Abstract
We developed a modified ChIP-chip method, designated ChAP-chip (Chromatin Affinity Precipitation coupled with tiling chip). The binding sites of Bacillus subtilis Spo0J determined using this technique were consistent with previous findings. A DNA replication initiator protein, DnaA, formed stable complexes at eight intergenic regions on the B. subtilis genome. Characterization of the binding sequences suggested that two factors—the local density of DnaA boxes and their affinities for DnaA—are critical for stable binding. We further showed that in addition to autoregulation, DnaA directly modulate the expression of sda in a positive, and ywlC and yydA in a negative manner. Examination of possible stable DnaA-binding sequences in other Bacillus species suggested that DnaA-dependent regulation of those genes is maintained in most bacteria examined, supporting their biological significance. In addition, a possible stable DnaA-binding site downstream of gcp is also suggested to be conserved. Furthermore, potential DnaA-binding sequences specific for each bacterium have been identified, generally in close proximity to oriC. These findings suggest that DnaA plays several additional roles, such as control of the level of effective initiator, ATP-DnaA, and/or stabilization of the domain structure of the genome around oriC for the proper initiation of chromosome replication.
Key words: ChIP-chip analysis, Bacillus subtilis, DnaA protein, DnaA box, high-density tiling chip
1. Introduction
The replication initiation protein of eubacteria, DnaA, binds to an asymmetrical 9 bp consensus sequence (TTATNCACA, DnaA box) that is repeated several times in the replication origin (oriC) region, and plays a central role in the formation of the initiation complex for chromosome replication (reviewed by Leonard and Grimwade,1 Kaguni,2 Mott and Berger,3 and Zakrzewska-Czerwinska et al.4). Additionally, the protein autogenously represses its own expression to maintain the cellular level through binding to DnaA boxes downstream of the dnaA promoter in Escherichia coli,5 Bacillus subtilis,6 Streptomyces lividance,7 Mycobacterium smegmatis, and M. bovis.8 The timing of chromosome replication initiation is tightly regulated, and involves the cellular DnaA level in E. coli (reviewed by Kaguni2), B. subtilis6 and M. smegmatis.9 However, the DnaA box sequence is not restricted to the oriC and dnaA promoter regions. A search for the consensus sequence, TTATNCACA, allowing a one base mismatch along the complete genome sequence of E. coli and B. subtilis, reveals the existence of 3742 and 4342 sites, respectively.
Indeed, several genes are possibly or conclusively regulated directly by DnaA in E. coli (reviewed by Messer and Weigel).10 DnaA negatively regulates mioC, rpoH, uvrB, and proS,5,11,12 and positively regulates polA expression.13 Levels of mioC and polA are controlled in a growth phase-dependent manner, and DnaA possibly contributes to the coupling of expression to growth rate.13–15 A recent study proposes that repression of nrdAB expression by ATP-bound DnaA is associated with coupling of the initiation frequency to elongation rate of chromosome replication,16 although DnaA has been demonstrated previously to regulate positively nrdAB promoter.17 DnaA might have a dual role in modulating nrdAB expression; low levels of DnaA-ATP stimulate the transcription whereas high levels repress it.18 In addition, a high-affinity DnaA-binding sequence containing five DnaA boxes, dat (DnaA Titration), has been identified.19 Multicopy plasmids harboring the dat sequence interfere with the normal replication cycle, although its exact role is unclear.20 A similar high-affinity DnaA-binding region is involved in regulating the initiation of chromosome replication in S. coelicolor.21 In B. subtilis, DnaA functions in repressing the initiation of sporulation in actively replicating cells by controlling the expression of Sda that blocks the signal transduction cascade leading to sporulation.22,23 The sequence upstream of the sda promoter includes five DnaA boxes, and DnaA binding to these is essential for sda expression. Transcriptome analysis of replication-inhibited cells and chromatin immunoprecipitation assays reveal that yllB (ftsL), dnaB, ywlC, and yydA are negatively regulated by DnaA binding to their promoter regions,23 suggesting that the protein is involved in tolerance against replication stress as a transcriptional regulator. Furthermore, DnaA acts as a transcriptional regulator in Caulobacter crescentus.24,25 In this case, DnaA stimulates the expression of a master regulator of cell cycle progression, gcrA, which in turn activates multiple genes, including those involved in chromosome replication and segregation, coordinating the initiation of chromosome replication and cell cycle progression.
Thus, in addition to the initiation of replication and autoregulation of its expression, DnaA appears to participate in the coordination between initiation of chromosome replication and cell cycle progression. However, genome-wide identification of DnaA-binding sites in vivo has not yet been attempted. ChIP-chip analysis, a powerful tool for monitoring the distribution of genome-associated proteins in vivo, employs chromatin immunoprecipitation in combination with a high-resolution oligonucleotide tiling chip.26 For instance, we and other groups recently reported the distribution of nucleoid-associated protein, H-NS, on the E. coli and Salmonella genomes, and found that H-NS preferentially binds to and represses horizontally acquired genes.27–29 However, ChIP-chip analysis requires a specific antibody that efficiently recognizes the target protein in protein–DNA complexes, which may limit its applicability. Previously, we developed a method to isolate protein complexes with high purity, using a 12 × Histidine-tag under denatured conditions.30 To improve versatility, this procedure was adapted for the purification of protein–DNA complexes, denoted ChAP-chip (Chromatin Affinity Precipitation coupled with high density tiling chip). The binding sites of Spo0J determined with the modified technique were consistent with previous reports.31 Clearly, DnaA forms stable complexes at eight intergenic regions on the B. subtilis genome, including oriC, oriC1, and oriC2. Characterization of the DnaA-binding sequences revealed that two factors—local density of the DnaA boxes and their affinities to DnaA—are critical for stable binding. Thus, filament formation of multiple ATP-bound DnaA proposed for the bacterial oriC–DnaA complex32 might occur at the stable-binding sites. In combination with transcriptome analysis of cells containing increased or decreased DnaA amounts, our data show that DnaA proteins stably associated with target DNA directly regulate the expression of sda in a positive manner, and ywlC and yydA in a negative manner. Examination of possible stable DnaA-binding sequences in genome sequences of other Bacillus species suggested that DnaA-dependent regulation of those genes is maintained in most of bacteria examined, supporting their biological significance. In addition, a possible stable DnaA-binding site downstream of gcp is also suggested to be conserved. Furthermore, potential DnaA-binding sequences specific for each bacterium have been identified, generally in close proximity to the oriC region. These findings suggest that DnaA plays several additional roles, such as control of the level of effective initiator, ATP-DnaA, and/or stabilization of the domain structure of the genome around oriC for the proper initiation of chromosome replication.
2. Materials and methods
2.1. Design of the B. subtilis tiling chip
We retrieved the B. subtilis genome sequence and other genetic information from the SubtiList database (http://genolist.pasteur.fr/SubtiList/) for the design of probes, according to the Affymetrix guidelines. Probe sequences were selected for protein coding and intergenic regions using different criteria.27 Apart from the rrnA operon, rRNA genes were excluded from probe design. In addition, skin element, two prophage (PNSX and spβ), and seven prophage-like sequences33 were excluded. The tRNA genes and those encoding proteins with lengths of <100 amino acids were taken as intergenic regions. As a result, 55 430 25-mer sequences on the coding strand were selected for protein coding regions at 25–30 bp intervals, and 72 218 sequences on both strands were selected for intergenic regions at 2–3 bp intervals. In total, 127 648 probe sets (perfect match and mismatch probes) were synthesized on the chip. Chip probe information is available on our website (http://genome.naist.jp/bacteria/array/ecol.html).
2.2. Construction of strains expressing C-terminally histidine-tagged proteins
Primers used for generating plasmids used in this study are presented in Supplementary Table S1.
To construct the pMUTinHis-▵spo0J plasmid, a 250 bp fragment encompassing the 3′-region of the spo0J gene (except the stop codon) was amplified by PCR from B. subtilis 168 genomic DNA using the primer set, spo0J.f–spo0J.r, and cloned between EcoRI–XhoI sites of pMUTinHis.30 The resultant plasmid was integrated into the B. subtilis chromosome by single crossover to generate B. subtilis strains expressing C-terminal histidine-tagged Spo0J (strain SI002).
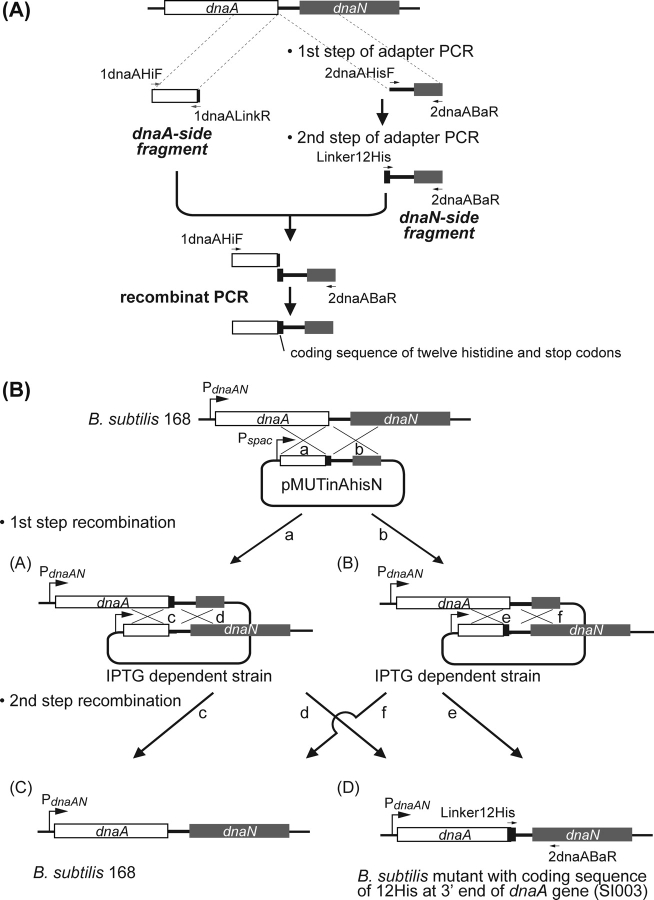
The dnaA gene constitutes an operon with the downstream dnaN gene and the intergenic region (oriC2) acts as the B. subtilis oriC, together with a sequence upstream of dnaA (oriC1).34 We generated a strain expressing C-terminal histidine-tagged DnaA without plasmid integration between dnaA and dnaN (strain SI003), as schematically shown in Fig. 1. A DNA fragment containing the 3′-region of dnaA, except the stop codon, was amplified using the primers, 1dnaAHiF and 1dnaALinkR, with a partial 12 × Histidine (His-tag) coding sequence (dnaA-side fragment). A fragment containing the His-tag, followed by the stop codon of dnaA and its 500 bp downstream sequence (dnaN-side fragment) was generated by two-step adaptor PCR, using the primer sets, 2dnaAHisF–2dnaABaR, for initial amplification, and Linker12His–2dnaABaR, for the second PCR. The two fragments were ligated by recombinant PCR using the 1dnaAHiF–2dnaABaR primer set, and cloned within HindIII and BamHI sites of pMUTinNC.35 Since the dnaN gene essential for cell growth was placed under control of the IPTG-inducible spac promoter in the resultant transformants, pMUTinAhisN was used for transformation of B. subtilis 168 by single crossover with selection on LB medium containing 0.5 µg/ml erythromycin and 1 mM IPTG (first step in Fig. 1B). To induce a second recombination event for removal of the integrated plasmid, the resultant strain was cultured in competent medium containing 1 mM IPTG. Cells that grew in the absence of IPTG were selected on LB medium without erythromycin and IPTG (second step in Fig. 1B). Cells with insertion of the His-tag sequence were selected from IPTG-independent and erythromycin-sensitive colonies by examining amplification using primers complementary to the His-tag sequence (Linker12His) and a dnaN coding sequence (2dnaAbaR), and chromosomal DNA extracted from these colonies. Finally, we confirmed the addition of the 12 × His coding sequence at the 3′ end of dnaA and removal of integrated plasmid by sequencing and the resultant strain expressing the His-tagged DnaA was designated SI003. Negligible changes if any in synchrony of initiation of chromosome replication in SI003 cells were confirmed by flow cytometory (Supplementary Fig. S1).
Figure 1.
Schematic representation of the generation of strain SI003. (A) Adapter PCR and recombinant PCR products used to create the plasmid, pMUTinAhisN. (B) Transformation of 168 cells with pMUTinAhisN and removal of the integrated plasmid to generate strain SI003. Integration of the plasmid into the chromosome through recombination at region a or b resulted in cells with structure 1 or 2, respectively. Induction of recombination at region c or d in cells with structure 1 resulted in cells with structure 3 or 4, respectively. On the other hand, recombination at region e or f in cells with structure 2 resulted in cells having structure 4 or 3, respectively. Cells displaying structure 4 were selected for ChAP-chip analysis.
2.3. ChAP-chip analysis
An overnight culture of B. subtilis cells expressing histidine-tagged protein in LB liquid medium at 37°C was inoculated into 400 mL LB medium to obtain an initial OD600 value of 0.01. During SI002 culture at 37°C, 0.5 µg/mL erythromycin and 1 mM IPTG were added at every step. At OD600 of 0.4, the culture was treated with formaldehyde (1% final concentration) for 30 min. Cells were washed with TBS buffer (pH 7.5), and stored at −80°C until use. Next, cells were disrupted by sonication on ice in 3 mL of UT buffer (100 mM HEPES, 50 mM imidazole, 8 M urea, 0.5 M NaCl, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, pH 7.4). After centrifugation at 8000 rpm for 10 min, 200 µL of MagneHis (Promega) was added to the supernatant, followed by overnight incubation at room temperature with gentle shaking. MagneHis was washed five times with UT buffer, and bound proteins eluted with 400 µL of elution buffer (100 mM Tris–HCl, pH 7.5, 0.5 M imidazole, 1% SDS, 10 mM DTT). The eluate was passed through Microcon-100 (Millipore) to remove non-specifically bound uncrossed-linked proteins with a molecular mass lower than 100 kDa. Protein complexes retained on the membrane were washed three times with wash buffer (100 mM Tris–HCl, pH 7.5, 1% SDS, 10 mM DTT), and recovered by the addition of 50 µL buffer. Cross-links were dissociated by heating at 65°C overnight, and DNA purified using Qiaquick (QIAGEN).
Terminal labeling of purified DNA fragments and hybridization to the oligonucleotide chip were performed essentially as described previously.27 Signal intensities of mismatch probes were subtracted from those of perfect match probes. The signal intensities of DNA in the affinity-purified fraction and those of DNA isolated from the whole cell extract fraction before purification (control DNA) were adjusted to confer a signal average of 500. Signal intensities of DNA in the affinity-purified fraction were subtracted by those of control DNA to obtain protein-binding signals. The distribution of protein-binding signals along the genome coordinate was visualized with the In Silico Molecular Cloning program, Array Edition (In Silico Biology).
2.4. High-resolution transcriptome analysis
NIS2022 cells harboring the IPTG-inducible dnaA–dnaN operon at the purA locus6 were pre-cultured overnight at 30°C on a PAB plate containing 5 µg/ml tetracycline and 10 µg/ml neomycin, with or without 10 µM IPTG. Cells were inoculated into PAB liquid medium with or without 0.1 M IPTG to obtain an initial OD600 value of 0.01, followed by culture at 30°C. At OD600 of 0.4, cells were harvested from 40 mL of culture, and total RNA extracted, as described previously.36 Expression of DnaA protein in wild-type CRK6000 cells and NIS2022 cells was confirmed by western blotting using a specific antibody, based on an earlier protocol.6 Synthesis of cDNA, terminal labeling, and hybridization to the oligonucleotide chip were performed using established methods.27 Amplification of cDNA was avoided to allow the coding strands of each transcript to be distinguished. To compensate for the differences in hybridization efficiency of each 25-mer probe on the chip, we divided the hybridization intensities of cDNA synthesized from total RNA by those of total genome DNA. The distribution of transcriptional signals along the genome coordinate was visualized with the In Silico Molecular Cloning program, Array Edition (In Silico Biology).
3. Results
3.1. Accurate mapping of known Spo0J-binding sites by ChAP-chip analysis
Previously, we developed a method to purify in vivo complexes of cell division proteins using a 12 × histidine- tag under denatured conditions.30 We anticipated that the procedure is similarly effective for the purification of in vivo DNA–protein complexes. To verify the sensitivity of the tag-based chromatin precipitation protocol, we initially examined the binding sites of Spo0J involved in chromosome partitioning in B. subtilis. Spo0J binds at eight specific positions (parS sites) on the genome, as observed from the identified binding consensus sequences and chromatin immunoprecipitation analysis.31 We expressed the Spo0J protein fused to a histidine tag at the C-terminus using SI002 cells. During exponential growth in LB medium under aerobic conditions, Spo0J-12 × His-expressing cells were treated with formaldehyde to cross-link protein and genome DNA, and disrupted by sonication in 8 M urea buffer to generate an average DNA fragment size of ∼500 bp. Protein–DNA complexes were purified with the Ni2+-resin under denatured conditions, and the cross-links removed by heat treatment. DNA fragments co-purified with Spo0J were hybridized with a high-density tiling chip. The hybridization intensities of Spo0J-associated DNA to each oligonucleotide probe on the chip are presented in Fig. 2 (see also detailed map in Supplementary Fig. S2). Our ChAP-chip analysis clearly shows that Spo0J forms a stable complex at eight regions, consistent with previous results.31 Spo0J-binding signals are more broadly distributed than those of DnaA, as described below (Fig. 1B and Supplementary Fig. S2). This result is consistent with the recent finding that Spo0J is associated with several kilobases of DNA flanking its specific binding sites (parS) through a parS-dependent nucleation event that promotes lateral spreading along the chromosome.37 Thus, it appears that our ChAP-chip method is sufficiently sensitive to detect the in vivo interaction sites of DNA-binding proteins.
Figure 2.
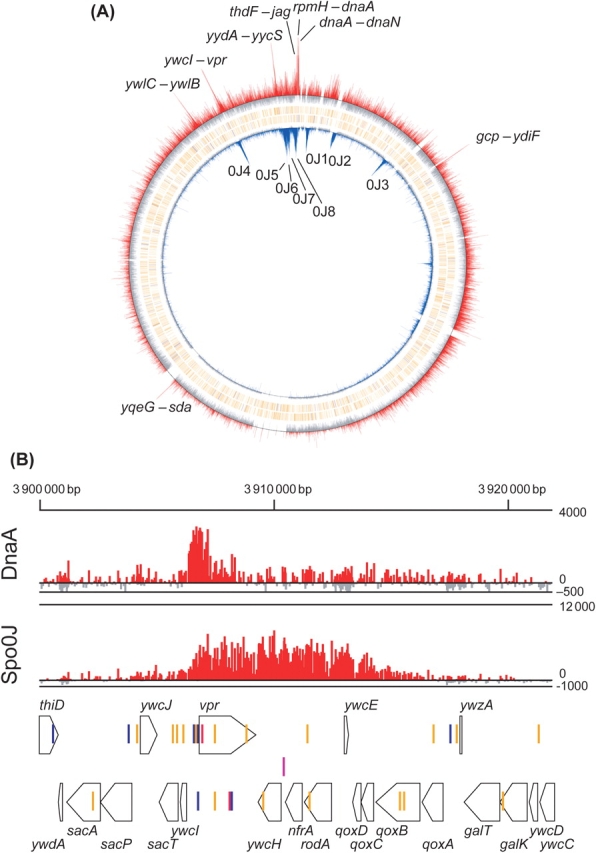
DNA-binding profiles of Spo0J and DnaA proteins on the B. subtilis genome revealed using ChAP-chip analysis. (A) Spo0J (inner blue bars) and DnaA-binding (outer red bars) signals for each 25-mer probe on the chip were calculated by subtracting signal intensities of control DNA from those of DNA in the affinity-purified fraction, and shown at their corresponding genome coordinates. 0J1–0J8 correspond to the sites previously determined by Lin and Grossman.31 DnaA boxes are specified as colored vertical lines under DnaA-binding signals: TTATCCACA: red, TTATACACA, TTATAGACA, and TTATATACA: blue, other DnaA boxes having one base mismatch: orange. (B) Detailed DnaA and Spo0J binding profiles around the ywcI–vpr region, where binding of both proteins was observed, are shown for comparison of binding profiles of two proteins. The arrangement of genes (thick arrows), DnaA-boxes (as in A), and Spo0J binding site (parS site, purple vertical line) are shown schematically at the bottom.
3.2. Identification of high-affinity binding sites of DnaA
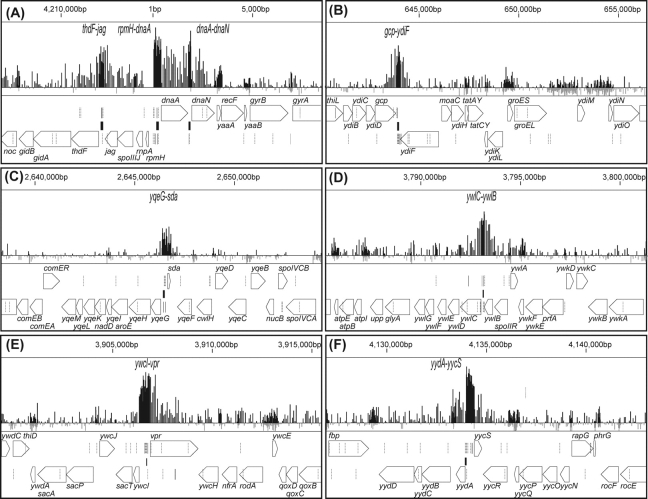
To identify the DnaA-binding sites on the B. subtilis genome, DnaA was expressed as a fusion protein with a 12 × histidine-tag by the authentic promoter using SI003 cells, and ChAP-chip analysis performed. The hybridization profiles of DnaA-associated DNA clearly disclose eight stable binding sites for DnaA on the B. subtilis genome (Fig. 2 and Supplementary Fig. S2). Interestingly, binding sites are mapped only in the intergenic regions (between rpmH–dnaA, dnaA–dnaN, yqeG–sda, ywlB–ywlC, jag–thdF, yycS–yydA, ywcI–vpr, and gcp–ydiF). Gene organization surrounding the eight binding sites, location of the DnaA boxes, and detailed DnaA-binding signals are depicted in Fig. 3, and the functions of the surrounding genes are summarized in Table 1. The regions upstream and downstream of dnaA, rpmH–dnaA (oriC1), and dnaA–dnaN (oriC2) comprise cis-acting sequences essential for the initiation of chromosome replication,34 and the upstream sequence contains the dnaA promoter that is repressed by the corresponding translated protein.6 Expression of sda is dependent on DnaA binding to the upstream sequence,22,23 and promoters of ywlC and yydA were reported to be negatively regulated by DnaA.23 Multiple DnaA boxes upstream of thdF were identified 17 years ago,38 but with no evidence of actual DnaA binding. Our results additionally suggest a possibility that DnaA regulates the expression of vpr and/or ywcI–secT. Binding to the gcp–ydiF region is uncharacteristic for transcriptional regulation, as it is located at the 3′ end of convergent gcp and ydiF genes.
Figure 3.
Local DNA-binding profiles of DnaA. Local DNA binding profiles of DnaA are presented, together with genes and DnaA boxes. DnaA boxes are specified as vertical lines under DnaA-binding signals: TTATCCACA: bold lines, TTATACACA, TTATAGACA and TTATATACA: gray lines, other DnaA boxes having one base mismatch: dotted lines. The top, middle, and bottom lines of ChAP-chip signals indicate signal intensities of 4000, 0, and −500, respectively. Closed boxes represent regions having high-affinity successive DnaA box clusters, as described in Table 2.
Table 1.
Genes in transcriptional units adjacent to the DnaA-binding sites
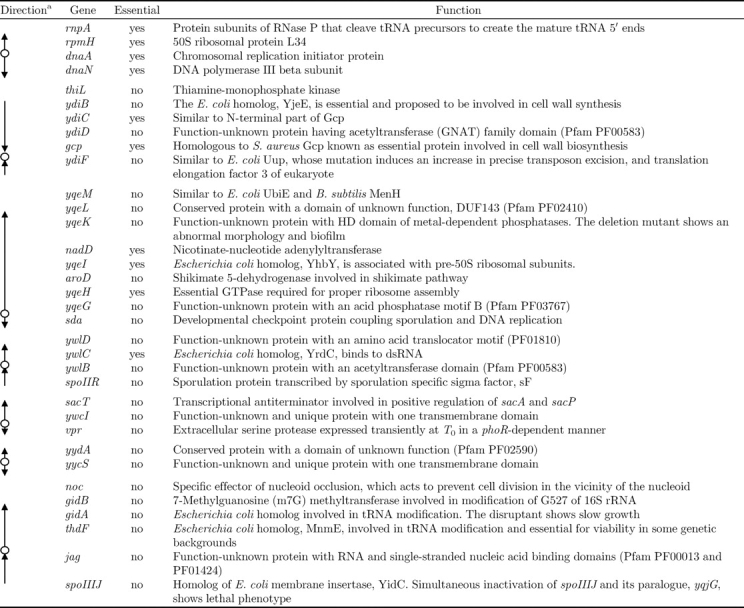
aThe location of DnaA-binding site is indicated by circle and direction of the adjacent transcriptional units is indicated by arrow.
DnaA protein binds to single DnaA box in vitro.39,40 However, the DnaA-binding regions identified here contain multiple DnaA boxes. These results suggest that the ChAP-chip analysis can be effectively used to detect stable DnaA–DNA complexes, but not transient or weak binding to dispersed DnaA boxes.
3.3. Effect of DnaA overproduction and depletion on transcription of genes surrounding the stable binding sites
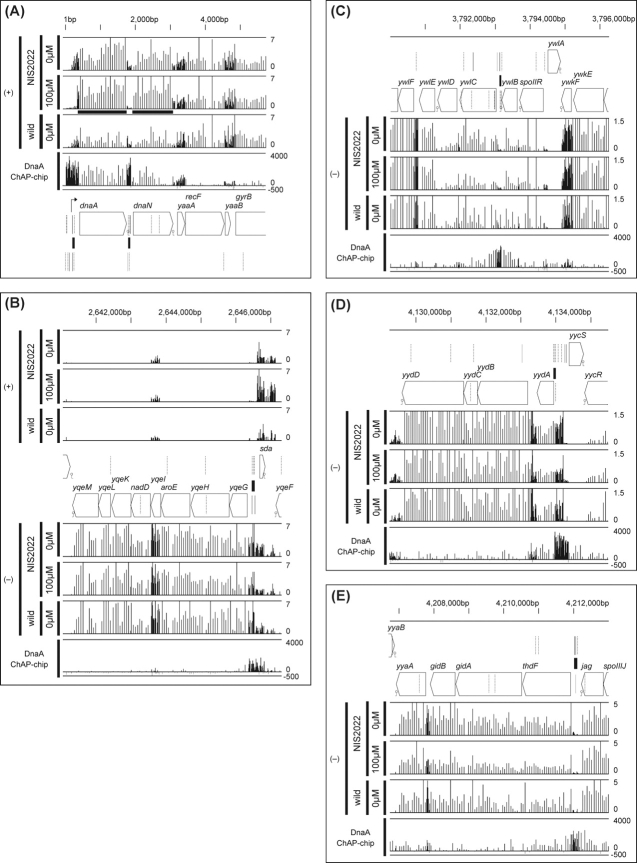
To establish the function of DnaA binding to sequences other than oriC, we examined transcriptional profiles in DnaA-overproducing and -depleted cells using the high-density tiling chip employed for ChAP-chip analysis. To this end, we employed NIS2022 cells with an IPTG-inducible dnaA–dnaN operon at the purA locus, and inactivated the authentic dnaA gene by introducing orcher mutations.6 Expression of the DnaA protein in NIS2022 cells is reduced to One-fifth that in non-induced conditions (0 µM IPTG), and increases fivefold upon induction with 100 µM IPTG, compared with the DnaA concentration in wild-type cells.6 Notably, under both non-induced and induced conditions, no apparent growth defects were observed, and the overall profiles of gene expression remained unchanged (Supplementary Fig. S3). Transcriptional initiation from the authentic promoter of dnaA is clearly repressed upon induction of DnaA expression, and stimulated slightly under non-induced conditions (Fig. 4A). This finding is in agreement with a previous study reporting autoregulation of dnaA.6 A similar change in transcription was observed for the intergenic region between dnaA and dnaN. The dependence of sda expression on DnaA is well characterized.22,23 We observed sda expression and repression in DnaA-induced and non-induced conditions, respectively (Fig. 4B). Interestingly, we identified another transcript covering the sda gene, but on the non-coding strand. The newly identified transcript was weakly, but significantly repressed by overexpressed DnaA at the DnaA-binding site. We additionally observed weak, but significant repression of ywlC, yydA, and thdF-gidA-gidB-noc transcription in DnaA-induced cells (Fig. 4C–E), although their de-repression in non-induced cells was not apparent. A sufficient amount of DnaA is present in cells to support growth at a normal rate, even under non-induced conditions, thus those genes may still be repressed by DnaA. The suppression of ywlC and yydA expression by DnaA is in accordance with reported microarray data.23 We have demonstrated DnaA-regulated expression of thdF-gidA-gidB-noc by northern blot analysis using NIS2022 cells (Ogura et al., unpublished result). These transcriptional alterations were reproducible in two independent experiments (Supplementary Fig. S3). Additionally, the DnaA level affected rocA, rocD, and yvdCD expression positively, and ykuNOP and dhbA expression negatively (Supplementary Fig. S3 and Supplementary Table S2). The molecular mechanisms and biological significance of these changes are currently unclear. On the other hand, the effects of DnaA on the expression of genes around ywcI–vpr and gcp–ydiF could not be distinguished due to weak expression.
Figure 4.
Transcriptional changes around the DnaA-binding regions in dnaA-induced and repressed cells. Transcriptional activity of NIS2022 cells in the absence and presence of 100 µM IPTG and wild-type cells were analyzed using tiling chip. Transcriptional signals for each probe were calculated by dividing hybridization intensities of cDNA synthesized from total RNA by those of total genome DNA, and shown by vertical blue bars at their corresponding genome coordinates, for regions surrounding the DnaA-binding sites. Signal intensities of probes on Watson (+) and Crick (−) strands were shown separately to indicate the direction of transcripts. The DnaA-binding signals (DnaA ChAP-chip) and location of genes and DnaA boxes are also shown, as in Fig. 2B. The transcriptional start sites, dnaA and sda, determined previously, are highlighted in the genetic maps with arrows. Possible termination signals of transcription are additionally specified. Signals marked by a bold underline in panel A are due to the expression of dnaA–dnaN placed at the purA locus in NIS2022 cells.
3.4. Sequence characteristics of stable DnaA-binding sites on the chromosome
Next, we explored the characteristics defining the stable DnaA-binding sites in the B. subtilis genome. Bacillus subtilis DnaA recognizes DnaA boxes in E. coli oriC, and vice versa, in vitro. Although the numbers and arrangement of DnaA boxes in oriC are different in B. subtilis and E. coli, the oriC regions were incompatible with each other in vivo.39,40 Recently, the crystal structure of the DNA-binding domain of E. coli DnaA complexed with a DnaA box (TTATCCACA) was resolved.41 In the complex, amino acids R399, P423, D433, H434, T435, and H439 form base-specific interactions, and are essential for sequence recognition by DnaA. Interestingly, these residues are conserved in B. subtilis DnaA, indicating that DNA-binding specificities are similar for B. subtilis and E. coli DnaA. Thus, we assume that the consensus sequence of the B. subtilis DnaA box is TTATNCACA, similar to the E. coli protein, and allow one base mismatch for identifying DnaA boxes. As shown in Fig. 3, stable DnaA-binding sites contain multiple boxes, as observed in oriC sequences of eubacteria.42 Initially, we evaluated the density of DnaA boxes along the genome, using a modified method of0 Mackiewicz et al.42 The density of DnaA box distribution was presented on charts as b = 1/d, calculated for each box, where d is the average distance between adjacent DnaA boxes. Mackiewicz and colleagues calculated the b value for three successive DnaA boxes (average of two distances), whereas we additionally calculated for four and five successive boxes, with no clear correlation with the identified binding sites. Recently, the DnaA-binding affinity to DnaA boxes with different fifth nucleotides was evaluated in E. coli cells and the affinity was found to decrease in the order: C > A = G > T.43 Accordingly, we assigned the binding affinities as 4, 2, 1.5, 1, and 1 for TTATCCACA, TTATACACA, TTATAGACA, TTATATACA, and other DnaA boxes (having one base mismatch from TTATNCACA), and the binding affinity of DnaA for four successive DnaA boxes was expressed as A = (sum of affinities of four successive DnaA boxes)/d. As the result, we found that the A value is well correlated with the DnaA-binding sites; eight binding sites identified here exclusively have A value >0.3 (Fig. 5A). Sequence characteristics of the eight DnaA-binding regions, including number of DnaA boxes and the A value, are summarized in Table 2. Our results suggest that two factors, specifically, closely clustered DnaA boxes of probably four or more, and the affinity of DnaA boxes for the protein, are essential for stable DnaA-binding in B. subtilis cells.
Figure 5.
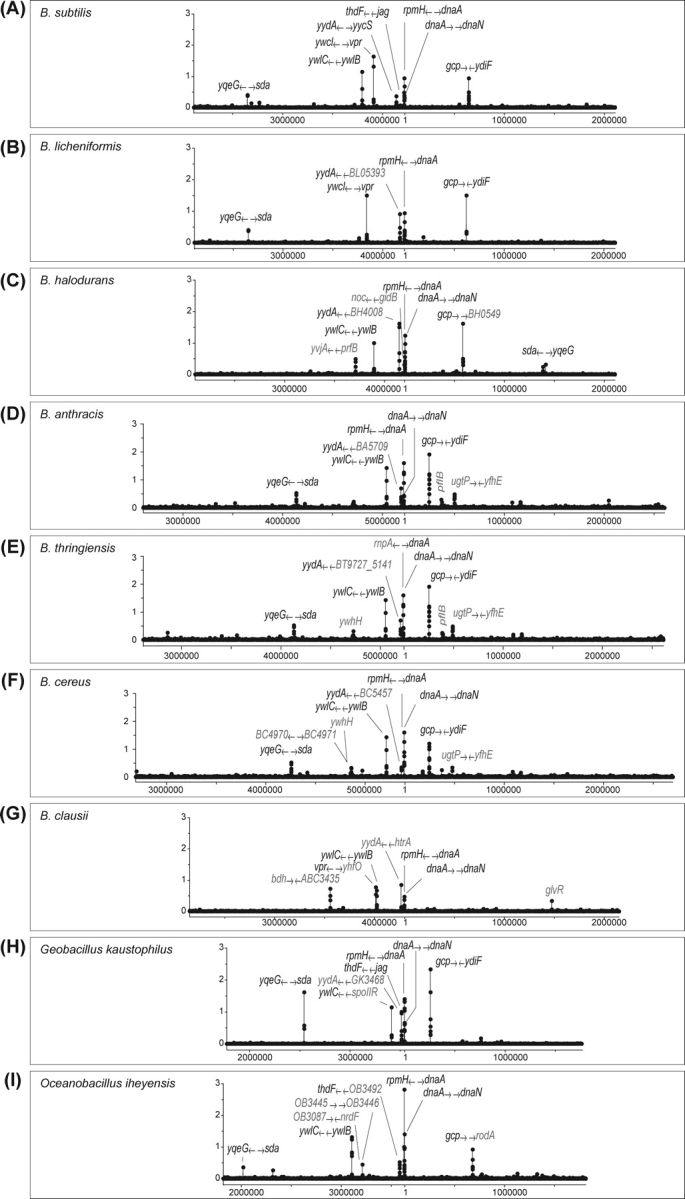
Estimation of the distribution of the possible DnaA-binding regions in genome sequences of the Bacillus species. The A value [(sum of affinities of four successive DnaA boxes)/d (average distance of adjacent DnaA boxes)] was calculated for all DnaA boxes, and plotted (closed circles joined by line) at corresponding genome coordinates in B. subtilis (A), B. licheniformis (B), B. halodurans (C), B. anthracis (D), B. thuringiensis (E), B. cereus (F), B. clausii (G), Geobacillus kaustophilus (H), and Oceanobacillus iheyensis (I). Genome sequences, except that of B. subtilis, were retrieved from the NCBI GeneBank database. The genes adjacent to the possible DnaA-binding regions are specified, and “ → ” indicates their orientations (5′ to 3′ direction) relative to DnaA-binding site located between them. Genes or possible DnaA-binding regions that are not conserved in B. subtilis are colored gray.
Table 2.
Sequence characteristics of the DnaA-binding regions identified by ChAP-chip analysis
| Regiona |
db | Sum of affinity | Ac | Number of DnaA boxesd | Number of perfect matchese | ||
|---|---|---|---|---|---|---|---|
| Surrounding genes | Start | End | |||||
| rpmH–dnaA | 193 | 257 | 10.7 | 10 | 0.94 | 8 | 2 (2) |
| dnaA–dnaN | 1797 | 1860 | 10.3 | 7 | 0.68 | 5 | 1 (1) |
| gcp–ydiF | 643 951 | 643 999 | 5.3 | 5 | 0.94 | 8 | 4 (3) |
| yqeG–sda | 2 646 452 | 2 646 536 | 17.3 | 7 | 0.40 | 5 | 2 (2) |
| ywlC–ywlB | 3 793 128 | 3 793 181 | 7.0 | 8 | 1.14 | 5 | 2 (1) |
| ywcI–vpr | 3 906 700 | 3 906 743 | 3.7 | 6 | 1.64 | 5 | 3 (0) |
| yydA–yycS | 4 133 932 | 4 133 997 | 11.0 | 4 | 0.36 | 4 | 0 (0) |
| thdF–jag | 4 212 013 | 4 212 108 | 21.0 | 10 | 0.48 | 4 | 3 (2) |
aRegions with A value >0.3 are shown. When DnaA box clusters overlap, those with the highest A value are indicated.
bAverage distance between four adjacent DnaA boxes.
cA = (sum of affinities of four successive DnaA boxes)/d. Only the highest value in each DnaA box cluster is shown. See text for detail.
dNumber of DnaA boxes allowing one base mismatch of the consensus DnaA box, TTATNCACA, in successive DnaA box clusters with an A value >0.3.
eThe number of perfectly matched sequences to the consensus DnaA box, TTATNCACA. The number of DnaA boxes with highest affinity (TTATCCACA) is indicated in parentheses.
3.5. Distribution of probable DnaA-binding sites in other Bacillus species
We estimated the stable DnaA-binding sites in the genome sequences of other Bacillus species, using the defined A value. Interestingly, regions with high A values locate in a similar arrangement on the genome in all the bacteria examined (Fig. 5B–I). However, close examination of possible DnaA-binding regions in various Bacillus species revealed their conserved and diverse characteristics (Table 3). As expected, multiple DnaA-binding sequences upstream and downstream of dnaA (oriC) are essentially conserved in all bacteria. Notably, in B. licheniformis, the dnaA–dnaN region contains only three DnaA boxes, and its A value is below the threshold. DnaA-binding sequences upstream of sda, ywlC, and yydA are also present in most bacteria, supporting the biological significance of DnaA-mediated regulation of these genes. Interestingly, a DnaA box cluster downstream of gcp is highly conserved, although its biological role is not clear at present. On the other hand, DnaA box cluster upstream of the thdF-gidA-gidB-noc operon is not conserved, although those genes are conserved in all bacteria examined (Table 3), thus the DnaA-dependent expression of the operon appears not to be general. Similarly, DnaA box clusters upstream of vpr are present in limited bacteria. In addition, other possible DnaA-binding regions (colored red in Fig. 5B–I) that are absent in B. subtilis have been identified. Interestingly, DnaA-binding regions with a high A value are generally located near oriC in all the bacteria examined.
Table 3.
Conservation of DnaA-binding sequences identified in B. subtilis and adjacent genes in genome sequences of Bacillus spices
| Genus | Species | rpmH← | →dnaA→ | →dnaN | yydA← | →yycS | gcp→ | ←ydiF | thdF← | ←jag | ywlC← | ←ywlB | yqeG← | →sda | ywcI← | →vpr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus | B. anthracis | + | + | + | + | + | + | + | (+) | (+) | + | + | + | + | ▵ | |
| B. cereus | + | + | + | + | + | + | ▵ | ▵ | + | + | + | + | ▵ | |||
| B. thuringiensis | + | + | + | + | + | + | ▵ | ▵ | + | + | + | + | ▵ | |||
| B. clausii | + | + | + | + | ▵ | ▵ | ▵ | ▵ | + | + | ▵ | ▵ | + | |||
| B. halodurans | + | + | + | + | + | + | ▵ | ▵ | + | + | + | + | ▵ | |||
| B. licheniformis | + | + | ▵ | + | + | ▵ | (+) | (+) | (+) | (+) | + | + | + | + | ||
| B. subtilis | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Geobacillus | G. kaustophilus | + | + | + | + | + | + | + | + | + | + | + | ||||
| Oceanobacillus | O. iheyensis | + | + | + | ▵ | ▵ | + | (+) | + | + | + | ▵ | ▵ |
Symbols indicate conservation of genes. When DnaA box clusters with A ≥ 0.3 and A < 0.3 exist in the similar arrangement with that in B. subtilis, cells are indicated as “+” and “(+)”, respectively. When DnaA box clusters do not exit, cells are indicated as “(Δ)”. “ → ” indicates orientation of genes (5′ to 3′) relative to DnaA-binding site located between them.
4. Discussion
In this study, we developed a ChAP-chip method, based on our original protocol for the purification of protein complexes.30 The binding sites of Spo0J determined using this technique were consistent with those reported earlier,31,37 clearly indicating that the ChAP-chip methods are sufficiently sensitive for the genome-wide determination of specific binding sites of DNA-binding proteins. Using this method, we have successfully visualized the distribution of RNA polymerase core enzyme and associated factors, SigA, NusA, and GreA (unpublished results). Thus, the ChAP-chip assay should be effective for proteins that are both stably and dynamically associated with genome DNA.
The initiator protein of chromosomal replication, DnaA, forms stable complexes at eight intergenic regions on the B. subtilis genome, including the oriC sequences, oriC1 and oriC2. In vitro assays demonstrate that DnaA protein binds to a single DnaA box.39,40 However, characterization of the DNA sequences bound to DnaA in vivo reveals that the presence of densely clustered DnaA boxes and affinity of these boxes for DnaA protein are critical factors for stable DNA–protein interactions in vivo. The recent crystal structure of Aquifex aeolicus DnaA bound to the non-hydrolyzable ATP analog, AMP-PCP, disclosed that ATP-DnaA forms a right-handed filament defined by specific protein–ATP interactions.3,32 This finding strongly suggests that through filament formation of multiple ATP-DnaA bound to oriC, a right-handed oriC DNA wrap is formed around the initiation nucleoprotein complex. It is plausible that this type of higher-order structure of the DnaA–DNA complex is formed at the stable DnaA-binding sites detected by our group.
Notably, genes related to basic cellular functions are over-represented in neighboring genes. These include chromosome replication (dnaA and dnaN), translation (yqeI, yqeH: ribosome assembly; gidA, thdF: modification of tRNA; gidB: modification of 16S rRNA), and unknown essential functions (gcp, ywlC). In combination with transcriptome analysis of cells containing increased or decreased amounts of DnaA, we demonstrated that in addition to autoregulation, DnaA proteins stably associated with DNA directly regulate sda expression in a positive and ywlC and yydA expression in a negative manner. Furthermore, DnaA-dependent regulation of these genes was suggested to be maintained in other Bacillus species, indicating their biological significance. The DnaA-dependent control of dnaA and sda is well documented. In contrast, the functions of ywlC and yydA and the rationale for their DnaA-dependent regulation await further investigation. We additionally show that although thdF-gidA-gidB-noc expression is under control of DnaA in B. subtilis, this regulation is not universal in the Bacillus species.
DnaA generally acts as a repressor of transcription in B. subtilis, probably through blocking the formation of initiation complex of transcription and/or a transcriptional roadblock. However, the location of DnaA boxes and transcriptional organization of the yqeG–sda region suggests an alternative mechanism of transcriptional regulation by DnaA. Interestingly, sda transcription is positively regulated by DnaA. It is possible that DnaA interacts with RNA polymerase as a transcriptional activator, although experimental evidence of these interactions is yet to be obtained. We identified a novel transcript negatively regulated by DnaA, which traverses the sda gene in the opposite orientation. RNA polymerase collisions would occur if both transcriptions were active simultaneously. If these collisions negatively regulate sda transcription, negative regulation of the opposite reaction would have a positive effect on sda expression. Additionally, upregulation of the sda promoter may be mediated through changes in superhelicity of the DNA template. If DnaA forms helical filaments and bound DNA is wrapped around the DnaA helix, as discussed earlier, positive superhelicity will be stabilized, as demonstrated by Erzberger et al.32 Superhelicity of template DNA is an important factor in determining transcriptional activity.44,45
Estimation of stable DnaA-binding sequences using the A value in genome sequences of Bacillus species suggests that in addition to the upstream regions of dnaA, dnaN, sda, yydA, and ywlC, DnaA binds downstream of gcp in most bacteria examined. In addition, specific DnaA-binding sequences for each bacterium may be present in close proximity to oriC. These observations suggest that DnaA plays several additional roles, such as control of the amount of effective initiator, ATP-DnaA, as proposed for E. coli,46 and/or stabilization of domain structure of genome around oriC for the proper initiation of chromosome replication.
However, although the importance of orientation of adjacent DnaA boxes for stable binding of DnaA has been investigated in E. coli,47 we did not take into account this factor to evaluate the DnaA-binding regions. Further improvements of bioinformatic methods to estimate the DnaA-binding regions are necessary.
In conclusion, we have analyzed the distribution of the DnaA protein on the B. subtilis genome in actively growing cells, and confirmed its role as a direct regulator of several genes. Moreover, we propose that DnaA has additional functions related to chromosome replication. Thus, genome-wide analysis of DnaA-binding sites should facilitate our understanding of the role of B subtilis DnaA as a coordinator of bacterial cell growth.
Supplementry Data
Supplementry data are available online at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas Systems Genomics from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Material
References
- 1.Leonard A. C., Grimwade J. E. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol. Microbiol. 2005;55:978–985. doi: 10.1111/j.1365-2958.2004.04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaguni J. M. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 3.Mott M. L., Berger J. M. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 4.Zakrzewska-Czerwinska J., Jakimowicz D., Zawilak-Pawlik A., Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 2007;31:378–387. doi: 10.1111/j.1574-6976.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- 5.Braun R. E., O'Day K., Wright A. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell. 1985;40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 6.Ogura Y., Imai Y., Ogasawara N., Moriya S. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakimowicz D., Majka J., Lis B., Konopa G., Wegrzyn G., Messer W., Schrempf H., Zakrzewska-Czerwinska J. Structure and regulation of the dnaA promoter region in three Streptomyces species. Mol. Gen. Genet. 2000;262:1093–1102. doi: 10.1007/pl00008652. [DOI] [PubMed] [Google Scholar]
- 8.Salazar L., Guerrero E., Casart Y., Turcios L., Bartoli F. Transcription analysis of the dnaA gene and oriC region of the chromosome of Mycobacterium smegmatis and Mycobacterium bovis BCG, and its regulation by the DnaA protein. Microbiology. 2003;149:773–784. doi: 10.1099/mic.0.25832-0. [DOI] [PubMed] [Google Scholar]
- 9.Greendyke R., Rajagopalan M., Parish T., Madiraju M. V. Conditional expression of Mycobacterium smegmatis dnaA, an essential DNA replication gene. Microbiology. 2002;148:3887–3900. doi: 10.1099/00221287-148-12-3887. [DOI] [PubMed] [Google Scholar]
- 10.Messer W., Weigel C. DnaA initiator—also a transcription factor. Mol. Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- 11.Nozaki N., Okazaki T., Ogawa T. In vitro transcription of the origin region of replication of the Escherichia coli chromosome. J. Biol. Chem. 1988;263:14176–14183. [PubMed] [Google Scholar]
- 12.Wang Q. P., Kaguni J. M. dnaA protein regulates transcriptions of the rpoH gene of Escherichia coli. J. Biol. Chem. 1989;264:7338–7344. [PubMed] [Google Scholar]
- 13.Quinones A., Wandt G., Kleinstauber S., Messer W. DnaA protein stimulates polA gene expression in Escherichia coli. Mol. Microbiol. 1997;23:1193–1202. doi: 10.1046/j.1365-2958.1997.2961658.x. [DOI] [PubMed] [Google Scholar]
- 14.Theisen P. W., Grimwade J. E., Leonard A. C., Bogan J. A., Helmstetter C. E. Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 1993;10:575–584. doi: 10.1111/j.1365-2958.1993.tb00929.x. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa T., Okazaki T. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J. Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gon S., Camara J. E., Klungsoyr H. K., Crooke E., Skarstad K., Beckwith J. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25:1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Augustin L. B., Jacobson B. A., Fuchs J. A. Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd-lac fusion. J. Bacteriol. 1994;176:378–387. doi: 10.1128/jb.176.2.378-387.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrick J., Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 2007;63:22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa T., Yamada Y., Kuroda T., Kishi T., Moriya S. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 2002;44:1367–1375. doi: 10.1046/j.1365-2958.2002.02969.x. [DOI] [PubMed] [Google Scholar]
- 20.Morigen , Molina F., Skarstad K. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. Bacteriol. 2005;187:3913–3920. doi: 10.1128/JB.187.12.3913-3920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smulczyk-Krawczyszyn A., Jakimowicz D., Ruban-Osmialowska B., Zawilak-Pawlik A., Majka J., Chater K., Zakrzewska-Czerwinska J. Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: from in silico to in vivo studies. J. Bacteriol. 2006;188:6184–6194. doi: 10.1128/JB.00528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkholder W. F., Kurtser I., Grossman A. D. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 23.Goranov A. I., Katz L., Breier A. M., Burge C. B., Grossman A. D. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hottes A. K., Shapiro L., McAdams H. H. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 25.Collier J., Murray S. R., Shapiro L. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 2006;25:346–356. doi: 10.1038/sj.emboj.7600927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katou Y., Kaneshiro K., Aburatani H., Shirahige K. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 2006;409:389–410. doi: 10.1016/S0076-6879(05)09023-3. [DOI] [PubMed] [Google Scholar]
- 27.Oshima T., Ishikawa S., Kurokawa K., Aiba H., Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 28.Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., Fang F. C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 29.Lucchini S., Rowley G., Goldberg M. D., Hurd D., Harrison M., Hinton J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathogens. 2006;2:746–752. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa S., Kawai Y., Hiramatsu K., Kuwano M., Ogasawara N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 2006;60:1364–1380. doi: 10.1111/j.1365-2958.2006.05184.x. [DOI] [PubMed] [Google Scholar]
- 31.Lin D. C., Grossman A. D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 32.Erzberger J. P., Mott M. L., Berger J. M. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 33.Kunst F., Ogasawara N., Moszer I., et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 34.Moriya S., Imai Y., Hassan A. K., Ogasawara N. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid. 1999;41:17–29. doi: 10.1006/plas.1998.1381. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto T., Loh P. C., Hirai T., Asai K., Kobayashi K., Moriya S., Ogasawara N. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology. 2002;148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa S., Yamane K., Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J. Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray H., Ferreira H., Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 2006;61:1352–1361. doi: 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- 38.Yoshikawa H., Ogasawara N. Structure and function of DnaA and the DnaA-box in eubacteria: evolutionary relationships of bacterial replication origins. Mol. Microbiol. 1991;5:2589–2597. doi: 10.1111/j.1365-2958.1991.tb01967.x. [DOI] [PubMed] [Google Scholar]
- 39.Fukuoka T., Moriya S., Yoshikawa H., Ogasawara N. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 1990;107:732–739. doi: 10.1093/oxfordjournals.jbchem.a123117. [DOI] [PubMed] [Google Scholar]
- 40.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 41.Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackiewicz P., Zakrzewska-Czerwinska J., Zawilak A., Dudek M. R., Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004;32:3781–3791. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen F. G., Christensen B. B., Nielsen C. B., Atlung T. Insights into the quality of DnaA boxes and their cooperativity. J. Mol. Biol. 2006;355:85–95. doi: 10.1016/j.jmb.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 44.Peter B. J., Arsuaga J., Breier A. M., Khodursky A.B., Brown P. O., Cozzarelli N. R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blot N., Mavathur R., Geertz M., Travers A., Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen F. G., Christensen B. B., Atlung T. The Initiator titration model: computer simulation of chromosome and minichromosome control. Res. Microbiol. 1991;142:161–167. doi: 10.1016/0923-2508(91)90025-6. [DOI] [PubMed] [Google Scholar]
- 47.Hansen F. G., Christensen B. B., Atlung T. Sequence characteristics required for cooperative binding and efficient in vivo titration of the replication initiator protein DnaA in E. coli. J. Mol. Biol. 2007;367:942–952. doi: 10.1016/j.jmb.2007.01.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.