Abstract
The 26 amino acid hemolytic melittin peptide was converted into a gene transfer peptide that binds to DNA and polymerized through disulfide bond formation. Melittin analogues were synthesized by addition of one to four Lys repeats at either the C or N-subterminal end along with terminal Cys residues. Melittin analogues were able to bind and polymerize on plasmids resulting in the formation of DNA condensates. In the absence of DNA, melittin analogues retained their red blood cell hemolytic potency but were inactive when bound to plasmid DNA. The in vitro gene transfer efficiency mediated by poly-melittin analogues was equivalent to PEI in HepG2 cells. Attempts to truncate portions of either of the two melittin α-helices resulted in concurrent loss of hemolytic potency and gene transfer efficiency. The results demonstrate the ability to transform melittin into a gene transfer peptide by transiently masking its membrane lytic activity by the addition of Lys and Cys residues to promote DNA binding and polymerization.
Keywords: gene delivery, gene therapy, melittin, peptides, transfection
Introduction
Non-viral gene delivery systems depend upon endosomal lysis to allow DNA to enter the cytosol (1–3). Two main strategies have emerged to achieve endosomal lysis. The first relies upon endosomal lytic peptides that respond to lowering of the pH, leading to fusion with the endosomal membrane and lysis (3–7). The most widely used fusogenic peptides used to enhance gene delivery include the hemolytic peptide (HA2), GALA and melittin. The second strategy to achieve endosomal lysis relies upon the use of endosomal buffering agents that resist changes in pH leading to osmotic lysis. This is the reported mechanism by which chloroquine, PEI and polyhistidine achieve enhanced gene delivery (8–10).
Naturally occurring melittin from bee venom is a 26 amino acid peptide composed of two α helices joined by an interrupting proline (11–13). A number of prior studies have examined melittin and its analogues to elucidate the amino acid sequences that are essential to its potent hemolytic and antimicrobial activity (14–16). Likewise, one prior study examined the hemolytic activity of dimeric melittin formed by disulfide bond formation between terminal Cys residues (17). Unlike HA2 which is primarily hemolytic at pH 4 but not 7, the hemolytic activity of melittin is not pH dependent. Consequently, because melittin remains hemolytic at pH 7, its utility for enhancing gene delivery necessitates masking to avoid unintended cell lysis.
Despite this potential limitation, melittin has been examined as an in vitro gene transfer agent to increase endosomal escape of DNA (3, 5, 18, 19). Several groups have incorporated a Cys containing melittin into either a lipid or PEI resulting in a significant improvement in the gene transfer efficiency while simultaneously diminishing melittin’s toxicity (3, 5, 19). An alternate strategy involved masking the hemolytic activity of melittin by reversible derivatization of Lys residues (18).
We previously reported that the gene transfer efficiency of short polylysine peptides can be dramatically increased by installing terminal Cys residues resulting in peptide polymerization (20, 21). The multiplication of cationic residue in sulfhydryl polymerized peptides relative to a monomeric peptide, results in much greater DNA binding affinity. Likewise, sulfhydryl-polymerized peptides are postulated to undergo intra-cellular reduction to trigger the release of DNA (22).
In the present study we have transformed melittin into a sulfhydryl polymerized peptide and investigated its in vitro gene transfer activity. One of the goals of this study was to investigate how structural modifications in the melittin sequence influence its hemolysis of red blood cells and gene transfer efficiency. The results establish a relationship between melittin hemolytic activity and gene transfer efficiency and provide a means to transiently mask the membrane lytic activity of melittin to enhance gene transfer.
Experimental Procedures
Substituted Wang resin for peptide synthesis, N-terminal Fmoc protected amino acids, 9-hydroxybenzotriazole, diisopropylcarbodiimide, and diisopropylethylamine were obtained from Advanced ChemTech (Lexington, KY). N,N-Dimethylformamide, trifluoroacetic acid (TFA), acetic anhydride, acetonitrile and piperidine were purchased from Fisher Scientific (Pittsburgh, PA). Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) and thiazole orange were obtained from Sigma Chemical Co. (St. Louis, MO). Polyethylene amine (PEI) 25 KDa was purchased from Aldrich (Milwaukee, WI). D-luciferin and luciferase from Photinus pyralis (EC 1.13.12.7) were obtained from Roche Applied Science (Indianapolis, IN). HepG2, CHO and COS-7 cells were acquired from the American Type Culture Collection (Manassas, VA). Inactivated “qualified” fetal bovine serum (FBS) was from Life Technologies, Inc. (Carlsbad, CA). BCA reagent was purchased from Pierce (Rockford, IL).
Peptide synthesis was carried out on an Apex 396 Advanced ChemTech solid phase peptide synthesizer. Peptide purification was performed using a semi-preparative (10 μm) C18 RP-HPLC column from Vydac (Hesperia, CA). Preparative HPLC was performed using a computer-interfaced HPLC and fraction collector from ISCO (Lincoln, NE). Electrospray ionization mass spectrometry (ESI-MS) was performed using an Agilent 1100 Ion Trap LC-MS system.
Synthesis and Characterization of Melittin Analogues
Melittin and its analogues were synthesized using standard Fmoc procedures with 9-hydroxybenzotriazole and diisopropylcarbodiimide double couplings on 30 μmol scale. Peptides were cleaved from the resin and side chain protecting groups were removed by reaction with TFA/ethane dithiol/water (95:2.5:2.5 v/v/v) for 3 hr. Cleaved peptides were precipitated with cold ether. The precipitate was washed twice with cold ether and dissolved in 0.1% TFA for purification. Peptides were purified to homogeneity on RP-HPLC by injecting 2 μmol onto a semi-preparative column (2 × 25 cm) eluted at 10 ml/min with 0.1 v/v% TFA and a Vydac C18 gradient of acetonitrile of 20–55 v/v% over 30 min while monitoring tryptophan absorbance at 280 nm. The major peak was collected and pooled from multiple runs, concentrated by rotary evaporation, lyophilized, and stored dry at −20 °C. Purified peptides were reconstituted in 0.1 v/v% TFA (degassed with argon) and quantified by tryptophan (ε280 = 5600 M−1 cm−1) absorbance to determine the isolated yield, which was typically 10–25%.
Purified peptides were characterized by LC-MS by injecting 1 nmol onto a Vydac C18 analytical column (0.47 × 25 cm) eluted at 0.7 ml/min with 0.1 v/v% TFA and an acetonitrile gradient of 5 to 65 v/v% over 30 min. The mass spectral data were obtained in the positive mode.
Circular Dichroism (CD) Spectrometry Melittin Analogues
CD spectra were recorded on a JASCO-J-715 spectra polarimeter in 1-cm path length cells under nitrogen at 25°C. The spectra were recorded between 200 and 240 nm at a peptide concentration of 10 μM in either 10 mM Na2HPO4 pH 7.4 or 40 v/v% TFE (trifluoroethanol) in 10 mM Na2HPO4 pH 7.4. The percentage of α-helix structure was calculated as follows: α-helix (%) = ([θ]222 − [θ]0222)/[θ]100222, where [θ]222 is the experimentally observed absolute mean residue ellipticity at 222 nm. Values for [θ]0222 and [θ]100222, corresponding to 0 and 100% helix content at 222 nm, were estimated to be −2000 and −28400 degrees cm2/dmol, respectively (16). The absolute mean residue ellipticity at 222 nm was calculated using the formula [θ]222 = 100 (θ)/(lcN), where (θ) is the observed ellipticity in degrees, l is the optical path length in centimeters, c is the molar concentration of the peptide, and N is the number of residues in the peptide.
RBC Hemolysis by Melittin Analogues
Whole blood samples were obtained from male ICR mice by heart puncture with heparinized 22G needles and collected in conical tubes containing 10 ml of 0.15 M PBS (pH 7.4) pre-warmed to 37°C. The erythrocytes were immediately separated from plasma by centrifugation at 2,000 rpm for 2 min, washed three times with 10 ml of PBS, and then diluted to 1.5 × 108 cells/ml.
Peptide stock solutions of 15 μM were prepared and serially diluted in a total volume of 100 μl in a MultiScreenHTS BV 96 well plate. Erythrocytes (50 μl, 7.5 × 106 cells) were added to the peptide and incubated at 37°C for 1 hr followed by filtration on a Multiscreen vacuum manifold (Millipore Corporation, Billerica, MA U.S.A.). The filtrate was measured for Abs415nm on a plate reader and the percent hemolysis calculated relative to complete hemolysis caused by replacing PBS with water. Peptide DNA condensates (5 μg DNA, 1nmol peptide in 100 μl PBS) were also assayed for hemolysis as described above.
Formulation of Melittin DNA Condensates
Peptide DNA condensates were prepared by combining 50 μg of DNA in 500 μl of Hepes-buffered mannitol (HBM) (0.27 M mannitol, 5 mM Hepes, pH 7.5) with 5–75 nmol of peptide in 500 μl of HBM while vortexing to create DNA condensates possessing a calculated for charge ratio (NH4+:PO4−) ranging from 0.1 to 2.4. Peptide DNA condensates were incubated 1 hr at room temperature to allow peptide polymerization.
The particle size of each peptide DNA condensate was determined by quasi-elastic light scattering at a scatter angle of 90° on a Brookhaven ZetaPlus particle sizer. Condensates were analyzed at a DNA concentration of 50 μg/ml in HBM at a stoichiometry of 0.3–0.8 nmol of peptide/μg of DNA corresponding to a charge ratio of approximately 1.5:1 for each. The mean diameter was computed from the diffusion coefficient using a unimodal cumulate analysis supplied by the manufacturer.
Stability of Poly-Melittin DNA Condensates
DNA condensates (50 μl of 50 μg/ml) were formed at 0.9 nmol peptide per μg pCMVL and incubated at RT for 30 min and then combined with 0–50 μl of 4 M sodium chloride and normalized to 100 μl with HBM to achieve a final sodium chloride concentration of 0, 0.2, 0.4, 0.8, 1.0 and 2.0 M. Each sample was sonicated for 30 s with a 100 W Microson XL-2000 ultrasonic probe homogenizer (Kontes, Vineland, NJ) with a vibrational amplitude of 6 to fragment uncondensed DNA (23). DNA condensates were digested with 2.5 μg of proteinase K for 18 hrs at 37°C and then electrophoresed on a 1 % agarose gel and visualized by ethidium bromide staining.
In Vitro Gene Transfer Using Poly-Melittin DNA Condensates
HepG2 cells (5 × 105) were plated on 6 × 35 mm wells and grown to ca. 50 % confluency. Transfections were performed in MEM (2 ml/35 mm well) supplemented with 2 % FBS, sodium pyruvate (1 mM) and penicillin/streptomycin (100 U and 100 μg/ml). Melittin analogues were combined with pCMVL in HBM to form condensates at 1.5:1 (N:P) charge ratio. Peptide-DNA condensates (10 μg of DNA in 0.2 ml of HBM) were added drop wise to wells in triplicate. After 6 hrs incubation at 37°C, the transfection media were replaced with 10 % FBS supplemented culture media.
After 24 hrs, cells were washed twice with 2 ml of ice-cold phosphate-buffered saline (Ca2+- and Mg2+-free) and then treated with 0.5 ml of lysis buffer (25 mM Tris chloride, pH 7.8, 1 mM EDTA, 8 mM magnesium chloride, 1% Triton X-100) for 10 min at 4 °C. The cell lysates were scraped, transferred to 1.5-ml microcentrifuge tubes, and centrifuged for 7 min at 13,000 × g at 4 °C to pellet cell debris. Luciferase relative light units were measured by a Lumat LB 9501 (Berthold Systems, Germany) with 10 s integration after automatic injection of 100 μl of 0.5 mM D-luciferin. The relative light units were converted to fmols using a standard curve generated by adding a known amount of luciferase to 35-mm wells containing 50% confluent HepG2 cells. The resulting standard curve had an average slope of 8.4 × 104 relative light units/fmol of enzyme. Protein concentrations were measured by BCA assay using bovine serum albumin as a standard (24). The amount of luciferase recovered in each sample was normalized to milligrams of protein and reported as the mean and standard deviation obtained from triplicate transfections.
PEI pCMVL complexes were prepared by mixing 50 μg of DNA in 500 μl of HBM with 60 μg PEI in 500 μl of HBM while vortexing to create DNA complexes possessing a charge ratio (NH4+:PO4−) of 9:1. HepG2 cells were transfected with 10 μg of PEI-DNA condensates as described above. COS-7 and CHO cells (1 × 105) were plated on 6 × 35-mm wells and grown to ca. 50 % confluency, and then transfected as described above.
The toxicity of melittin analogues were evaluated by MTT assay (25). Briefly, HepG2, COS-7, and CHO cells were plated on 6 × 35-mm wells at 3×105 cells/well and grown to 40 – 70% confluency. The culture media was replaced with 2 ml of fresh MEM supplemented with 2% FBS and 3 μM of melittin analogues. After 6 hrs of incubation, the media was replaced with fresh culture media and grown for an additional 18 hrs. The media was replaced with 2 ml of fresh media and 500 μl of 0.5 % (w/v) 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) in PBS solution, and then incubated for 2 hrs to let formazan crystals form. The crystals were dissolved by adding 2 ml of dimethylsulfoxide (DMSO) then measured spectrophotometrically at 595 nm on a microplate reader (Bio-Rad, Bethesda, MD, USA). The percent viability was determined relative to untreated cells.
Results
Melittin is a well characterized α-helical membrane disrupting peptide derived from bee venom. Its utility in enhancing PEI mediated gene transfer has been previously established (3, 19). To investigate its potential utility for gene transfer, the hemolytic activity of melittin has been transiently masked by reversible N-acetylation (18). Based on these results, we proposed the transient masking of melittin’s hemolytic activity by controlling its reversible binding to plasmid DNA. This would potentially mask the hemolytic activity of melittin until released intracellularly following disulfide bond reduction.
Natural melittin possesses six positively charged amino groups that are insufficient to mediate strong binding to DNA to form stable condensates. Therefore, we prepared a series of melittin analogues supplemented with additional Lys residues to endow it with the ability to bind and condense plasmid DNA. To facilitate DNA binding, one to four Lys residues were added to the N-terminus or C- terminus of melittin (Table 1). In addition, each melittin analogue was equipped with N and C terminal Cys residues to encourage melittin disulfide polymerization on DNA. We anticipated that polymeric melittin should possess higher DNA binding affinity due to the larger number of amines per molecule.
Table 1.
Structure and Properties of Melittin Analogues and DNA Condensates
| Amino Acid Sequence of Synthetic Melittin Analogues | Massa | HL50 (μM)b pH 7.4: 4.5 | NaClc (M) | Particle Sized (μm) | Zeta Potentiale (+ mV) | |
|---|---|---|---|---|---|---|
| M0 | GIGA VLKV LTTG LPAL ISWI KRKR QQ | 2847.5/2850.0 | 1.5: 6 | 0.1 | >1 | 5.8 |
| M1 | C IGA VLKV LTTG LPAL ISWI KRKR QQ C | 2996.7/2998.8 | 3.0: 1.5 | 0.4 | >1 | 5.0 |
| M2 | CK IGA VLKV LTTG LPAL ISWI KRKR QQ C | 3124.9/3126.9 | 3.5: >10 | 0.4 | >1 | 5.0 |
| M3 | CKK IGA VLKV LTTG LPAL ISWI KRKR QQ C | 3253.0/3255.4 | 5.0: >10 | 0.4 | >1 | 5.0 |
| M4 | CKKK IGA VLKV LTTG LPAL ISWI KRKR QQ C | 3381.2/3383.7 | 7.5: >10 | 0.4 | >1 | 5.0 |
| M5 | CKKKK IGA VLKV LTTG LPAL ISWI KRKR QQ C | 3509.4/3509.7 | 7.5: >10 | 0.4 | >1 | 5.5 |
| M6 | CKK IGA VLKV LTTG LPAL ISWI KRKR QQ KKC | 3509.4/3509.7 | 2.5: >10 | 0.4 | >1 | 17.2 |
| M7 | CKKK IGA VLKV LTTG LPAL ISWI KRKR QQ KKKC | 3765.3/3765.9 | 2.5: >10 | 0.4 | >1 | 22.3 |
| M8 | CKKKK IGA VLKV LTTG LPAL ISWI KRKR QQ KKKKC | 4022.1/4022.4 | 2.5: >10 | 0.8 | 0.326 | 32.0 |
| M9 | CKKKK A VLKV LTTG LPAL ISWI KRKR QQ KKKKC | 3851.9/3852.3 | 2.5: >10 | 0.8 | 0.233 | 24.2 |
| M10 | CKKKK LKV LTTG LPAL ISWI KRKR QQ KKKKC | 3681.7/3682.3 | 5.0: >10 | 0.8 | 0.341 | 22.4 |
| M11 | CKKKK V LTTG LPAL ISWI KRKR QQ KKKKC | 3440.3/3440.9 | >10: >10 | 0.8 | 0.275 | 27.6 |
| M12 | CKKKK TTG LPAL ISWI KRKR QQ KKKKC | 3228.0/3228.3 | >10: >10 | 0.8 | 0.229 | 32.2 |
| M13 | CKKKK G LPAL ISWI KRKR QQ KKKKC | 3025.8/3026.0 | >10: >10 | 0.8 | 0.206 | 20.5 |
| M14 | CKKKK IGA VLKV LTTG LPAL ISWI KKKKC | 3197.1/3198.4 | 8: >10 | 0.8 | 0.253 | 29.6 |
| M15 | CKKKK IGA VLKV LTTG LPAL ISW KKKKC | 3083.9/3086.4 | >10: >10 | 0.8 | 0.332 | 12.9 |
| M16 | CKKKK IGA VLKV LTTG LPAL W KKKKC | 2883.7/2885.7 | >10: >10 | 0.8 | 0.145 | 15.4 |
| M17 | CKKKK IGA VLKV LTTG LP W KKKKC | 2699.5/2700.4 | >10: >10 | 0.4 | 0.252 | 5.9 |
| M18 | CKKKK IGA VLKV LTTG W KKKKC | 2489.2/2490.6 | >10: >10 | 0.4 | 0.159 | 6.5 |
Calculated and observed mass.
RBC hemolytic activity at pH 7.4 and 4.5.
Salt concentration above which peptide dissociates from DNA determined by gel electrophoresis as illustrated in Fig. 2.
Mean particle size of peptide DNA condensates as determined by QELS.
Mean zeta potential determined in 5 mM Hepes pH 7.4.
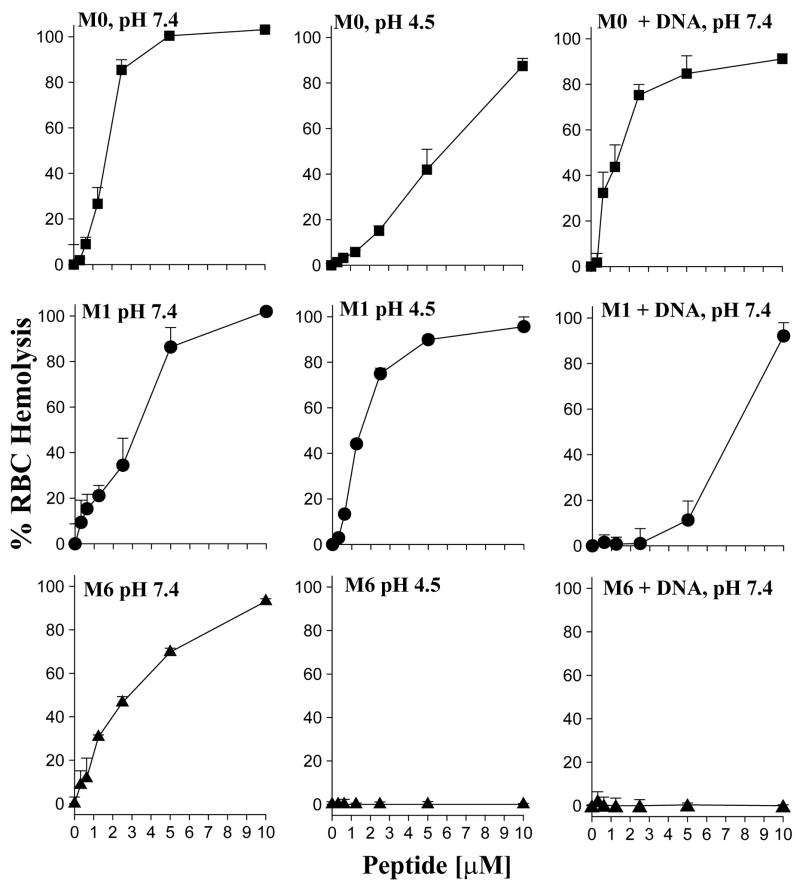
The hemolytic activity of melittin analogues were evaluated using mouse blood cells as a primary screen. Dose-response curves revealed that natural melittin (M0) possessed an HL50 (hemolytic activity of 50%) of approximately 1.5 μM at pH 7.4 (Fig. 1) whereas M0 was 4-fold less active at pH 4.5. Importantly, the combination of M0 and DNA resulted in hemolytic activity at pH 7.4 that was indistinguishable from M0 alone (Fig. 1) suggesting that M0 failed to bind to DNA.
Figure 1. Hemolysis Activity of Melittin Analogues and Melittin DNA Condensates.
The RBC hemolytic activity of melittin (M0) and melittin analogues (M1 and M6) are compared at pH 7.4, pH 4.5 and when bound to DNA at pH 7.4. The results illustrated in the left panels indicate that M0 is able to lyse RBCs with HL50 of 1.5 μM at pH 7.4, compared to M1 and M6 with HL50 of 3.0 and 2.5 μM. At pH 4.5 (middle panels), M0 is less potent (HL50 of 6 μM) whereas M1 has an HL50 of 1 μM and M6 is inactive. When combined with plasmid DNA at pH 7.4 (right panels), M0 retains its hemolytic activity, M1 is reduced to HL50 of 8 μM and M6 is completely inactivated.
Comparison of the results of M0 with a melittin peptide endowed with C and N-terminal Cys residues (M1) established an HL50 = 3 μM at pH 7.4 and 1 μM at pH 4.5. In contrast to M0, there was nearly a complete loss of hemolytic activity in the presence of DNA up to 5 μM peptide, followed by a recovery of activity at higher concentrations of peptide (Fig. 1). This result suggested that M1 was able to weakly bind and polymerize on DNA, resulting in its lack of hemolytic activity below 5 μM.
To enhance the ability of melittin to bind DNA, analogues were prepared by adding additional Lys residues to the N-terminal side of the peptide, considering that the endogenous KRKR sequence in melittin would likely be sufficient to promote DNA binding near the C-terminal and encourage disulfide bond formation with a neighboring peptide. The addition of one to four N-terminal Lys residues (Table 1, M2–M5) resulted in up to a 5-fold decrease in hemolytic activity relative to M0 but simultaneously afforded increased DNA binding affinity as determined by gel electrophoresis as described below.
Melittin analogues were also prepared with two to four-Lys residues added to both the N and C-subterminal end of melittin, along with terminal Cys residues. Comparison of the hemolytic activity for melittin analogues possessing 2, 3, or 4 N and C-subterminal Lys residues (M6, M7 and M8) established that each was slightly less potent than M0 (Table 1).
Comparison of the hemolytic activity of peptides M0–M8 at pH 4.5 revealed that only M0 and M1 possessed significant activity at low pH whereas M2–M8 were inactive. Interestingly, M1, which possessed terminal Cys residues, was more potent in hemolysis at pH 4.5 compared to native melittin (M0) (Table 1).
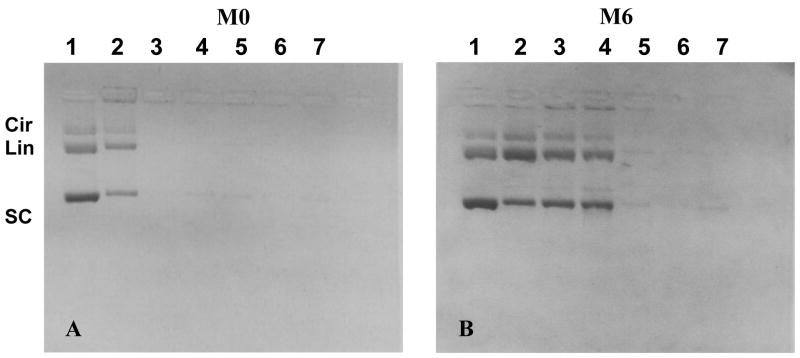
The hemolytic activity of M2–M8 at pH 7.4 was masked when the peptides were combined with DNA as illustrated for M6 (Fig. 1). This result can be rationalized by considering the binding properties of M0–M8 with DNA using a gel based assay as illustrated in figure 2 (20, 21). Agarose gel electrophoresis revealed peptide protection of the DNA from sonication until a critical salt concentration, at which the peptide dissociates and DNA is fragmented. The results established that M0 dissociates from DNA between 0–0.2 M sodium chloride (Fig. 2A, lane 2), whereas M6 binds and polymerizes on DNA resulting in condensates that are stable up to 0.4 M sodium chloride (Fig. 2B, lane 6). A similar result was observed for peptides M2–M8, all of which demonstrated the ability to bind and polymerize on DNA and thereby protect it from sonication in 0.4 M sodium chloride or higher (Table 1). These data establish that melittin analogues with four or more additional Lys and terminal Cys residues bind and polymerize on DNA, thereby masking their hemolytic activity.
Figure 2. Stability of Melittin DNA condensates.
The stability of melittin DNA condensates were analyzed by gel electrophoresis following 30 second sonication in the presence of increasing concentration of sodium chloride. Lanes 1–7 represent DNA standard (1), and peptide-DNA in 0 (2), 0.2 (3), 0.4 (4), 0.8 (5), 1.0 (6) and 2.0 M (7) sodium chloride, respectively. In panel A, M0 was only able to protect DNA during sonication at 0 M sodium chloride (lane 2) but not at 0.2 M (lane 3), whereas in panel B, M6 was able to polymerize on DNA and stabilized condensates up to 0.4 M sodium chloride (lane 4).
A second series of shorter melittin analogues were prepared in an attempt to truncate either the C or N-terminal α-helix while retaining hemolytic activity. To evaluate which of the two α-helices was most active, analogues were prepared that eliminated either 2, 4, 6, 8 or 10 amino acids from the N-terminal α-helix of M8. The hemolytic potency of M9 and M10 established that most of the activity could be retained after deletion of 4 residues but that further deletion of 6, 8 or 10 N-terminal melittin residues (M 11, M12, M13) abolished hemolytic activity (Table 1).
A melittin analogue (M14) possessing a full length N-terminal α-helix sequence with a deletion of 6 amino acids from the C-terminal α-helix retained modest hemolytic activity (Table 1). However, deletion of 7, 8, 9 or 10 amino acids from the C-terminal α-helix (M15, M16, M17, M18) resulted in complete loss of hemolytic activity (Table 1). In addition, the hemolytic activity of melittin analogues was not influenced by the addition of 1 mM DTT, suggesting that the monomeric peptides were the primary form active in hemolysis.
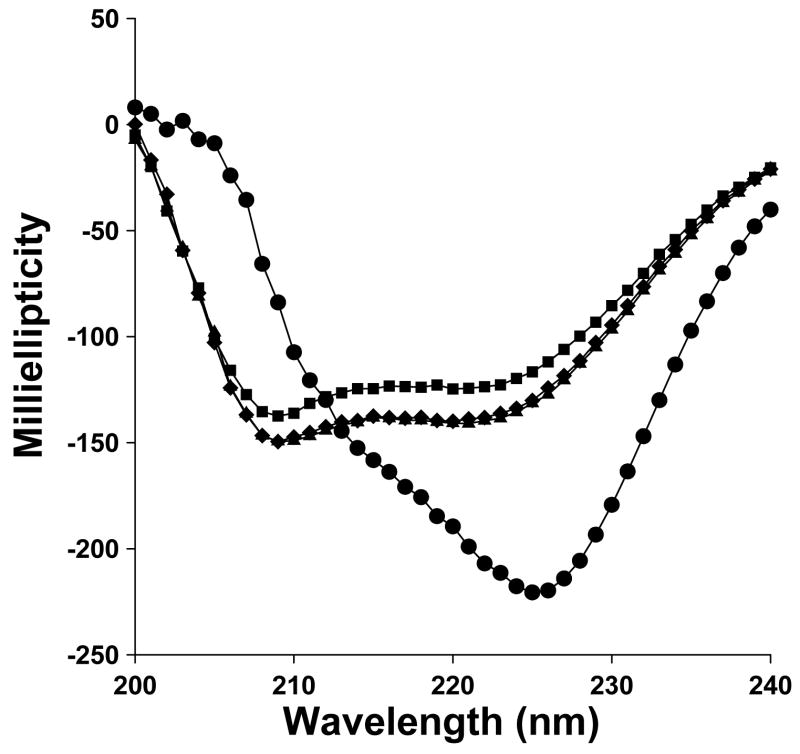
The conformation of melittin peptides were examined by CD spectroscopy acquired in 10 mM sodium phosphate pH 7.4 containing 40%TFE in an attempt to mimic a membrane environment (16). Representative hemolytic peptides M0, M6 and M14 each produced coincident CD spectra calculated to be 80% α-helical (Fig. 3). In contrast, M11, which was devoid of hemolytic activity, produced a CD spectrum consistent with a random coil (Fig. 3). These results are representative of the data obtained in each of the peptides in the series.
Figure 3.
Circular Dichroism of Melittin Analogues. The CD spectra of M0 (
 ), M6 (◆), M11 (●) and M14 (■) acquired in 40% trifluoroethanol (v/v) in 10 mM pH 7.4 phosphate buffer at 25 °C are compared. The results establish that M11 was unable to form an α-helix, thereby accounting for its lack of hemolytic activity and gene transfer efficiency.
), M6 (◆), M11 (●) and M14 (■) acquired in 40% trifluoroethanol (v/v) in 10 mM pH 7.4 phosphate buffer at 25 °C are compared. The results establish that M11 was unable to form an α-helix, thereby accounting for its lack of hemolytic activity and gene transfer efficiency.
The particle size of each poly-melittin DNA condensate was evaluated using quasi-elastic light scattering at a constant charge ratio of 1.5:1 (N:P). M0–M7 formed large DNA condensates >1 μm in mean diameter (Table 1). Alternatively, M8–M18 condensed DNA to form smaller condensates of mean diameter of approximately 200–300 nm.
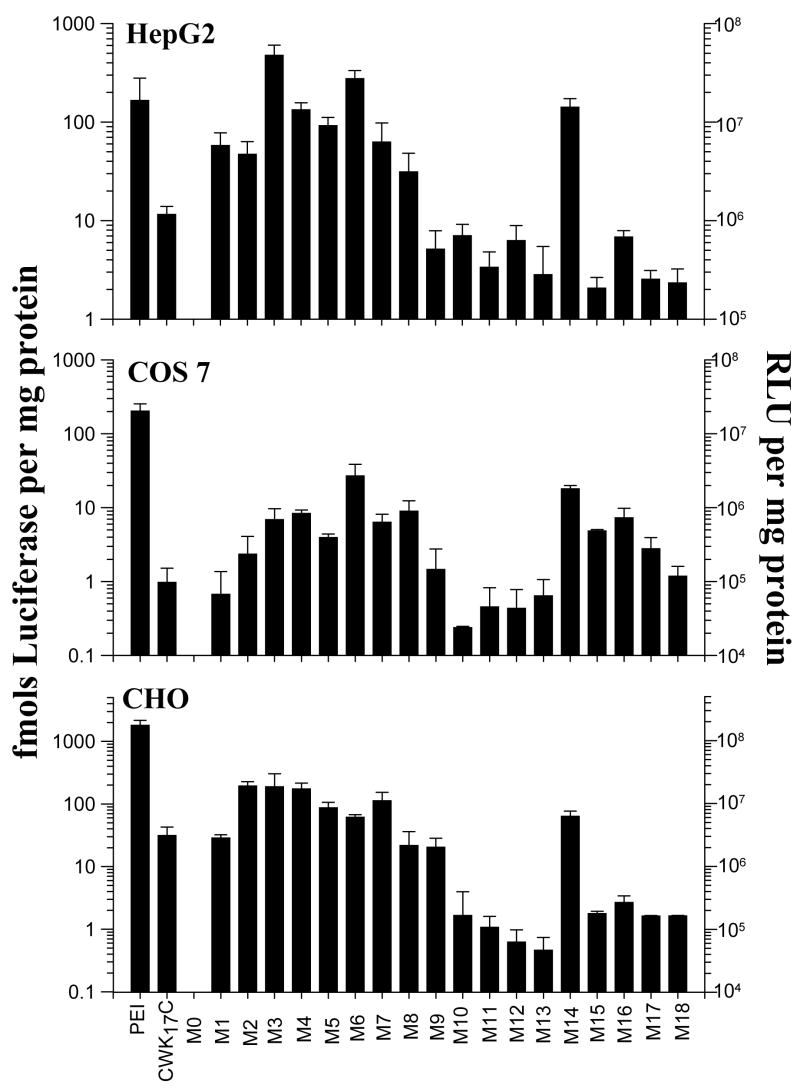
Poly-melittin DNA condensates were evaluated for their in vitro transfection activity in HepG2, COS-7 and CHO cells using PEI and CWK17C (20) to bracket the level of gene expression with (PEI) and without (CWK17C) endosomal escape. In all three cell lines, M0 DNA failed to produce measurable luciferase expression, which was due to its cellular toxicity as demonstrated by MTT assay. In contrast, M1 mediated equivalent gene expression as CWK17C in CHO and COS 7 and was 10-fold more efficient in mediating gene expression in HepG2 (Fig. 4). The cellular toxicity of M0 DNA (<20% cell viability) by MTT assay, in comparison to that determined for M1–M18 DNA (>70% cell viability), supports the hypothesis that DNA binding transiently masks the membrane lytic activity of melittin analogues.
Figure 4. In Vitro Gene Transfer of Melittin Analogue DNA Condensates.
Transfections were performed with peptide DNA charge ratio of 1.5:1. The luciferase reporter gene expression was measured in HepG2 (A), COS-7(B) and CHO (C) cells at 24 hrs. PEI and CWK17C mediated gene delivery were used as positive controls. The results represent the mean and standard deviation for three independent transfections.
Comparison of the relative luciferase gene expression mediated by M1–M8 in all three cell lines established a general trend in which melittin analogues were either equivalent or 10-fold more potent than CWK17C. Within the series, M3 and M6 demonstrated the most potent activity in HepG2 cells which was equivalent to the gene transfer mediated by PEI (Fig. 4).
Comparison of the gene transfer efficiency of M1–M8 with deletion peptides M9–M13 established a loss of activity for M9–M13 in all three cell lines that was coincident with the loss of hemolytic potency (Fig. 4). The partial recovery of gene transfer activity for M14 was coincident with its ability to produce hemolysis (Fig. 4 and Table 1). Likewise, the decreased gene transfer activity for deletion peptides M15–M18 was coincident with the lack of hemolytic activity for these truncated melittin peptides (Fig. 4 and Table 1).
Discussion
One of the rate limiting steps in gene delivery is the efficient escape of DNA from targeting lysosomes. To facilitate this process, fusogenic peptides have been incorporated into gene delivery carriers in an attempt to mimic the process used by viruses. A recent example emphasizes an important directionality in linking melittin into a gene delivery system. In this study it was found that linking melittin to PEI through its C-terminus improves its activity relative to linking through the N-terminus (19).
In the present study we attempted to evolve melittin into a sulfhydryl polymerized peptide, considering that its potent membrane lytic properties could potentially be transiently while bound to DNA. We further considered that a sulfhydryl polymerized melittin analogue could potentially be adapted for in vivo use by incorporating it into a non-viral gene delivery system through co-polymerization with Cys containing peptides (26). One potential difficulty in using melittin for this purpose would be its cytotoxicity, since unlike HA2, it is highly hemolytic at pH 7.
In order for a peptide to be active in non-viral gene delivery, it must bind and condense DNA, usually through cationic interactions. Although melittin has six cationic residues, we have previously shown that polycations of a minimum 13–18 are usually necessary to fully condense DNA and mediate gene transfer in vitro (27). Fewer cationic groups result in DNA condensates that are large (>1 μm) and dissociate at physiological salt concentration (0.15 M) (23). The results of the gel electrophoresis assay (Fig. 3) demonstrate this effect for M0 binding to DNA to form large condensates that dissociate between 0–0.2 M sodium chloride. Dissociation at this salt concentration would release M0 from ionic binding to DNA and explains why M0 DNA results in red blood cell lysis (Fig. 1). These results also explain the lack of gene transfer activity and the cytotoxicity observed by M0 DNA in three cell lines (Fig. 4).
It was possible to decrease the cellular toxicity of M0 by the addition of C and N terminal Cys residues resulting in M1. This modification leads to the improved salt stability of M1 DNA condensates (Table 1) indicating that the peptide is able to polymerize on DNA. Polymeric M1 displays weaker hemolysis activity when bound to DNA and an increase in gene transfer potency. The further addition of 1-4 Lys residues on the N-terminal side of melittin (M2–M5) or 2-3 Lys residues on both the C and N-terminal (M6 and M7) did not increase polymerization as measured by gel electrophoresis (Table 1). DNA condensates prepared with M1–M7 were large (>1 μm) and possessed a relatively low zeta potential (+ 5 mV). The large particle size was most likely the result of particle coalescences initiated through interaction of hydrophobic domains on melittin peptides. Nonetheless, M1–M7 possessed significant hemolytic activity and M3 proved to be one of the most potent gene transfer peptides in the series. The addition of 4 Lys residues to the C and N-subterminal to form M8 led to a more complete polymerization on DNA as revealed by dissociation above 0.8 M sodium chloride (Table 1). Despite a decrease in particle size for M8 DNA to approximately 0.3 μm (Table 1), the gene transfer efficiency was not increased (Fig. 4).
Attempts to truncate a portion of either the N or C terminal α-helix revealed a relationship between hemolytic activity and gene transfer activity. Only M14, in which KRKR QQ was deleted and replaced with a tetra Lys and terminal Cys, was able to mediate gene transfer activity comparable to full length melittin analogues. This was likely because the QQ was unnecessary and the KRKR as substituted for by KKKK.
The results of this study demonstrate the conversion of a hemolytic peptide into a DNA condensing peptide. We hypothesize that melittin analogues function by depolymerizing in the reducing environment inside the cell resulting in the release of DNA. The neutralization of hemolytic activity when melittin analogues bind and polymerize on DNA appears to protect cells from cytotoxicity and facilitate gene transfer. This strategy should be useful to evolve many different fusogenic peptides into sulfhydryl polymerized peptides to positively influence non-viral gene delivery.
Acknowledgments
The authors gratefully acknowledge support for this work from NIDDK 063196.
References
- 1.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The Influence of Endosome-disruptive Peptides on Gene Transfer Using Synthetic. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 2.Van Dyke RW. Acidification of lysosomes and endosomes. Sub-Cellular Biochemistry. 1996;27:331–60. doi: 10.1007/978-1-4615-5833-0_10. [DOI] [PubMed] [Google Scholar]
- 3.Ogris M, Carlisle RC, Bettinger T, Seymour LW. Melittin Enables Efficient Vesicular Escape and Enhanced Nuclear Access of Nonviral Gene Delivery Vectors. J Biol Chem. 2001;276:47550–47555. doi: 10.1074/jbc.M108331200. [DOI] [PubMed] [Google Scholar]
- 4.Wagner E, Plank C, Zatloukal K, Cotten M, Birnstiel ML. Influenza virus hemagglutinin HA-2 N-terminal fusogenic peptides augment gene transfer by transferrin-polylysine-DNA complexes: Toward a synthetic virus-like gene-transfer vehicle. Proc Natl Acad Sci USA. 1992;89:7934–7938. doi: 10.1073/pnas.89.17.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legendre JY, Bohrmann TB, Deuschle U, Kitas E, Supersaxo A. Dioleoymelittin as a Novel Serum-Insensitive Reagent for Efficient Transfections of Mammalian Cells. Bioconjug Chem. 1997;8:57–63. doi: 10.1021/bc960076d. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Nicol F, Szoka FC., Jr GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 7.Balicki D, Putnam CD, Scaria PV, Beutler E. Structure and function correlation in histone H2A peptide-mediated gene transfer. Proc Natl Acad Sci USA. 2002;99:7467–7471. doi: 10.1073/pnas.102168299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconj Chem. 1999;10:406–11. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 9.Wolfert MA, Seymour LW. Chloroquine and amphipathic peptide helices show synergistic transfection in vitro. Gene Therapy. 1998;5:409–14. doi: 10.1038/sj.gt.3300606. [DOI] [PubMed] [Google Scholar]
- 10.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Contr Rel. 1999;60:149–60. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 11.Goto Y, Hagihara Y. Mechanism of the conformational transition of melittin. Biochemistry. 1992;31:732–8. doi: 10.1021/bi00118a014. [DOI] [PubMed] [Google Scholar]
- 12.Subbalakshmi C, Nagaraj R, Sitaram N. Biological activities of C-terminal 15-residue synthetic fragment of melittin: design of an analog with improved antibacterial activity. FEBS Letters. 1999;448:62–6. doi: 10.1016/s0014-5793(99)00328-2. [DOI] [PubMed] [Google Scholar]
- 13.Terwilliger TC, Eisenberg D. The structure of melittin. II Interpretation of the structure. J Biol Chem. 1982;257:6016–22. [PubMed] [Google Scholar]
- 14.Blondelle SE, Houghten RA. Hemolytic and Antimicrobial Activities of the Twenty-Four Individual Omission Analogues of Melittin. Biochemistry. 1991;30:4671–4678. doi: 10.1021/bi00233a006. [DOI] [PubMed] [Google Scholar]
- 15.Juvvadi P, Vunnam S, Merrifield RB. Synthetic Melittin, Its Enantio, Retro, and Retroenantio Isomers, and Selected Chimeric Analogs: Their Antibacterial, Hemolytic, and Lipid Bilayer Action. JACS. 1996;118:8989–8997. [Google Scholar]
- 16.Yan H, Li S, Sun X, Mi H, He B. Individual substitution analogs of Mel(12–26), melittin’s C-terminal 15-residue peptide: their antimicrobial and hemolytic actions. FEBS Letters. 2003;554:100–4. doi: 10.1016/s0014-5793(03)01113-x. [DOI] [PubMed] [Google Scholar]
- 17.Takei J, Remenyi A, Clarke AR, Dempsey CE. Self-association of disulfide-dimerized melittin analogues. Biochemistry. 1998;37:5699–708. doi: 10.1021/bi9729007. [DOI] [PubMed] [Google Scholar]
- 18.Rozema DB, Ekena K, Lewis DL, Loomis AG, Wolff JA. Endosomolysis by masking of a membrane-active agent (EMMA) for cytoplasmic release of macromolecules. Bioconj Chem. 2003;14:51–7. doi: 10.1021/bc0255945. [DOI] [PubMed] [Google Scholar]
- 19.Boeckel S, Wagner E, Ogris M. C-Versus N-terminally Linked Melittin-Polyethylenimine Conjugates: The Site of Linkage Strongly Influences Activity of DNA Polyplexes. J Gene Medicine. 2005;7:1335–1347. doi: 10.1002/jgm.783. [DOI] [PubMed] [Google Scholar]
- 20.McKenzie DL, Kwok KY, Rice KG. A potent new class of reductively activated peptide gene delivery agents. J Biol Chem. 2000;275:9970–9977. doi: 10.1074/jbc.275.14.9970. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie D, Smiley B, Kwok KY, Rice KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconj Chem. 2000;11:901–911. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 22.Oupicky D, Carlisle RC, Seymour LW. Triggered intracellular activation of disulfide crosslinked polyelectrolyte gene delivery complexes with extended systemic circulation in vivo. Gene Ther. 2001;8:713–24. doi: 10.1038/sj.gt.3301446. [DOI] [PubMed] [Google Scholar]
- 23.Adami RC, Rice KG. Metabolic stability of glutaraldehyde cross-linked peptide DNA condensates. J Pharm Sci. 1999;88:739–746. doi: 10.1021/js990042p. [DOI] [PubMed] [Google Scholar]
- 24.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujumoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 25.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 26.Kwok KY, Park Y, Yongsheng Y, McKenzie DL, Rice KG. In Vivo Gene Transfer using Sulfhydryl Crosslinked PEG-peptide/Glycopeptide DNA Co-Condensates. J Pharm Sci. 2003;92:1174–1185. doi: 10.1002/jps.10384. [DOI] [PubMed] [Google Scholar]
- 27.Wadhwa MS, Collard WT, Adami RC, McKenzie DL, Rice KG. Peptide-mediated gene delivery: influence of peptide structure on gene expression. Bioconj Chem. 1997;8:81–8. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]