Abstract
NagA is a member of the amidohydrolase superfamily and catalyzes the deacetylation of N-acetyl-d-glucosamine-6-phosphate. The catalytic mechanism of this enzyme was addressed by the characterization of the catalytic properties of metal-substituted derivatives of NagA from Escherichia coli with a variety of substrate analogs. The reaction mechanism is of interest since NagA from bacterial sources is found with either one or two divalent metal ions in the active site. This observation indicates that there has been a divergence in the evolution of NagA and suggests that there are fundamental differences in the mechanistic details for substrate activation and hydrolysis. NagA from E. coli was inactivated by the removal of the zinc bound to the active site and the apo-enzyme reactivated upon incubation with one equivalent of Zn2+, Cd2+, Co2+, Mn2+, Ni2+ or Fe2+. In the proposed catalytic mechanism the reaction is initiated by the polarization of the carbonyl group of the substrate via a direct interaction with the divalent metal ion and His-143. The invariant aspartate (Asp-273) found at the end of β-strand 8 in all members of the amidohydrolase superfamily abstracts a proton from the metal-bound water molecule (or hydroxide) to promote the hydrolytic attack on the carbonyl group of the substrate. A tetrahedral intermediate is formed and then collapses with cleavage of the C-N bond after proton transfer to the leaving group amine by Asp-273. The lack of a solvent isotope effect by D2O and the absence of any changes to the kinetic constants with increases in solvent viscosity indicate that net product formation is not limited to any significant extent by proton transfer steps or the release of products. N-trifluoroacetyl-d-glucosamine-6-phosphate is hydrolyzed by NagA 26-fold faster than the corresponding N-acetyl derivative. This result is consistent with the formation or collapse of the tetrahedral intermediate as the rate limiting step in the catalytic mechanism of NagA.
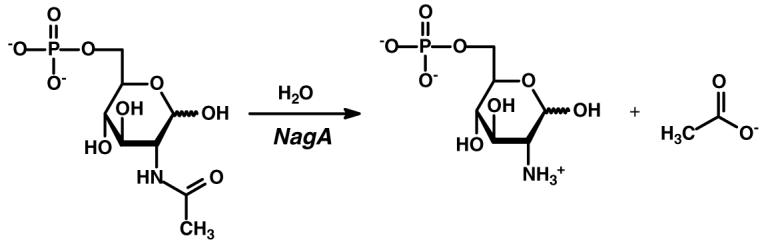
NagA1 catalyzes the hydrolytic cleavage of N-acetyl-d-glucosamine-6-phosphate as illustrated in Scheme 1. The deacetylation of this compound provides a source of carbon and nitrogen by preparing this substrate for entry into the glycolytic pathway. This reaction is a key step in the catabolism of N-acetyl-d-glucosamine, derived from the degradation of chitin, and is an essential component for the biosynthesis of lipopolysaccharides and peptidoglycans. More recently, the reaction catalyzed by NagA has been shown to be an important step in the recycling of cell wall murein (1-3).
Scheme 1.
The purification of NagA from Escherichia coli was originally reported by White and Pasternak (4). The enzyme oligomerizes as a tetramer and each subunit contains a reactive sulfhydryl group near the active site (5). In the forward reaction, inhibition occurs at high substrate concentrations while the products, d-glucosamine-6-phosphate and acetate, are noncompetitive and competitive inhibitors, respectively. A phosphonate analogue of the putative tetrahedral intermediate has been synthesized and it is a potent, tight-binding inhibitor (6). The structure of apo-NagA from E. coli was first reported by the team led by Steve Almo (pdb code: 1ymy) and later by Ferreira et al. (7) (pdb code: 1yrr). The protein folds as a (β/α)8-barrel and is a member of the amidohydrolase superfamily (8, 9). The enzyme binds up to 1.4 equivalents of zinc and the metal can be removed with a chelating agent, resulting in the loss of activity that can be restored after reconstitution with Zn, Co, Mn or Fe (7).
The precise role of the metal cofactor is of particular interest for NagA because the two crystal structures available for the enzyme from E. coli have no metal bound in the active site. However, the crystal structures of NagA from B. subtilis and T. maritima have been determined with divalent cations bound within the active site (10). The B. subtilis enzyme (pdb code: 1un7) binds two equivalents of Fe2+ (10). One of the metals is coordinated to the H×H motif at the end of β-strand 1 and an aspartic acid from the end of β-strand 8. This site has been designated as the Mα-position (8). The second metal (Mβ-site) is coordinated to two histidine residues from the ends of β-strands 5 and 6, respectively. The two metal ions are bridged to one another by a glutamate from the end of β-strand 3 and a hydroxide (or water) from solvent.
The active site of the T. maritima enzyme is quite similar to that of the B. subtilis structure except that a single metal ion is bound to the Mβ-site (pdb code: 1o12). The metal ligation scheme in the NagA from E. coli must be significantly different since the H×H motif from the end of β-stand 1 is replaced by Q×N and thus it is highly likely that this enzyme can bind only a single divalent cation in the active site. This fundamental divergence in the active sites of NagA from these sources suggests that the mechanism for the activation of the hydrolytic water and substrate required for catalysis must differ to a significant degree.
NagA has been characterized as a member of the amidohydrolase superfamily based on an exhaustive set of sequence and structural comparisons (8, 9, 11). This superfamily is a diverse group of enzymes that catalyzes the hydrolytic cleavage of amide bonds to nucleic acids, amino acids, and sugars (8, 9). Within this superfamily, four metal ligation schemes have been identified. The most prevalent is a binuclear metal center where the two divalent cations are bridged by a hydroxide from solvent. An additional bridging ligand from the protein may include a carboxylated lysine from the end of β-strand 4, a glutamate from the end of β-strand three or four, or a cysteine from the end of β-strand 2 (8, 9). Enzymes of this type include dihydroorotase (12), the phosphotriesterase homology protein (13), renal dipeptidase (14) and d-amino acid deacetylase (15). Many members of the amidohydrolase superfamily bind a single divalent cation, including cytosine (16) and adenosine deaminase (17). This class of enzymes binds one metal at the α-position via ligation to the H×H motif from β-strand 1, a histidine from the end of β-strand 5 and an invariant asparate from β-strand 8. The third ligation scheme, which may include NagA from E. coli and T. maritima, utilizes a single metal bound exclusively to the β-site. The most extreme example within the amidohydrolase superfamily is uronate isomerase from E. coli. This enzyme catalyzes the isomerization of d-glucuronate to d-fructuronate and a metal ion is not required for catalytic activity (18).
There are a significant number of issues regarding the mechanism for substrate hydrolysis by NagA that are unresolved. It is unclear how the divalent cation binds within the active site of NagA and how the water molecule is activated for nucleophilic attack on the amide bond. The disparity in the metal ligation schemes between the E. coli and B. subtilis forms of NagA suggests a divergence in the evolution of this enzyme at a significant locale within the active site. In this paper, we have systematically interrogated the mechanism of substrate hydrolysis by NagA from E. coli with a battery of substrate analogs and active site mutants using a host of metal-substituted forms of the enzyme. We conclude from these studies that NagA from E. coli can bind up to one divalent cation for the activation of the hydrolytic water molecule. This water molecule is further activated by general base catalysis through the abstraction of a proton by Asp-273. There is no evidence to suggest that Glu-131 plays any role in catalysis other than to help coordinate the single divalent cation. The enzyme is rate limited by the cleavage of the amide bond.
MATERIALS and METHODS
Materials
N-Acetyl-d-glucosamine-6-phosphate, N-acetyl-d-glucosamine-6-sulfate, and all buffers and purification reagents were purchased from Sigma-Aldrich. Chromatographic columns and resins were purchased from G. E. Healthcare. Chelex 100 resin was purchased from BioRad. ICP standards were obtained from Inorganic Ventures Inc.
Preparation of N-thioacetyl-d-glucosamine-6-phosphate
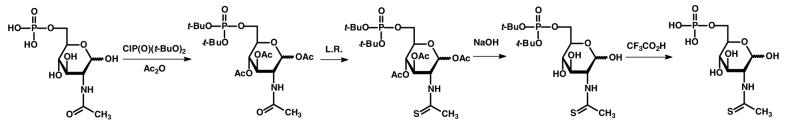
The N-thioacetyl derivative of D-glucosamine-6-phosphate (4) was synthesized according to the procedure outlined in Scheme 2. In a 250 mL flask was added 2.0 g of N-acetyl-d-glucosamine (9.0 mmol) and 150 mL of pyridine. The mixture was stirred at 4 °C and 2.1 g of di-t-butyl chlorophosphate (9.0 mmol) was added. For the next 4 days 1.0 g of di-t-butyl chlorophosphate (4.3 mmol) was added every 12 h. The temperature was subsequently raised to 25 °C and 5 mL of acetic anhydride was added and allowed to react overnight. After removal of the solvent, the residue was dissolved in 200 mL of chloroform and washed with aqueous HCl. The organic phase was dried and the product purified by column chromatography on silica gel using ethyl acetate as the eluent. The yield of 1,3,4-tri-O-acetyl-N-acetyl-d-glucosamine-6-di-t-butylphosphate was 2.5 g (51%).
Scheme 2.
The carbonyl group of the N-acetyl substituent was replaced with sulfur using Lawesson's reagent. In a 25 mL flask was added 0.60 g of 1,3,4-tri-O-acetyl-N-acetyl-d-glucosamine-6-di-t-butylphosphate (1.2 mmol), 0.5 g of Lawesson's reagent (1.2 mmol) and 10 mL of THF. The mixture was stirred overnight at room temperature. After removal of the solvent, the residue was purified on silica gel through elution with ethyl acetate. The yield of 1,2,4-tri-O-acetyl-N-thioacetyl-d-glucosamine-6-di-t-butylphosphate was 60 mg (10%). The O-acetyl groups of this product were subsequently removed with hydroxide. In a 25 mL flask was added 55 mg of 1,2,4-tri-O-acetyl-N-thioacetyl-d-glucosamine-6-di-t-butylphosphate (0.1 mmol), 13 mg of NaOH (0.3 mmol) and 5 mL of aqueous methanol. The mixture was stirred in an ice-water bath for 1 hour. After removal of the solvent, N-thioacetyl-d-glucosamine-6-di-t-butylphosphate was purified on silica gel by elution with a mixture of EtOAc/MeOH (ratio of 10/1).
The t-butyl protecting groups were removed from the phosphate substituent with trifluroacetic acid. In a 25 mL flask was added the entire amount of N-thioacetyl-d-glucosamine-6-di-t-butylphosphate from the previous step, 5 mL of CH2Cl2 and 0.5 mL of CF3CO2H. The mixture was stirred at room temperature for 1 h. The solvent was removed to obtain 20 mg of N-thioacetyl-d-glucosamine-6-phosphate in an overall yield of 64% from the previous step. 1H NMR (300 MHz, CD3OD): 5.92 and 5.21 ppm (1H, s, d, J = 5.4 and 6.6 Hz, CH), 4.41-4.38 ppm (1H, m, CH), 4.04-4.02 ppm (3H, m, CH2, CH), 3.81-3.77 ppm (1H, m, CH), 3.43-3.25 ppm (1H, m, CH), 2.59 and 2.35 ppm (3H, s, CSCH3). 31P NMR (121 MHz, CD3OD): −0.03, −0.05 ppm. 13C NMR (75 MHz, CD3OD): 203.20, 90.85, 72.19, 72.12, 71.84, 67.08, 61.96 and 33.37 ppm. MS (ESI negative mode) found: 315.9 (M–H)− ; calculated for C8H15NO8PS (M–H)−: 316.0.
Preparation of N-formyl-d-glucosamine-6-phosphate
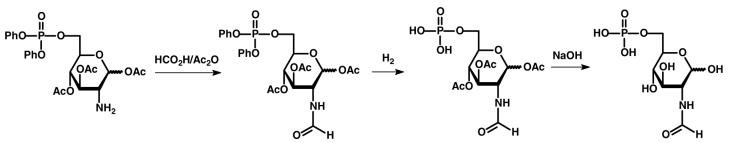
The synthesis of N-formyl-d-glucosamine-6-phosphate (5) was conducted according to the procedure illustrated in Scheme 3. In a 100 mL flask, 1.16 g (2.0 mmol) of 1,3,4-tri-O-acetyl-6-diphenylphospho-d-glucosamine hydrochloride, and 0.56 mL triethylamine (3.0 mmol) was mixed with 30 mL of chloroform and allowed to stir for 30 min before the removal of the solvent. The residue was dissolved in 40 mL of formic acid and 7.0 mL of acetic anhydride was added drop wise at reduced temperature. After 2 h the solvent was removed under reduced pressure and the solid residue dissolved in 150 mL of chloroform. This solution was washed with 100 mL of dilute HCl, saturated NaHCO3 solution and then dried over Na2SO4. The product, diphenyl N-formyl-1,3,4-tri-O-acetyl-d-glucosamine-6-phosphate was obtained by flash chromatography in a yield of 90% (1.02 g).
Scheme 3.
The compound from the previous step (0.85 g, 1.5 mmol) was mixed with PtO2 (80 mg) and 60 mL of acetic acid in a 100 mL flask and allowed to stir at room temperature for 14 days under an atmosphere of hydrogen. Every 24 h 20-40 mg of PtO2 was added and the progress of the reaction monitored by 31P NMR spectroscopy. After removal of the catalyst by filtration and the solvent by evaporation, the solid residue was washed with ethyl ether to obtain N-formyl-1,3,4-tri-O-acetyl-d-glucosamine-6-phosphate in a yield of 95%. The acetyl groups from this compound were removed with base. In a 100 mL flask was added 0.50 g (1.2 mmol) of Nformyl-1,3,4-tri-O-acetyl-d-galactosamine-6-phosphate and 10 mL of water, and cooled in an ice-water bath. To this solution was added 0.29 g (7.2 mmol) of NaOH in 5 mL of water and allowed to stir for 1 h before neutralization of the reaction mixture with acetic acid. After removal of the water and washing with MeOH/CH3COCH3, the disodium salt of N-formyl-d-glucosamine-6-phosphate was obtained in quantitative yield. 1H NMR (300 MHz, D2O): 8.04, 7.98, 7.82, and 7.80 ppm (1H, s, CHO), 5.08-5.04 and 4.55-4.47 ppm(1H, m, CH), 3.92-3.27 ppm (6H, m, 4CH, OCH2). 31P NMR (121.4 MHz, D2O): 6.48ppm. MS (ESI negative mode): found: 286.04 (M-2Na+H)− ; calculated for C7H13NO9P: 286.03.
Preparation of N-trifluoroacetyl-d-glucosamine-6-phosphate
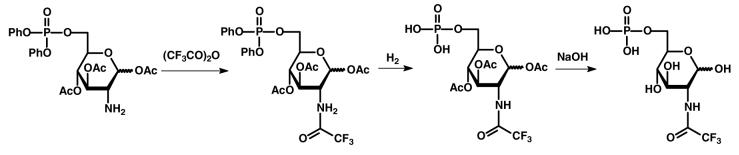
The preparation of the trifluoroacetate derivative of d-glucosamine-6-phosphate (6) was conducted as outlined in Scheme 4. In a 100 mL flask was added 0.58 g of 1,3,4-tri-O-acetyl-d-glucosamine-6-diphenylphosphate (1.0 mmol), 0.5 mL of Et3N (5.0 mmol), 30 mL CHCl3 and 0.63 g of trifluoroactic anhydride (3.0 mmol). The mixture was stirred for 2 h at room temperature. After removal of the solvent, the residue was eluted from a column of silica gel with a mixture of EtOAc/hexane (5/4). The yield of 1,3,4-tri-O-acetyl-N-trifluoroacetyl-d-glucosamine-6-diphenylphosphate was 0.60 g. The phenyl groups were removed from this compound by hydrogenation. In a 100 mL flask was added 0.59 g of 1,3,4-tri-O-acetyl N-trifluoroacetyl-d-glucosamine-6-diphenylphosphate (0.93 mmol), PtO2 (20 mg) and acetic acid (30 mL). H2 was bubbled into this mixture overnight at room temperature. After removal of the solvent, the residue was washed with ether to obtain 0.43 g of 1,3,4-tri-O-acetyl-N-trifluoroacetyl-d-glucosamine-6-phosphate in a yield of 98%.
Scheme 4.
The disodium salt ofN -trifluoroacetyl-d-glucosamine-6-phosphate was prepared by mixing 0.23 g of 1,3,4-tri-O-acetyl-N-trifluoroacetyl-d-glucosamine-6-phosphate (0.47 mmol), NaOMe (76 mg, 1.41 mmol) and methanol (8 mL). The solution was stirred in an ice-water bath for 2 h and then the pH adjusted to 7.0 using 0.1N HCl. After removal of the solvent the residue was dissolved in water (8 mL) and the pH adjusted to 7.0 using 0.1N NaOH. The solvent was removed to obtain the disodium salt of N-trifluoroacetyl-d-glucosamine-6-phosphate. 1H NMR (300 MHz, D2O): 5.20 and 4.28 ppm (1H, d, J = 5.7 and 8.1 Hz, CH), 4.02-3.41 ppm (6H, m, CH2, CH). 31P NMR (121 MHz, D2O): 6.27 ppm. 13C NMR (75 MHz, D2O): 160.04, 159.54, 117.91, 114.11, 94.41, 90.61, 75.54, 75.45, 72.80, 71.31, 71.22, 69.93, 69.72, 69.48, 63.08, 63.03, 57.40, 54.94 and 49.22 ppm. 19F NMR (282 MHz, D2O): 100.37 and 100.09 ppm. MS (ESI positive mode) found: 399.98 (M+H)+: calculated for C8H12F3NNa2O9P (M+H)+: 400.00. MS (ESI negative mode) found: 354.03 (M−2Na+H)− and 376.01 (M−Na)−; calculated for C8H12F3NO9P (M−2Na+H)−: 354.02 and C8H11F3NNaO9P (M−Na)−: 376.00.
Preparation of N-acetyl-d-galactosamine-6-phosphate
The synthesis of N-acetyl-d-galactosamine-6-phosphate (2) was conducted according to the procedure that is presented in Scheme 5. In a 100 mL flask was added 0.5 g (2.3 mmol) of N-acetyl-d-galactosamine to 40 mL of pyridine. The mixture was cooled to −40 °C and 0.91 g (3.4 mmol) of diphenylchlorophosphate was added drop wise. The mixture was stirred at −23 °C for 2-4 days. At this time 1.5 mL of acetic anhydride was added and allowed to react at room temperature overnight. After removal of the solvent, the residue was dissolved in 100 mL of chloroform and washed with dilute HCl, saturated NaHCO3 and then dried over Na2SO4. The product, diphenyl N-acetyl-1,3,4-tri-O-acetyl-d-galactosamine-6-phosphate was isolated by flash chromatography with a yield of 50% (0.65 g). The phenyl protecting groups were removed by hydrogenation. In a 100 mL flask was added 0.58 g (1.0 mmole) of diphenyl N-acetyl-1,3,4-tri-O-acetyl-d-galactosamine-6-phosphate, 40 mg PtO2 and 40 mL of acetic acid. The mixture was stirred at room temperature overnight under an atmosphere of hydrogen. After removal of the solvent, the solid residue was washed with ethyl ether to obtain N-acetyl-1,3,4-tri-O-acetyl-d-galactosamine-6-phosphate in a yield of 95%.
Scheme 5.
The acetyl protecting groups were removed by base. In a 100 mL flask was added 0.4 g (0.94 mmol) of N-acetyl-1,3,4-tri-O-acetyl-d-galactosamine-6-phosphate in 10 mL of water. The solution was cooled with an ice-water bath and 5 mL of a solution of sodium hydroxide (0.24 g, 6 mmol) was added drop wise and allowed to stir for 1h before being neutralized with acetic acid. After removal of the solvent the residue was washed with MeOH/CH3COCH3 to obtain the sodium salt of N-acetyl-D-galactosamine-6-phosphate (2) in quantitative yield. 1H NMR (300 MHz, D2O): 5.16 ppm (0.6H, d, J = 3.6 Hz, CH), 4.61 ppm (0.4H, d, J = 8.1 Hz, CH), 4.20-3.72 ppm (5H, m, 3CH, OCH2), 3.74-3.65 ppm (1H, m, NCH), 2.04 and 2.03 ppm (3H, s, NCOCH3). 31P NMR (121.4 MHz, D2O): 7.19ppm. MS (ESI negative mode) found: 300.05 (M−2Na+H)− , 322.04 (M−Na)− ; calculated for C8H15NO9P: 300.05 and C8H14NNaO9P: 322.03.
Cloning, Expression and Purification
The gene from E. coli encoding NagA was cloned into a PET-30a(+) expression vector through the use of the NdeI and EcoRI restriction sites. Mutants of NagA were prepared in accordance with procedures published in the Quikchange Site-Directed Mutagenesis Kit. Gene sequences were verified by the Gene Technologies Lab at Texas A &M University. The expression plasmids were inserted into BL21(DE3) cells through electroporation. The transformation solution was then transferred into 1 mL of LB growth medium for incubation at 37 °C for 30 minutes, which was followed by plating onto LB agar containing 50 μg/mL kanamycin. The cells were allowed to grow overnight at the same temperature. Single colonies were selected for inoculation into 50 mL of LB/kanamycin for use as overnight starter cultures for inoculation into 2 L of LB/kanamycin. Prior to induction with 1.0 mM IPTG at an A600 nm of 0.6, 1.0 mM ZnCl2 was added to supplement the growth medium. The cells grew overnight at a temperature of 30 °C, after which they were centrifuged at 3,400g for 12 min. The cells were then re-suspended and disrupted by sonication in 10 × (v/w) 50 mM Tris buffer, pH 7.5, containing 1.0 mM DTT and 100 μg/mL of the protease inhibitor phenylmethanesulfonylfluoride. Insoluble cell debris was removed by centrifugation at 13,900g for 12 minutes, after which 1% w/v protamine sulfate from salmon sperm was used to precipitate the nucleic acids. The precipitate was removed by centrifugation and the protein fractionated with ammonium sulfate to 50% of saturation. The precipitated protein was isolated by centrifugation at 13,900g for 12 minutes. The protein was then re-suspended in a minimal amount of 50 mM Tris buffer, pH 7.5, containing 1.0 mM DTT and then passed through a 0.2 μM pore filter prior to loading onto a pre-equilibrated HiLoad 26/60 Superdex 200 prep grade gel filtration column. The pooled fractions containing NagA were diluted to 50 mL in the same buffer and loaded onto a 6 mL Resource Q anion exchange column. The column was washed with several column volumes of buffer and the protein eluted with a linear gradient of 1 M NaCl, 50 mM Tris, pH 7.5. The appropriate fractions were pooled after analysis with SDS PAGE for purity. Protein concentrations were determined using the calculated extinction coefficient of 18,490 M−1 cm−1 at 280 nm (www.scripps.edu/~cdputnam/protcalc.html).
Measurement of Enzymatic Activity
Kinetic assays were performed at 30 °C using a 96 well quartz plate in conjunction with a SpectraMax 384-Plus spectrophotometer from Molecular Devices. The data were analyzed using Softmax Pro version 4.7.1. Extinction coefficients for the different substrates were determined from plots of absorbance versus substrate concentration before and after total enzymatic hydrolysis.
Metal Analysis
Metal determination and quantification were performed with an Elan DRC II ICP-MS from Perkin Elmer. An analog detection mode was used with three averaged replicates per reading. External calibration standards were prepared through the serial dilution of a single 10 ppm stock mixture of Zn, Cd, Co, Cu, Mn, Ni and Fe in 2% nitric acid. Freshly prepared standards generally contained 2, 20 and 200 ppb of the metal ions in 1% Trace Select nitric acid from Fluka, diluted in MilliQ deionized water. The masses of the isotopes detected were 55Mn, 57Fe, 59Co, 60Ni, 66Zn and 111Cd. 115In was used as an internal standard for 111Cd whereas 69Ga was used as an internal standard for all other isotopes.
Metal Chelation and Reconstitution
Apo-enzyme was prepared by dialysis of NagA against buffer A (20 mM dipicolinate, 10 mM β-mercaptoethanol, 50 mM MES, pH 6.0). After 3 × 300-fold buffer changes with buffer A over two days, the chelator was removed by dialysis with Chelex-treated HEPES buffer, pH 8.0. For the metal reconstitution studies, 100 μL of apo-NagA at 8.23 mg/mL was mixed with 3 equivalents of various metal solutions. The metal solutions consisted of freshly prepared NiCl2, CdCl2, CoCl2, MnCl2, or ZnCl2 in water, and Fe(NH4)2(SO4)2 in 1% HCl. Reconstitution of the apo-enzymes was conducted overnight at 4 °C before removal of unbound metal by passage through a PD-10 column. The PD-10 column was pre-treated with dipicolinate to remove traces of unbound metal and then washed with five column volumes of metal free HEPES buffer, pH 8.0. After elution, the concentration of the metal-reconstituted enzyme was determined by UV absorbance and the metal content of the samples determined by ICP-MS.
pH Studies
The pH dependence of the kinetic constants, kcat and kcat/Km, was determined at intervals of ∼0.25 pH units from pH 5 to 10 using 20 mM piperazine (pH 5-6.25), phosphate (pH 6.25-8), and borate (pH 8-10) with varying amounts of substrate. The pH values of the assays were determined after the reactions were completed.
Solvent Viscosity Analysis
Solvent viscosity effects were analyzed for Zn-NagA with N-acetyl-d-glucosamine-6-phosphate as the substrate using sucrose as the micro-viscogen in 20 mM phosphate buffer, pH 7.5. The viscosity effects were measured using 0, 10, 14, 20, 24, 27.5, 32.5 and 35% (w/w) sucrose and the corresponding relative viscosities were 1, 1.32, 1.5, 1.88, 2.2, 2.48, 3.06, and 3.42, respectively (19, 20).
Data Analysis
All kinetic data were fit to the corresponding equations using the nonlinear least-squares curve fitting program SigmaPlot 9.0. Simple substrate saturation curves were fit to equation 1 where A is the substrate concentration, v is the velocity of the reaction, kcat is the turnover number and Km is the Michaelis constant. When substrate saturation was not achieved, kcat/Km values were determined from fits of the data to a straight line. Saturation curves showing substrate inhibition were fit to equation 2 were Kis is the apparent inhibition constant for the substrate inhibition. For the analysis of pH-rate profiles, plots of log kcat and log kcat/Km vs. pH that indicated the deprotonation of a single group required for maximum activity were fit to equation 3, where c is the maximum activity and Ka is the acid dissociation constant. pH-rate profiles that showed the loss of catalytic activity at high and low values of pH were fit to equation 4, where Kb is the dissociation constant of the group that must be protonated for full activity. The pH profiles which showed that the deprotonation of two acid ic groups and the protonation of one basic group are required for maximum activity were fit to equation 5 where Ka is the average dissociation constant of two acidic groups and Kb is the dissociation constant of the basic group. Equation 6 was used to determine the average Ka for two ionizable acidic groups and the average Kb for two ionizable basic groups.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
RESULTS and DISCUSSION
Purification and Properties of NagA
The wild type NagA was over-expressed in E. coli. From 3 g of cell paste about 100 mg of homogeneous protein were isolated. Enzymatic activity was dependent upon the presence of disulfide reducing reagents such as dithiothreitol or β-mercaptoethanol during storage and purification. The native molecular weight of the purified enzyme was estimated to be 180 kDa based upon the elution volume through a calibrated gel filtration column (data not shown). The molecular weight of a single subunit from the derived amino acid sequence of NagA is 40,951 Da. These results confirm that NagA adopts a tetrameric quaternary structure in the presence of disulfide reducing agents (5, 7).
Metal Dependence of Enzyme Activity
The initial purification of NagA resulted in enzyme with a turnover number of 35 s−1. This protein contained 0.4 equivalents of Zn2+ and 0.05 equivalents of Fe2+ as determined by ICP-MS. NagA was subsequently expressed in LB medium supplemented with 1.0 mM ZnCl2 and the protein purified from these cells contained 0.95 equivalents of Zn per subunit with a specific activity of 96 s−1. The role of the bound metal on the enzymatic reaction rate was addressed by characterization of the apo-enzyme. The metal was removed from the enzyme via dialysis against 20 mM dipicolinate and the resulting protein solution was verified to be metal-free by ICP-MS. However, during the time course for measurement of catalytic activity by the apo-enzyme, the rate of substrate turnover increased slowly with time. The activation of the apo-enzyme was apparently due to the binding of trace metals in the assay solution since the activity of the apo-enzyme in the presence of 500 μM EDTA was less than 2% of the activity exhibited by the native enzyme. These results are consistent with an absolute requirement for a bound divalent cation for the expression of catalytic activity by NagA.
Incubation of the apo-enzyme with one equivalent of Zn2+ for 30 minutes fully restored catalytic activity. The titration of apo-enzyme with varying amounts of ZnCl2 is shown in Figure 1. These results demonstrate that NagA from E. coli has a maximum catalytic activity with a single divalent cation bound to the active site. These results are consistent with the absence of the two histidines at the end of β-strand one and the determination of the x-ray structure of the Zn-NagA that shows a single Zn bound to the Mβ-site (21). The apo-enzyme was incubated with three equivalents of Fe2+, Mn2+, Ni2+, Cd2+, Co2+ or Zn2+ and the metal-substituted forms of NagA were found to contain approximately one equivalent of metal per protein subunit after removal of the excess metal by passage through a PD-10 column. The metal content and the corresponding kinetic parameters for the metal-substituted variants of NagA are listed in Table 1.
Figure 1.
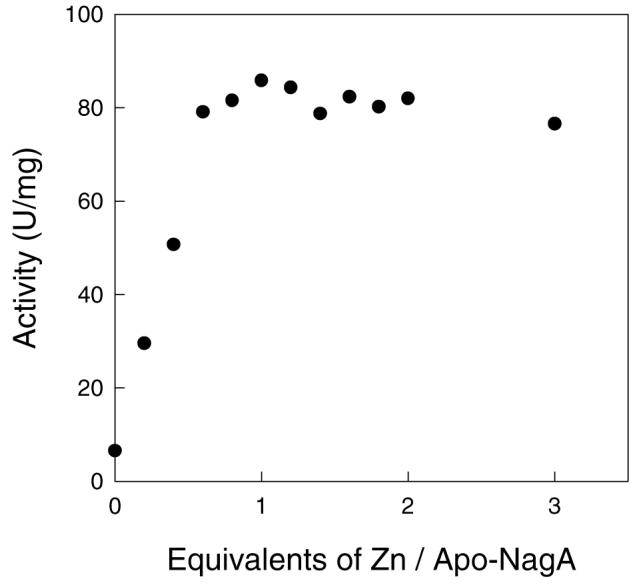
Reconstitution of apo-NagA (0.5 μM) with varying amounts of ZnCl2. The rate of substrate hydrolysis was determined after diluting the enzyme to a concentration of 30 nM in 1.0 mM N-acetyl-d-glucosamine-6-phosphate, 1.0 μM EDTA and 25 mM Tris at pH 7.5 and 30 °C.
Table 1.
Kinetic Parameters for Metal-Reconstituted Forms of NagAa
| Metal | Metal/subunit | kcat (s−1) | Km (mM) | kcat /Km (M−1s−1) |
|---|---|---|---|---|
| Zn | 0.9 | 102 ± 2 | 0.08 ± 0.01 | (1.3 ± 0.2) × 106 |
| Cd | 0.8 | 163 ± 3 | 0.20 ± 0.01 | (8.2 ± 0.4) × 105 |
| Co | 1.0 | 177 ± 5 | 0.15 ± 0.01 | (1.2 ± 0.1) × 106 |
| Ni | 0.8 | 41 ± 3 | 0.64 ± 0.12 | (6.4 ± 1.3) × 104 |
| Mn | 0.7 | 92 ± 2 | 0.10 ± 0.01 | (9.2 ± 0.9) × 105 |
| Fe | 0.5 | 58 ± 2 | 0.23 ± 0.03 | (2.5 ± 0.3) × 105 |
The kinetic parameters were determined at pH 7.5, 30 °C from fits of the data to equation 1 using N-acetyl-d-glucosamine-6-phosphate (1) as the substrate.
Kinetic Constants for NagA
The effect of metal substitution on the catalytic constants for NagA can be utilized to evaluate the contributions made by these divalent cations on enzymatic activity (22, 23). The relationship on the value of kcat/Km observed for the different metals follow the trend: Zn = Co > Mn > Cd > Fe > Ni . The Fe containing enzyme was not fully reconstituted with metal and thus the kinetic constants may be larger if the metal site was fully occupied. The values for kcat follow the trend Co > Cd > Zn > Mn > Fe > Ni. The Ni enzyme has the highest Km while the Zn enzyme has the lowest. Similar trends in the range of the catalytic constants were reported previously for the Co, Zn, Mn and Fe reconstitutions of apo-NagA from E. coli (7).
Substrate Specificity
The structures of the compounds tested as substrates for NagA are presented in Scheme 6. The substitution of sulfur for oxygen can be used in conjunction with hard and soft metals to probe the potential for metal-ligand interaction during the activation of electrophilic reactions (24, 25). Cd- and Zn-NagA were used to address the occurrence of direct interactions of the carbonyl oxygen/sulfur and the metal ion through a comparison of the kinetic constants for N-acetyl-d-glucosamine-6-phosphate (1) and N-thioacetyl-d-glucosamine-6-phosphate (4) as substrates. The Cd-NagA catalyzes the hydrolysis of the thioacetyl substrate about an order of magnitude better than does the Zn-substituted enzyme (Table 2). The ratio of (kcat/Kthio)/(kcat/Kacetyl) is 0.78 (6.4 × 105 M−1s−1/8.2 × 105 M−1s−1) for the Cd-enzyme whereas the corresponding ratio for the Zn-NagA is only 0.036 (4.3 × 104 M−1s−1/1.2 × 106 M−1s−1). These results are consistent with the preferred interactions between hard and soft metals and ligands, since the relatively soft thio-carbonyl group is a better ligand for the softer cadmium ion than it is for zinc (26).
Scheme 6.
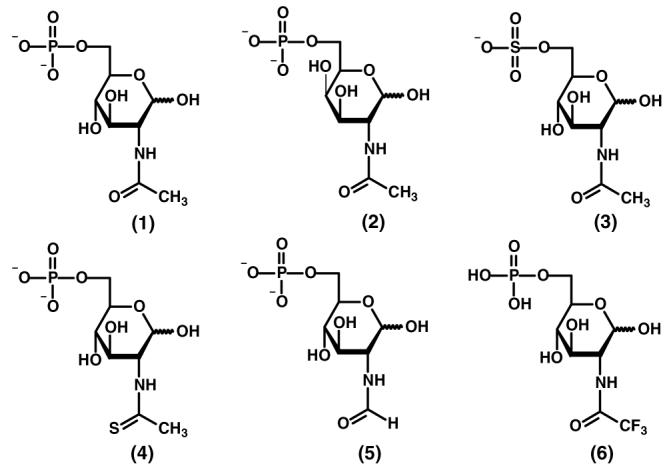
Table 2.
Kinetic Parameters for Substrates with Metal and Mutant Variant Forms of NagAa.
| Substrate | Enzyme | kcat (s−1) | Km (mM) | kcat/Km (M−1s−1) |
|---|---|---|---|---|
| 1 | Zn | 102 ± 2 | 0.08 ± 0.01 | (1.3 ± 0.2) × 106 |
| 1 | Cd | 163 ± 3 | 0.20 ± 0.01 | (8.2 ± 0.4) × 105 |
| 1 | Mn | 92 ± 2 | 0.10 ± 0.01 | (9.2 ± 0.9) × 105 |
| 2 | Zn | 154 ± 15 | 1.24 ± 0.16 | (1.2 ± 0.2) × 105 |
| 3 | Zn | 64 ± 4 | 4.9 ± 1.0 | (1.3 ± 0.3) × 104 |
| 3 | Cd | 23 ± 1 | 11 ± 1 | (2.1 ± 0.2) × 103 |
| 3 | Zn-K139M | 60 ± 4 | 9.7 ± 1.3 | (6.2 ± 0.9) × 10 |
| 4 | Zn | 10 ± 0.2 | 0.24 ± 0.02 | (4.2 ± 0.4) × 104 |
| 4 | Cd | 128 ± 5 | 0.20 ± 0.03 | (6.4 ± 1.0) × 105 |
| 4 | Mn | 11 ± 0.3 | 0.23 ± 0.03 | (4.8 ± 0.6) × 104 |
| 5 | Zn | 22 ± 0.2 | 0.29 ± 0.01 | (7.6 ± 0.3) × 104 |
| 6 | Zn | 2610 ± 60 | 0.40 ± 0.04 | (6.5 ± 0.7) × 106 |
The kinetic parameters were determined at pH 7.5, 30 °C from fits of the data to equation 1.
These results are consistent with those previously observed for carboxypeptidase A, where thio-amide substrate analogs were compared to the corresponding oxo-amides with the Zn-, and Cd-substituted enzymes (27). For carboxypeptidase A, the Zn-enzyme was the best catalyst for the hydrolysis of oxo-amides, while the Cd-enzyme was the less efficient. For the thio-amide substrates, the Cd-enzyme was the best enzyme and the Zn-enzyme was poorer. The catalytic constants for the Zn- and Cd-substituted forms of NagA support the conclusion that the carbonyl group of the substrate is polarized by a direct interaction with the single divalent cation bound to the Mβ-position in the active site.
In addition to N-acetyl-d-glucosamine-6-phosphate (1) and the thioacetyl analogue (4), NagA can hydrolyze N-trifluoroacetyl-d-glucosamine-6-phosphate (6), N-acetyl-d-glucosamine-6-sulfate (3), N-acetyl-d-galactosamine-6-phosphate (2) and N-formyl-d-glucosamine-6-phosphate (5). The kinetic constants for NagA with these substrate analogs are listed in Table 2 The N-trifluoroacetyl substituted substrate is hydrolyzed 26 times faster than the natural substrate but the N -formyl substrate is hydrolyze more slowly by a factor of 5. The value of Km for the galactosamine derivative (2) is about an order of magnitude higher than it is for the physiological substrate (1).
pH Rate Profiles
The effect of pH on the kinetic constants for the hydrolysis of substrates by NagA was measured in an attempt to identify those functional groups in the substrate and active site that are required to be in a specific state of protonation. The log kcat vs. pH-profile shown in Figure 2A for Zn-NagA was fit to equation 3 and these results indicate that the deprotonation of a single acid with a kinetic pKa of 5.6 is required for catalytic activity in the enzyme-substrate complex. The log kcat/Km vs. pH-profile for Zn-NagA is shown in Figure 2B. In this profile, catalytic activity is lost at high and low values of pH and the data were fit to equation 4 where two groups, in either the substrate or free enzyme, with identical pKa values of 6.4 are required to be ionized. An additional group must be protonated with a pKa of ∼9.3. Similar results were obtained for the effect of pH on the kinetic constants for the hydrolysis of compound 1 by Cd-NagA (data not shown). From the log kcat vs. pH profile, a single group must be ionized with a kinetic pKa of 6.4. From the log kcat/Km pH-rate profile activity is lost at low and high pH with limiting slopes of 2 and −2, respectively, and the data were fit to equation 5.
Figure 2.
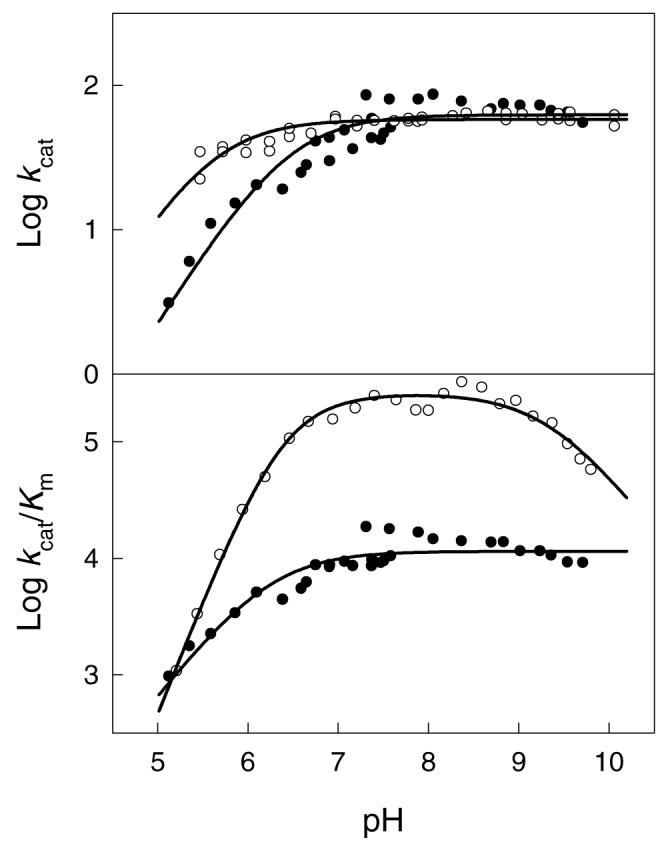
pH-rate profiles for the hydrolysis of substrates by NagA (A) log kcat vs. pH profiles for Zn-NagA with N-acetyl-d-glucosamine-6-phosphate ( ○ ) and N-acetyl-d-glucosamine-6-sulfate ( ● ). The solid lines represent fits of the data to equation 3. (B) logk cat/Km vs. pH profiles for Zn-NagA with N-acetyl-d-glucosamine-6-phosphate ( ○ ) and N-acetyl-d-glucosamine-6-sulfate ( ● ). The solid lines represent fits of the data to equations 5 and 3, respectively. Additional details are provided in the text and the dissociation constants from fits of the data to the appropriate equations are listed in Table 3.
For the log kcat vs. pH profile, a single ionization is observed for either Zn- or Cd-NagA. This ionization must represent the protonation of the metal-bound hydroxide that is utilized for substrate hydrolysis or, alternatively, the general base that is utilized to deprotonate the metal-bound water molecule prior to substrate hydrolysis. The pKa value is slightly higher for Cd-NagA than for Zn-NagA, as expected from the tendency of zinc to lower the pKa of water relative to cadmium (28). For the log kcat/Km profiles, two ionizations are observed at low pH for both metal-substituted forms of the enzyme. One of these ionizations must be from the free enzyme and likely reflects the state of protonation for the group that must activate the metal-bound water molecule (or hydroxide). From the x-ray crystal structure of NagA and homologous active site structures for other members of the amidohydrolase superfamily, this residue must be the invariant carboxylate group, Asp-273, from the end of β-strand 8 (8, 21). The second ionization observed in the log kcat/Km profile could originate from other residues within the active site of NagA but it could also arise from the protonation of the substrate itself since the phosphate moiety has a pKa of ∼6.1 (29). If the enzyme requires (or highly prefers) a doubly ionized phosphate substituent for binding then a diminution in activity will be observed in the kcat/Km plot but not the kcat vs. pH profile (30).
To address this issue experimentally, the pH-rate profiles for the hydrolysis of the sulfate analog of the substrate (3) were measured. The sulfate-derivative is a mono-anion above pH 5 and thus no ionizations can originate from the protonation of this compound in the pH range available for the characterization of NagA (pH 5 – 10). At saturating substrate the sulfate analogue (3) is hydrolyzed at approximately half the rate of the physiological substrate (1), although the Km value is significantly higher (Table 2). For the pH-rate profiles (Figures 2A and 2B) a single ionization of pKa 6.5 is observed in the log kcat vs. pH profiles and a single ionization of pKa 6.2 is observed in the log kcat/Km profile (Table 3). These results are fully consistent with the conclusion that the second ionization observed in the pH-rate profile for log kcat/Km with compound 1 is due to the protonation of the phosphate moiety to a mono-anion. It is also curious to note that the ionization that is observed at high pH in the log kcat/Km vs. pH-rate profiles for the natural substrate is not observed during the hydrolysis of the sulfate derivative. These results suggest that the mono-anionic sulfate derivative does not require the interaction with an enzyme group that apparently ionizes in this pH range. The likely candidates for this group include Lys-139 and Tyr-223, based on the structure of d-glucosamine-6-phosphate bound to the enzyme from B. subtilis (10), and the inhibitor bound form of the D273N mutant from E. coli (21).
Table 3.
Kinetic pKa Values from pH – Rate Profiles of Metal- Substituted Variants and Mutant.
| Enzyme | Metal | Substrate |
kcat vs. pH pKa |
kcat/Km vs. pH pKa |
kcat/Km vs. pH pKb |
|---|---|---|---|---|---|
| Wild type | Zn | 1 | 5.6 ± 0.1 | 6.4 ± 0.1a | 9.3 ± 0.4 |
| Wild type | Cd | 1 | 6.4 ± 0.1 | 6.1 ± 0.1a | 9.1 ± 0.2b |
| K139M | Zn | 1 | 5.4 ± 0.2 | 7.0 ± 0.1 | 9.1 ± 0.4 |
| Wild type | Zn | 3 | 6.5 ± 0.1 | 6.2 ± 0.1 | - |
Average value of pKa for two ionizations.
Average value of pKb for two ionizations.
Mutation of Active Site Residues
Site specific mutants of NagA were constructed in order to ascertain the roles of specific residues in substrate recognition and catalytic function. The metal content and the kinetic parameters for the mutants of NagA constructed for this investigation are presented in Table 4. Residues Q59 and N61 were mutated together to a pair of histidine residues to create an HxH motif that is analogous to that found in NagA from B. subtilis and T. maritima. This alteration resulted in a decrease in kcat, a large increase in Km, and an increase in the average amount of Zn bound to the protein, suggesting that an additional metal ion can bind to this mutant. The drop in catalytic activity may result from the binding of this second metal ion or to steric crowding within the confines of an active site that has apparently evolved to operate with a single divalent cation. For the single histidine substitutions, the N61H mutant is similar in catalytic activity to the double mutant whereas the Q59H mutant is diminished by less than an order of magnitude relative to the wild type enzyme. For the substitutions at Asn-61, the replacement with an alanine is significantly less disruptive than the change to a histidine.
Table 4.
Metal Content and Kinetic Parameters of NagA and Mutantsa.
| Mutant | Zn/Subunit | kcat (s−1) | Km (mM) | kcat/Km (M−1s−1) |
|---|---|---|---|---|
| Q59H/N61H | 1.5 | 1.2 ± 0.1 | 5.4 ± 0.5 | (2.2 ± 0.3) × 102 |
| Q59H | 1.3 | 32 ± 3 | 0.31 ± 0.06 | (1.0 ± 0.2) × 105 |
| Q59A | 0.9 | 10 ± 0.4 | 0.08 ± 0.01 | (1.3 ± 0.2) × 105 |
| N61H | 1.0 | 2.6 ± 0.1 | 1.7 ± 0.2 | (1.5 ± 0.2) × 103 |
| N61A | 0.9 | 24 ± 1 | 0.80 ± 0.05 | (3.0 ± 0.2) × 104 |
| E131Q | 0.1 | 0.7 ± 0.1 | 3.6 ± 1.1 | (1.9 ± 0.7) × 103 |
| E131A | 0.2 | 1.9 ± 0.3 | 0.15 ± 0.06 | (1.3 ± 0.5) × 104 |
| K139M | 0.9 | 49 ± 2 | 2.0 ± 0.2 | (2.5 ± 0.3) × 104 |
| H143N | 0.6 | 0.43 ± 0.02 | 2.1 ± 0.1 | (2.0 ± 0.1) × 102 |
| H143Q | 0.3 | 2.4 ± 0.1 | 0.33 ± 0.04 | 7.3 ± 0.9 × 103 |
| Y223F | 0.8 | 167 ± 15 | 0.76 ± 0.11 | (2.2 + 0.4) × 105 |
| H251N | 0.9 | 6.8 ± 0.4 | 2.7 ± 0.4 | (2.5 + 0.4) × 103 |
| D273N | 0.7 | < 0.02 | - | - |
| D273A | 0.8 | < 0.02 | - | - |
Kinetic parameters were determined at pH 7.5 and 30 °C from fits to equation 1 using compound 1 as the substrate.
The x-ray structures of NagA from E. coli (21), T. maritima (pdb code: 1o12), and B. subtilis (10) indicate in each case that a glutamate from the end of β-strand 3 of the (β/α)8-barrel interacts with one or both of the divalent cations bound within the active site. Mutation of this residue to either glutamine or alanine results in mutant enzymes that have lost a significant amount of catalytic activity. The value of kcat is reduced approximately two orders of magnitude and the Km for the E131Q mutant is increased to 3.6 mM. A significant portion of this reduction in catalytic efficiency has arisen because the metal content of the purified proteins is relatively low. Incubation of 25 μM E131A mutant with 15 equivalents of ZnCl2 enhanced the catalytic activity of this enzyme by a factor of three.
In the crystal structure of the D273N mutant of NagA, the side chain of His-143 is 3.0 Å away from the phosphonate oxygen of the mimic of the tetrahedral intermediate (21). This result suggests that this group could facilitate the activation of the carbonyl group of the substrate in conjunction with an interaction with the metal ion bound within the active site. Mutation of H143 to an asparagine resulted in the dramatic loss of catalytic activity and a moderate increase in the Km, producing a 6,000-fold decrease in kcat/Km. The H143Q mutant was more active with a decrease in the value of kcat/Km of greater than two orders of magnitude. The H143Q mutant was isolated with 0.6 equivalents of Zn, while the H143N mutant contained only 0.3 equivalents. Overall, these results are consistent with the participation of H143 in the polarization of the substrate carbonyl group.
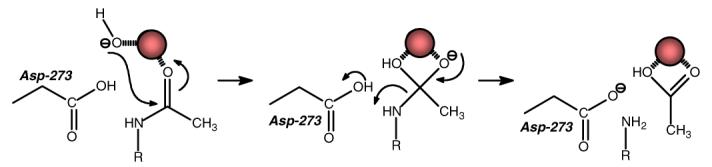
The most significant reduction in catalytic activity occurs with the mutation of the invariant aspartate (Asp-273) from the end of β-strand 8. In the x-ray crystal structure of NagA from E. coli this residue is hydrogen bonded to the lone water molecule (or hydroxide) that is coordinated to the metal ion bound to the Mβ-site (21). Therefore, it is expected that this residue will function in catalysis by abstraction of a proton from water prior to, or concomitant with, the attack of hydroxide on the amide bond of the substrate. Within the detection limits of our assay for product formation, we were unable to measure any catalytic activity for either the D273N or D273A mutant. The loss of activity is not due to a diminished capacity to bind divalent cations since the metal content of the purified mutants was identical to the wild type enzyme. The catalytic properties of these mutants are fully consistent with the proposed role of this residue in the chemical mechanism as the primary general base for activation of the hydrolytic water molecule and the subsequent protonation of the leaving group amine.
The crystal structure of NagA from B. subtilis bound with the product, d-glucosamine-6-phosphate, indicates that an invariant arginine residue (equivalent to Arg-227 in the E. coli enzyme) from the adjacent subunit ion-pairs with the phosphate moiety of the substrate (10). It is unlikely that this arginine residue could contribute to the diminution in kcat/Km at high pH since the pKa value for the ionization of the guanidino group is expected to exceed 12. However, in NagA from E. coli there are tyrosine (Y223) and lysine (K139) residues that may also be contributing to the interaction with the phosphate moiety of the substrate. Deprotonation of either of these residues at high pH could result in a drop in the value of kcat/Km. The mutation of Lys-139 to methionine reduces the value of kcat by a factor of two and the value of kcat/Km by a factor of ∼50 when the phosphate derivative (1) is used as the substrate, relative to the wild type enzyme. However, there is only a 2-fold drop in kcat/Km relative to the wild type enzyme when K139M is used to hydrolyze N-acetyl-d-glucosamine-6-sulfate (3). However, the pH-rate profile for K139M still exhibits a drop in activity at high pH when N-acetyl-d-glucosamine-6-phosphate (1) is used as the substrate and thus it is unlikely that the ionization state of this lysine is critical for catalytic activity. Mutation of Y223 to a phenylalanine residue resulted in a mutant with nearly the same catalytic activity as the wild type enzyme and thus this residue is not significant for the binding of substrate in NagA.
The structure of the D273N mutant of the E. coli NagA in the presence of the phosphonate inhibitor indicates that H251 interacts with the anomeric hydroxyl group at carbon 1 (21). The H251N mutant has a kcat of 6.8 s−1 and a Km of 2.7 mM. The value for kcat/Km is 1/400 of that measured for the wild type enzyme. These results confirm the importance for the role of H251 in the binding of substrate within the active site of NagA. The homologous residue was found to interact with d-glucosamine-6-phosphate in the structure of NagA from B. subtilis. This histidine residue (H258) was found to be 2.7 Å from the oxygen atom on the anomeric carbon in the α-conformation (10).
Rate Limiting Steps
Three experimental probes were marshaled with NagA to unveil the source of the rate limitation for this enzyme. The first of these experiments compared the rates of substrate hydrolysis in H2O and D2O. Using Zn-NagA with N-acetyl-d-glucosamine-6-phosphate as the substrate, initial velocity kinetics were measured at a pH/pD of 7.5. At concentrations of substrate up to 4 mM there is evidence for substrate inhibition and the data were therefore fit to equation 2 to yield kinetic constants of kcat (78 s−1), Km (0.14 mM) and kcat/Km (5.7 × 105 M−1s−1) for the results in H2O. The corresponding values in D2O under identical conditions with the same enzyme stock were the following: kcat = 69 s−1, Km = 0.12 mM and kcat/Km = 5.6 × 105 M−1s−1. The solvent isotope effect for the substitution of D2O for H2O on kcat/Km is 1.02 and the value on kcat is 1.1. These relatively modest solvent isotope effects indicate that proton transfer is not a significant rate determining step in this transformation.
The rate limitation imposed by the chemical cleavage of the amide bond was tested by employing the trifluoroacetate derivative of d-glucosamine-6-phosphate (6) as a substrate. Viola et al. have demonstrated that the trifluoroacetate analog of N-acetyl-l-aspartate is a very good substrate for aspartoacylase from human brain (31). The kinetic parameters for the hydrolysis of 6 with NagA are listed in Table 2 This substrate is hydrolyzed at a significantly faster rate than is the corresponding N-acetyl derivative. Since the pKa of trifluoroacetic acid (0.23) is significantly lower than that of acetic acid (4.76), the carbon-nitrogen bond is weaker and the carbonyl group more electrophilic. Therefore, the rate of hydrolysis of 6 is expected to be faster than the hydrolysis of 1 if NagA is limited by the chemical step in the steady state. This assessment does not differentiate whether the rate limiting step is the formation or cleavage of the putative tetrahedral intermediate. It could be argued, however, that if NagA were limited by product release from the active site, then the dissociation of trifluoroacetate could be inherently faster than the release of acetate. This specific scenario was addressed by the utilization of changes in solvent viscosity to systematically alter the rate constants for the association and dissociation of products and substrates with NagA (19, 20).
The effects of solvent viscosity on the catalytic rate constants for the hydrolysis of 1 by Zn-NagA are shown in Figures 4A and 4B using sucrose as the micro-viscogen. The plot of relative kcat vs. relative viscosity was fit to a linear equation yielding a slope of 0.008 ± 0.02. The corresponding fit of data for the effect of solvent viscosity on kcat/Km gave a slope of 0.065 ± 0.049. These results indicate quite clearly that the rate of product dissociation does not limit kcat for the hydrolysis of substrates by NagA. These experiments also indicate that the value of kcat/Km is not limited by the association rate constant for the formation of the NagA-substrate complex. These results (together with the enhanced rate of reaction of compound 6 relative to compound 1) are only consistent with the conclusion that NagA is limited by either the formation or breakdown of the tetrahedral intermediate.
Figure 4.
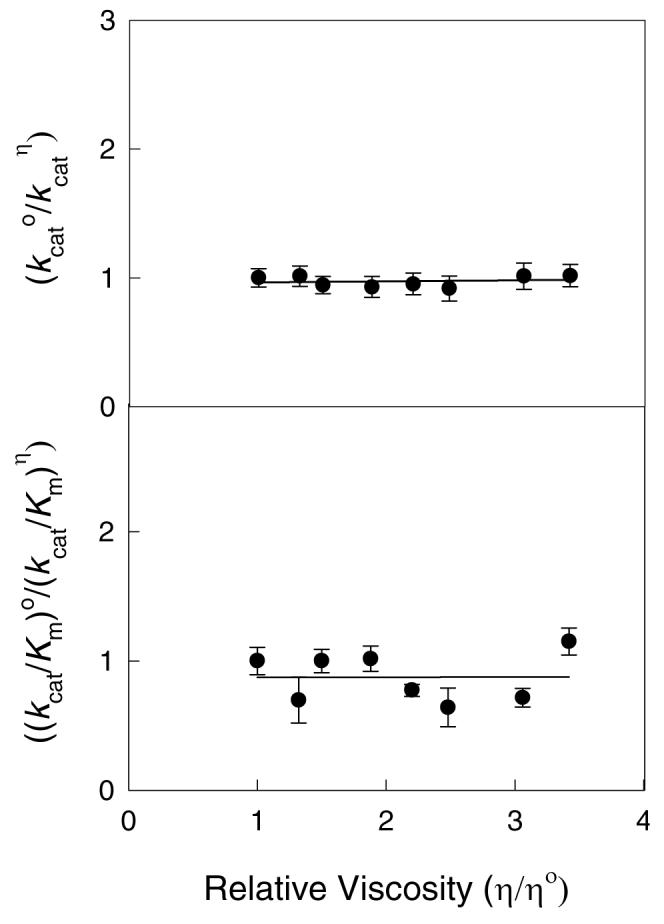
The effect of viscosity on the relative values of kcat (A) and kcat/Km (B) using sucrose as the micro-viscogen. Additional details are available in the text.
Mechanism of Action
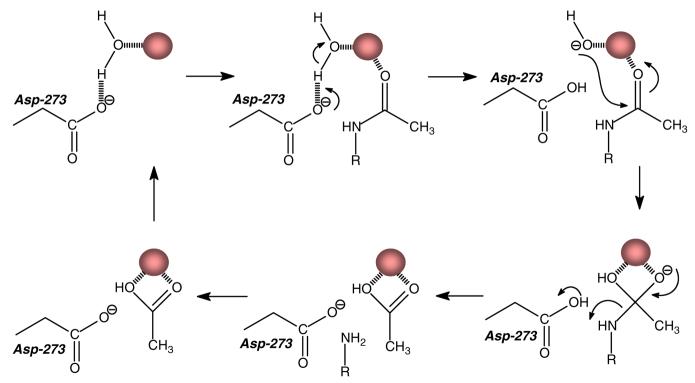
A model for the mechanism of amide bond hydrolysis by NagA from E. coli is presented in Figure 5. In this scheme the resting state of the enzyme has a single divalent cation bound to the active site through ligation with the two histidines at the end of β-stands 5 and 6 (His-195 and His-216), and the glutamate (Glu-131) from the end of β-stand 3 (21). The remaining ligand is a bound water molecule that is, in turn, hydrogen bonded to the invariant aspartate (D273) that originates from the end of β-strand 8. The reaction is initiated by the binding of the substrate in the active site and polarization of the carbonyl oxygen via ligation to the divalent cation. There is a proton transfer from the bound water molecule to Asp-273 and this is followed by nucleophilic attack on the carbonyl carbon and formation of the tetrahedral intermediate. The collapse of the tetrahedral intermediate is facilitated by proton transfer from Asp-273 to the leaving group amine of the product, d-glucosamine-6-phosphate. The products depart the active site and another round of catalysis commences with the charging of the metal center with a molecule of water from the solvent.
Figure 5.
Proposed mechanism for the substrate hydrolysis by NagA from E. coli.
The mechanism of substrate and water activation by NagA from E. coli must be different than the mechanism utilized by the same enzyme from B. subtilis. The B. subtilis enzyme contains a binuclear metal center whereas the E. coli enzyme can bind but a single divalent cation in the active site. For a binuclear metal center, the activation of the amide bond and the activation of the solvent water can be distributed separately between the two metal ions. However, in the mononuclear metal center, both functions must be borne by a single divalent cation. These results highlight the significant diversity for the evolution of function within the amidohydrolase superfamily. It is not intuitively obvious whether a binuclear or mononuclear metal center represents a more “advanced” or sophisticated site of catalytic power. However, it is tempting to speculate that the binuclear metal center currently found in NagA in some organisms is in the process of shedding one of the divalent cations to create a fully functional active site that operates with a single divalent cation.
Figure 3.
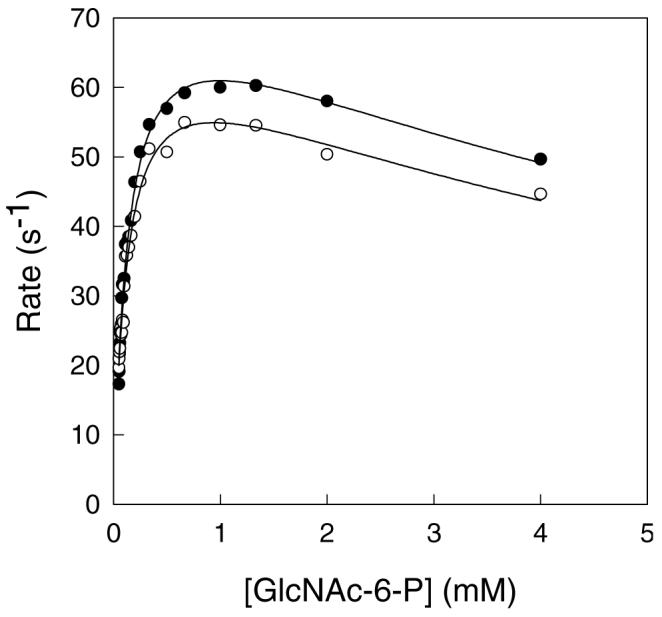
Solvent isotope effects for reactions performed in H2O ( ● ) and D2O ( ○ ) at pH/pD 7.5. The solid lines represent fits of the data to equation 2. Additional details are available in the text.
For Table of Contents Use Only
ACKNOWLEDGEMENT
We are indebted to Jessica A. DiGirolamo for the initiation of the preliminary experiments with NagA (CHE-0243829). We thank Dr. Ricardo Marti-Arbona for advice and experimental assistance.
Footnotes
This work was supported in part by the NIH (GM 71790) and the Robert A. Welch Foundation (A-840). RSH was supported by a Chemical Biology Interface Training Grant (GM 08523).
Abbreviations: NagA, N-acetyl-d-glucosamine-6-phosphate deacetylase; ICP-MS, inductively coupled plasma mass spectrometry.
REFERENCES
- 1.Uehara T, Park JT. The N-acetyl-d-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 2004;186:7273–7279. doi: 10.1128/JB.186.21.7273-7279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uehara T, Suefuji K, Valbuena N, Meehan B, Donegan M, Park JT. Recycling of the anhydro-n-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 2005;187:3643–3649. doi: 10.1128/JB.187.11.3643-3649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uehara T, Suefuji K, Jaeger T, Mayer C, Park JT. MurQ etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J. Bacteriol. 2006;188:1660–1662. doi: 10.1128/JB.188.4.1660-1662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White RJ, Pasternak CA. The purification and properties of N-acetylglucosamine 6-phosphate deacetylase from Escherichia coli. Biochem. J. 1967;105:121–125. doi: 10.1042/bj1050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Souza JM, Plumbridge JA, Calcagno ML. N-acetyl-glucosamine-6-phosphate deacetylase from Escherichia coli: purification and molecular and kinetic characterization. Arch. Biochem. Biophys. 1997;340:338–346. doi: 10.1006/abbi.1997.9780. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Hall R, Cummings J, Raushel FM. Tight binding inhibitors of N-acyl amino sugar and n-acyl amino acid deacetylases. J. Am. Chem. Soc. 2006;128:4244–4245. doi: 10.1021/ja0600680. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira FM, Mendoza-Hernandez G, Castaneda-Bueno M, Aparicio R, Fischer H, Calcagno ML, Oliva G. Structural analysis of N-acetyl-d-glucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli. J. Mol. Biol. 2006;359:308–321. doi: 10.1016/j.jmb.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 8.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 9.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 10.Vincent F, Yates D, Garman E, Davies GJ, Brannigan JA. The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily. J. Biol. Chem. 2004;279:2809–2816. doi: 10.1074/jbc.M310165200. [DOI] [PubMed] [Google Scholar]
- 11.Pegg SC-H, Brown S, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 12.Thoden JB, Phillips GN, Jr., Neal TM, Raushel FM, Holden HM. Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center. Biochemistry. 2001;40:6989–6997. doi: 10.1021/bi010682i. [DOI] [PubMed] [Google Scholar]
- 13.Buchbinder JL, Stephenson RC, Dresser MJ, Pitera JW, Scanlan TS, Fletterick RJ. Biochemical characterization and crystallographic structure of an Escherichia coli protein from the phosphotriesterase gene family. Biochemistry. 1998;37:10860. doi: 10.1021/bi985043v. [DOI] [PubMed] [Google Scholar]
- 14.Nitanai Y, Satow Y, Adachi H, Tsujimoto M. Crystal structure of human renal dipeptidase involved in beta-lactam hydrolysis. J. Mol. Biol. 2002;321:177–184. doi: 10.1016/s0022-2836(02)00632-0. [DOI] [PubMed] [Google Scholar]
- 15.Liaw SH, Chen SJ, Ko TP, Hsu CS, Chen CJ, Wang AH, Tsai YC. Crystal structure of d-aminoacylase from Alcaligenes faecalis DA1. A novel subset of amidohydrolases and insights into the enzyme mechanism. J. Biol. Chem. 2003;278:4957–4962. doi: 10.1074/jbc.M210795200. [DOI] [PubMed] [Google Scholar]
- 16.Ireton GC, McDermott G, Black ME, Stoddard BL. The structure of Escherichia coli cytosine deaminase. J. Mol. Biol. 2002;315:687–697. doi: 10.1006/jmbi.2001.5277. [DOI] [PubMed] [Google Scholar]
- 17.Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 18.Williams L, Nguyen T, Li Y, Porter TN, Raushel FM. Uronate isomerase: a nonhydrolytic member of the amidohydrolase superfamily with an ambivalent requirement for a divalent metal ion. Biochemistry. 2006;45:7453–7462. doi: 10.1021/bi060531l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brouwer AC, Kirsch JF. Investigation of diffusion-limited rates of chymotrypsin reactions by viscosity variation. Biochemistry. 1982;21:1302–1307. doi: 10.1021/bi00535a030. [DOI] [PubMed] [Google Scholar]
- 20.Maurice M, Bearne SL. Kinetics and thermodynamics of mandelate racemase catalysis. Biochemistry. 2002;41:4048–4058. doi: 10.1021/bi016044h. [DOI] [PubMed] [Google Scholar]
- 21.Hall RS, Brown S, Federov A, Federov L, Babbitt PC, Almo SC, Raushel FM. “Structural Diversity within the Mononuclear and Binuclear Active Sites of N-acetyl-D-Glucosamine-6-Phosphate Deacetylase”. Biochemistry. 2007;46:0000–0000. doi: 10.1021/bi700544c. [DOI] [PubMed] [Google Scholar]
- 22.Omburo GA, Kuo JM, Mullins LS, Raushel FM. Characterization of the zinc binding site of bacterial phosphotriesterase. J. Biol. Chem. 1992;267:13278–13283. [PubMed] [Google Scholar]
- 23.Marti-Arbona R, Fresquet V, Thoden JB, Davis ML, Holden HM, Raushel FM. Mechanism of the reaction catalyzed by isoaspartyl dipeptidase from Escherichia coli. Biochemistry. 2005;44:7115–7124. doi: 10.1021/bi050008r. [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, Liu Y, Bruzik KS, Tsai MD. A novel calcium-dependent bacterial phosphatidylinositol-specific phospholipase C displaying unprecedented magnitudes of thio effect, inverse thio effect, and stereoselectivity. J. Am. Chem. Soc. 2003;125:22–23. doi: 10.1021/ja029019n. [DOI] [PubMed] [Google Scholar]
- 25.Hougland JL, Kravchuk AV, Herschlag D, Piccirilli JA. Functional identification of catalytic metal ion binding sites within RNA. PLoS Biology. 2005;3:e277. doi: 10.1371/journal.pbio.0030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson RG. Acids and bases. Science. 1966;151:172–177. doi: 10.1126/science.151.3707.172. [DOI] [PubMed] [Google Scholar]
- 27.Bond MD, Holmquist B, Valee BL. Thioamide substrate probes of metal-substrate interactions in carboxypeptidase A catalysis. J. Inorg. Biochem. 1986;28:97–105. doi: 10.1016/0162-0134(86)80074-5. [DOI] [PubMed] [Google Scholar]
- 28.Barnum DW. Hydrolysis of cations - formation-constants and standard free-energies of formation of hydroxy complexes. J. Inorg. Chem. 1983;22:2297–2305. [Google Scholar]
- 29.Jencks WP, Westheimer FH. pKa Data Compiled by R. Williams. http://research.chem.psu.edu/brpgroup/pKa_compilation.pdf (accessed 11/06/06), part of B. R. Peterson's web page at Penn State University (accessed 11/06/06). http://research.chem.psu.edu/brpgroup/grouphomepage.html (accessed 11/06/06)
- 30.Cleland WW. The use of pH studies to determine chemical mechanisms of enzyme-catalyzed reactions. Methods Enzymol. 1982;87:390–405. doi: 10.1016/s0076-6879(82)87024-9. [DOI] [PubMed] [Google Scholar]
- 31.Le Coq J, An HJ, Lebrilla C, Viola RE. Characterization of human aspartoacylase: The brain enzyme responsible for Canavan disease. Biochemistry. 2006;45:5878–5884. doi: 10.1021/bi052608w. [DOI] [PMC free article] [PubMed] [Google Scholar]