AcrB-rhodamine 6G reached 10.3 μM. As NorM shows a pH-dependent profile very different from the profiles of the PMF-dependent pumps AcrB and EmrE, it is very likely that NorM does not use H+ as an energy-coupling ion. Since NorM tends to bind to positively charged drugs, we expect that its drug-binding pocket should consist of at least one acidic residue. Indeed, the protein sequence of NorM suggests that this transporter consists of two negatively charged residues, Glu 261 and Asp 377, in the transmembrane region. These acidic residues could potentially form the multidrug binding site. Thus, the modest pH effect on NorM could be accounted for by the protonation state of the acid residue(s) in the binding pocket.
FIG. 7.
Effect of pH on the KD of rhodamine 6G binding to NorM. The resulting KDs were plotted against the pH.
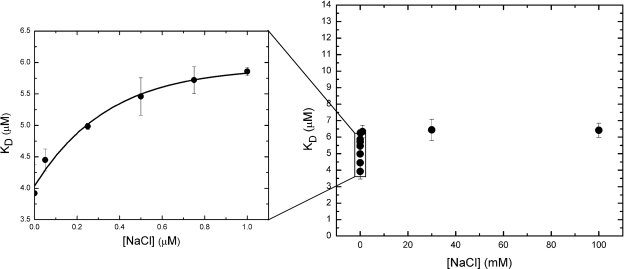
Na+ dependence of drug binding to NorM.
As the results of in vivo studies of drug accumulation and efflux suggest, NorM is a Na+-dependent transporter, and the presence of Na+ ions may affect the drug binding affinity in vitro. We therefore investigated the Na+ dependence of drug binding using fluorescence polarization. As shown in Fig. 8, the KD for rhodamine 6G is Na+ dependent when the concentration of Na+ is in the submicromolar range. In this region, the KD for rhodamine 6G was a simple hyperbolic function of the Na+ concentration. The curve showed saturability over 0.01 to 100 mM. Like the pH effect, the change in KD for rhodamine 6G due to the presence of the Na+ ion is quite small. Within 0 to 100 mM Na+, the change in KD reached only 2.5 μM (Fig. 8). We also added various concentrations of NaCl ranging from 100 to 400 mM (data not shown) and observed no significant change in the KD.
FIG. 8.
Effect of Na+ concentration on the KD of rhodamine 6G binding to NorM. The resulting KDs were plotted against the NaCl concentration.
Competition binding of different drugs in YdhE.
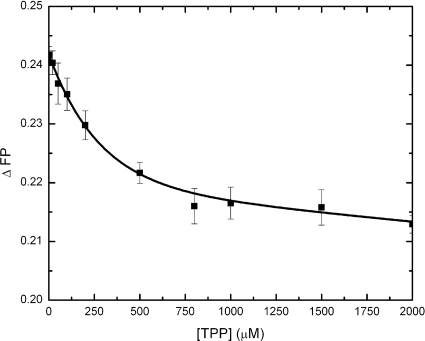
To confirm that the drug molecules bind specifically in the YdhE transporter, we performed competition experiments in which TPP was titrated into a solution containing the preformed YdhE-rhodamine 6G complex. In this case, TPP was chosen as a second ligand to knock off the bound rhodamine 6G from YdhE. The absorption spectra of TPP (from 200 to 600 nm) showed that this molecule absorbs light at wavelengths of 224.9, 268.0, and 275.9 nm. At a λ value of 527 nm, which is the λex for rhodamine 6G, the energy is too low to excite TPP. Thus, TPP was treated as a nonfluorescent ligand in the “knock-off” experiments. The data revealed that TPP was able to bind to YdhE and replace the bound rhodamine 6G molecule from the protein, as demonstrated by the release of rhodamine 6G observed from the reduction of polarization (Fig. 9). This binding assay provides direct evidence that TPP interferes with the binding of rhodamine 6G, possibly by a direct competitive binding process for the same binding site. Alternatively, the release of rhodamine 6G by TPP may be due to an allosteric interaction between distinct binding sites of these two ligands. Regardless, the titrations demonstrate that rhodamine 6G is bound specifically in the YdhE transporter.
FIG. 9.
YdhE binding competition experiment between rhodamine 6G and TPP. YdhE (5 μM) was preincubated with rhodamine 6G (1 μM) for 2 h before titration. The change in the fluorescence polarization signals (ΔFP) of rhodamine 6G was measured at an emission wavelength of 550 nm. TPP was nonfluorescent under the experimental conditions used. The decrease in the change in the fluorescence polarization signals showed that the bound rhodamine 6G was knocked off by TPP.
We have determined the binding affinities of five different drugs to the purified, detergent-solubilized NorM and YdhE proteins using fluorescence polarization assays. We note that fluorescence polarization has been widely used to study protein-DNA interactions (12, 14, 18) and protein-ligand interactions in transcriptional regulators (4, 34, 38). To our knowledge, this is the first attempt to use this methodology to investigate the interaction between MATE proteins and their transported substrates. This approach also allows us, for the first time, to quantify the strength of the transporter-drug interaction among the MATE family of transporters.
Supplementary Material
Acknowledgments
We thank Hiroshi Nikaido for providing us with the E. coli AG100AX strain.
This work was supported by PHS grants AI-21150 (to W.M.S.) and GM-074027 (to E.W.Y.) from the NIH. W. M. Shafer is the recipient of a senior research career scientist award from the VA Medical Research Service.
Footnotes
Published ahead of print on 30 June 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Begum, A., M. Rahaman, W. Ogawa, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. Gene cloning and characterization of four MATE family multidrug efflux pumps from Vibrio cholerae non-O1. Microbiol. Immunol. 49:949-957. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkette III, C. A. Block, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis, H., D. Molenaar, G. Polarends, H. W. van Veen, B. Poolman, A. J. M. Driessen, and W. N. Konings. 1994. Proton motive force-driven and ATP-dependent drug extrusion systems in multidrug-resistant Lactococcus lactis. J. Bacteriol. 176:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, B. E., K. M. Piro, and R. G. Brennan. 2007. Multidrug-binding transcription factor QacR binds the bivalent aromatic diamidines DB75 and DB359 in multiple positions. J. Am. Chem. Soc. 129:8389-8395. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., Y. Morita, M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2002. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delahay, R. M., B. D. Robertson, J. T. Balthazar, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127-2133. [DOI] [PubMed] [Google Scholar]
- 8.Griffith, J. K., M. E. Baker, D. A. Rouch, M. G. Page, R. A. Skurray, I. T. Paulsen, K. F. Chater, S. A. Baldwin, and P. J. Henderson. 1992. Membrane transport proteins: implications of sequence comparisons. Curr. Opin. Cell Biol. 4:684-695. [DOI] [PubMed] [Google Scholar]
- 9.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 10.Hagman, K. E., C. E. Lucas, J. T. Balthazar, L. A. Snyder, M. Nilles, R. C. Judd, and W. M. Shafer. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117-2125. [DOI] [PubMed] [Google Scholar]
- 11.He, G.-X., T. Kuroda, T. Mima, Y. Morita, T. Mizushima, and T. Tsuchiya. 2004. An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186:262-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyduk, T., and J. C. Lee. 1990. Application of fluorescence energy transfer and polarization to monitor Escherichia coli cAMP receptor protein and lac promoter interaction. Proc. Natl. Acad. Sci. USA 87:1744-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, K. M., D. Williams, W. M. Shafer, and R. G. Brennan. 2005. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J. Bacteriol. 187:5008-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper, D. C. 2000. New uses for new and old quinolones and the challenge of resistance. Clin. Infect. Dis. 30:243-254. [DOI] [PubMed] [Google Scholar]
- 16.Huda, M. N., Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2001. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 203:235-239. [DOI] [PubMed] [Google Scholar]
- 17.Lee, E.-H., and W. M. Shafer. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839-845. [DOI] [PubMed] [Google Scholar]
- 18.LeTilly, V., and C. A. Royer. 1993. Fluorescence anisotropy assays implicate protein-protein interactions in regulating trp repressor DNA binding. Biochemistry 32:7753-7758. [DOI] [PubMed] [Google Scholar]
- 19.Lewinson, O., and E. Bibi. 2001. Evidence for simultaneous binding of dissimilar substrates by the Escherichia coli multidrug transporter MdfA. Biochemistry 40:12612-12618. [DOI] [PubMed] [Google Scholar]
- 20.Li, L., Z. He, G. K. Pandey, T. Tsuchiya, and S. Luan. 2002. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277:5360-5368. [DOI] [PubMed] [Google Scholar]
- 21.Maness, M. J., and P. F. Sparling. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128:321-330. [DOI] [PubMed] [Google Scholar]
- 22.Marger, M. D., and M. H. Saier, Jr. 1993. A major superfamily of transmembrane facilitators that catalyse uniporter, symporter and antiporter. Trends Biochem. Sci. 18:13-20. [DOI] [PubMed] [Google Scholar]
- 23.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morita, Y., A. Kataoka, S. Shiota, T. Mizushima, and T. Tsuchiya. 2000. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug effux pump. J. Bacteriol. 182:6694-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muth, T. R., and S. Schuldiner. 2000. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 19:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nazmul Huda, N., J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Gene cloning and characterization of VcrM, a Na+-coupled multidrug efflux pump from Vibrio cholerae non-O1. Microbiol. Immunol. 47:419-427. [DOI] [PubMed] [Google Scholar]
- 28.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otsuka, M., M. Yasuda, Y. Morita, C. Ostuka, T. Tsuchiya, H. Omote, and Y. Moriyama. 2005. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 187:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouquette-Loughlin, C., S. A. Dunham, M. Kuhn, J. T. Balthazar, and W. M. Shafer. 2003. The NorM efflux pump of Neisseria gonorrhoeae and Neisseria meningitides recognizes antimicrobial cationic compounds. J. Bacteriol. 185:1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouquette-Loughlin, C. E., J. T. Balthazar, and W. M. Shafer. 2005. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 56:856-860. [DOI] [PubMed] [Google Scholar]
- 34.Schumacher, M. A., M. C. Miller, and R. G. Brennan. 2004. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J. 23:2923-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shafer, W. M., W. L. Veal, E.-H. Lee, L. Zarentonelli, J. T. Balthazar, and C. Rouquette. 2001. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitides. J. Mol. Microbiol. Biotechnol. 3:219-225. [PubMed] [Google Scholar]
- 36.Shafer, W. M., X.-D. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh, A. K., R. Halder, D. Mandal, and M. Kundu. 2006. Analysis of the topology of Vibrio cholerae NorM and identification of amino acid residues involved in norfloxacin resistance. Antimicrob. Agents Chemother. 50:3717-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su, C.-C., D. J. Rutherford, and E. W. Yu. 2007. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem. Biophys. Res. Commun. 361:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su, C.-C., and E. W. Yu. 2007. Ligand-transporter interaction in the AcrB multidrug efflux pump determined by fluorescence polarization assay. FEBS Lett. 581:4972-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su, X.-Z., J. Chen, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49:4362-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 42.Yang, S., S. R. Clayton, and E. L. Zechiedrich. 2003. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 51:545-556. [DOI] [PubMed] [Google Scholar]
- 43.Yu, E. W., G. McDermott. H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.