Abstract
Genome-wide analysis of sequence divergence patterns in 12 024 human–mouse orthologous pairs reveals, for the first time, that the trends in nucleotide and amino acid substitutions in orthologs of high and low GC composition are highly asymmetric and polarized to opposite directions. The entire dataset has been divided into three groups on the basis of the GC content at third codon sites of human genes: high, medium, and low. High-GC orthologs exhibit significant bias in favor of the replacements, Thr → Ala, Ser → Ala, Val → Ala, Lys → Arg, Asn → Ser, Ile → Val etc., from mouse to human, whereas in low-GC orthologs, the reverse trends prevail. In general, in the high-GC group, residues encoded by A/U-rich codons of mouse proteins tend to be replaced by the residues encoded by relatively G/C-rich codons in their human orthologs, whereas the opposite trend is observed among the low-GC orthologous pairs. The medium-GC group shares some trends with high-GC group and some with low-GC group. The only significant trend common in all groups of orthologs, irrespective of their GC bias, is (Asp)Mouse → (Glu)Human replacement. At the nucleotide level, high-GC orthologs have undergone a large excess of (A/T)Mouse → (G/C)Human substitutions over (G/C)Mouse → (A/T)Human at each codon position, whereas for low-GC orthologs, the reverse is true.
Key words: high-GC orthologs, low-GC orthologs, amino acid replacement matrix, nucleotide replacement matrix, sequence divergence
1. Introduction
Mammalian genomes are highly heterogeneous in base composition. These are composed of long stretches of DNA with distinct GC composition, commonly known as the isochore structures1–4 or GC-content domains.5 The local GC composition correlates with a number of important genomic features such as gene density, gene length, patterns of gene expression, repeat element distribution, recombination rate etc.6–11 Evolutionary stability of the GC-content distribution has been demonstrated for mice and humans on a genome-wide level.12 The GC-rich sequences from one genome were demonstrated to be GC rich in the other genome and vice versa. Finding such one-to-one correspondence between the local GC distribution patterns in mouse and human was, however, not trivial. Since the divergence of the rodent and primate lineages at around 84–121 million years ago,13,14 multiple substitutions might have occurred at the same sites of a pair of mouse–human orthologs independently in two lineages and if there had not been a strong directionality of the selection process(es) prevailing over the random mutation and fixation, such multiple substitutions should have randomized the local GC distribution patterns in two genomes. Invariance of the overall patterns of GC distribution along the chromosomes of mouse and human, therefore, suggests that there might be some well-defined trends in the nucleotide and/or amino acid substitution patterns across these two species. The present study was designed to determine such trends, if any.
A number of efforts have been made earlier to determine the evolutionary trends in mammalian genomes, but no definite conclusion could be reached. On the basis of the analysis of orthologous gene sequences from closely related species, it has been proposed that GC-rich regions of primate and cetartiodactyl genomes are becoming GC poorer, i.e. GC-rich isochores are now vanishing in these lineages.15–18 Alvarez-Valin et al.,19 however, described the ‘vanishing isochores’ effect as an artifact created due to inaccurate reconstruction of ancestral GC levels in such studies,15 offering an evidence for an AT substitution bias within the repetitive elements of mammals. On the contrary, the maximum parsimony analysis conducted by Gu and Li20 advocated for recent enrichment of the GC content of GC-rich genes in some genomes, e.g. the rabbit. Therefore, the direction(s) of evolution of mammalian genes is a matter of conjecture. Did mammalian genes of varying GC bias follow distinct evolutionary trajectories, and if yes, to what extent could they influence the evolution of encoded proteins? In an attempt to address these questions, the present study carried out a genome-scale analysis of the trends in nucleotide and amino acid substitutions between human and mouse orthologous pairs of varying GC content. The analysis showed that indeed there exist definite trends not only in nucleotide, but also in amino acid substitution patterns between mouse and human orthologous pairs, and that these trends are, in general, highly asymmetric and polarized to the reverse directions in high-GC and low-GC sets of orthologs in such a way that in course of evolution, the compositional heterogeneity has been significantly enhanced in coding regions in human compared with that in mouse.
2. Materials and methods
2.1. Sequence retrieval
Nucleotide sequences of 12 405 pairs of orthologous coding regions of human and mouse were extracted from the Searchable Prototype Experimental Evolutionary Database (SPEED)21 (http://www.bioinfobase.umkc.edu/speed/), using an in-house program developed in Perl. To minimize the sampling errors, a total of 174 sequences, which were shorter than 100 codons in either organism, were excluded from the analysis. The remaining sequences were subjected to a codon integrity check using a freely available program, CodonW22 (http://www.molbiol.ox.ac.uk/cu/), and the dataset was further screened for removing redundant sequences. The final dataset of human mouse orthologs contains 12 024 nonredundant sequences. We generated corresponding nonredundant protein sequence using C program developed in-house.
2.2. Classification of orthologs in three compositional groups
The pairs of orthologous sequences under study exhibited significant correlations, not only between the overall GC contents, but also between the GC contents at third codon sites (GC3) as shown in Fig. 1A and B. These orthologs were then classified into three groups according to the GC3 contents of the human genes: the low-GC group with (GC3)Human < 50%, the medium-GC group with 50% ≤ (GC3)Human ≤ 70%, and the high-GC group with (GC3)Human > 70%. The numbers of pairs of orthologous genes in these three groups were comparable with one another (3896, 3960, and 4168 numbers in high-, medium-, and low-GC groups, respectively). The sequences in these three groups were used to examine the trends in amino acid and nucleotide substitution patterns.
Figure 1.
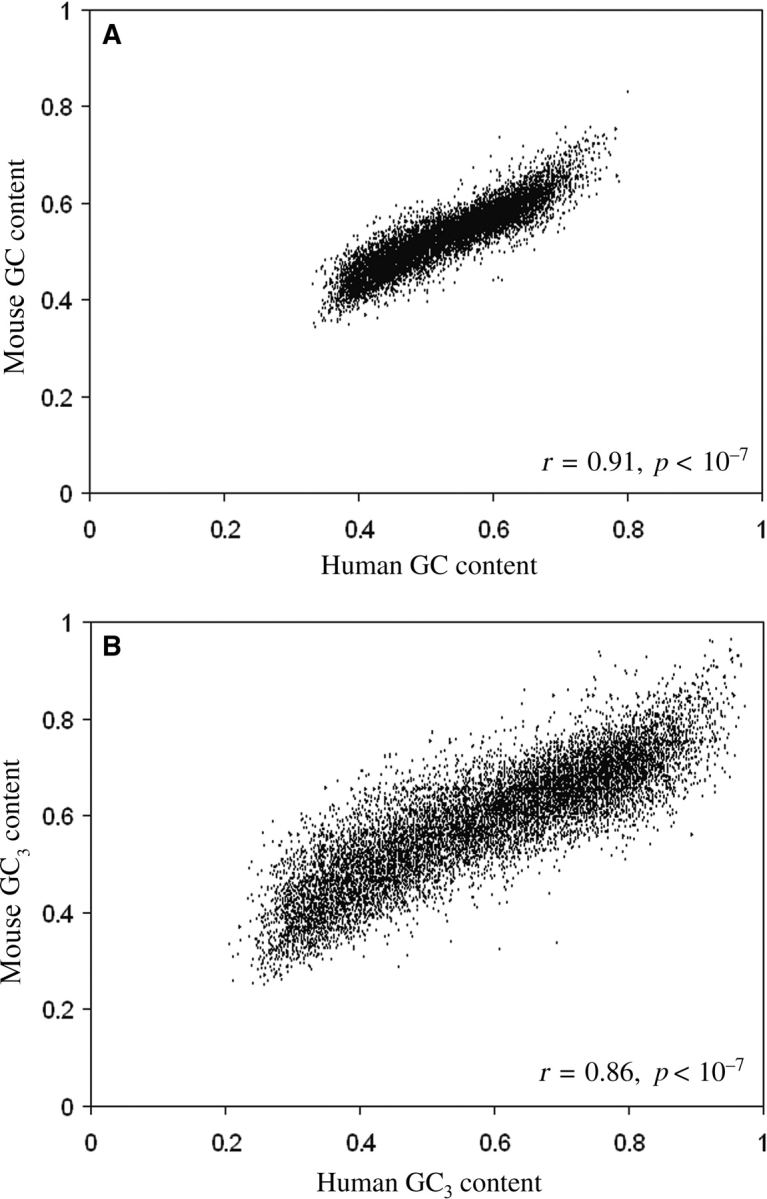
Scatter plot of (A) overall GC content (%) and (B) GC content at third codon sites (%) of 12 024 orthologous genes of human and mouse with their correlation coefficient values.
The total dataset was also classified into another three groups on the basis of GC3 contents of the mouse genes: the new low-GC group with (GC3)Mouse< 50%, the new medium-GC group with 50% ≤ (GC3)Mouse ≤ 70%, and the new high-GC group with (GC3)Mouse > 70%. The entire study carried out with the high-, medium-, and low-GC groups were re-checked with these new high-, new medium-, and new low-GC groups of orthologs.
We classified the datasets on the basis of the GC3 of coding sequences rather than the overall GC, because the GC3 contents of mammalian genes are known to exhibit strong correlation with the GC content of the genomic region, where the genes are located.23,24
2.3. Analysis of amino acid substitution patterns: evaluation of amino acid replacement matrix (AARM) for different groups of orthologs
The alignments of orthologous sequences of three groups were created separately using the pairwise alignment program ClustalW25 and only the gap-free aligned regions of length >100 residues were considered to avoid any spurious short-alignment regions. The numbers of pairs of aligned orthologous genes in three groups of sequences with gap-free regions of length >100 residues were less than the previous set (3659, 2669, and 3291 numbers in high-, medium-, and low-GC groups, respectively). Amino acid replacements were calculated for all gap-free alignment regions of >100 residues and also for fully aligned sequences including gaps. The replacement data are represented in a 20 × 20 matrix, designated as amino acid replacement matrix (AARM), as shown in Tables 1, 2, and 3 for gap-free alignment regions of >100 residues. (To avoid confusion with the standard amino acid substitution matrices like PAM or BLOSUM, we have used the term ‘Replacement’ matrix.) The elements of AARM represent the ratio between the number of forward replacements and the number of backward replacements for any specific pairs of residues, i.e. the value of any element Rij of the AARM represents the ratio of the total number of [i]Mouse → [j]Human replacements to the number of [j]Mouse → [i]Human replacements. If Rij > 1, then there will be an overall gain in the amino acid residue j at the cost of the amino acid residue i in human compared with that in mouse. If Rij < 1, the reverse will be true. The actual number of forward and backward replacements for all possible pairs of amino acid residues for high-, medium-, and low-GC groups are given in Supplementary Table S1a–c. Other than diagonal positions of the matrices (representing the identical substitutions), all other elements represent the nonidentical substitutions. The replacement values for the gap-free alignment regions were not changed significantly from the result obtained by alignment of full sequences including gaps. In order to test whether there were any significant intra-group variations in the replacement values, subsets of 500 pairs of sequences were taken sequentially from start to end and also randomly from the entire dataset of a specific group of orthologs (i.e. high-/medium-/low-GC group) and 20 × 20 AARM was determined for each subset of sequence pairs. Comparison of the replacement values obtained from different subsets of any particular group was then carried out, and no significant variations in substitution values were found for individual residue pairs within a group. All these computations were done using Substitution Pattern Analysis Software Tool (SPAST), a program in C, developed in-house.
Table 1.
Amino Acid Replacement Matrix (AARM) for high-GC group in human mouse orthologs
| Human |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gly | Ala | Arg | Pro | Phe | Tyr | Met | Ile | Asn | Lys | Ser | Thr | Cys | Trp | Val | Leu | Glu | Asp | His | Gln | ||
| Mouse | Gly | – | 0.93 | 0.84 | 1.02 | 0.62 | 0.35 | 0.84 | 0.40 | 0.47 | 0.47 | 0.67 | 0.55 | 0.76 | 0.86 | 0.72 | 0.76 | 0.77 | 0.73 | 0.69 | 0.83 |
| Ala | 1.08 | – | 0.85 | 1.04 | 0.69 | 0.40 | 0.62 | 0.47 | 0.49 | 0.55 | 0.59 | 0.58 | 0.67 | 1.00 | 0.70 | 0.78 | 0.71 | 0.64 | 1.03 | 0.79 | |
| Arg | 1.19 | 1.17 | – | 0.87 | 0.45 | 0.47 | 0.76 | 0.72 | 0.56 | 0.74 | 0.85 | 1.00 | 0.82 | 0.87 | 1.06 | 0.77 | 0.86 | 0.94 | 0.71 | 0.72 | |
| Pro | 0.98 | 0.96 | 1.15 | – | 0.44 | 0.48 | 0.66 | 0.49 | 0.81 | 0.81 | 0.58 | 0.66 | 0.59 | 0.74 | 0.63 | 0.79 | 1.00 | 0.82 | 0.72 | 0.82 | |
| Phe | 1.62 | 1.44 | 2.21 | 2.25 | – | 1.13 | 0.73 | 0.82 | 1.00 | 0.67 | 1.07 | 0.67 | 1.22 | 1.50 | 1.20 | 1.27 | 1.14 | 0.78 | 1.82 | 1.06 | |
| Tyr | 2.82 | 2.50 | 2.12 | 2.09 | 0.88 | – | 1.00 | 0.85 | 1.13 | 1.71 | 1.31 | 1.53 | 1.25 | 1.33 | 0.70 | 1.08 | 1.83 | 1.68 | 1.50 | 1.61 | |
| Met | 1.20 | 1.60 | 1.32 | 1.51 | 1.38 | 1.00 | – | 0.81 | 0.81 | 0.99 | 0.89 | 0.90 | 0.62 | 1.00 | 1.21 | 1.45 | 1.83 | 1.07 | 0.5 | 1.25 | |
| Ile | 2.51 | 2.12 | 1.39 | 2.02 | 1.22 | 1.18 | 1.24 | – | 1.38 | 1.07 | 1.42 | 1.40 | 1.00 | 0.33 | 1.22 | 1.48 | 1.47 | 0.65 | 0.91 | 1.64 | |
| Asn | 2.13 | 2.03 | 1.77 | 1.24 | 1.00 | 0.89 | 1.23 | 0.73 | – | 1.12 | 1.34 | 1.11 | 1.76 | – | 1.24 | 1.10 | 1.42 | 1.26 | 1.30 | 1.48 | |
| Lys | 2.15 | 1.81 | 1.36 | 1.24 | 1.50 | 0.58 | 1.01 | 0.94 | 0.89 | – | 1.39 | 1.00 | 0.80 | 3.67 | 1.76 | 1.30 | 1.60 | 1.22 | 1.61 | 1.47 | |
| Ser | 1.49 | 1.70 | 1.18 | 1.72 | 0.94 | 0.76 | 1.13 | 0.71 | 0.75 | 0.72 | – | 1.03 | 0.93 | 1.10 | 1.00 | 1.01 | 1.36 | 1.06 | 1.15 | 1.31 | |
| Thr | 1.83 | 1.71 | 1.00 | 1.52 | 1.50 | 0.65 | 1.11 | 0.72 | 0.90 | 1.00 | 0.97 | – | 1.16 | 0.89 | 1.08 | 1.06 | 1.37 | 1.09 | 0.91 | 0.99 | |
| Cys | 1.31 | 1.50 | 1.22 | 1.69 | 0.82 | 0.80 | 1.60 | 1.00 | 0.57 | 1.25 | 1.07 | 0.86 | – | 1.17 | 1.04 | 0.98 | 0.75 | 1.17 | 1.01 | 0.63 | |
| Trp | 1.16 | 1.00 | 1.15 | 1.35 | 0.67 | 0.75 | 1.00 | 3.00 | – | 0.27 | 0.91 | 1.12 | 0.85 | – | 1.05 | 0.94 | 1.00 | 0.33 | 0.59 | 0.76 | |
| Val | 1.40 | 1.43 | 0.94 | 1.60 | 0.84 | 1.43 | 0.82 | 0.82 | 0.8 | 0.57 | 1.00 | 0.93 | 0.96 | 0.95 | – | 1.08 | 0.81 | 0.86 | 1.17 | 1.07 | |
| Leu | 1.31 | 1.28 | 1.31 | 1.26 | 0.79 | 0.92 | 0.69 | 0.67 | 0.91 | 0.77 | 0.99 | 0.94 | 1.02 | 1.07 | 0.92 | – | 1.51 | 1.04 | 1.16 | 0.88 | |
| Glu | 1.30 | 1.41 | 1.16 | 1.00 | 0.88 | 0.55 | 0.55 | 0.68 | 0.71 | 0.62 | 0.74 | 0.73 | 1.33 | 1.00 | 1.23 | 0.66 | – | 0.85 | 0.79 | 0.86 | |
| Asp | 1.38 | 1.57 | 1.06 | 1.22 | 1.29 | 0.59 | 0.93 | 1.53 | 0.80 | 0.82 | 0.95 | 0.92 | 0.86 | 3.00 | 1.16 | 0.96 | 1.17 | – | 0.94 | 1.26 | |
| His | 1.45 | 0.97 | 1.41 | 1.38 | 0.55 | 0.67 | 2.00 | 1.10 | 0.77 | 0.62 | 0.87 | 1.10 | 0.99 | 1.70 | 0.85 | 0.86 | 1.27 | 1.06 | – | 1.02 | |
| Gln | 1.20 | 1.27 | 1.38 | 1.21 | 0.94 | 0.62 | 0.80 | 0.61 | 0.68 | 0.68 | 0.76 | 1.01 | 1.58 | 1.31 | 0.93 | 1.13 | 1.17 | 0.79 | 0.98 | – | |
Each element Rij in the AARM represents the ratio of number of replacements of the residue i by the residue j in the forward direction (mouse → human) to that in the reverse direction. This means that if Rij >1, the number of (i)Mouse → (j)Human replacements is higher than the number of (j)Mouse → (i)Human replacements and if Rij <1, the reverse is true. Bold and Bold-italics ratios signifies the directional bias at p < 0.0001 and 0.001 respectively.
Table 2.
Amino Acid Replacement Matrix (AARM) for medium-GC group in human mouse orthologs
| Human |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gly | Ala | Arg | Pro | Phe | Tyr | Met | Ile | Asn | Lys | Ser | Thr | Cys | Trp | Val | Leu | Glu | Asp | His | Gln | ||
| Mouse | Gly | – | 0.92 | 0.90 | 1.09 | 0.93 | 0.9 | 1.04 | 0.76 | 0.84 | 0.83 | 0.92 | 0.73 | 0.89 | 0.90 | 1.10 | 1.20 | 0.97 | 0.88 | 0.69 | 0.80 |
| Ala | 1.09 | – | 1.19 | 1.08 | 1.19 | 1.00 | 0.9 | 0.85 | 0.97 | 1.05 | 0.85 | 0.85 | 0.8 | 0.59 | 0.95 | 0.96 | 0.90 | 0.74 | 1.06 | 1.21 | |
| Arg | 1.11 | 0.84 | – | 1.02 | 0.91 | 0.84 | 0.85 | 0.76 | 1.04 | 1.09 | 1.10 | 1.07 | 1.07 | 1.23 | 0.91 | 0.92 | 1.12 | 0.98 | 0.95 | 0.89 | |
| Pro | 0.92 | 0.93 | 0.98 | – | 0.76 | 0.61 | 1.21 | 0.82 | 0.79 | 1.03 | 0.82 | 0.97 | 0.62 | 1.75 | 0.77 | 0.85 | 1.05 | 0.78 | 1.11 | 1.15 | |
| Phe | 1.08 | 0.84 | 1.10 | 1.32 | – | 1.19 | 0.72 | 0.95 | 1.41 | 1.67 | 0.98 | 0.93 | 0.97 | 1.38 | 0.82 | 1.01 | 1.80 | 0.93 | 1.62 | 1.15 | |
| Tyr | 1.11 | 1.00 | 1.19 | 1.64 | 0.84 | – | 1.25 | 0.78 | 0.88 | 0.94 | 0.94 | 1.12 | 0.91 | 1.32 | 1.04 | 0.84 | 1.12 | 0.86 | 1.12 | 0.88 | |
| Met | 0.97 | 1.11 | 1.17 | 0.82 | 1.38 | 0.80 | – | 1.09 | 0.96 | 0.86 | 0.77 | 0.86 | 1.11 | 0.75 | 0.98 | 1.11 | 0.91 | 0.65 | 0.93 | 1.46 | |
| Ile | 1.32 | 1.18 | 1.32 | 1.21 | 1.05 | 1.29 | 0.92 | – | 1.13 | 1.02 | 1.03 | 0.95 | 1.28 | 0.25 | 0.85 | 0.92 | 1.33 | 0.88 | 0.83 | 1.29 | |
| Asn | 1.19 | 1.04 | 0.96 | 1.27 | 0.71 | 1.13 | 1.04 | 0.88 | – | 1.00 | 0.89 | 0.88 | 0.76 | 1.33 | 0.89 | 0.92 | 1.07 | 1.01 | 0.98 | 0.72 | |
| Lys | 1.20 | 0.95 | 0.92 | 0.97 | 0.60 | 1.06 | 1.16 | 0.98 | 1.00 | – | 0.89 | 0.84 | 0.73 | 1.94 | 1.05 | 1.35 | 1.13 | 0.87 | 0.81 | 1.04 | |
| Ser | 1.09 | 1.18 | 0.91 | 1.22 | 1.02 | 1.06 | 1.30 | 0.97 | 1.13 | 1.13 | – | 1.17 | 0.91 | 1.10 | 1.08 | 1.05 | 1.14 | 1.04 | 1.05 | 1.22 | |
| Thr | 1.38 | 1.17 | 0.93 | 1.03 | 1.08 | 0.89 | 1.16 | 1.06 | 1.14 | 1.20 | 0.85 | – | 0.86 | 0.57 | 1.06 | 0.80 | 1.05 | 0.93 | 0.80 | 1.15 | |
| Cys | 1.12 | 1.24 | 0.93 | 1.62 | 1.03 | 1.10 | 0.90 | 0.78 | 1.31 | 1.36 | 1.09 | 1.16 | – | 0.93 | 0.84 | 1.18 | 1.00 | 0.76 | 0.93 | 1.00 | |
| Trp | 1.11 | 1.70 | 0.81 | 0.57 | 0.72 | 0.76 | 1.33 | 4.00 | 0.75 | 0.52 | 0.91 | 1.75 | 1.08 | – | 1.30 | 0.89 | 0.76 | 0.75 | 0.55 | 0.83 | |
| Val | 0.91 | 1.06 | 1.10 | 1.30 | 1.21 | 0.96 | 1.02 | 1.17 | 1.12 | 0.96 | 0.92 | 0.94 | 1.19 | 0.77 | – | 1.05 | 0.91 | 0.72 | 0.90 | 1.02 | |
| Leu | 0.83 | 1.04 | 1.08 | 1.18 | 0.99 | 1.19 | 0.90 | 1.09 | 1.09 | 0.74 | 0.95 | 1.25 | 0.84 | 1.12 | 0.95 | – | 0.98 | 1.39 | 1.10 | 0.94 | |
| Glu | 1.03 | 1.11 | 0.89 | 0.95 | 0.56 | 0.89 | 1.10 | 0.75 | 0.93 | 0.88 | 0.87 | 0.95 | 1.00 | 1.32 | 1.10 | 1.02 | – | 0.86 | 0.69 | 0.91 | |
| Asp | 1.14 | 1.35 | 1.03 | 1.29 | 1.07 | 1.17 | 1.55 | 1.14 | 0.99 | 1.15 | 0.96 | 1.08 | 1.32 | 1.33 | 1.38 | 0.72 | 1.16 | – | 0.81 | 0.83 | |
| His | 1.45 | 0.95 | 1.06 | 0.90 | 0.62 | 0.90 | 1.08 | 1.20 | 1.02 | 1.23 | 0.95 | 1.25 | 1.07 | 1.83 | 1.11 | 0.91 | 1.46 | 1.23 | – | 1.06 | |
| Gln | 1.25 | 0.83 | 1.13 | 0.87 | 0.87 | 1.13 | 0.69 | 0.78 | 1.38 | 0.96 | 0.82 | 0.87 | 1.00 | 1.21 | 0.98 | 1.06 | 1.09 | 1.21 | 0.94 | – | |
Each element Rij in the AARM represents the ratio of number of replacements of the residue i by the residue j in the forward direction (mouse → human) to that in the reverse direction. This means that if Rij >1, the number of (i)Mouse → (j)Human replacements is higher than the number of (j)Mouse → (i)Human replacements and if Rij <1, the reverse is true. Bold and Bold-italics ratios signifies the directional bias at p < 0.0001 and 0.001 respectively.
Table 3.
Amino Acid Replacement Matrix (AARM) for low-GC group in human mouse orthologs
| Human |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gly | Ala | Arg | Pro | Phe | Tyr | Met | Ile | Asn | Lys | Ser | Thr | Cys | Trp | Val | Leu | Glu | Asp | His | Gln | ||
| Mouse | Gly | – | 0.88 | 0.98 | 0.91 | 2.10 | 1.50 | 1.60 | 2.08 | 1.77 | 1.22 | 1.15 | 1.09 | 1.32 | 0.97 | 1.80 | 1.56 | 1.29 | 1.35 | 1.23 | 1.08 |
| Ala | 1.13 | – | 0.97 | 1.24 | 1.61 | 1.32 | 1.36 | 1.67 | 1.97 | 1.97 | 1.24 | 1.25 | 0.97 | 3.00 | 1.28 | 1.37 | 1.31 | 1.24 | 1.62 | 1.45 | |
| Arg | 1.02 | 1.03 | – | 0.97 | 1.32 | 1.38 | 1.45 | 1.50 | 1.89 | 1.63 | 1.18 | 1.14 | 1.11 | 0.97 | 1.04 | 1.23 | 1.40 | 1.69 | 1.41 | 1.09 | |
| Pro | 1.10 | 0.81 | 1.03 | – | 1.34 | 2.43 | 1.40 | 1.64 | 2.63 | 2.00 | 1.26 | 1.51 | 0.77 | 1.07 | 1.10 | 1.15 | 1.45 | 1.32 | 1.56 | 1.69 | |
| Phe | 0.48 | 0.62 | 0.76 | 0.74 | – | 1.25 | 0.68 | 0.95 | 1.21 | 1.54 | 0.76 | 0.75 | 0.60 | 0.68 | 0.64 | 0.78 | 0.76 | 0.50 | 0.92 | 0.85 | |
| Tyr | 0.67 | 0.76 | 0.72 | 0.41 | 0.80 | – | 0.73 | 1.12 | 0.86 | 0.93 | 0.67 | 0.70 | 0.74 | 1.38 | 0.76 | 0.91 | 1.00 | 0.80 | 0.77 | 0.83 | |
| Met | 0.62 | 0.73 | 0.69 | 0.71 | 1.46 | 1.38 | – | 1.48 | 1.23 | 1.05 | 0.83 | 0.88 | 0.50 | 0.38 | 0.81 | 0.84 | 0.60 | 0.96 | 0.87 | 1.04 | |
| Ile | 0.48 | 0.60 | 0.67 | 0.61 | 1.05 | 0.89 | 0.68 | – | 1.17 | 1.00 | 0.65 | 0.66 | 0.50 | 0.89 | 0.67 | 0.68 | 0.65 | 0.70 | 0.96 | 0.57 | |
| Asn | 0.57 | 0.51 | 0.53 | 0.38 | 0.83 | 1.16 | 0.81 | 0.86 | – | 1.04 | 0.57 | 0.60 | 0.54 | 0.14 | 0.72 | 0.68 | 0.90 | 0.81 | 0.57 | 0.74 | |
| Lys | 0.82 | 0.51 | 0.61 | 0.50 | 0.65 | 1.08 | 0.95 | 1.00 | 0.96 | – | 0.62 | 0.62 | 0.75 | 0.65 | 0.77 | 0.70 | 0.93 | 0.89 | 0.57 | 0.73 | |
| Ser | 0.87 | 0.80 | 0.85 | 0.79 | 1.32 | 1.48 | 1.20 | 1.53 | 1.74 | 1.60 | – | 1.26 | 0.76 | 0.66 | 1.04 | 1.15 | 1.33 | 1.39 | 0.99 | 1.10 | |
| Thr | 0.91 | 0.80 | 0.88 | 0.66 | 1.33 | 1.43 | 1.13 | 1.51 | 1.67 | 1.60 | 0.79 | – | 1.00 | 1.8 | 0.91 | 0.91 | 1.19 | 1.08 | 0.91 | 1.05 | |
| Cys | 0.76 | 1.03 | 0.90 | 1.30 | 1.67 | 1.36 | 2.00 | 2.00 | 1.84 | 1.33 | 1.31 | 1.00 | – | 1.00 | 0.52 | 1.11 | 0.57 | 1.44 | 0.94 | 1.31 | |
| Trp | 1.03 | 0.33 | 1.03 | 0.93 | 1.48 | 0.73 | 2.67 | 1.12 | 7.00 | 1.54 | 1.52 | 0.56 | 1.00 | – | 1.22 | 1.16 | 1.19 | 0.8 | 0.94 | 1.11 | |
| Val | 0.55 | 0.78 | 0.96 | 0.91 | 1.56 | 1.32 | 1.23 | 1.48 | 1.39 | 1.31 | 0.96 | 1.10 | 1.91 | 0.82 | – | 1.13 | 0.79 | 0.89 | 1.16 | 0.97 | |
| Leu | 0.64 | 0.73 | 0.82 | 0.87 | 1.28 | 1.09 | 1.19 | 1.48 | 1.47 | 1.42 | 0.87 | 1.09 | 0.9 | 0.86 | 0.89 | – | 0.92 | 0.58 | 1.13 | 0.92 | |
| Glu | 0.78 | 0.76 | 0.71 | 0.69 | 1.31 | 1.00 | 1.67 | 1.53 | 1.12 | 1.08 | 0.75 | 0.84 | 1.75 | 0.84 | 1.27 | 1.09 | – | 0.84 | 1.06 | 0.95 | |
| Asp | 0.74 | 0.81 | 0.59 | 0.76 | 2.00 | 1.25 | 1.04 | 1.43 | 1.24 | 1.12 | 0.72 | 0.93 | 0.69 | 1.25 | 1.12 | 1.72 | 1.19 | – | 0.78 | 1.00 | |
| His | 0.81 | 0.62 | 0.71 | 0.64 | 1.09 | 1.30 | 1.15 | 1.04 | 1.76 | 1.75 | 1.01 | 1.10 | 1.07 | 1.06 | 0.86 | 0.89 | 0.94 | 1.28 | – | 1.09 | |
| Gln | 0.93 | 0.69 | 0.92 | 0.59 | 1.170 | 1.20 | 0.96 | 1.77 | 1.36 | 1.37 | 0.91 | 0.95 | 0.77 | 0.90 | 1.03 | 1.08 | 1.05 | 1.00 | 0.91 | – | |
Each element Rij in the AARM represents the ratio of number of replacements of the residue i by the residue j in the forward direction (mouse → human) to that in the reverse direction. This means that if Rij >1, the number of (i)Mouse → (j)Human replacements is higher than the number of (j)Mouse → (i)Human replacements and if Rij <1, the reverse is true. Bold and Bold-italics ratios signifies the directional bias at p < 0.0001 and < 0.001 respectively.
2.4. Analysis of nucleotide substitution patterns: evaluation of nucleotide replacement matrix (NRM) for three codon sites of different groups of orthologs
We created the nucleotide sequence alignments on the basis of amino acid alignments and calculated the nucleotide replacements in the form of 4 × 4 NRM, individually for three codon positions for three different groups of orthologs under study. The elements rij of NRM represent the ratio of the number of forward replacements to that of backward replacements for any specific pairs of nucleotides. Comparison of the nucleotide replacement values obtained from different subsets of 500 orthologous sequences (taken sequentially from start to end and also randomly) of any particular group was then carried out and no significant variations in replacement values were found for individual nucleotide pairs within a group.
2.5. Tests for statistical significance of different elements (Rij/rij) of AARM and NRM
For a given pair of amino acids or nucleotides, the mouse to human replacement was taken as the forward direction and human to mouse as the reverse direction, and each Rij in AARM or rij in NRM represents the ratio of number of replacements of the residue i by the residue j in the forward direction (mouse to human) to that in the reverse direction (human to mouse). This means that if Rij (or rij) > 1, the number of (i)Mouse → (j)Human replacements is higher than the number of (j)Mouse → (i)Human replacements, and if Rij (or rij) < 1, the reverse is true.
For each pair of replacements, the ratio of forward to reverse replacements was expected to be 1:1 under unbiased conditions. To test this hypothesis, the observed and expected numbers (based on a 1:1 ratio) were recorded for each pair of a particular group. In all cases, the Chi-square test was used to assess the significance of the directional bias, if any, at p = 0.001 and 0.0001. For each pair of replacements, the first and second rows of the 2 × 2 contingency table represented the number of replacements from one particular residue (say, i) to another (say, j) of the pair and the total count of the remaining replacements (say, k) from the residue i (where k ≠ j), respectively. The procedure was repeated also for orthologous replacements of 500 sequences taken sequentially from start to end and randomly. The significant (at p < 0.001 or 0.0001) trends for the whole dataset are also consistent with sequences taken sequentially from start to end and randomly.
2.6. Correspondence analysis (COA) on relative synonymous codon usage (RSCU) and estimation of synonymous and nonsynonymous substitution
Correspondence analysis on RSCU26 was performed using the CodonW 1.4.222 program to identify the major factors influencing the variation in synonymous codon usage in three groups of orthologous sets. These analyses generate a series of orthogonal axes to identify trends that explain the variation within a dataset, with each subsequent axis explaining a decreasing amount of the variation.
To examine the nucleotide substitution patterns, we estimated the number of synonymous substitutions per synonymous site, dS, and the number of nonsynonymous substitutions per nonsynonymous site, dN, of randomly chosen 500 pairs of ten sets of orthologs in each three groups using the MEGA program (version 2.1), as described by Nei and Gojobori.27 The values of dS, dN, and dN/dS of three orthologous groups were compared by t-test.
3. Results
3.1. Specific trends in amino acid substitution patterns between mouse and human orthologs
In order to investigate whether the mouse and human proteins of high-, medium-, and low-GC composition followed the same or different evolutionary trajectories since the divergence of the two species, trends in amino acid substitution between the human–mouse orthologous pairs were studied individually in three groups of genes, using the program SPAST developed in-house. Tables 1, 2, and 3 represents the AARMs for aligned regions of orthologous pairs (gap-free regions of length >100 residues at a stretch) in high-, medium-, and low-GC groups, respectively. As already mentioned, the mouse to human replacements was taken by convention as the forward direction and human to mouse as the reverse direction.
For each group of orthologs, some specific amino acid pairs exhibit significant bias in the replacement patterns. For instance, in high-GC group, the value of RIG, i.e. the ratio of [Ile]Mouse → [Gly]Human to [Gly]Mouse → [Ile]Human, is 2.51 (p < 0.0001), implying that the frequency of replacement of Ile in mouse sequence with Gly in human is >2.5-fold higher than that in reverse direction, i.e. the frequency of replacement of Gly of mouse sequence with Ile in human. On the contrary, in low-GC group, RIG = 0.48, indicating that in low-GC orthologs, the frequency of substitution of Ile of mouse sequence by Gly in human is more than two-fold lower than the frequency of the reverse substitution. For the medium-GC group, the value of RIG is not statistically significant, suggesting that the frequencies of substitution of Ile by Gly and of Gly by Ile are comparable in cases of medium-GC orthologs of mouse and human.
As Rij = 1/Rji, out of the 380 off-diagonal elements of an AARM (Tables 1, 2, and 3), only 190 are independent. Out of these 190 AARM elements, 53 are significantly biased in a specific direction for high-GC group (46 at p < 0.0001 and seven at p < 0.001), whereas for low-GC group, 67 elements are found to be significantly biased (56 and 11 at p < 0.0001 and 0.001, respectively). In the medium-GC group, only 15 AARM elements are statistically significant (ten at p < 0.0001 and five at p < 0.001). Some of them are shared with the high-GC group and some with the low-GC group.
3.2. Significant trends in amino acid substitution in high-GC and low-GC groups are, in general, opposite to one another
A careful examination of Tables 1, 2, and 3 reveals that when i represents a residue encoded by A/U-rich codons and j represents a residue encoded by relatively G/C-rich codons, the AARM element, Rij, in most cases (but not in all), is >1 in the high-GC group, <1 in the low-GC group, and nearly equal to ∼1 in the medium-GC group. Reverse situation occurs, in general, when i represents a residue encoded by G/C-rich codons and j by relatively A/U-rich codons. For instance, for i = Ala (A) (encoded by GCN), RAj is significantly <1 in high-GC group and significantly >1 in low-GC group for j = Ile, Asn, Lys, Ser, Thr, Val, Glu etc (encoded, respectively, by AUH, AAY, AAR, UCN/AGY, ACN, GUN, and GAR). On the contrary, for i = Asn (N) (encoded by AAY), RNj > 1 for high-GC group and <1 for low-GC group, when j = Gly or Ala or Arg (encoded by GGN, GCN, and CGN/AGR, respectively). There are altogether 33 AARM elements, which are polarized to the opposite directions (>1 and <1) in high- and low-GC groups and are found to be statistically significant in both groups.
Table 4 provides the lists of the 15 amino acid pairs having the largest differences in total number of forward (mouse to human) and backward (human to mouse) substitutions between them for three different groups of orthologs under study. There are eight pairs of residues (marked with ±) that appear among the top 15 trends in both high- and low-GC groups, but with opposite directionality (Table 4). There are four other pairs of residues among the top 15 of the high-GC group (marked with +), which exhibit significant, but opposite, bias in the low-GC group (Table 4), but did not come among the top 15 in the later group. Similarly, there are also four pairs (marked with –) among the top 15 of the low-GC group, which are opposite and significant, but not among the top 15 in the high-GC group. Thus, the trends in amino acid substitutions between the mouse and human orthologs follow reverse directionality, in general, in the high- and low-GC groups. Among the top 15 trends in the medium-GC groups, some are common in directionality with high-GC group and some with the low-GC group.
Table 4.
Top 15 amino acid pairs of three orthologous groups according to differences in number of forward (mouse to human) and reverse (human to mouse) replacements in AARM
| Pair | Forward no. | Reverse no. | Difference | Ratio | Codon changes | Trends |
|---|---|---|---|---|---|---|
| High-GC group | ||||||
| Thr → Ala* | 6873 | 4020 | 2853 | 1.71 | ACN → GCN | ± |
| Ser → Ala* | 4160 | 2447 | 1713 | 1.70 | UCN/AGY → GCN | ± |
| Ser → Pro* | 3603 | 2097 | 1506 | 1.72 | UCN/AGY → CCN | ± |
| Val → Ala* | 4484 | 3144 | 1340 | 1.43 | GUN → GCN | ± |
| Ser → Gly* | 3749 | 2516 | 1233 | 1.49 | UCN/AGY → GGN | + |
| Gln → Arg* | 3389 | 2448 | 941 | 1.38 | CAR → CGN/AGR | |
| Lys → Arg* | 3412 | 2518 | 894 | 1.36 | AAR → CGN/AGR | ± |
| Ile → Val* | 4572 | 3756 | 816 | 1.22 | AUH → GUN | ± |
| Asn → Ser* | 2878 | 2145 | 733 | 1.34 | AAY → UCN/AGY | ± |
| Asp → Glu* | 3799 | 3237 | 562 | 1.17 | GAY → GAR | • |
| His → Arg* | 1899 | 1343 | 556 | 1.41 | CAY → CGN/AGR | + |
| Met → Leu* | 1753 | 1211 | 542 | 1.45 | AUG → UUR/CUN | + |
| Ile → Leu* | 1500 | 1012 | 488 | 1.48 | AUN → UUR/CUN | ± |
| Lys → Glu* | 1257 | 785 | 472 | 1.60 | AAR → GAR | |
| Leu → Pro* | 2060 | 1634 | 426 | 1.26 | YYR/CUN → CCN | + |
| Medium-GC group | ||||||
| Val → Ile* | 5839 | 4982 | 857 | 1.17 | GUN → AUH | □ |
| Thr → Ala* | 5566 | 4753 | 813 | 1.17 | ACN → GCN | ○ |
| Ser → Pro* | 3804 | 3113 | 691 | 1.22 | UCN/AGY → CCN | ○ |
| Asp → Glu* | 4443 | 3833 | 610 | 1.16 | GAY → GAR | • |
| Ser → Thr* | 3652 | 3113 | 539 | 1.17 | UCN/AGY → ACN | □ |
| Ser → Ala* | 3507 | 2972 | 535 | 1.18 | UCN/AGY → GCN | ○ |
| Ser → Asn* | 3626 | 3213 | 413 | 1.13 | UCN/AGY → AAY | □ |
| Arg → Lys** | 4017 | 3701 | 316 | 1.08 | CGN/AGR → AAR | □ |
| Leu → Pro* | 1999 | 1691 | 308 | 1.18 | UUR/CUN → CCN | ○ |
| Gln → Arg* | 2676 | 2372 | 304 | 1.13 | CAR → CGN/AGR | ○ |
| Ser → Gly | 2782 | 2560 | 222 | 1.09 | UCN/AGY → GGN | ○ |
| Val → Ala | 3759 | 3561 | 198 | 1.06 | GUN → GCN | ○ |
| Lys → Glu** | 1514 | 1336 | 178 | 1.13 | AAR → GAR | ○ |
| Asp → Ala* | 661 | 488 | 173 | 1.35 | GSN → GCN | |
| Met → Leu | 1648 | 1483 | 165 | 1.11 | AUG → UUR/CUN | ○ |
| Low GC group | ||||||
| Val → Ile* | 9402 | 6335 | 3067 | 1.48 | GUN → AUH | ± |
| Ser → Asn* | 6508 | 3733 | 2775 | 1.74 | UCN/AGY → AAY | ± |
| Arg → Lys* | 6603 | 4059 | 2544 | 1.63 | CGN/AGR → AAR | ± |
| Ala → Thr* | 6393 | 5117 | 1276 | 1.25 | GCN → ACN | ± |
| Leu → Ile* | 3003 | 2035 | 968 | 1.48 | UUR/CUN → AUH | ± |
| Asp → Glu* | 5952 | 5014 | 938 | 1.19 | GAY → GAR | • |
| Ser → Thr* | 4498 | 3563 | 935 | 1.26 | UCN/AGY → ACN | |
| Ala → Val* | 4193 | 3264 | 929 | 1.28 | GCN → GUN | ± |
| Thr → Ile* | 2612 | 1733 | 879 | 1.51 | CAN → AUH | – |
| Pro → Ser* | 4178 | 3304 | 874 | 1.26 | CCN → UCN/AGY | ± |
| Ala → Ser* | 3975 | 3195 | 780 | 1.24 | GCN → UCN/AGY | ± |
| Leu → Phe* | 2990 | 2330 | 660 | 1.28 | UUR/CUN → UUY | – |
| Thr → Asn* | 1630 | 975 | 655 | 1.67 | CAN → AAY | |
| Met → Ile* | 1900 | 1288 | 612 | 1.48 | AUG → AUH | – |
| His → Asn* | 1258 | 715 | 543 | 1.76 | CAY → AAY | – |
*,**Replacements of pairs are significant at p < 0.0001 and 0.001, respectively.
Symbols used in codons: R, A/G; Y, C/T; S, G/C; W, A/T; H, A/C/T; N, any nucleotide. Symbols used in trends: opposite trends among top 15 amino acid pairs of both high- and low-GC groups (±); opposite trends in high- and low-GC group, but not present in top 15 of low-GC group (+); opposite trends in high- and low-GC group, but not present in top 15 of high-GC group (–); same trend in high-, medium-, and low-GC groups (•); same trend in high- and medium-GC groups (○); same trend in low- and medium-GC groups (□).
In the high-GC group, although amino acids of mouse proteins encoded by A/U-rich codons tend to be replaced by the amino acids encoded by G/C-rich codons in their human orthologs, not all amino acid residues encoded by A/U-rich codons exhibit equal bias in replacement patterns. There are six residues, viz. Phe, Tyr, Met, Ile, Asn, and Lys, which are encoded by A/U-rich codons and four residues, viz. Gly, Ala, Arg, and Pro, encoded by G/C-rich codons. As can be seen from Tables 1, 2, and 3, among the amino acid residues encoded by A/U-rich codons, Ile, Asn, and Lys have a more number of significantly biased replacement ratios (AARM elements) both in the high-GC group and in the low-GC group. Replacement ratios of Phe and Tyr, though follow the general trend, are not statistically significant in most cases. Rather, some other residues like Ser, Thr, Val, Leu etc., which are not necessarily encoded by A/U codons, exhibit significant bias in the replacement ratios (Tables 1 and 3). Similarly, among Gly, Ala, Arg, and Pro, the former two have more number of significant Rij values. Previous analysis of many prokaryotic genomes28 and high-GC rice genes with their Arabidopsis homologs29 showed that proteins encoded by GC-rich sequences are characterized by increased levels of Gly, Ala, Arg, and Pro residues and a corresponding decrease in Phe, Tyr, Met, Ile, Asn, and Lys residues. It is, therefore, intriguing to examine to what extent the overall usage of the residues Gly/Ala/Arg/Pro and that of Phe/Tyr/Met/Ile/Asn/Lys vary within the mouse and human orthologs of high-, medium- and low-GC groups. Our analysis indicates that the mouse and human orthologs of three groups are indeed characterized by distinct usage profile of these residues (Supplementary Fig. S1). In the high-GC group, the human orthologs have higher usage of Gly, Ala, Arg, and Pro and lower usages of Phe, Tyr, Met, Ile, Asn, and Lys compared with their mouse orthologs, but the differences are not as pronounced as shown previously for homologous gene pairs from the rice and Arabidopsis, having large difference in GC content.29 In the low-GC group, the reverse is true, whereas in the medium-GC group, there is no significant difference between mouse and human orthologous pairs in the usage of these two groups of residues.
3.3. (Asp)Mouse → (Glu)Human trend in all groups of orthologs irrespective of their GC content
There is only one replacement ratio RDE, which exhibits same directionality and almost the same value in all three AARMs (Tables 1, 2, and 3), indicating that in all groups of orthologs, the frequency of (Asp)Mouse→ (Glu)Human replacements is slightly higher than the replacement in the opposite direction. As the Asp → Glu replacement is among the top 15 trends in substitution in all three groups (Table 4), it is one of the most common trends in amino acid replacement in mouse–human orthologs. These observations suggest that irrespective of the GC content of the encoding genes, there has been a consistent increase in glutamic acid in human proteins at the cost of aspartic acid compared with their mouse orthologs. The structural and/or functional implications of this unique evolutionary trend is, however, not clear. There are two other substitution trends, Ser → Thr and Phe → Tyr, which also exhibit same directionality in all three groups under study, but the replacement values are not statistically significant for the high-GC group.
3.4. High-GC orthologs are biased towards (A/T)Mouse → (G/C)Human replacements, whereas in low-GC orthologs, (G/C)Mouse → (A/T)Human replacements prevail
As already emphasized, the major trends in amino acid replacements between mouse and human orthologs (Tables 1–4) indicate that in the high-GC group, the amino acid residues encoded by relatively GC-rich codons tend to increase in human proteins compared with mouse orthologs, and in the low-GC group, the reverse trends prevail. In the medium-GC-group, however, there is no such specific directionality in codon substitution patterns. These observations have prompted us to examine the trends in nucleotide substitution patterns individually in three codon positions in three groups of orthologs. As can be seen from the NRMs shown in Table 5, in the high-GC dataset, rij, is significantly greater than 1, when i = A or T and j = G or C. On the contrary, rij is significantly less than 1, when i = G or C and j = A or T. These trends are valid in all three codon positions, although the deviation of rij from 1 (for a particular set of m and n) is highest in third codon positions, followed by the first and second codon positions. Therefore, in the high-GC group, there has been an excess of (A/T)Mouse → (G/C)Human replacements over (G/C)Mouse → (A/T)Human at each codon position individually. For the low-GC group, the reverse situation has been encountered (Table 5), i.e. there is a tendency for G and C in mouse genes to be replaced by A or T in their human orthologs, the bias being maximal at the third codon positions. For the medium-GC group, however, no significant difference between (A/T)Mouse → (G/C)Human and (G/C)Mouse → (A/T)Human replacements could be observed at the first and second codon sites, whereas for the third codon sites, the (A)Mouse → (G)Human and (T)Mouse → (C)Human replacements dominate over the reverse replacements. These observations imply that for the high-GC group, either the GC content tends to increase in human genes relative to mouse or tends to decrease in mouse genes relative to human, whereas for low-GC group, either there is a trend in relative decrease in GC content in human compared with mouse or there is a trend in relative increase in GC content in mouse compared with human. This suggests that with time, there is a relative increase in compositional heterogeneity within human genes compared with that within mouse genes or decrease in compositional heterogeneity within mouse genes compared with that within human genes.
Table 5.
Nucleotide replacement matrices at (NRMs) three codon positions for human mouse orthologs of high-, medium-, and low-GC groups under study
| Human |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | First codon position | Second codon position | Third codon position | |||||||||||
| Mouse | A | T | G | C | A | T | G | C | A | T | G | C | ||
| High-GC group | A | ― | 1.00 | 1.13 | 1.16 | ― | 1.01 | 1.08 | 1.06 | ― | 1.00 | 1.86 | 1.71 | |
| T | 1.00 | ― | 1.11 | 1.19 | 0.99 | ― | 1.06 | 1.06 | 1.00 | ― | 1.69 | 1.86 | ||
| G | 0.88 | 0.90 | ― | 1.01 | 0.92 | 0.95 | ― | 0.98 | 0.54 | 0.59 | ― | 1.01 | ||
| C | 0.86 | 0.84 | 0.99 | ― | 0.94 | 0.95 | 1.02 | ― | 0.58 | 0.54 | 0.99 | ― | ||
| Medium-GC group | A | ― | 1.01 | 1.02 | 1.03 | ― | 0.99 | 1.01 | 1.01 | ― | 1.00 | 1.04 | 1.01 | |
| T | 0.99 | ― | 1.03 | 1.02 | 1.01 | ― | 1.02 | 1.02 | 1.00 | ― | 1.02 | 1.03 | ||
| G | 0.98 | 0.97 | ― | 1.00 | 0.99 | 0.98 | ― | 1.00 | 0.97 | 0.98 | ― | 1.00 | ||
| C | 0.97 | 0.98 | 1.00 | ― | 0.99 | 0.98 | 1.00 | ― | 0.99 | 0.97 | 1.00 | ― | ||
| Low-GC group | A | ― | 1.01 | 0.93 | 0.90 | ― | 0.99 | 0.90 | 0.94 | ― | 0.99 | 0.64 | 0.65 | |
| T | 0.99 | ― | 0.93 | 0.83 | 1.01 | ― | 0.94 | 0.95 | 1.01 | ― | 0.70 | 0.62 | ||
| G | 1.08 | 1.07 | ― | 0.97 | 1.11 | 1.06 | ― | 1.02 | 1.56 | 1.42 | ― | 0.96 | ||
| C | 1.11 | 1.20 | 1.03 | ― | 1.07 | 1.06 | 0.98 | ― | 1.53 | 1.62 | 1.04 | ― | ||
Each element rij in the NRM represents the ratio of number of replacements of the nucleotide i by the nucleotide j in the forward direction (mouse to human) to that in the reverse direction. This means that if rij > 1, the number of (i)Mouse → (j)Human replacements is higher than the number of (j)Mouse → (i)Human replacements and if rij < 1, the reverse is true. Bold and bold-italics ratios signifies the directional bias at p < 0.0001 and 0.001, respectively.
Our next task was to see to what extent the observed trends in nucleotide substitution patterns have affected the relative GC divergence between mouse and human orthologs. To this end, the number of genes was plotted against their GC12 and GC3 values both for mouse and for human in all three groups of orthologs (Fig. 2) using STATISTICA (version 6.0). In all cases, normal distributions were obtained (Fig. 2). In high-GC group, both GC12 and GC3 distributions in human are skewed towards right (increasing GC contents) compared with mouse (Fig. 2A and B), but for low-GC group, the reverse is true (Fig. 2E and F). The extent of inter-species divergence in GC distribution is much more apparent in case of third codon positions (Fig. 2B, D, and F) compared with the first and second positions (Fig. 2A, C, and E). For medium-GC orthologs, medians of both GC12 and GC3 distributions are almost same in both species under study (Fig. 2C and D). These observations imply that the intra-species divergence in base composition is higher in case of human genes than that in their mouse orthologs such that among the GC-rich pairs of orthologs, human coding sequences are usually higher in GC content than their mouse counterparts, but among the GC-poor orthologous pairs, human coding sequences are, in general, lower in GC content than the respective mouse sequences.
Figure 2.
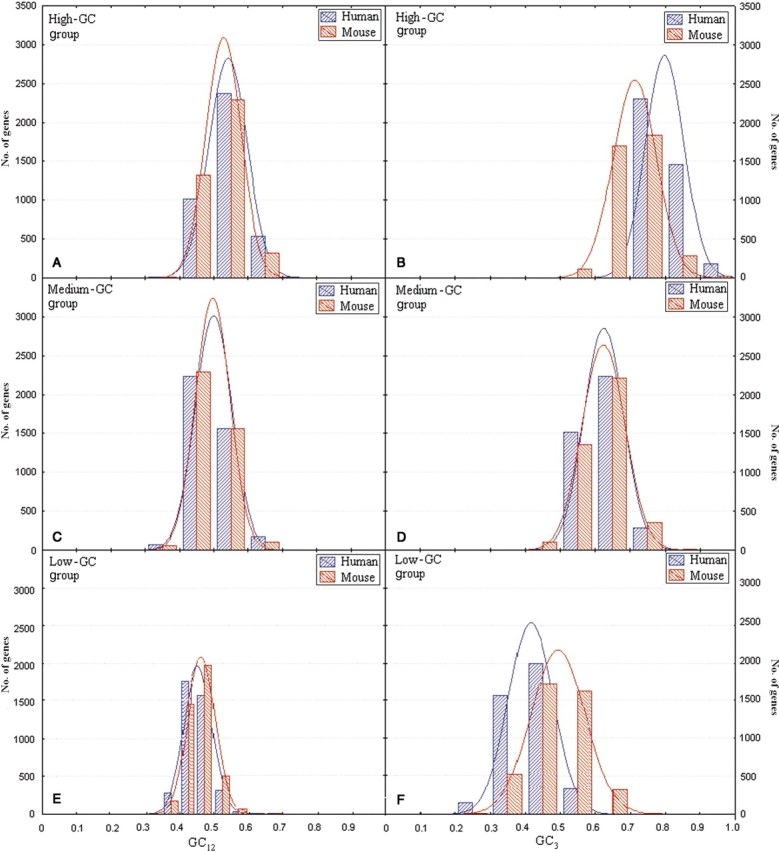
Left panel: distribution of GC content at first and second codon positions among human (blue) and mouse (red) orthologous genes for (A) high-, (C) medium-, and (E) low-GC group with their normal distributions. Right panel: distribution of GC content at third codon position among human and mouse orthologous genes for (B) high-, (D) medium-, and (F) low-GC group with their normal distributions.
3.5. Multivariate analysis of synonymous codon usage confirms opposite trends in high-GC and low-GC groups of orthologs
The skewness of GC3 in human genes towards increasing GC in high-GC group and decreasing GC in low-GC group compared with mouse orthologs (Fig. 2B, D, and F) has also been reflected in the COA of RSCU. Fig. 3A–C represents axis-1 versus axis-2 plot of the COA on RSCU of genes in three different groups. In all cases, axis-1 exhibits strong negative correlation with GC content at synonymous substitution sites (GC3S). The distribution of human and mouse genes along axis-1 confirms that in high-GC group (Fig. 3A), human genes exhibit higher usage of G or C ending synonymous codons compared with their mouse orthologs, whereas for low-GC group, the reverse trend dominates (Fig. 3C). For medium-GC group, as expected, usage of G/C-ending codons is comparable in mouse and human (Fig. 3B).
Figure 3.
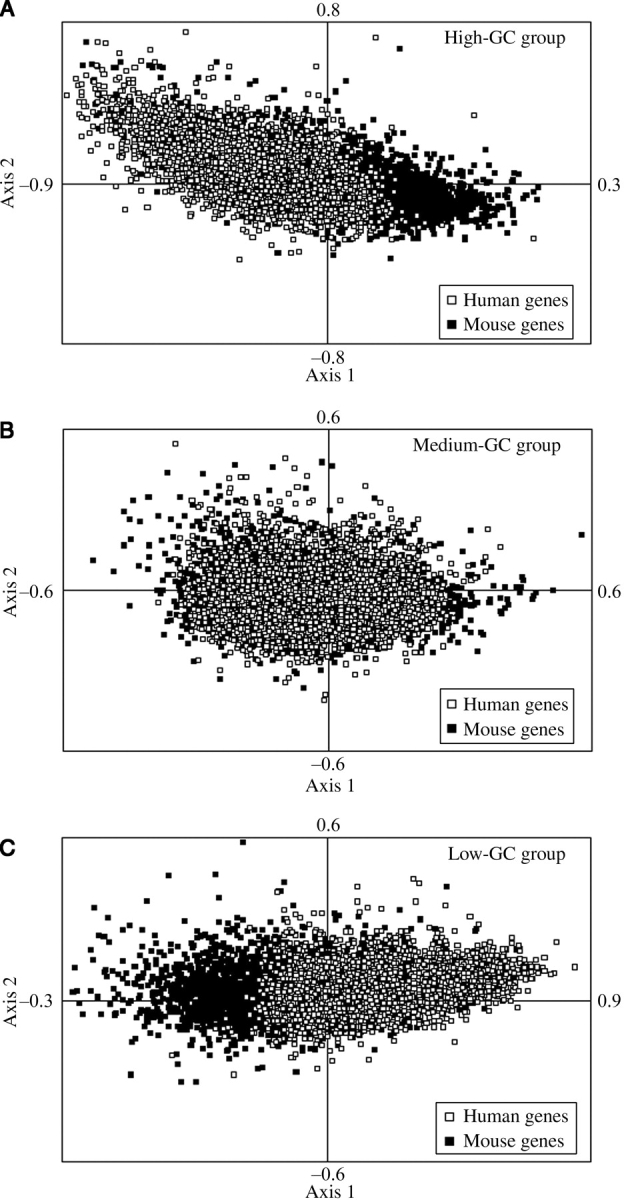
Positions of orthologous gene pairs between human and mouse along the first two identical principal axes generated by COA on RSCU values of (A) 3896 genes from high-GC group, (B) 3960 genes from medium-GC group, and (C) 4168 genes from low-GC group. The filled quadrangle and open quadrangle represent genes from mouse and human orthologous genes, respectively.
It is worth mentioning that the GC contents of the synonymous substitution sites in the mouse and human orthologous pairs exhibit negative correlations in all three groups (supplement-I). These observations are in accordance with the previous report by Takahashi and Nakashima.30
3.6. Rate of synonymous and nonsynonymous substitutions are same in all three groups of orthologs
In order to examine whether the rate of nucleotide substitution between mouse and human orthologs varies in three different groups, the number of synonymous substitutions per synonymous site, dS, and the number of nonsynonymous substitutions per nonsynonymous site, dN, were estimated for randomly selected 500 pairs of the orthologs from each group. The value of dS remains almost same in all three groups (data not shown). The value of dN apparently seems to be lower in the medium-GC group, but its difference with its values for the other two groups is not statistically significant. These observations indicate that although the trends in nucleotide substitutions are polarized to the opposite directions in the high- and low-GC groups of orthologs, the rates of synonymous or nonsynonymous substitutions did not vary with the GC bias of the genes.
4. Discussion
Since the divergence of the rodent and primate lineages, multiple substitutions might have occurred at the same site of a pair of mouse–human orthologs independently in two lineages. Had there not been a strong directionality of selection process(es) prevailing over the random mutational events, such multiple hits should have obscured the true pattern of substitution, if any, between such orthologous pairs. However, the present study has revealed that the nucleotide and amino acid substitution patterns in mouse–human orthologs have followed definite trends that are highly asymmetric and polarized to opposite directions in high- and low-GC groups, suggesting that indeed there has been a definite directionality in gene/protein evolution towards increasing compositional divergence in human protein-coding regions compared with that in mouse protein-coding regions or towards decreasing compositional divergence in mouse protein-coding regions compared with that in human protein-coding regions. It is true that the GC content shows evolutionary stability between mouse and human, i.e. orthologs have similar GC contents in two species, but among the high-GC orthologs, human proteins are slightly higher in GC content than their mouse orthologs, whereas among the low-GC orthologs, human proteins are slightly higher in AT content than their mouse counterparts.
A question may be raised at this point: why, of all mammalian species, only mouse and human were chosen as the species of study in the present report. The reason is as follows: initially we intended to analyze the sequence divergence patterns between the orthologous coding regions of human, chimpanzee, and rhesus monkey. However, the numbers of nonsynonymous substitutions between two orthologs of any two primate species were often too low to reveal any significant statistical trend. Therefore, we have decided to analyze the trends in substitution patterns between a rodent and a primate species, mouse and human have been chosen as the representative species of the two lineages.
As already mentioned in Section 2, the trends reported here are robust enough to be valid for any subset of the total datasets of orthologous sequences. Any trend in amino acid/nucleotide replacement between the pairs of orthologs of a particular dataset remains invariant, in general, when a subset of sequences are chosen randomly from that particular dataset. This indicates that same trends are usually followed individually by each pair of orthologs in a particular group (high-, medium-, or low-GC group).
The trends in amino acid and nucleotide replacement patterns also remained same when the orthologous sequences were classified in high-, medium-, and low-GC groups on the basis of the GC3 content of mouse genes instead of human genes. The same previous directionality was observed for high- or low-GC groups, i.e. GC content either increase in human genes relative to mouse or decrease in mouse genes relative to human for the high-GC group, whereas for low-GC group, either there is relative decrease in GC content in human genes compared with mouse gene or relative increase in GC content in mouse genes compared with human gene. This was, however, expected as the two genome sequences exhibit a one-to-one correspondence in their local GC content.
The only significant trend common in all three groups of orthologs is (Asp)Mouse → (Glu)Human. Surprisingly, the value of RDE is almost same in all three groups and the trend has also been exhibited by the subsets chosen randomly from the whole dataset of any particular compositional group. This indicates that this trend, in general, does not alter with the compositional bias or functional characteristics of the genes. In accordance with this, average frequency of Glu (7.01% for mouse and 7.11% for human) is significantly higher in human (p < 10−5) and that of Asp (4.90% for mouse and 4.81% for human) is significantly higher in mouse (p < 10−5). The structural consequence of this trend is, however, not clear.
No significant differences could be observed between the synonymous or nonsynonymous substitution rates in three groups of orthologs under study. This suggests that although the directionality of evolution in orthologs of two extreme GC compositions is oppositely polarized, the rate at which they evolve is almost same in both cases.
In a nutshell, the present study indicates that in comparison with mouse, the coding regions of the human genome have experienced an expansion, not shrinkage, in intra-species heterogeneity in local GC content. This observation, however, does not warrant the relative expansion of the human GC islands as a whole, since it would depend not only on the evolutionary trends of the coding region, but also on those of the noncoding regions. One should also remember that a relative increase in GC heterogeneity in human orthologs compared with mouse orthologs not necessarily implies an absolute increase in GC heterogeneity in human coding regions with evolution. In absolute sense, both human and mouse might have evolved towards decreasing compositional heterogeneity, the rate of decrease in heterogeneity being less in human than in mouse, or alternatively, both the species might be evolving towards increasing intra-species inhomogeneity, the rate of increase being higher in human relative to mouse.
Funding
Council of Scientific and Industrial Research (Project No. CMM 0017 to C.D and S.G); Department of Biotechnology, Government of India (BT/BI/04/055-2001 to S.K.B and S.P).
Supplementary Material
Acknowledgements
We are grateful to Dr. A. Pan, Indian Association for the Cultivation of Science, Kolkata, India, for critical reading of the manuscript.
Supplementary data
Supplementary data are available online at http://www.dnaresearch.oxfordjournals.org
References
- 1.Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- 2.Eyre-Walker A, Hurst L. D. The evolution of isochores. Nat. Rev. Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- 3.Filipski J, Thiery J. P, Bernardi G. An analysis of the bovine genome by Cs2SO4-Ag density gradient centrifugation. J. Mol. Biol. 1973;80:177–197. doi: 10.1016/0022-2836(73)90240-4. [DOI] [PubMed] [Google Scholar]
- 4.Hughes S, Zelus D, Mouchiroud D. Warm-blooded isochore structure in Nile crocodile and turtle. Mol. Biol. Evol. 1999;16:1521–1527. doi: 10.1093/oxfordjournals.molbev.a026064. [DOI] [PubMed] [Google Scholar]
- 5.Lander E. S, Linton L. M, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Caron H, van Schaik B, van der Mee M, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 7.Fullerton S. M, Bernardo Carvalho A, Clark A. G. Local rates of recombination are positively correlated with GC content in the human genome. Mol. Biol. Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- 8.Kong A, Gudbjartsson D. F, Sainz J, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 9.Lercher M. J, Urrutia A. O, Pavlicek A, Hurst L. D. A unification of mosaic structures in the human genome. Hum. Mol. Genet. 2003;12:2411–2415. doi: 10.1093/hmg/ddg251. [DOI] [PubMed] [Google Scholar]
- 10.Mouchiroud D, D'Onofrio G, Aissani B, Macaya G, Gautier C, Bernardi G. The distribution of genes in the human genome. Gene. 1991;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- 11.Saccone S, De Sario A, Wiegant J, Raap A. K, Della Valle G, Bernardi G. Correlations between isochores and chromosomal bands in the human genome. Proc. Natl. Acad. Sci. USA. 1993;90:11929–11933. doi: 10.1073/pnas.90.24.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterston R. H, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 13.Nei M, Glazko G. V. The Wilhelmine E. Key 2001 Invitational Lecture. Estimation of divergence times for a few mammalian and several primate species. J. Hered. 2002;93:157–164. doi: 10.1093/jhered/93.3.157. [DOI] [PubMed] [Google Scholar]
- 14.Glazko G. V, Koonin E. V, Rogozin I. B. Molecular dating: ape bones agree with chicken entrails. Trends Genet. 2005;21:89–92. doi: 10.1016/j.tig.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Arndt P. F, Petrov D. A, Hwa T. Distinct changes of genomic biases in nucleotide substitution at the time of Mammalian radiation. Mol. Biol. Evol. 2003;20:1887–1896. doi: 10.1093/molbev/msg204. [DOI] [PubMed] [Google Scholar]
- 16.Duret L, Semon M, Piganeau G, Mouchiroud D, Galtier N. Vanishing GC-rich isochores in mammalian genomes. Genetics. 2002;162:1837–1847. doi: 10.1093/genetics/162.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith N. G, Webster M. T, Ellegren H. Deterministic mutation rate variation in the human genome. Genome Res. 2002;12:1350–1356. doi: 10.1101/gr.220502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster M. T, Smith N. G, Ellegren H. Compositional evolution of noncoding DNA in the human and chimpanzee genomes. Mol. Biol. Evol. 2003;20:278–286. doi: 10.1093/molbev/msg037. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez-Valin F, Clay O, Cruveiller S, Bernardi G. Inaccurate reconstruction of ancestral GC levels creates a ‘vanishing isochores’ effect. Mol. Phylogenet. Evol. 2004;31:788–793. doi: 10.1016/j.ympev.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Gu J, Li W. H. Are GC-rich isochores vanishing in mammals? Gene. 2006;385:50–56. doi: 10.1016/j.gene.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 21.Vallender E. J, Paschall J. E, Malcom C. M, Lahn B. T, Wyckoff G. J. SPEED: a molecular-evolution-based database of mammalian orthologous groups. Bioinformatics. 2006;22:2835–2837. doi: 10.1093/bioinformatics/btl471. [DOI] [PubMed] [Google Scholar]
- 22.Penden J, Sharp P. M. CodonW. 1997;v. 1.4.2:1.4.2. [Google Scholar]
- 23.Aissani B, D'Onofrio G, Mouchiroud D, Gardiner K, Gautier C, Bernardi G. The compositional properties of human genes. J. Mol. Evol. 1991;32:493–503. doi: 10.1007/BF02102651. [DOI] [PubMed] [Google Scholar]
- 24.Bernardi G, Olofsson B, Filipski J, et al. The mosaic genome of warm-blooded vertebrates. Science. 1985;228:953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J. D, Higgins D. G, Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp P. M, Li W. H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 28.Singer G. A, Hickey D. A. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 2000;17:1581–1588. doi: 10.1093/oxfordjournals.molbev.a026257. [DOI] [PubMed] [Google Scholar]
- 29.Wang H. C, Singer G. A, Hickey D. A. Mutational bias affects protein evolution in flowering plants. Mol. Biol. Evol. 2004;21:90–96. doi: 10.1093/molbev/msh003. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi N, Nakashima H. Negative correlation of G + C content at silent substitution sites between orthologous human and mouse protein-coding sequences. DNA Res. 2006;13:135–140. doi: 10.1093/dnares/dsl007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


