Abstract
Background and Aims
Cedrus (true cedars) is a very important horticultural plant group. It has a disjunct distribution in the Mediterranean region and western Himalaya. Its evolution and biogeography are of great interest to botanists. This study aims to investigate the phylogeny and biogeography of Cedrus based on sequence analyses of seven cytoplasmic DNA fragments.
Methods
The methods used were PCR amplification and sequencing of seven paternal cpDNA and maternal mtDNA fragments, parsimony and maximum likelihood analyses of the DNA dataset, and molecular clock estimate of divergence times of Cedrus species.
Key Results
Phylogenies of Cedrus constructed from cpDNA, mtDNA and the combined cp- and mt-DNA dataset are identical in topology. It was found that the Himalayan cedar C. deodara diverged first, and then the North African species C. atlantica separated from the common ancestor of C. libani and C. brevifolia, two species from the eastern Mediterranean area. Molecular clock estimates suggest that the divergence between C. atlantica and the eastern Mediterranean clade at 23·49 ± 3·55 to 18·81 ± 1·25 Myr and the split between C. libani and C. brevifolia at 7·83 ± 2·79 to 6·56 ± 1·20 Myr.
Conclusions
The results, combined with palaeogeographical and palaeoecological information, indicate that Cedrus could have an origin in the high latitude area of Eurasia, and its present distribution might result from vicariance of southerly migrated populations during climatic oscillations in the Tertiary and further fragmentation and dispersal of these populations. It is very likely that Cedrus migrated into North Africa in the very late Tertiary, while its arrival in the Himalayas would not have been before the Miocene, after which the phased or fast uplift of the Tibetan plateau happened.
Key words: Cedrus, molecular phylogeny, biogeography, Mediterranean, Himalayas, molecular clock
INTRODUCTION
Mountain regions between the fortieth parallels of latitude are of interest to biogeographers due to their importance in the survival of many plant species during the Ice Ages (Hewitt, 2004). The Mediterranean is one of 18 world hotspots of biodiversity, where approx. 25 000 plant species, of which 50 % are endemics, are harboured (Cowling et al., 1996; Myers et al., 2000; Scarascia-Mugnozza et al., 2000; Comes, 2004). Phylogeographical studies have provided strong evidence that some refugia were available in montane Mediterranean during the Quaternary, and also shaped the distribution of modern biota (Svenning, 2003; Comes, 2004; Hellwig, 2004). Unfortunately, the origin of the Mediterranean flora is still obscure (Valcárcel et al., 2003). In Asia, the uplift of the Himalayas and the onset of the Asian monsoon changed the world climate (Griffin, 2002; Liu and Yin, 2002), and made the Tibetan plateau, especially its south-eastern part, an important centre for not only the survival of many arctic plants, when they migrated southwards, but also the speciation of various organisms (Axelrod et al., 1996; Myers et al., 2000). It is of great interest to investigate the floristic relationship between the Mediterranean and the Tibetan plateau. Moreover, the significant geological and climatic changes in the two regions during the Cenozoic (Mai, 1989; LePage, 2003; Hellwig, 2004) produced many disjunct distributions (Sanmartín, 2003; Comes, 2004; Hellwig, 2004), and thus give us opportunities to test the dispersal and vicariance theories of biogeography.
Cedrus (true cedar) is one of 11 commonly accepted genera in Pinaceae, first described by Trew in 1757 (Farjón, 2001). It comprises four species with a highly disjunct distribution in circum-Mediterranean and western Himalayas (Farjón, 1990, 2001), i.e. Cedrus deodara (Roxb.) G. Don in the Hindu Kush, Karakoram and Indian Himalayas, Cedrus libani A. Rich. in Turkey, Lebanon and Syria, Cedrus brevifolia (Hook. f.) Henry in Cyprus, and Cedrus atlantica (Endl.) Manetti ex Carriére in North Africa (Algeria, Morocco). The true cedars occurring around the Mediterranean basin were once treated as one species C. libani, including four subspecies, namely ssp. atlantica, ssp. brevifolia, ssp. stenocoma in Turkey and ssp. libani in Lebanon and Syria (Davis, 1965). Some authors divided Cedrus into three species (cf. Scaltsoyiannes, 1998), regarding C. brevifolia as a subspecies of C. libani (Meikle, 1977). The great debate in species delimitation and phylogeny of Cedrus remains unresolved, although previous cytological (Li and Fu, 1995; Bou Dagher-Kharrat et al., 2001), biochemical (Scaltsoyiannes, 1998) and gene flow analyses (Fady et al., 2003) contributed greatly to the interpretation of interspecific relationships of true cedars.
In recent years, sequence analysis of multiple genes from different genomes has been successfully used in reconstructing complex phylogenies, meanwhile providing important information to the inference of biogeographical histories of many plant groups (e.g. Baum et al., 1998; Kusumi et al., 2002; Schneeweiss et al., 2004; Winkworth and Donoghue, 2005; Xiang et al., 2005). The combined gene analysis has great advantages in some conifer groups such as Pinaceae (Wang et al., 2000), where chloroplast and mitochondria are paternally and maternally inherited, respectively (Hipkins et al., 1994; Birky, 1995; Mogensen, 1996; Ahuja, 2001). The joint cp- and mt-DNA analysis can detect interspecific gene flow and reticulate evolution. However, no DNA sequence-based phylogeny has been constructed for Cedrus, a very important horticultural plant group. On the other hand, the systematic position of Cedrus in Pinaceae is also quite controversial, like the difficulty in species delimitation of the genus. Based on leaf arrangement, the presence of short shoots, and the petiole and pulvinus form, Cedrus was placed in the subfamily Laricoideae, with Larix and Pseudolarix (Cheng and Fu, 1978; Krüssmann, 1985). This classification was not supported by the subsequent anatomical (Hart, 1987; Frankis, 1988; Wu and Hu, 1997), immunological (Price et al., 1987) and cytological studies (Li, 1995). In particular, Cedrus does not have the same position in molecular phylogenies constructed by different markers such as PCR–RFLP (Tsumura et al., 1995) and DNA sequence (Quinn et al., 2002), and may represent a very distinctive lineage (Wang et al., 2000). The reconstruction of a robust phylogeny of Cedrus comprising all species could also shed some light on the origin of the genus.
To resolve interspecific relationships of plants, sequences of intergenic spacers and gene introns are most widely used because of their fast evolutionary rates (Wang et al., 1999; Wei and Wang, 2003; Shaw et al., 2005). Here, the phylogeny of true cedars based on sequences of seven cytoplasmic DNA fragments, including five from the paternal chloroplast genome and two from the maternal mitochondrial genome, is reconstructed, and the intrageneric classification, divergence times and biogeography of Cedrus are discussed.
MATERIALS AND METHODS
Plant materials
All four species of Cedrus were sampled, and each species was represented by two individuals. To investigate whether the molecular markers we used have infraspecific variation, six individuals of C. libani that are at least 100 m apart were further analysed. Pinus armandii Franchet and Cathaya argyrophylla Chun et Kuang, each represented by one individual, were chosen as outgroups based on molecular phylogenies of Pinaceae (Wang et al., 1998, 2000; Quinn et al., 2002). The origins of materials are shown in Table 1.
Table 1.
Sources of material
| Taxa | Origins | Individuals/vouchers |
|---|---|---|
| Ingroups | ||
| Cedrus atlantica (Endl.) Manetti ex Carriére | Natural History Museum, Budapest, Hungary (cultivated) | 2/Folly acb 1905, Debreczy-ca1 |
| Cedrus brevifolia (Hook. f.) Henry | Natural History Museum, Budapest, Hungary (cultivated) | 1/Debreczy-cb1 |
| Cyprus | 1/Debreczy-cb2 | |
| Cedrus deodara (Roxb.) G. Don | Botanic Garden, Institute of Botany, Beijing, China (cultivated) | 2/Wang-cd1, Wang-cd2 |
| Cedrus libani A. Rich. | Elmali, Antalya, Turkey | 7/Wang-cl-15 |
| Taurus Mt, Turkey | 1/Debreczy-cl1 | |
| Outgroups | ||
| Pinus armandii Franchet | Botanic Garden, Institute of Botany, Beijing, China (cultivated) | 1/Wang 6102 |
| Cathaya argyrophylla Chun et Kuang | Jinfo Mountain, Nanchuan, Sichuan, China | 1/WangXQ94513 |
DNA sequences of these materials determined in the present study have been deposited in GenBank under accession numbers DQ983599–DQ983604 (nad1), DQ983605–DQ983610 (nad5), DQ983611–DQ983616 (psbB-psbH), DQ983617–DQ983622 (rpL16), DQ983623–DQ983628 (5′rps12-rpL20), DQ983629–DQ983634 (trnC-trnD) and DQ983635–DQ983640 (trnT-trnF). All vouchers are deposited in PE (Herbarium, Institute of Botany, the Chinese Academy of Sciences, Beijing).
DNA extraction, PCR amplification and sequencing
Total DNA was extracted from silica gel-dried needles using the CTAB method following the protocol of Rogers and Bendich (1988) and used as the template in the polymerase chain reaction. The seven cytoplasmic DNA fragments used were chloroplast trnT-trnF (Wei and Wang, 2003), trnC-trnD (Lee and Wen, 2004), 5′rps12-rpL20, psbB-psbH and rpL16 intron (Shaw et al., 2005), and mitochondrial nad1 intron 2 (Song et al., 2002; Won and Renner, 2003) and nad5 intron 1 (Jaramillo-Correa et al., 2003). These gene regions were amplified using the primers listed in Table 2. The PCR reaction was carried out in a volume of 25 µL containing 5–50 ng of DNA template, 6·25 pmol of each primer, 0·2 mm of each dNTP, 2 mm MgCl2, and 0·75 U of Taq DNA polymerase. Amplification was conducted in a Tpersonal Thermocycle and T1 Thermocycle (Biometra, Goettingen, Germany). PCR cycles were as follows: one cycle of 4 min at 70 °C, four cycles of 40 s at 94 °C, 20 s at 52–58 °C, and 2–2·5 min at 72 °C, followed by 36 cycles of 20 s at 94 °C, 20 s at 50–56 °C, and 2–2·5 min at 72 °C, with a final extension step for 10 min at 72 °C. PCR products were separated by 1·5 % agarose gel electrophoresis. The band with the right size was cut out and purified using the GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Amersham, UK). Sequencing reactions were performed with the two PCR primers listed above and several internal primers using the DYEnamic Energy Transfer (ET) Terminator Reagent Premix Kit (Amersham Biosciences). The internal primers included c (Wei and Wang, 2003) for the trnT-trnF region, petN2G (5′-CTTGGGCTGCTTTAATGGT AG-3′, forward), psbM2GF (5′-GTAGAGCAGCAATAAATGCAAG-3′, forward), psbM2GR (5′-CTTGCATTTATTGCTGCTCTAC-3′, reverse) and petN3G (5′-ATGGTACGAGGTCCTTCATCC-3′, forward) for the trnC-trnD region, and nad5-IF (5′-GGCTTTAGGGGGCCTTATG-3′, forward) for the nad5 intron 1. petN2G, psbM2GF and psbM2GR were modified from petN2, psbM2 and psbM2R of Lee and Wen (2004), respectively. After precipitation in 95 % EtOH and 3 m NaAc (pH 5·2), the sequencing products were separated on a MegaBACE 1000 automatic sequencer (Amersham Biosciences).
Table 2.
Sequences and references for chloroplast and mitochondrial DNA primers used in the present study
| Region | Forward primer | Sequence (5′–3′) | Reserve primer | Sequence (5′–3′) | Reference |
|---|---|---|---|---|---|
| cpDNA | |||||
| psbB–psbH | psbB(F) | TCCAAAAACTGGGAGATCCAAC | psbH(R) | TCAATGGTCTGTGTAGCCAT | Shaw et al., 2005* |
| rpL16 | rpL16F | GCTATGCTTAGTGTGCGACTCGTTG | rpL16R | CCYTTCATTCTTCCCCTATGTTG | Small et al., 1998* |
| 5′rpS12–rpL20 | 5′rpS12(F) | ATTAGAAATGCAAGACAGCCAAT | rpL20(R) | CGTTTTCGRGCTATA/GTATCC | Shaw et al., 2005* |
| trnC–trnD | trnC | CCAGTTCGAATCCGGGTGTC | trnD | GGGATTGTAGCTCAATTGGT | Demesure et al., 1995* |
| trnT–trnF | trnT-F(F) | CATTACAAATGCGATGCTCT | trnT-F(R) | ATTTGAACTGGTGACACGAG | Taberlet et al., 1991* |
| mtDNA | |||||
| nad1 intron2 | nad1F1 | GATCGGCCATAAATGTACTCC | nad1R1 | CCCCATATATTCCCGGAGC | Song et al., 2002 |
| nad5 inton 1 | nad5-aF | GGAAATGTTTGATGCTTCTTGGG | nad5-bR | CTGATCCAAAATCACCTACTCG | Wang et al., 2000 |
* Primers that have been modified in this study.
Data analysis
Sequence alignments were made with CLUSTAL X (Thompson et al., 1997) and refined manually for the maximization of sequence homology using BioEdit 5.0.9 (Hall, 1999). Maximum parsimony analysis was performed using PAUP version 4.0b10 (Swofford, 2002) with Pinus armandii and Cathaya argyrophylla as outgroups (Wang et al., 2000). Branch-and-bound searches were conducted with the MULTREES option. Indels in the alignment induced by the ingroups were coded as 1/0 binary characters, and gaps of different lengths were all treated as single events. All character states were specified as unordered and equally weighted. To evaluate relative robustness of the clades found in the most-parsimonious trees, the non-parametric bootstrap analysis (Felsenstein, 1985) was performed with 1000 replicates using the same search settings. The incongruence length difference test (Farris et al., 1995) was conducted, using the partition homogeneity test in PAUP ver. 4.0b10 (Swofford, 2002), to examine the congruence between different datasets. Test settings were 100 random stepwise additions and 1000 replicates of heuristic search with TBR branch swapping using two outgroups.
Owing to the large length variation in the mitochondrial nad1 intron 2, only the combined chloroplast dataset was used to estimate divergence times of Cedrus species. To estimate branch lengths, sequences of the ingroups were fitted using maximum likelihood (ML) models estimated on the topology of the parsimony trees. Optimal ML models for the combined data were selected using Modeltest 3.06 (Posada and Crandall, 1998). To estimate ages of nodes, first the hypothesis of a molecular clock was tested for the dataset using a Likelihood Ratio Test (LRT) that compared likelihood scores of the ML phylogenetic hypothesis with (L1) and without (L0) the molecular clock enforced. Significance was assessed by comparing two times the difference in log likelihoods to a chi-square distribution with n – 2 degrees of freedom, where n was the number of ingroups. When LRT showed that the data did not adhere to a molecular clock, non-parametric rate smoothing (NPRS) (Sanderson, 1997) in the program TreeEdit (http://evolve.zoo.ox.ac.uk/software/TreeEdit/; Rambaut and Charleston, 2002) transformed ML trees into ultrametric topologies. All node heights, sum of the branch lengths from a node to a tip, were transformed from units of molecular change into units of time by calibration to a node of known age. In addition, standard errors for estimates of node age were calculated for two nodes by using the non-parametric bootstrap resampling method (100 replicates) in PAUP*.
Since the NPRS method does not use rate smoothing, penalized likelihood (PL) analysis (Sanderson, 2002) was used additionally to estimate the divergence times of Cedrus. The ages of the nodes in the tree were estimated using penalized likelihood rate smoothing under TN algorithm with the program r8s version 1.7 (Sanderson, 2003, available at http://ginger.ucdavis.edu/r8s). A cross-validation analysis was performed to obtain the most likely smoothing parameter. Standard errors of the divergence times were estimated by a three-step nonparametric bootstrap procedure (Efron and Tibshirani, 1993): (1) 100 data sets were simulated by using the SEQBOOT program in PHYLIP Version 3.6a2 (Felsenstein, 1993); (2) the matrices were imported into PAUP version 4.0b10 and ML trees were generated; (3) the tree file was processed by the program r8s, which summarizes the bootstrap distribution of divergence times for each node that were used to calculate standard errors.
Although fossil and geological event can both be selected as a calibration point for time estimation, the latter as the calibration point may prove to be circular. The earliest identifiable cedar fossil dates back to the Paleocene (Blokhina, 1998), and therefore a minimum date of 54.8 Myr was applied to the node connecting Cedrus to the outgroup in the present study.
RESULTS
The five cpDNA fragments, psbB-psbH, rpL16 intron, 5′rps12-rpL20, trnC-trnD and trnT-trnF, are 404–409, 799, 705, 2253–2259 and 1336 bp in length, respectively (Table 3). No intraspecific sequence variation was detected. In the trnC-trnD region, three 5-bp gaps, one shared by C. libani, C. brevifolia and C. atlantica, one shared by C. deodara and C. atlantica and another one unique to C. deodara, were found. In addition, a 5-bp gap occurred in the psbB-psbH region of C. atlantica that included a 256-bp coding sequence. The partition-homogeneity test indicated that data sets of these cpDNA fragments were significantly congruent (P = 0·996). The combined cpDNA data set had 5875 characters, of which 697 were variable and 198 were parsimony-informative.
Table 3.
Summary of sequence variation in cp- and mt-DNA regions of the genus Cedrus
| Region | Ingroups length (bp) | Alignment length (bp) | Substitution sites (informative) |
Indels in ingroups (informative) | |
|---|---|---|---|---|---|
| Ingroups + outgroups | Ingroups | ||||
| psbB-psbH | 404–409 | 415 | 24 (4) | 0 (0) | 1 (0) |
| rpL16 | 799 | 824 | 82 (30) | 1 (0) | 0 (0) |
| 5′rpS12-rpL20 | 705 | 706 | 52 (16) | 1 (0) | 0 (0) |
| trnC-trnD | 2253–2259 | 2513 | 368 (90) | 13 (2) | 3 (1) |
| trnT-trnF | 1336 | 1417 | 171 (58) | 3 (0) | 0 (0) |
| cp regions combined | 5497–5508 | 5875 | 697 (198) | 18 (2) | 4 (1) |
| nad1 intron 2 | 530–852 | 622* | 54 (8) | 8 (0) | 2 (2) |
| nad5 intron 1 | 1252 | 1266 | 59 (28) | 10 (1) | 0 (0) |
| mt regions combined | 1782–2104 | 1888 | 113 (36) | 18 (1) | 2 (2) |
* Some regions that could not be aligned have been removed.
The two mtDNA fragments, nad1 intron 2 (partial sequence) and nad5 intron 1, are 530–852 and 1252 bp in length, respectively (Table 3). No intraspecific sequence variation was detected. In the nad1 intron 2, two deletions, relative to C. deodara and outgroups, were found. One of 116 bp occurred in C. atlantica, and the other one of 322 bp was shared by C. libani and C. brevifolia. The partition-homogeneity test indicated that data sets of the two mtDNA regions were fully congruent (P = 1). The combined mtDNA data set was 1888 bp in length after removing some sequences that could not be aligned, including 113 variable sites, of which 36 were parsimony-informative.
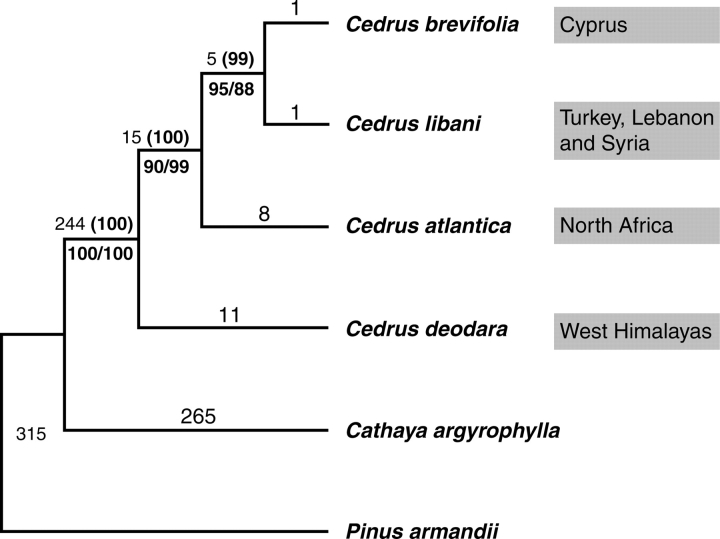
Parsimony analysis of the combined cpDNA dataset generated only one most-parsimonious tree with a tree length of 743, a consistency index (CI) of 0·9960 and a retention index (RI) of 0·9851 (tree not shown). Phylogenetic analysis of the combined mtDNA dataset also found single most-parsimonious tree (tree length = 122, CI = 0·9590, RI = 0·8780) (tree not shown). A further partition-homogeneity test between cpDNA and mtDNA data sets suggested that they were completely congruent (P = 1). Parsimony analysis of the combined cp- and mt-DNA data set yielded a single most-parsimonious tree (tree length = 865, CI = 0·9908, RI = 0·9671) topologically the same as the cp- and mt- DNA trees, and all clades of the tree were robustly supported by the bootstrap analysis (Fig. 1). It was found that the Himalayan cedar C. deodara diverged first, and then the North African species C. atlantica separated from the common ancestor of C. libani and C. brevifolia, two species from the eastern Mediterranean area.
Fig. 1.
The single most-parsimonious tree obtained from sequence analysis of combined cp- and mt-DNA regions (length = 865, CI = 0·9908, RI = 0·9671). Numbers above the branches denote branch lengths and bootstrap values (in parentheses), respectively. Numbers below the branches are bootstrap values above 50 % yielded in the cp- (left) and mt- (right) DNA trees, respectively. The present distribution of each species was indicated on the right.
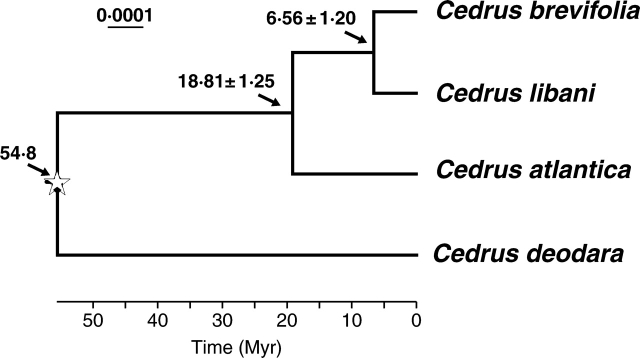
When an LRT compared likelihood scores under F81 (the best model given by Akaike Information Criterion), with and without the clock assumption on the ML tree obtained from the combined chloroplast data, the molecular clock was rejected in favour of the null hypothesis of no clock (δ = 2·62614, d.f. = 2, P > 0·05). Thus, a time-scaled NPRS ultrametric tree estimated under TreeEdit, using the topology with initial branch lengths estimated under maximum likelihood (Ribera et al., 2004), was generated and shown in Fig. 2. On the basis of a crown group age estimate of 54·8 Myr, molecular clock estimates suggested the divergence between the North African C. atlantica and the eastern Mediterranean clade comprising C. libani and C. brevifolia at 18·81 ± 1·25 Myr and the split between C. libani from Turkey, Lebanon and Syria and C. brevifolia from Cyprus at 6·56 ± 1·20 Myr. In the PL analysis, divergence times of the above nodes were estimated to be 23·49 ± 3·55 and 7·83 ± 2·79 Myr, respectively.
Fig. 2.
Cedar phylogeny and time scale of evolution determined by NPRS based on the combined chloroplast DNA data, with a calibration point of 54·8 Myr at the root node (indicated by a star) where Cedrus diverged from the outgroups. Standard errors for estimates of node ages are given for two nodes.
DISCUSSION
Intrageneric classification and phylogeny of Cedrus
Based on morphological and anatomical characteristics, all taxonomists accept the Himalaya cedar C. deodara, but classification of the circum-Mediterranean cedars is quite controversial (Davis, 1965; Meikle, 1977; Farjón, 1990, 2001; Scaltsoyiannes, 1998). Davis (1965) treated the circum-Mediterranean cedars as a single species, C. libani. Tutin et al. (1964) and Meikle (1977) recognized the North African cedar as a separate species C. atlantica based on its downy young twigs and small female cones. According to a combination of morphological characters such as angle of branch, cone size, male strobili length, leaf length, number of leaves in pseudo-whorls, together with native distributions, Farjón (1990, 2001) divided the circum-Mediterranean cedars into three closely related species, i.e. C. atlantica, C. brevifolia and C. libani. The external morphology is often affected by biotic and environmental factors (Davis, 1965; Meikle, 1977; Farjón, 1990; Pijut, 2000), and many characters vary continuously among cedar species, which could be responsible for the difficulty of intrageneric division of Cedrus.
Through terpene composition analysis, Canard et al. (1997) found a clear differentiation between C. libani and C. atlantica. This finding is further supported by the isozyme analysis of Scaltsoyiannes (1998), in which the dendrogram of genetic distances of 21 cedar populations comprises five distinct groups corresponding to C. libani ssp. libani, C. libani ssp. stenocoma, C. libani ssp. brevifolia, C. deodara and C. atlantica, respectively. Furthermore, fluorochrome banding patterns indicate that C. atlantica could be recognized as a separate species, and C. brevifolia as a subspecies of C. libani, although the three species are closely related (Bou Dagher-Kharrat et al., 2001). Based on open- and controlled-pollinations, as well as analyses using nuclear and cytoplasmic markers, Fady et al. (2003) found that gene flow occurs naturally between Cedrus taxa in plantation forests, and there are no strong pre-zygotic reproductive isolating barriers among Mediterranean cedars, when sympatric. Therefore, they proposed that Mediterranean Cedrus should be considered as units of a single collective species comprising two regional groups, North Africa and the Middle East.
The distinctness of C. deodara and the close relationships among the Mediterranean cedars are further corroborated by the present cp- and mt-DNA analyses. We found that the Himalaya cedar C. deodara has a basal position, and C. atlantica is sister to the clade comprising C. libani and C. brevifolia (Fig. 1). The tree topology, together with branch length, also supports North African C. atlantica as a separate species, and the reciprocal monophyly of Cedrus species could be the result of geographical isolation, which prevents interspecific gene flow from occurring.
Biogeography of Cedrus
Distribution patterns of organisms are greatly influenced by climatic and geological changes as well as their evolutionary histories. Climate tolerance is a crucial factor for plant distribution, in particular cold and aridity for trees (Pither, 2003; Svenning, 2003). From the beginning of the Tertiary, the global climate changed from extremes of expansive warmth with ice-free poles to extremes of cold with massive continental ice-sheets and polar ice caps, including several warm periods during the mid-Paleocene to early Eocene, and mid-Miocene (Zachos et al., 2003). The climatic oscillation provided opportunities for biota dispersal and subdivision (Mai, 1989; LePage, 2003; Sanmartín, 2003; Spicer et al., 2003; Comes, 2004; Hellwig, 2004; Vasiliev et al., 2004).
Farjón (1990) proposed that the highly disjunct distribution of apparently very closely related taxa of Cedrus could come from contraction of a formerly more extensive range of possibly one or two species, and a possible migration route that lay across the so called Thetys Fold Belt, a series of late Tertiary mountain uplifts along the brink of the old Thetys Sea, once extending from south-east Asia to north-west Africa. However, true cedars have reliable fossil records from the Oligocene of western Kazakhstan, the Miocene of south-west Europe, the Pliocene of south-east Europe, and the early Pleistocene of the Ahaggar Massif in the central Sahara (Florin, 1963; Farjón, 1990), in addition to fossil wood from the Paleocene of Kamchatka in Russian Far East (Blokhina, 1998, 2005). The fact that the fossils are progressively younger from north to south suggests an origin of Cedrus in the high latitude area of Eurasia. Its present distribution in several isolated regions could result from vicariance of southerly migrated populations during climatic oscillation in the Tertiary and further fragmentation and dispersal of these populations. This inference is robustly supported by the molecular phylogenies constructed in this study (Fig. 1), in which the Himalaya cedar is sister to the clade comprising Mediterranean cedars that was further divided into two subclades, i.e. the North African subclade and the subclade consisting of C. libani and C. brevifolia, two species from the eastern Mediterranean area.
The ancestor of Mediterranean cedars might have reached south Europe in the Miocene, which is supported by the fossil evidence mentioned above and the molecular clock estimate of 23·49 ± 3·55 Myr (PL) to 18·81 ± 1·25 Myr (NPRS) for the divergence between the North African C. atlantica and the eastern Mediterranean clade comprising C. libani and C. brevifolia (Fig. 2). The eastward dispersal of this lineage could be finally stopped by aridity in Asia Minor (Wulff, 1944; Griffin, 2002; Liu and Yin, 2002). On the other hand, biological communications between Europe and north Africa could have occurred through the Iberian connection during the Messinian (7–5 Mya) as a consequence of the desiccation of the Tethys, the Italian connection and the Arabian connection during the Miocene (Mai, 1989; Fici, 1991; Geraads, 1998; Krijgsman, 2002; Sanmartín, 2003; Hellwig, 2004). The last two connections, however, were unavailable to Cedrus for the presence of Saharan and Arab-Syrian deserts (Griffin, 2002; Liu and Yin, 2002; Sanmartín, 2003). It is very likely that Cedrus migrated into North Africa in the very late Tertiary, although C. atlantica was isolated from the eastern Mediterranean clade as early as the Miocene. The arrival of Cedrus in the Himalayas should not have been before the Miocene, whenafter the phased or fast uplift of the Tibetan plateau happened as suggested by recent studies, although there are a lot of debates on the time of the uplift (Dettman et al., 2003; LePage, 2003; Spicer et al., 2003). This inference is also corroborated by fossil pollen records from the late Miocene and Pliocene (8.10–4.07 Mya) of the central Loess Plateau in China (Ma et al., 2005) and from the Pliocene of the central Himalayas (Hsü et al., 1973).
It is obvious that more fossil evidence is still needed to retrieve the place of origin of Cedrus. In addition, the present molecular clock estimation of divergence times of true cedars is based on the fossil wood from the Paleocene, the earliest one recorded (Blokhina, 2005). The time values obtained in the present study could be younger than the real divergence times of the group, considering that Cedrus represents a deep or basal branch in the phylogeny of Pinaceae, a family with an origin dating back to the early Cretaceous (Wang et al., 2000). To investigate in detail the differentiation process and dispersal routes of Cedrus, it may be necessary to conduct a phylogeographical study on all of the four cedar species in the future, with extensive population sampling.
ACKNOWLEGEMENTS
The authors thank the handling editor and the two anonymous reviewers for their insightful comments and suggestions on the manuscript. We also thank Prof. Zsolt Debreczy of the International Dendrological Research Institute, Wellesley, MA, USA for providing some leaf samples of Cedrus, especially C. brevifolia, a species endemic to Cyprus; Dr S. Tugrul Koruklu in University of Ankara for his kind help in collecting population samples of C. libani; Drs Xiao-Xin Wei, Xian-Zhao Kan, Dan Peng, Fu-Sheng Yang, Wei Wang and Rui-Qi Li for their help in the laboratory or data analysis. This study was supported by the National Natural Science Foundation of China (grant no. 30425028) and the Chinese Academy of Sciences (grant KZCX2-YW-415 and the special fund to Xiao-Quan Wang).
LITERATURE CITED
- Ahuja MRV. Recent advances in molecular genetics of forest trees. Euphytica. 2001;121:173–195. [Google Scholar]
- Axelrod DI, Al-Shehbaz I, Raven PH. History of the modern flora of China. In: Zhang AL, Wu SG, editors. Floristic characteristics and diversity of East Asian plants. Beijing: Higher Education Press; 1996. [Google Scholar]
- Baum DA, Small RL, Wendel JF. Biogeography and floral evolution of baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Systematic Biology. 1998;47:181–207. doi: 10.1080/106351598260879. [DOI] [PubMed] [Google Scholar]
- Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: mechanisyms and evolution. Proceedings of the National Academy of Sciences of the USA. 1995;92:11331–1338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina NI. Fossil wood of Cedrus (Pinaceae) from the Paleogene of Kamchatka. Paleontological Journal. 1998;32:532–538. [Google Scholar]
- Blokhina NI. Fossil wood from the Paleogene of Russian Far East. XVII International Botanical Congress – Abstract 6.6.2 (P99).2005. [Google Scholar]
- Bou Dagher-Kharrat M, Grenie G, Bariteau M, Brown S, Siljak-Yakolev S, Savouré A. Karyotype analysis reveals interspecific differentiation in the genus Cedrus despite genome size and base composition constancy. Theoretical and Applied Genetics. 2001;103:846–854. [Google Scholar]
- Canard D, Perru O, Tauzin V, Devillard C, Bonhoure JP. Terpene composition variations in diverse provenances of Cedrus libani (A.) Rich. and Cedrus atlantica Manet. Trees: Structure and Function. 1997;11:504–510. [Google Scholar]
- Cheng WC, Fu LK. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; 1978. [Google Scholar]
- Comes HP. The Mediterranean region – a hotspot for plant biogeographic research. New Phytologist. 2004;164:11–14. doi: 10.1111/j.1469-8137.2004.01194.x. [DOI] [PubMed] [Google Scholar]
- Cowling RM, Rundel PW, Lamont BB, Arroyo MK, Arianoutsou M. Plant diversity in Mediterranean-climate regions. Trends in Ecology and Evolution. 1996;11:362–366. doi: 10.1016/0169-5347(96)10044-6. [DOI] [PubMed] [Google Scholar]
- Davis PH. Flora of Turkey and the East Aegean islands. Edinburgh: Edinburgh University Press; 1965. [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Dettman DL, Fang XM, Garzione CN, Li JJ. Uplift-driven climate change at 12 Ma: a long delta O-18 record from the NE margin of the Tibetan plateau. Earth and Planetary Science Letters. 2003;214:267–277. [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. New York, NY: Chapman and Hall; 1993. Monographs on Statistics and Applied Probability No. 57. [Google Scholar]
- Fady B, Lefèvre F, Reynaud M, Vendramin GG, Bou Dagher-Kharrat M, Anzidei M, et al. Gene flow among different taxonomic units: evidence from nuclear and cytoplasmic markers in Cedrus plantation forests. Theoretical and Applied Genetics. 2003;107:1132–1138. doi: 10.1007/s00122-003-1323-z. [DOI] [PubMed] [Google Scholar]
- Farjón A. Königstein: Koeltz Scientific Books; 1990. Pinaceae: drawings and descriptions of the genera Abies, Cedrus, Pseudolarix, Keteleeria, Nothotsuga, Tsuga, Cathaya, Pseudotsuga, Larix and Picea. [Google Scholar]
- Farjón A. World checklist and bibliography of conifers. 2nd edn. London: Royal Botanic Gardens, Kew; 2001. [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Seattle: Department of Genome Sciences, University of Washington; 1993. PHYLIP (Phylogeny Inference Package) Version 3.6a2. Distributed by the author. Available at http://evolution.gs.washington.edu/phylip.html . [Google Scholar]
- Fici S. Floristic relations between eastern Africa and the Mediterranean region with special references to northern Somalia. Flora Mediterranea. 1991;I:175–185. [Google Scholar]
- Florin R. The distribution of conifer and taxad genera in time and space. Acta Horti Bergianizo. 1963;20:194–256. [Google Scholar]
- Frankis MP. Generic inter-relationships in Pinaceae. Notes from the Royal Botanic Garden, Edinburgh. 1988;45:527–548. [Google Scholar]
- Geraads D. Biogeography of circum-Mediterranean Miocene-Pliocene rodents; a revision using factor analysis and parsimony analysis of endemicity. Palaeogeography, Palaeoclimatology, Palaeoecology. 1998;137:273–288. [Google Scholar]
- Griffin DL. Aridity and humidity: two aspects of the late Miocene climate of North Africa and the Mediterranean. Palaeogeography, Palaeoclimatology, Palaeoecology. 2002;182:65–91. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hart JA. A cladistic analysis of conifers: preliminary results. Journal of the Arnold Arboretum. 1987;68:269–307. [Google Scholar]
- Hellwig FHA. Centaureinae (Asteraceae) in the Mediterranean-history of ecogeographical radiation. Plant Systematics and Evolution. 2004;246:137–162. [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London. Series B. Biological Sciences. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipkins VD, Krutovskii KV, Strauss SH. Organelle genomes in conifers: structure, evolution, and diversity. Forest Genetics. 1994;1:179–189. [Google Scholar]
- Hsü J, Tao JR, Sun XJ. On the discovery of a Quercus semicarpifolia bed in mount Shisha Pangma and its significance in botany and geology. Acta Botanica Sinica. 1973;15:103–119. [Google Scholar]
- Jaramillo-Correa JP, Bousquet J, Beaulieu J, Isabel N, Perron M, Bouillé M. Cross-species amplification of mitochondrial DNA sequence-tagged-site markers in conifers: the nature of polymorphism and variation within and among species in Picea. Theoretical and Applied Genetics. 2003;106:1353–1367. doi: 10.1007/s00122-002-1174-z. [DOI] [PubMed] [Google Scholar]
- Krijgsman W. The Mediterranean: mare nostrum of earth sciences. Earth and Planetary Science Letters. 2002;205:1–12. [Google Scholar]
- Krüssmann G. Manual of cultivated conifers. 2nd edn. Prtland, OR: Timber Press; 1985. [Google Scholar]
- Kusumi J, Tsumura Y, Yoshimaru H, Tachida H. Molecular evolution of nuclear genes in Cupressaceae, a group of conifer. Molecular Biology and Evolution. 2002;19:736–747. doi: 10.1093/oxfordjournals.molbev.a004132. [DOI] [PubMed] [Google Scholar]
- Lee C, Wen J. Phylogeny of Panax using chloroplast trnC-trnD intergenic region and the utility of trnC-trnD in interspecific studies of plants. Molecular Phylogenetics and Evolution. 2004;31:894–903. doi: 10.1016/j.ympev.2003.10.009. [DOI] [PubMed] [Google Scholar]
- LePage BA. The evolution, biogeography and palaeoecology of the Pinaceae based on fossil and extant representatives. Acta Horticulturae. 2003;615:29–52. [Google Scholar]
- Li LC. Studies on the karyotype and phylogeny of the Pinaceae. Acta Phytotaxonomica Sinica. 1995;33:417–432. [Google Scholar]
- Li LC, Fu YX. Studies on the cytotaxonomy and the historical phytogeography of Cedrus (Pinaceae) Acta Botanica Yunnanica. 1995;17:41–47. [Google Scholar]
- Liu X, Yin ZY. Sensitivity of East Asian monsoon climate to the uplift of the Tibetan Plateau. Palaeogeography, Palaeoclimatology, Palaeoecology. 2002;183:223–245. [Google Scholar]
- Ma Y, Wu F, Fang X, Li J, An Z, Wang W. Pollen record from red clay sequence in the central Loess Plateau between 8.10 and 2.60 Ma. Chinese Science Bulletin. 2005;50:2234–2242. [Google Scholar]
- Mai DHZ. Development and regional differentiation of the European vegetation during the Tertiary. Plant Systematics and Evolution. 1989;162:79–91. [Google Scholar]
- Meikle RD. Flora of Cyprus. Vol. I. London: Royal Botanic Gardens, Kew; 1977. [Google Scholar]
- Mogensen HL. The hows and ways of cytoplasmic inheritance in seed plants. American Journal of Botany. 1996;83:383–404. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Pijut PM. Cedrus – the true cedars. Journal of Arboriculture. 2000;26:218–224. [Google Scholar]
- Pither J. Climate tolerance and interspecific variation in geographic range size. Proceeding of the Royal Society of London. Series B. Biological Sciences. 2003;270:475–481. doi: 10.1098/rspb.2002.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Price RA, Olsen-Stojkovich J, Lowenstein JM. Relationships among the genera of Pinaceae: an immunological comparison. Systematic Botany. 1987;12:91–97. [Google Scholar]
- Quinn CJ, Price RA, Gadek PA. Familial concepts and relationships in the conifers based on rbcL and matK sequence comparisons. Kew Bulletin. 2002;57:513–531. [Google Scholar]
- Rambaut A, Charleston MA. TreeEdit. 2002. 1.0a10, ed. Available at http://www.evolve.zoo.oxford.ac.uk .
- Ribera I, Nilsson AN, Vogler AP. Phylogeny and historical biogeography of Agabinae diving beetles (Coleoptera) inferred from mitochondrial DNA sequences. Molecular Phylogenetics and Evolution. 2004;30:545–562. doi: 10.1016/S1055-7903(03)00224-0. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. Plant Molecular Biology (Manual) 1988;A6:1–10. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Molecular Biology and Evolution. 1997;14:1218–1231. [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. R8s: analysis of rates (“r8s”) of evolution (and other stuff), version 1.60. 2003. http://ginger.ucdavis.edu/r8s/
- Sanmartín I. Dispersal vs. vicariance in the Mediterranean: historical biogeography of the Palearctic Pachydeminae (Coleoptera, Scarabaeoidea) Journal of Biogeography. 2003;30:1883–1897. [Google Scholar]
- Scaltsoyiannes A. Allozyme differentiation and phylogeny of cedar species. Silvae Genetica. 1998;48:61–68. [Google Scholar]
- Scarascia-Mugnozza G, Oswald H, Piussic P, Radoglou K. Forests of the Mediterranean region: gaps in knowledge and research needs. Forest Ecology and Management. 2000;132:97–109. [Google Scholar]
- Schneeweiss G, Schonswetter P, Kelso S, Niklfeld H. Complex biogeographic patterns in Androsace (Primulaceae) and related genera: evidence from phylogenetic analyses of nuclear internal transcribed spacer and plastid trnL-F sequences. Systematic Biology. 2004;53:856–876. doi: 10.1080/10635150490522566. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Mille J, et al. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Small RL, Ryburn JA, Cronn RC, Seelanan T, Wendel JF. The tortoise and the hare: choosing between noncoding plastome and nuclear Adh sequences for phylogeny reconstruction in a recently diverged plant group. American Journal of Botany. 1998;85:1301–1315. [PubMed] [Google Scholar]
- Song BH, Wang XQ, Wang XR, Sun LJ, Hong DY, Peng PH. Maternal lineages of Pinus densata, a diploid hybrid. Molecular Ecology. 2002;11:1057–1063. doi: 10.1046/j.1365-294x.2002.01502.x. [DOI] [PubMed] [Google Scholar]
- Spicer RA, Harris NBW, Widdowson M, Herman AB, Guo S, Valdes PJ, et al. Constant elevation of southern Tibet over the past 15 million years. Nature. 2003;421:622–624. doi: 10.1038/nature01356. [DOI] [PubMed] [Google Scholar]
- Svenning JC. Deterministic Plio-Pleistocene extinctions in the European cool-temperate tree flora. Ecology Letters. 2003;6:646–653. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y, Yoshimura K, Tomaru N, Ohba K. Molecular phylogeny of conifers using PCR-RFLP analaysis of PCR-amplified specific chloroplast genes. Theoretical and Applied Genetics. 1995;91:1222–1236. doi: 10.1007/BF00220933. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, et al. Flora Europaea I. Cambridge: Cambridge University Press; 1964. [Google Scholar]
- Valcárcel V, Fiz O, Vargas P. Chloroplast and nuclear evidence for multiple origins of polyploids and diploids of Hedera (Araliaceae) in the Mediterranean basin. Molecular Phylogenetics and Evolution. 2003;27:1–20. doi: 10.1016/s1055-7903(02)00364-0. [DOI] [PubMed] [Google Scholar]
- Vasiliev I, Krijgsman W, Langereis CG, Panaiotu CE, Matenco L, Bertotti G. Towards an astrochronological framework for the eastern Paratethys Mio-Pliocene sedimentary sequences of the FocYani basin (Romania) Earth and Planetary Science Letters. 2004;227:231–247. [Google Scholar]
- Wang XQ, Han Y, Hong DY. A molecular systematic study of Cathaya, a relic genus of the Pinaceae in China. Plant Systematics and Evolution. 1998;213:165–172. [Google Scholar]
- Wang XQ, Tank DC, Sang T. Phylogeny and divergence times in Pinaceae: evidence from three genomes. Molecular Biology and Evolution. 2000;17:773–781. doi: 10.1093/oxfordjournals.molbev.a026356. [DOI] [PubMed] [Google Scholar]
- Wang XR, Tsumura Y, Yoshimaru H, Nagasaka K, Szmidt AE. Phylogenetic relationships of Eurasian pines (Pinus, Pinaceae) based on chloroplast rbcL, matK, rpl20-rps18 spacer and trnV intron sequences. American Journal of Botany. 1999;86:1742–1753. [PubMed] [Google Scholar]
- Wei XX, Wang XQ. Phylogenetic split of Larix: evidence from paternally inherited cpDNA trnT-trnF region. Plant Systematics and Evolution. 2003;239:67–77. [Google Scholar]
- Winkworth RC, Donoghue MJ. Viburnum phylogeny based on combined molecular data: implications for taxonomy and biogeography. American Journal of Botany. 2005;92:653–666. doi: 10.3732/ajb.92.4.653. [DOI] [PubMed] [Google Scholar]
- Won H, Renner SS. Horizontal gene transfer from flowering plants to Gnetum. Proceedings of the National Academy of Sciences of the USA. 2003;100:10824–10829. doi: 10.1073/pnas.1833775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Hu ZH. Comparative anatomy of resin ducts of the Pinaceae. Trees: Structure and Function. 1997;11:135–143. [Google Scholar]
- Wulff EV. Historical plant geography: history of the world flora. Waltham, MA: Chronica Botanica; 1944. [Google Scholar]
- Xiang (J) QY, Manchester SR, Homas DT, Zhang WH, Fan CZ. Phylogeny, biogeography, and molecular dating of cornelian cherries (Cornus, Cornaceae): tracking Tertiary plant migration. Evolution. 2005;59:1685–1700. [PubMed] [Google Scholar]
- Zachos JC, Wara MW, Bohaty S, Delaney ML, Petrizzo MR, Brill A, et al. A transient rise in tropical sea surface temperature during the Paleocene-Eocene thermal maximum. Science. 2003;302:1551–1554. doi: 10.1126/science.1090110. [DOI] [PubMed] [Google Scholar]