Abstract
To study the role of carbohydrate in lysosomal protein transport, we engineered two novel glycosylation signals (Asn-X-Ser/Thr) into the cDNA of human procathepsin L, a lysosomal acid protease. We constructed six mutant cDNAs encoding glycosylation signals at mutant sites Asn-138, Asn-175, or both sites together, in the presence or absence of the wild-type Asn-204 site. We stably transfected wild-type and mutant cDNAs into NIH3T3 mouse fibroblasts and then used species-specific antibodies to determine the glycosylation status, phosphorylation, localization, and transport kinetics of recombinant human procathepsin L containing one, two, or three glycosylation sites. Both novel glycosylation sites were capable of being glycosylated, although Asn-175 was utilized only 30–50% of the time. Like the wild-type glycosylation at Asn-204, carbohydrates at Asn-138 and Asn-175 were completely sensitive to endoglycosidase H, and they were phosphorylated. Mutant proteins containing two carbohydrates were capable of being delivered to lysosomes, but there was not a consistent relationship between the efficiency of lysosomal delivery and carbohydrate content of the protein. Pulse-chase labeling revealed a unique biosynthetic pattern for proteins carrying the Asn-175 glycosylation sequence. Whereas wild-type procathepsin L and mutants bearing carbohydrate at Asn-138 appeared in lysosomes by about 60 min, proteins with carbohydrate at Asn-175 were processed to a lysosome-like polypeptide within 15 min. Temperature shift, brefeldin A, and NH4Cl experiments suggested that the rapid processing did not occur in the endoplasmic reticulum and that Asn-175 mutants could interact with the mannose 6-phosphate receptor. Taken together, our results are consistent with the interpretation that Asn-175 carbohydrate confers rapid transport to lysosomes. We may have identified a recognition domain in procathepsin L that is important for its interactions with the cellular transport machinery.
INTRODUCTION
The transport of proteins to their proper intracellular and extracellular compartments requires a highly regulated set of processes in eukaryotic cells. One important intracellular compartment is the lysosome, an acidic organelle populated by soluble hydrolases that degrade proteins and glycans. The primary transport pathway for newly synthesized lysosomal hydrolases utilizes the mannose 6-phosphate receptor (MPR), which recognizes mannose 6-phosphate (man6P) residues present specifically on carbohydrates of lysosomal enzymes (reviewed by von Figura and Hasilik, 1986; Ludwig et al., 1995). MPR binds mannose 6-phosphorylated proteins in the trans-Golgi network, the receptor-ligand complex moves to a prelysosome/late endosome acidic compartment, the receptor dissociates from ligand, and the receptor recycles back to the trans-Golgi.
Under certain growth conditions, some lysosomal proteins are overproduced and predominantly secreted instead of being delivered to lysosomes (Gottesman, 1978; Sloane et al., 1982; Gal et al., 1985; Kane et al., 1988; Capony et al., 1989). The secretion of lysosomal proteins is specific, occurring predominantly for the protein that is overproduced in a particular cell (Dong et al., 1989; Achkar et al., 1990; Isidoro et al., 1991). The following mechanisms have been proposed for how proteins escape transport to lysosomes: 1) altered man6P content or structure of the carbohydrate, which prevents MPR binding (Hasilik and von Figura, 1981; Pagano et al., 1989; Pohlmann et al., 1995); 2) altered availability of MPR due to its saturation, down-regulation, or redistribution to the plasma membrane (Achkar et al., 1990; Prence et al., 1990); 3) altered binding to MPR due to noncarbohydrate effects on the lysosomal/secreted protein (Lazzarino and Gabel, 1990). There is growing evidence to suggest that lysosomal transport may be regulated (or dysregulated) in some manner in cells that overproduce and selectively secrete lysosomal hydrolases (Prence et al., 1990; Isidoro et al., 1991, 1995; Ulbricht et al., 1996). However, the exact mechanism by which proteins escape lysosomal delivery and the role that carbohydrate or protein determinants play in the distribution between intracellular and extracellular trafficking remain unknown.
A man6P-independent pathway for transporting lysosomal enzymes to their proper destination has been proposed for several cell types. Man6P-independent transport has been demonstrated directly for the lysosomal protein procathepsin D (proCD) and indirectly for a variety of lysosomal hydrolases (Owada and Neufeld, 1982; Waheed et al., 1982; Tsuji et al., 1988; Rijnboutt et al., 1991b; Glickman and Kornfeld, 1993; Ludwig et al., 1994; Pohlmann et al., 1995). Membrane-associated “receptors” appear to be involved in man6P-independent aspects of transport for at least two lysosomal proteins, proCD and procathepsin L (McIntyre and Erickson, 1993; Zhu and Conner, 1994). However, the exact mechanism of man6P-independent transport to lysosomes is not known. In addition, it is not known whether man6P-dependent and man6P-independent transport are completely distinct processes or whether they actually intersect during the normal transport of lysosomal enzymes. Recent reports claim that malignant cells utilize the man6P-independent pathway to a greater extent than do normal cells, suggesting that a defect or inefficiency of this mechanism may also play a role in secretion of lysosomal proteins by growth-stimulated cells (Capony et al., 1994; Isidoro et al., 1997).
Cathepsin L is a lysosomal cysteine protease that has been used as a model system for protein localization. Procathepsin L (proCL), the 42-kDa inactive proenzyme precursor of cathepsin L, contains a single carbohydrate at amino acid 204 (Gal and Gottesman, 1988). The most abundant form of this carbohydrate carries two man6P residues in phosphomonoester linkages (Dong and Sahagian, 1990; Lazzarino and Gabel, 1990). ProCL synthesized at basal levels by normal cells is efficiently transported to lysosomes and proteolytically processed to single-chain (32 kDa) and two-chain (25 + 5 kDa) cathepsin L, the enzymatically active forms of the protein (Gal et al., 1985).
ProCL synthesis is elevated in many cell types under conditions of growth stimulation and malignant transformation (Gottesman, 1978; Nielsen-Hamilton et al., 1982; Kane and Gottesman, 1990; Boyer and Tannock, 1993), in osteoclasts of remodeling bones (Kakegawa et al., 1993), in activated macrophages (Reddy et al., 1995), and in macrophages and synoviocytes of arthritic joints (Maciewicz et al., 1990). In all cases of its overexpression, the protein is abundantly secreted, but other lysosomal enzymes are not concomitantly secreted. The mechanism by which overexpressed proCL is selectively secreted is not entirely known. The secreted and intracellular forms of proCL appear to be identical in their carbohydrate structures and man6P content, and this glycosylation structure is not affected by the growth state of cells (Dong and Sahagian, 1990; Lazzarino and Gabel, 1990). Therefore, altered patterns of secretion and transport observed during the overexpression of proCL cannot be attributed to loss of an MPR-binding motif.
ProCL has low binding affinity for MPR (Dong et al., 1989; Lazzarino and Gabel, 1990). This observation has prompted the suggestion that when MPR is limiting, the low-affinity proCL is preferentially secreted by a default mechanism of protein trafficking. Two hypotheses have been set forth for why proCL binds MPR with low affinity: 1) the protein has only a single carbohydrate, whereas MPR has multiple man6P-binding domains (Dong and Sahagian, 1990); 2) there might be a sequence or structure in the protein that interferes with MPR recognition or binding (Lazzarino and Gabel, 1990). Neither of these models has been adequately tested. Whatever the cause, it is possible that low-affinity binding is an important component of the regulated transport of proCL.
In the current work, we explore the effects of carbohydrate and sequence on the transport of proCL. We engineered two novel glycosylation signals (Asn-138 and Asn-175) into the coding sequence of proCL. The novel sites were utilized for carbohydrate addition and man6P modification. We found that hyperglycosylation of proCL did not, in and of itself, confer enhanced transport to lysosomes. However, one of the novel glycosylations (Asn-175) appeared to confer the rapid appearance of a processed, lysosome-like form of proCL, both in the presence and absence of the wild-type carbohydrate. This suggests that Asn-175 may lie in a domain of proCL that is important for its interaction with cellular transport factors. In addition, we observed significant man6P-independent transport to lysosomes for recombinant human proCL proteins expressed in mouse NIH3T3 cells. We did not observe man6P-independent transport for endogenous human proCL expressed by human cells or for recombinant mouse proCL expressed in mouse (Kane, 1993) or human cells. This raises the possibility that there is species or cell type specificity to the MPR-independent pathway of lysosomal transport.
MATERIALS AND METHODS
Cell Lines
Parental and transfected NIH3T3 cells were maintained in DMEM containing 10% bovine serum (Irvine Scientific, Santa Ana, CA), 50 U of penicillin per ml, and 50 μg of streptomycin per ml. Transfected cells were maintained in medium containing colchicine, as indicated below.
Computer Modeling
Cathepsin L was modeled using a knowledged-based approach (Blundell et al., 1987) as implemented in the program HOMOLOGY (Molecular Simulations, San Diego, CA) running on a Silicon Graphics (Mountain View, CA) Indigo-2 R10000 high-impact workstation maintained by the Molecular Modeling Facility of the City of Hope. X-ray coordinates of actinidin (2ACT, Baker, 1980) and papain (9PAP, Kamphuis et al., 1984) were used as templates for modeling structurally conserved regions. Coordinates for nonconserved loops were extracted from a high-resolution subset of the Brookhaven Protein Databank (Bernstein et al., 1977). The model was refined using a combination of energy minimization and molecular dynamics (DISCOVER subroutines within HOMOLOGY). The consistent-valence forcefield was used throughout (Dauber-Osguthorpe et al., 1988). A ribbon model of the structure was created using MOLSCRIPT (Kraulis, 1991).
Site-directed Mutagenesis and Cloning
A cDNA fragment encoding human preprocathepsin L (nucleotides 127-1391; numbering according to Gal and Gottesman [1988], GenBank Accession No. X12451) was subcloned using SalI linkers into the SalI site of pGEM-3 cloning vector (Promega, Madison, WI). Site-directed mutagenesis was performed on this fragment using the PCR. A glycosylation signal at amino acid 138 (amino acid numbering relative to the amino terminus of proCL lacking the N-terminal signal sequence) was engineered by mutating Thr-Gly-Arg codons to Asn-Gly-Ser codons. The mutagenic oligonucleotide, 5′-ATGTTCCGGAAAAATGGGAGCCTTATCTCACTG (nucleotides 740–771; single underlines are mutated sites; double underline is a BspEI site in the proCL cDNA), was used as the upstream primer for PCR, and the downstream primer was complementary to the SP6 promoter in the cloning vector. The PCR product was digested with BspEI and HindIII and used to replace the corresponding fragment in the wild-type cDNA, creating the N1N3 mutant (see Figure 1A for nomenclature). For introducing a glycosylation signal at amino acid 175, Asp-Asn-Gly codons were mutated to Asn-Asn-Ser codons. The mutagenic oligonucleotide, 5′-CCACTATTATTCTGAACATACTG (complementary to nucleotides 872–850), was used as the downstream primer for PCR, and the upstream primer corresponded to the T7 primer in the cloning vector. The PCR product was digested with XbaI at the 5′-end and left blunt-ended at the 3′-end. A second PCR fragment from nucleotides 873 through the SP6 promoter was left blunt-ended at its 5′-end and digested with HindIII at its 3′-end. The two PCR fragments were cloned in a three-way ligation into the XbaI-HindIII site of pGEM·proCL, with the two blunt ends of the PCR fragments ligated together. This created the N2N3 mutant.
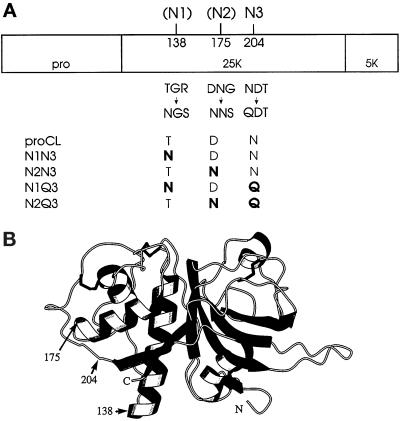
Figure 1.
Representations of (pro)cathepsin L. (A) Linear map of proCL showing the profragment (pro) and both segments of the mature enzyme (25K + 5K). On top of the map are the amino acid positions of the glycosylation sites used in this study: 204,N3 is the single wild-type site in human proCL; 138 (N1) and 175 (N2) are two novel sites introduced in this work. Directly below the map are the wild-type sequences at residues 138–140, 175–177, and 204–206 and the mutations at those residues that either create (138 and 175) or destroy (204) glycosylation signals. The bottom of the panel shows the names of the relevant mutants and their corresponding amino acids encoded at positions 138, 175, and 204, with mutations shown in boldface. (B) Drawing of a three-dimensional computer model of cathepsin L. The model was developed as described in MATERIALS AND METHODS. Numbers and arrows designate the positions of amino acids relevant to this study.
To mutate the wild-type glycosylation site, Asn-204 was mutated to a Gln codon. The mutagenic oligonucleotide, 5′-TGTAAGTACAATCCCAAGTATTCTGTTGCTCAGGACACCGGC (nucleotides 919–960; single underlines are mutations; double underline is a RsaI site in the proCL cDNA) was the upstream PCR primer, and the downstream primer was complementary to the SP6 promoter of pGEM-3. The PCR product was digested with RsaI (at the 5′-end) and ApaI (at nucleotide 1020 of proCL) and used to replace the corresponding fragment in N1N3 and N2N3 constructs, creating the N1Q3 and N2Q3 mutants, respectively. Finally, fragments encoding the Asn-138 and Asn-175 glycosylations were cloned together into the same cDNA in either a Asn-204 or Gln-204 background, creating N1N2N3 and N1N2Q3 mutants (not shown in Figure 1A). All nucleotide sequences at mutated sites were confirmed by the dideoxy chain termination method using a kit from United States Biochemical (Cleveland, OH). For expression in NIH3T3 cells, recombinant cDNAs were cloned as SalI fragments into the pSK1.MDR expression vector (Kane et al., 1989).
For construction of mouse proCL lacking both of its glycosylation signals, we modified the proCL.Q204 mutant that already contained a Asn → Gln mutation at residue 204 (Kane, 1993). Asn-251, a normally cryptic glycosylation site in mouse proCL, was mutated to Gln using the method of Kunkel (1985). The mutagenic oligonucleotide was 5′-TATGAACCCCAGTGTAGCAGC. After the mutation at residue 251 was confirmed, a EcoRV–SalI fragment containing that mutation was cloned into the corresponding region of proCL.Q204, creating the double mutant Q204Q251. Wild-type and mutant mouse cDNAs were cloned into the RcRSV expression vector (Invitrogen, San Diego, CA) for expression in human cell lines.
Stable Expression in Cell Lines
Vectors encoding mutant human proCL were transfected into NIH3T3 mouse fibroblasts by the DNA-CaPO4 coprecipitation method, and transfected cells were selected with colchicine (60 ng/ml) as described elsewhere (Kane et al., 1989). Individual colonies or populations of transfected cells were expanded and maintained in colchicine (80 ng/ml) during subsequent experiments. Control cells were transfected with vector encoding wild-type human proCL and grown in a high concentration of colchicine (640 ng/ml) to select for amplified expression of recombinant proCL, as described in Kane et al. (1988). RcRSV vectors encoding mouse cDNAs were transfected into SK-Hep human hepatoma cells using the same procedure except that selections were with G418 (1.5 mg/ml, GIBCO BRL).
Metabolic Labeling and Immunoprecipitation
For continuous labeling experiments, transfected cells were incubated in methionine-free medium (GIBCO, Grand Island, NY) in the presence of 200–500 μCi/ml [35S]methionine (EXPRE35S35S, DuPont-New England Nuclear, Boston, MA), for 0.5–5 h, depending on the experiment. Culture supernatants were collected, and cell lysates were harvested in SDS-buffer A (0.15 M NaCl, 50 mM Tris, pH 7.4, 0.5% Nonidet P-40, 0.05% SDS) containing protease inhibitors (1 mM PMSF, 1 μM E-64, 1 μM pepstatin). Cell lysates corresponding to 5–20 × 106 trichloroacetic acid-precipitable counts per min, and an equivalent volume of culture supernatants, were subjected to immunoprecipitation, SDS-PAGE, and fluorography as previously described (Kane, 1993). Polyclonal, species-specific antisera were used to immunoprecipitate either recombinant human (Smith et al., 1989) or endogenous mouse (Gottesman and Cabral, 1981) proCL synthesized by transfected cells (both antisera were gifts from M. Gottesman). For 32P-labeling, cells were incubated in phosphate-free medium containing 0.5 mCi/ml of [32P]orthophosphate (DuPont-New England Nuclear) for 4–5 h. Immunoprecipitations were performed on lysate and culture medium corresponding to 3–5 × 106 cpm of lysate. Immunoprecipitations were subjected to SDS-PAGE and dried gels were exposed to X-AR film (Kodak, Rochester, NY) at −80°C with an intensifying screen.
Pulse-Chase Analysis
For pulse-chase labeling, transfected cells were incubated in methionine-free medium for 1 h before labeling. Cells were labeled using the same conditions as in continuous labeling except labeling time was decreased to 15 min. After labeling, cells were thoroughly washed with PBS. Medium containing unlabeled methionine (2 mM) was added to cells for the chase. Culture supernatants and cell lysates were collected in SDS-Buffer A at 0, 15, 30, 60, 90, 120, 180, 240, and 360 min of chase. Equal volumes of each sample were immunoprecipitated and subjected to SDS-PAGE and fluorography.
Endoglycosidase Treatments
Endoglycosidase F (endoF) and endoglycosidase H (endoH) digestions were done by adding 4 mU of endoH or 170 mU of N-glycosidase F (both from Boehringer Mannheim) directly to cell lysates in SDS-Buffer A. Digestions were for 4 h at 37°C. Samples were then immunoprecipitated and subjected to SDS-PAGE and fluorography.
Ammonium Chloride, Brefeldin A, and Tunicamycin Experiments
For continuous labeling in the presence of NH4Cl, transfected cells were preincubated in methionine-free medium with or without NH4Cl (10 mM) for 1 h. Cells were then labeled for 5 h with 300 μCi per ml of [35S]methionine in methionine-free medium, either with or without 10 mM NH4Cl. Culture supernatant and cell lysates were collected, samples were immunoprecipitated separately with anti-human cathepsin L antiserum or anti-mouse cathepsin L antiserum and subjected to SDS-PAGE and fluorography. Brefeldin A (BFA) labeling experiments were done in the same manner except that 10 μg/ml BFA and 500 μCi/ml of [35S]methionine were used and the labeling time was for 3 h. For tunicamycin experiments, cells were incubated in standard growth medium in the presence or absence of tunicamycin (15 μg/ml) for 4 h before labeling. Cells were washed with PBS and labeled in methionine-free medium supplemented with 200–300 μCi/ml of [35S]methionine, with or without tunicamycin, for 3 h. Culture supernatants and cell lysates were collected and processed as above.
RESULTS
Novel Glycosylation Signals in ProCL are Utilized
Human proCL contains a single carbohydrate signal (Asn-Asp-Thr) with glycosylation at Asn-204. ProCL is efficiently transported to lysosomes in normal cells (<5% secretion), but secretion is enhanced when the protein is overexpressed in malignant cells or in immortalized cells transfected with a proCL expression vector (Gottesman, 1978; Kane et al., 1988).
To determine whether having multiple mannose-phosphorylated carbohydrates would affect proCL transport, we engineered two novel glycosylation signals into the primary sequence of human proCL. The novel signals were engineered alone or in combination into either wild-type proCL or proCL lacking the wild-type glycosylation signal at Asn-204. Figure 1A is a schematic representation of proCL indicating the positions of wild-type and novel glycosylation sites within the linear protein sequence. The nomenclature of mutants and their sequences at each of three potential glycosylation sites (two mutant sites [N1 and N2] and the wild type [N3] site) are shown below the linear map. Figure 1B is a computer-generated model of proCL illustrating the positions of wild-type and novel glycosylation sites within the folded protein. The novel sites were purposely engineered into an accessible loop and on the same face of the molecule as the wild-type site, as predicted by our computer model and consistent with the recently published crystal structure of proCL (Coulombe et al., 1996). We reasoned that these locations would most likely be utilized for carbohydrate addition and that the carbohydrates would be recognized for mannose phosphorylation (Cantor and Kornfeld, 1992).
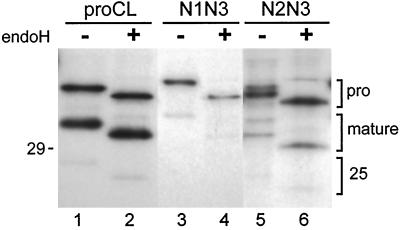
Mutant and wild-type cDNAs were transfected into NIH3T3 mouse fibroblasts, and cells expressing the corresponding recombinant proteins were isolated as described in MATERIALS AND METHODS. To determine the glycosylation status of the various recombinant proteins, transfected cells were metabolically radiolabeled with [35S]methionine; cell lysates were incubated with or without endoH and then subjected to immunoprecipitation with species-specific antiserum raised against human proCL. Results of the analysis are shown in Figure 2. A single species of recombinant N1N3 proenzyme (glycosylation sites at Asn-138 and Asn-204) was detected that migrated approximately 2 kDa slower than wild-type proCL in the absence of endoH (Figure 2, compare lane 1 with lane 3). After endoH treatment, wild-type and N1N3 proteins comigrated (Figure 2, lanes 2 and 4). These results suggest that N1N3 was fully glycosylated at both Asn-204 and the novel Asn-138.
Figure 2.
EndoH digestion of recombinant proCLs. Cell lines stably expressing wild type (proCL) or mutants encoding the wild-type glycosylation site plus a novel glycosylation site at Asn-138 (N1N3) or Asn-175 (N2N3). Cells were radiolabeled for 5 h with [35S]methionine, cell lysates were incubated in the presence (+) or absence (−) of endoH to remove high-mannose carbohydrate, and immunoprecipitations were performed with antiserum specific for human proCL. Immunoprecipitated proteins were run on SDS-PAGE and processed for fluorography as described in MATERIALS AND METHODS. Proenzyme (pro), lysosomal single-chain (mature), and two-chain (25) forms of the proteins are indicated. The 5-kDa carboxy-terminal fragment from the two-chain enzyme was run off the gel. The position of the 29-kDa size standard is shown on the left and lane numbers are provided on the bottom.
In contrast, two species of N2N3 proenzyme (glycosylation sites at Asn-175 and Asn-204) were detected in the absence of endoH, one that migrated slightly slower than glycosylated wild-type proCL and one that migrated slightly faster than proCL. After digestion with endoH, the N2N3 species were reduced to a single band that migrated slightly faster than endoH-digested proCL (Figure 2, lanes 5 and 6 compared with lanes 1 and 2). We interpret these results to mean that N2N3 was partially glycosylated at the novel site (Asn-175). Thus, the slower migrating protein contained two glycosylations, at Asn-204 and Asn-175; the faster migrating protein contained one glycosylation at the wild-type site, Asn-204. Utilization of Asn-175 was variable from experiment to experiment, but was always in the 30–50% range. The mutation creating the Asn-175 glycosylation signal (Asp-Asn-Gly → Asn-Asn-Ser) apparently altered the migration of the polypeptide to a slightly faster migrating species on SDS-PAGE, relative to wild-type proCL. This interpretation is reinforced by the relative migration patterns of the mature forms of the respective polypeptides (Figure 2). Results with mutants lacking the wild-type glycosylation signal (N1Q3 and N2Q3) were consistent with our interpretations that Asn-138 was fully utilized for carbohydrate addition, Asn-175 was partially utilized, and the mutation creating the Asn-175 signal affected migration of the resulting proteins (see below).
Mutant N1N2N3 encoding three glycosylation sites was not stably synthesized in large enough quantities to detect by metabolic labeling with [35S]methionine and SDS-PAGE. The N1N2Q3 mutant encoding two novel glycosylation sites and lacking the wild-type site was glycosylated in accord with the other mutants (i.e., completely at Asn-138, partially at Asn-175). However, it did not appear to be transported out of the endoplasmic reticulum, probably because it was misfolded or aggregated (our unpublished results). N1N2N3 and N1N2Q3 mutants were not analyzed further.
Novel Carbohydrates Are Modified with Man6P
It has previously been demonstrated that proCL is phosphorylated only in its man6P residues (Sahagian and Gottesman, 1982). To determine whether the novel carbohydrates at Asn-138 and Asn-175 were capable of being recognized for man6P modification, we metabolically radiolabeled transfected cells in parallel with [32P]orthophosphate or [35S]methionine, subjected cell lysates to immunoprecipitation, and analyzed the immunoprecipitates by SDS-PAGE. Figure 3 shows the results of this analysis for mutants lacking the wild-type glycosylation site and encoding only a single novel glycosylation signal at Asn-138 or Asn-175.
Figure 3.
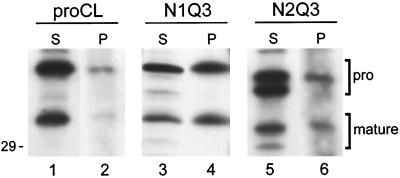
Phosphorylation of novel carbohydrates. Cell lines stably expressing wild type (proCL) or mutants containing only the novel glycosylation sites at Asn-138 (N1Q3) or Asn-175 (N2Q3). Cells were radiolabeled for 4–5 h with either [35S]methionine (S) or [32P]orthophosphate (P). Cell lysates were subjected to immunoprecipitation and SDS-PAGE, as previously, and processed for fluorography or autoradiography, respectively. The proenzymes (pro) and the processed, single-chain proteins (mature) are indicated. The position of the 29-kDa size standard is shown on the left and lane numbers are provided on the bottom.
For N1Q3 (Asn-138), we detected a single proenzyme species with both radioisotopes that migrated at a position consistent with glycosylation and phosphorylation at Asn-138 (Figure 3, lanes 3 and 4). Digestion of this protein (and of N1N3) with endoH completely eliminated the [32P] signal, indicating that the mutant protein was phosphorylated only in its carbohydrate moiety and that novel amino acid phosphorylation was not introduced by the Asn-138 mutation (our unpublished results). For N2Q3 (Asn-175), we detected two proenzyme bands with [35S]methionine. The upper band migrated slightly faster than wild-type proCL, and only this band was radiolabeled with [32P]orthophosphate (Figure 3, lanes 5 and 6). These results are consistent with the interpretation that Asn-175 was partially utilized for glycosylation (the upper band) and that the carbohydrate was modified with man6P. The faster form of the N2Q3 proenzyme contained no carbohydrate and thus was not radiolabeled with [32P]orthophosphate. This again confirmed carbohydrate-specific phosphorylation in mutants containing Asn-175. Also seen in Figure 3 is the fact that N1Q3 and both forms of N2Q3 were transported to lysosomes, as detected by the appearance of proteolytically processed, mature forms of the proteins.
Mutant ProCLs Can Be Transported to Lysosomes and Secreted
We now have the ability to distinguish between proCLs that have zero, one, or two carbohydrates. To determine whether mutant proCLs were secreted or localized to lysosomes, we performed immunoprecipitations from cell lysates and media collected from metabolically radiolabeled transfected cell lines. In this analysis, mature processed proteins represent transport to lysosomes (also see below). Secreted proteins are always full-length and glycosylated. As shown in Figure 4, all of the mutant proCLs were capable of being secreted into the medium (lanes marked M) and transported to lysosomes (labeled in the Figure as “mature”), regardless of their carbohydrate content. Figure 4 shows the results of one representative clone from each of the transfected cell lines. Similar results were obtained for populations of cells obtained from two separate transfections and for multiple independent clones of cells expressing each recombinant construct (our unpublished observations).
Figure 4.
Steady-state distribution of recombinant proteins. Individual clones of cells stably expressing wild-type (proCL) or mutant proteins, as indicated. Cells were radiolabeled for 4 h with [32P]orthophosphate (left-hand panel in A and B) or [35S]methionine (right-hand panel in A and B, both panels in C). Cell lysates (C) and culture media (M) were collected and subjected to immunoprecipitation, as previously described. The proenzymes (pro) and the processed lysosomal proteins (mature) are indicated by brackets. The positions of size markers are indicated on the left of each panel and lane numbers are provided on the bottom.
We were not able to quantify these results, due to the relatively low level of secreted proteins and the variable distributions of proteins from experiment to experiment, even for wild-type proCL (for example, see Figure 4B, lanes 1 and 2 compared with lanes 5 and 6; in this comparison proCL appeared to be secreted more efficiently in lane 2 than in lane 6). Although it appears in Figure 4 that double-glycosylated proteins were secreted less than single-glycosylated proCL, this was not a consistent result in all experiments. Furthermore, reduced secretion did not seem to correlate with an enhanced transport to lysosomes for proteins carrying two carbohydrates.
Secretion of the unglycosylated version of N2Q3 was significantly higher than that of the corresponding monoglycosylated version of the protein (Figure 4C, lanes 7 and 8). This is to be expected given that man6P-independent lysosomal transport of proCL is inefficient in mouse fibroblasts (Kane, 1993). Surprisingly, we also observed a significant amount of a mature, lysosome-like form of unglycosylated N2Q3 (see below).
ProCLs with Carbohydrate at Asn-175 Are Rapidly Processed to Lysosome-Like Forms
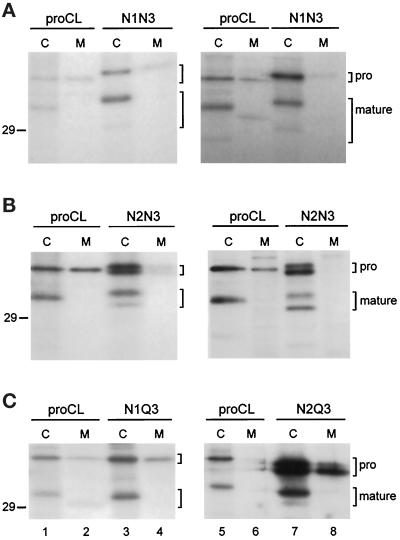
To determine whether novel carbohydrates might affect the rate of transport to lysosomes, we performed pulse-chase analysis on transfected cell lines. Cells were radiolabeled for 15 min with [35S]methionine and then chased for various times in the presence of excess unlabeled methionine. Immunoprecipitations were performed on cell lysates and media collected at each time point. Data for transport to lysosomes are shown in Figure 5. Wild-type proCL started to appear in lysosomes by 30–60 min of chase, as determined by the appearance of the mature, 32-kDa form of the protein (Figure 5A). This is consistent with previous reports of the transport kinetics for endogenous mouse proCL and with our own results using transfected recombinant proCL (Gal et al., 1985; Kane, 1993). N1N3 and N1Q3, containing two (Asn-138 + Asn-204) and one (Asn-138) carbohydrate, respectively, were transported to lysosomes with kinetics similar to wild-type proCL (Figure 5, B and C).
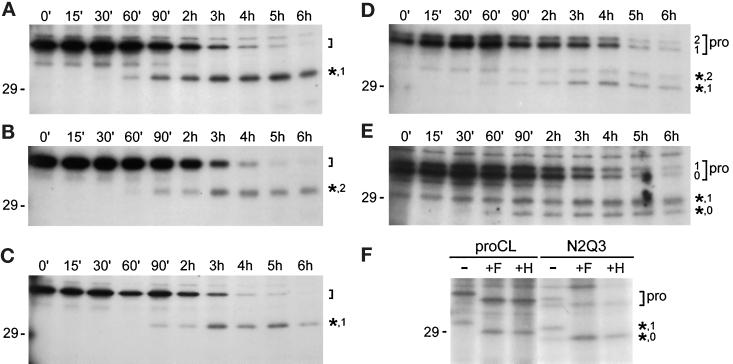
Figure 5.
Pulse-chase analysis. (A–E) Cell lines stably expressing recombinant proteins were pulse-labeled for 15 min with [35S]methionine and then chased with unlabeled methionine for the indicated times. Cell lysates were collected at each time point and subjected to immunoprecipitation as previously. (A) Wild-type proCL; (B) N1N3; (C) N1Q3; (D) N2N3; (E) N2Q3. (F) Lysates from the 2h time point of proCL and N2Q3 cells were treatd with endoF (+F), endoH (+H), or left untreated (−) before immunoprecipitation. Brackets on the right of each panel indicate the positions of proenzymes. The number of carbohydrates on each proenzyme are included in panels D and E. Asterisks indicate the processed, lysosome-like bands for the respective recombinants, along with the number of carbohydrates present on each of those proteins. Positions of the 29-kDa standard are shown.
We obtained a surprising result with the Asn-175 constructs. For the double-glycosylated form of N2N3, we detected the rapid appearance of a lysosome-like band within the 15-min pulse labeling time (Figure 5D, 0′ time point). The processed form of the monoglycosylated species appeared with normal kinetics (60 min). We next wanted to determine whether the rapid appearance of the lysosome-like form was due to the presence of two carbohydrates on N2N3 or was related to the location of carbohydrate at Asn-175. For this, we did pulse-chase analysis on N2Q3, which lacks the wild-type glycosylation site. Again, we detected a lysosome-like form of the monoglycosylated species (carbohydrate only at Asn-175) within the 15-min pulse-labeling time. Surprisingly, we also observed significant amounts of a mature form of the unglycosylated species of N2Q3 within 60 min (normal kinetics) (Figure 5E). Endoglycosidase digestions confirmed that the two lysosome-like species of N2Q3 were glycosylated and unglycosylated versions, respectively, of the mature protein (Figure 5F). Similar analysis of N2N3 confirmed the glycosylation patterns of its mature proteins (our unpublished results). Finally, although the relative levels of protein secretion were variable from cell line to cell line, kinetics of secretion was the same for wild-type and each of the mutant proteins, with initial appearance in the medium by 30–60 min of chase (our unpublished observations).
Processed Forms Are Not Produced in the Endoplasmic Reticulum
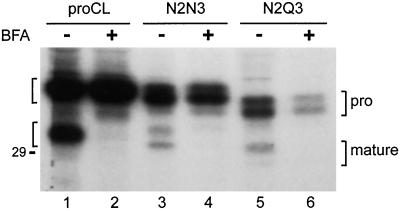
To try to determine whether the processing of N2N3 and N2Q3 was occurring in the endoplasmic reticulum rather than in lysosomes, we performed experiments with BFA, a fungal antibacterial agent that blocks secretory-pathway proteins in the endoplasmic reticulum (Lippincott-Schwartz et al., 1989). Transfected cells were incubated with or without BFA and radiolabeled with [35S]methionine, as described in MATERIALS AND METHODS, and cell lysates were subjected to immunoprecipitation. Results are shown in Figure 6. In the absence of BFA, results were consistent with previous experiments in which both species of N2N3 and N2Q3 appeared as processed, mature proteins (Figure 6, lanes 3 and 5). However, these proteins did not appear in the presence of BFA (Figure 6, lanes 4 and 6), suggesting that processing did not occur in the endoplasmic reticulum. Similar results were obtained when cells were pulse-labeled at 20°C to trap newly synthesized proteins early in the secretory pathway (Braulke et al., 1988) and then chased at 37°C to allow transport to lysosomes. Mature bands appeared only by 2 h after the shift in temperature to 37°C, and kinetics were in this case identical for all species of wild-type, N2N3, and N2Q3 proteins (our unpublished observations).
Figure 6.
BFA treatment. Cell lines stably expressing the indicated recombinants were radiolabeled for 3 h with [35S]methionine in the absence (−) or presence (+) of BFA (10 μg/ml) to block transport in the endoplasmic reticulum. Cell lysates were collected and subjected to immunoprecipitation, as previously. Proenzyme (pro) and processed (mature) proteins are indicated by brackets on both sides of the image. The position of the 29-kDa size standard is shown and lane numbers are provided on the bottom.
Mutant proCLs Interact with Man6P Receptor
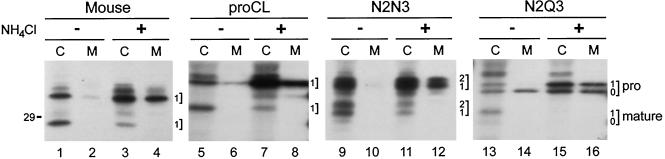
To determine whether Asn-175 proCLs were transported to lysosomes by MPR, we labeled cells in the presence and absence of NH4Cl, which raises the pH of endosomes, prevents the release of lysosomal proteins from MPR, and results in the predominant secretion of those proteins (Gonzalez-Noriega et al., 1980). Results of the steady-state labeling experiment are shown in Figure 7. Secretion of full-length proenzyme was enhanced by NH4Cl in the cases of endogenous mouse proCL (lane 4 vs. lane 2) and wild-type recombinant proCL (lane 8 vs. lane 6). Similarly, the secretion of both proenzyme species of N2N3, carrying one and two carbohydrates, respectively, were dramatically enhanced in NH4Cl-treated cells relative to control cells in the absence of NH4Cl (Figure 7, lane 12 vs. lane 10). For N2Q3, however, it appeared that enhanced secretion in NH4Cl-treated cells was specific for the monoglycosylated species and not the unglycosylated species (Figure 7, lane 16 vs. lane 14). These results suggest that MPR mediates transport of the glycosylated species but not the unglycosylated species, as expected.
Figure 7.
Ammonium chloride treatment. Cell lines stably expressing proCL, N2N3, or N2Q3 recombinants. Cells were radiolabeled for 5 h with [35S]methionine in the absence (−) or presence (+) of NH4Cl (10 mM). Cell lysates (C) and culture media (M) were collected and subjected to immunoprecipitation with anti-human proCL antiserum (last three panels). In the left panel (Mouse), anti-mouse proCL antiserum was used to immunoprecipitate endogenous protein from lysates of cells expressing the N2N3 recombinant, using 10-fold less sample than in the N2N3 panel. Positions of proenzymes and processed proteins (along with the number of carbohydrates on each protein) are indicated, as previously. The 29-kDa size standard is shown on the left and lane numbers are provided below each panel.
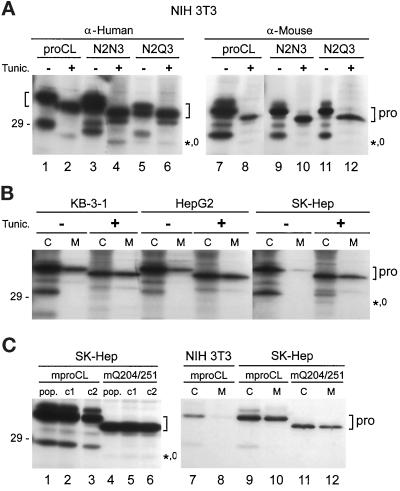
Recombinant, Unglycosylated Human proCLs Are Transported to Lysosomes by NIH3T3 Cells
To determine whether lysosomal transport of unglycosylated human proCL was specific for the N2Q3 mutant, we analyzed transport of wild-type, N2N3, and N2Q3 proteins in the presence of tunicamycin, which inhibits all glycosylation. Results are shown in Figure 8. NIH3T3 cells expressing any of the three human proteins transported a significant amount of those proteins to lysosomes in the presence of tunicamycin (Figure 8A, lanes 2, 4, and 6). This rules out the trivial explanation that mature, unglycosylated N2Q3 was derived from mature, glycosylated N2Q3 after transport to lysosomes. A low or undetectable amount of endogenous mouse proCL was delivered to lysosomes in the same tunicamycin-treated cells (Figure 8A, lanes 8, 10, and 12). The result with the mouse protein is similar to what we have previously observed for unglycosylated recombinant mouse proCL expressed in NIH3T3 cells (<5% transport to lysosomes) (Kane, 1993).
Figure 8.
Tunicamycin treatment. (A) Transfected cell lines were incubated in the presence (+) or absence (−) of tunicamycin (tunic.) for 4 h and then radiolabeled for 3 h with [35S]methionine (+/- tunic.). Cell lysates were immunoprecipitated with anti-human proCL antiserum or anti-mouse proCL antiserum, as indicated, followed by SDS-PAGE and fluorography. (B) Three human cell lines, as indicated, were treated with tunicamycin (+) or left untreated (−) and radiolabeled with [35S]methionine. Cell lysates (C) or media (M) were immunoprecipitated with anti-human proCL antiserum, followed by SDS-PAGE. (C) Human SK-Hep cells were transfected with wild-type mouse proCL (mproCL) or with a mutant lacking both of its normal glycosylation signals (mQ204/251). Left, Populations of cells (pop.) or two individual clones from each population (c1 and c2) were radiolabeled and cell lysates were immunoprecipitated with anti-mouse proCL antiserum. Right, NIH3T3 cells expressing authentic mouse proCL or populations of SK-Hep cells expressing wild-type or mutant recombinant mouse proCLs, as indicated, were radiolabeled with [35S]methionine. Cell lysates (C) and media (M) were immunoprecipitated with anti-mouse proCL antiserum. Brackets mark the positions of proenzymes (pro). The positions of the small amount of unglycosylated, lysosomal proteins are indicated (*,0). The 29-kDa size standard is shown on the left and lane numbers are provided below the panels.
We also wanted to determine whether unglycosylated proCL could be transported to lysosomes in other cell types. This did not appear to be the case. Thus, when we treated human epidermoid cells (KB-3–1) and human hepatoma cells (SK-Hep and HepG2) with tunicamycin, endogenous proCL was predominantly secreted and not transported to lysosomes (Figure 8B). Finally, we constructed a mouse proCL cDNA that lacked both of its wild-type glycosylation signals and we stably expressed this mutant in SK-Hep cells. The recombinant mouse protein was not appreciably transported to lysosomes by the human cells (Figure 8C, lanes 4–6), although it could be secreted (Figure 8C, lane 12). Taken together, these results suggest that carbohydrate-independent transport of proCL to lysosomes may be species-specific (for the human protein expressed in mouse cells) and possibly cell type-specific (for NIH3T3 cells).
DISCUSSION
In this paper we have analyzed the carbohydrate content, phosphorylation, localization, and transport kinetics of recombinant human proCLs containing novel glycosylation signals. One of those novel sites, Asn-138, was 100% utilized, while the other site, Asn-175, was utilized 30–50% of the time. This was true in N2N3, which has both the engineered site at Asn-175 and the wild-type site at Asn-204, and in N2Q3, which has only the Asn-175 site. This observation would seem to rule out steric hindrance from the wild-type carbohydrate as a reason for the partial utilization of Asn-175. Although we cannot rule out the possibility that Asn-175 was glycosylated more frequently and then lost carbohydrate due to instability of the site, this explanation does not seem likely given the fixed ratio of double- to monoglycosylated species of N2N3 and mono- to unglycosylated species of N2Q3 during the course of pulse-chase analyses (Figure 5).
The novel carbohydrates added at Asn-138 and Asn-175 were both phosphorylated (Figure 3). We expected this result, given the predicted positions of the two sites on the same face of proCL as Asn-204 carbohydrate (Figure 1, panel B). This is consistent with proCD studies, in which mannose phosphorylation of carbohydrates is rather promiscuous within the context of a lysosomal enzyme, but also depends on their proximity to a putative phosphorylation domain on those proteins (Cantor and Kornfeld, 1992). We should be able to extend our analysis of novel glycosylation sites to map the domain on proCL that is responsible for recognition of carbohydrate for mannose phosphorylation.
One purpose of our experiments was to test the hypothesis that extra man6P residues would enhance the affinity of proCL for MPR and thus increase its transport to lysosomes or reduce its secretion (Dong and Sahagian, 1990). We have not determined the affinity of our hyperglycosylated proteins for MPR, and we were not able to achieve high enough expression levels of recombinant proteins to adequately test the hypothesis. Nevertheless, our results suggest that proCL proteins carrying two carbohydrates were still capable of being secreted (Figure 4), even though both carbohydrates were modified with man6P and the proteins appeared to bind MPR (Figure 7). Furthermore, a direct comparison of double- and monoglycosylated protein expressed in the same cell (the two forms of N2N3 protein) suggested that extra carbohydrate was not sufficient to confer enhanced transport to lysosomes (Figure 4, panel B). Results with N1N3 (two carbohydrates), compared with N1Q3 and wild-type proCL (one carbohydrate each), were consistent with this interpretation (Figure 4, panels A and C). A similar study was performed with proCD, in which one or the other of its two glycosylation signals was selectively mutated. The conclusion of that study was that the number of carbohydrates does not correlate with efficiency of transport to lysosomes, although the man6P content of recombinant proteins (wild type or mutants) was not determined in that work (Fortenberry et al., 1995).
We made a surprising and novel observation using proteins that encoded the Asn-175 glycosylation signal: proteins with carbohydrate at Asn-175 appeared to be transported to lysosomes very rapidly. This conclusion is based on the very rapid appearance of low-molecular weight (mature) forms of newly synthesized protein in pulse-chase labeling experiments (Figure 5). The production of the mature forms did not seem to occur in the endoplasmic reticulum or Golgi, since BFA (Figure 6) and low temperature inhibited the appearance of those bands. In addition, proteolytic processing appeared to be accurate, since these forms migrated on SDS-PAGE with authentic lysosomal cathepsin L after digestion with endoF or endoH (Figure 5F). We have been unable to detect lysosomal appearance of mutant proCLs directly by subcellular fractionation, due to insufficient sensitivity of this assay, particularly with pulse-labeled lysates. Therefore, we cannot formally rule out the possibility that carbohydrate at Asn-175 increases the sensitivity of the protein to proteolytic processing and results in the appearance of the lysosome-like fragment in a prelysosomal compartment. Nevertheless, we believe our results with BFA, temperature shift, and NH4Cl are consistent with the interpretation that processing occurred in lysosomes.
The rapid kinetics required carbohydrate at Asn-175, since the double-glycosylated form of N2N3 (carbohydrate at Asn-175 + Asn-204) was processed rapidly, but its monoglycosylated form (Asn-204 only) was not. The rapid kinetics did not depend on the number of carbohydrates, however, since monoglycosylated N2Q3 (Asn-175 only) was also processed rapidly. Furthermore, double-glycosylated N1N3 (Asn-138 + Asn-204) and monoglycosylated N1Q3 (Asn-138 only) were not transported rapidly, again indicating that rapid kinetics did not correlate with the number of carbohydrates. Rather, rapid transport to lysosomes may depend on the position and/or sequence of the novel carbohydrate site.
We can speculate on a number of possible mechanisms by which a novel carbohydrate and/or sequence at residues 175–177 (Asp-Asn-Gly mutated to Asn-Asn-Ser) might affect the transport of proCL:
1. Asn-175 might lie in a domain that, when modified with carbohydrate, binds MPR with higher affinity than the wild-type Asn-204 domain. This would be consistent with observations that proCL normally has relatively low binding affinity for MPR (Dong et al., 1989; Lazzarino and Gabel, 1990). However, we have not measured the MPR binding affinity of our mutants, and it is not clear whether a higher affinity, if it exists, would necessarily alter the relative amounts of secreted versus lysosomal proCL, as the authors of those studies have suggested. Nevertheless, if it is MPR affinity that is affecting transport kinetics of our mutants, then a normally low affinity may be an essential feature of wild-type proCL localization, so that secretion under certain growth conditions can be achieved.
2. The new primary amino acid sequence at residues 175–177 might directly enhance recognition by MPR. This would be consistent with the hypothesis that the affinity of proCL for MPR is normally low because of a noncarbohydrate domain that interferes with binding (Lazzarino and Gabel, 1990). Thus Asn-Asn-Ser at residues 175–177 could either create a new motif that enhances MPR binding or disrupt a motif that normally inhibits binding. Our results indicate that carbohydrate at Asn-175 is crucial for the rapid transport kinetics, suggesting that primary amino acid sequence at residues 175–177 is not sufficient for conferring this phenotype. Nevertheless, sequence and carbohydrate could somehow work in concert to enhance MPR binding and transport. Again, this binding and transport effect need not inhibit proCL secretion if that trafficking pathway is regulated by a mechanism other than MPR binding.
3. The new sequence at residues 175–177 might enhance interaction with the proCL-specific lysosomal proenzyme receptor (LPR) proposed by McIntyre and Erickson (1993). LPR is a 43-kDa integral membrane protein that specifically recognizes a nine-amino acid sequence in the profragment of proCL (McIntyre et al., 1994). Binding to LPR is carbohydrate-independent (McIntyre and Erickson, 1991). Enhanced binding to LPR by our mutants could indirectly enhance their interaction with the MPR-dependent pathway, when carbohydrate is present, giving us the rapid-transport phenotype. According to our computer modeling of cathepsin L (Figure 1) and the published crystal structure of proCL (Coulombe et al., 1996), residues 175–177 are in a region that might interact with the profragment, although not within the nine-amino acid domain previously identified as important for LPR binding (McIntyre et al., 1994). Using our model of cathepsin L, we predict that the mutation at residues 175–177 would not cause any major changes to the overall conformation of the protein (Sherman and Kane, unpublished observations). Nevertheless, we cannot rule out an indirect effect of the mutation on the profragment and its interaction with LPR or a more direct effect on other protein–protein interactions via the 175–177 region. The fact that we eliminated a negatively charged amino acid in making the Asn-175 glycosylation signal might support a direct effect on protein–protein interactions.
4. The new primary sequence at residues 175–177 might affect interaction with the carbohydrate-dependent pathway as mediated by an unknown factor(s) in the cellular transport machinery. This will be the topic of future research.
A second, somewhat surprising observation was that completely unglycosylated human proCLs were transported to lysosomes by NIH3T3 mouse fibroblasts. This conclusion is based on the appearance of a mature form of unglycosylated N2Q3, both on steady-state labeling and pulse-chase labeling (Figures 3, 4, and 5), and the appearance of mature human proteins in transfected NIH3T3 cells treated with tunicamycin (Figure 8, panel A). Again, the production of this band seemed to be accurate (Figure 5, panel F), and it appeared to take place in lysosomes (Figure 6). Lysosomal delivery of unglycosylated N2Q3 protein was not rapid but occurred with the same kinetics as for wild-type, glycosylated proCL (Figure 5, panel E).
We have previously reported that recombinant mouse proCL that lacks carbohydrate is quantitatively secreted by NIH3T3 cells, even though it is properly folded, stable to incubation at 37°C (pH 7), and capable of being proteolytically processed during incubation at pH 4 (Kane, 1993). We also did not observe any lysosomal form of unglycosylated, endogenous human proCL in tunicamycin-treated cell lines (Figure 8, panel B) or of unglycosylated, recombinant mouse proCL expressed in human hepatoma cell lines (Figure 8, panel C). It is possible that unglycosylated protein was delivered to lysosomes in these experiments, but that the protein was rapidly degraded in the human cell lines. However, we think this is unlikely given our previous analysis of unglycosylated proCL (Kane, 1993, and this work) and the fact that recombinant cathepsin L purified from bacteria is stable and enzymatically active (Smith et al., 1989). Therefore, there may be species or cell type specificity to the MPR-independent transport of human proCL in NIH3T3 cells.
Other lysosomal enzymes appear to be transported quite efficiently by an MPR-independent mechanism in some cell types. For example, human hepatocytes and lymphocytes transport unglycosylated proCD to lysosomes (Glickman and Kornfeld, 1993; Rijnboutt et al., 1991b). In addition, fibroblasts isolated from knock-out mice lacking both the cation-independent MPR (MPR300) and the cation-dependent MPR (MPR46) still exhibit some residual lysosomal transport of proCD. Therefore, these fibroblasts must have a functional MPR-independent pathway, at least in the absence of MPR-mediated transport (Ludwig et al., 1994; Pohlmann et al., 1995).
ProCL and proCD are thought to associate with membranes in a carbohydrate-independent manner in fibroblasts, macrophages, and hepatocytes (Diment et al., 1988; McIntyre and Erickson, 1991; Rijnboutt et al., 1991a; Zhu and Conner, 1994). For proCL, membrane association is low pH-dependent and is mediated by the 43-kDa LPR (McIntyre and Erickson, 1991, 1993). For proCD, membrane association is also pH-dependent (McIntyre and Erickson, 1991) and appears to involve another lysosomal protein, prosaposin (Zhu and Conner, 1994). However, it is not known whether carbohydrate-independent membrane association is part of the normal MPR-mediated transport pathway or is related to an MPR-independent transport mechanism, or both. Further work with proCL glycosylation mutants showing altered transport phenotypes may help to clarify this issue.
ACKNOWLEDGMENTS
The authors wish to thank S.J. Currier, A.H. Erickson, S. Gal, J. Momand, and S. Pfeffer for their input into the design of experiments and the writing of the manuscript. This work was partially supported by a grant to S.E.K. from the American Cancer Society (JFRA-382).
REFERENCES
- Achkar C, Gong Q, Frankfater A, Bajkowski AS. Differences in targeting and secretion of cathepsins B and L by BALB/3T3 fibroblasts and Moloney murine sarcoma virus-transformed BALB/3T3 fibroblasts. J Biol Chem. 1990;265:13650–13654. [PubMed] [Google Scholar]
- Baker EN. Structure of actinidin, after refinement at 1.7 Å resolution. J Mol Biol. 1980;141:441–484. doi: 10.1016/0022-2836(80)90255-7. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The protein data bank: A computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blundell TL, Sibanda BL, Sternberg MJE, Thorton JM. Knowledged-based prediction of protein structures and the design of novel molecules. Nature. 1987;326:347–352. doi: 10.1038/326347a0. [DOI] [PubMed] [Google Scholar]
- Boyer MJ, Tannock IF. Lysosomes, lysosomal enzymes, and cancer. Adv Cancer Res. 1993;60:269–291. doi: 10.1016/s0065-230x(08)60828-3. [DOI] [PubMed] [Google Scholar]
- Braulke T, Hasilik A, von Figura K. Low temperature blocks transport and sorting of cathepsin D in fibroblasts. Biol Chem Hoppe-Seyler. 1988;369:441–449. doi: 10.1515/bchm3.1988.369.1.441. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Kornfeld S. Phosphorylation of Asn-linked oligosaccharides located at novel sites on the lysosomal enzyme cathepsin D. J Biol Chem. 1992;267:23357–23363. [PubMed] [Google Scholar]
- Capony F, Braulke T, Rougeot C, Roux S, Montcourrier P, Rochefort H. Specific mannose 6-phosphate receptor-independent sorting of pro-cathepsin D in breast cancer cells. Exp Cell Res. 1994;215:154–163. doi: 10.1006/excr.1994.1327. [DOI] [PubMed] [Google Scholar]
- Capony F, Rougeot C, Montcourrier P, Cavailles V, Salazar G, Rochefort H. Increased secretion, altered processing, and glycosylation of pro-cathepsin D in human mammary cancer cells. Cancer Res. 1989;49:3904–3909. [PubMed] [Google Scholar]
- Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Structure and energetics of ligand binding to proteins: E. coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1988;4:31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Diment S, Leech MS, Stahl PD. Cathepsin D is membrane-associated in macrophage endosomes. J Biol Chem. 1988;263:6901–6907. [PubMed] [Google Scholar]
- Dong J, Prence EM, Sahagian GG. Mechanism for selective secretion of a lysosomal protease by transformed mouse fibroblasts. J Biol Chem. 1989;264:7377–7383. [PubMed] [Google Scholar]
- Dong J, Sahagian GG. Basis for low affinity binding of a lysosomal cysteine protease to the cation-independent mannose 6-phosphate receptor. J Biol Chem. 1990;265:4210–4217. [PubMed] [Google Scholar]
- Fortenberry SC, Schorey JS, Chirgwin JM. Role of glycosylation in the expression of human procathepsin D. J Cell Sci. 1995;108:2001–2006. doi: 10.1242/jcs.108.5.2001. [DOI] [PubMed] [Google Scholar]
- Gal S, Gottesman MM. Isolation and sequence of a cDNA for human pro-(cathepsin L) Biochem J. 1988;253:303–306. doi: 10.1042/bj2530303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal S, Willingham MC, Gottesman MM. Processing and lysosomal localization of a glycoprotein whose secretion is transformation stimulated. J Cell Biol. 1985;100:535–544. doi: 10.1083/jcb.100.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman JN, Kornfeld S. Mannose 6-phosphate-independent targeting of lysosomal enzymes in I-cell disease B lymphoblasts. J Cell Biol. 1993;123:99–108. doi: 10.1083/jcb.123.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Noriega A, Grubb JH, Talkad V, Sly WS. Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. J Cell Biol. 1980;85:839–852. doi: 10.1083/jcb.85.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM. Transformation-dependent secretion of a low molecular weight protein by murine fibroblasts. Proc Natl Acad Sci USA. 1978;75:2767–2771. doi: 10.1073/pnas.75.6.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Cabral F. Purification and characterization of a transformation dependent protein secreted by cultured murine fibroblasts. Biochemistry. 1981;20:1659–1665. doi: 10.1021/bi00509a039. [DOI] [PubMed] [Google Scholar]
- Hasilik A, von Figura K. Oligosaccharides in lysosomal enzymes. Eur J Biochem. 1981;121:125–129. doi: 10.1111/j.1432-1033.1981.tb06440.x. [DOI] [PubMed] [Google Scholar]
- Isidoro C, Demoz M, de Stefanis D, Baccino FM, Bonelli G. Synthesis, maturation and extracellular release of procathepsin D as influenced by cell proliferation or transformation. Int J Cancer. 1995;63:866–871. doi: 10.1002/ijc.2910630619. [DOI] [PubMed] [Google Scholar]
- Isidoro C, Demoz M, de Stefanis D, Baccino FM, Hasilik A, Bonelli G. Differential targeting and processing of procathepsin D in normal and transformed murine 3T3 fibroblasts. Int J Cancer. 1997;70:310–314. doi: 10.1002/(sici)1097-0215(19970127)70:3<310::aid-ijc11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Isidoro C, Horst M, Baccino FM, Hasilik A. Differential segregation of human and hamster cathepsin D in transfected baby-hamster kidney cells. Biochem J. 1991;273:363–367. doi: 10.1042/bj2730363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa H, Nikawa T, Tagami K, Kamioka H, Sumitani K, Kawata T, Drobnic-Kosorok M, Lenarcic B, Turk V, Katunuma N. Participation of cathepsin L on bone resorption. FEBS Lett. 1993;321:247–250. doi: 10.1016/0014-5793(93)80118-e. [DOI] [PubMed] [Google Scholar]
- Kamphuis IG, Kalk KH, Swarte MBA, Drenth J. Structure of papain refined at 1.65 Å resolution. J Mol Biol. 1984;179:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- Kane SE. Mouse procathepsin L lacking a functional glycosylation site is properly folded, stable, and secreted by NIH 3T3 cells. J Biol Chem. 1993;268:11456–11462. [PubMed] [Google Scholar]
- Kane SE, Gottesman MM. The role of cathepsin L in malignant transformation. Semin Cancer Biol. 1990;1:127–136. [PubMed] [Google Scholar]
- Kane SE, Reinhard DH, Fordis CM, Pastan I, Gottesman MM. A new vector using the human multidrug resistance gene as a selectable marker enables overexpression of foreign genes in eukaryotic cells. Gene. 1989;84:439–446. doi: 10.1016/0378-1119(89)90518-0. [DOI] [PubMed] [Google Scholar]
- Kane SE, Troen BR, Gal S, Ueda K, Pastan I, Gottesman MM. Use of a cloned multidrug resistance gene for coamplification and overproduction of major excreted protein, a transformation-regulated secreted acid protease. Mol Cell Biol. 1988;8:3316–3321. doi: 10.1128/mcb.8.8.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarino D, Gabel CA. Protein determinants impair recognition of procathepsin L phosphorylated oligosaccharides by the cation-independent mannose 6-phosphate receptor. J Biol Chem. 1990;265:11864–11871. [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T, Le Borgne R, Hoflack B. Roles for mannose 6-phosphate receptors in lysosomal enzyme sorting, IGF-II binding and clathrin-coat assembly. Trends Cell Biol. 1995;5:202–206. doi: 10.1016/s0962-8924(00)89000-5. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Munier-Lehmann H, Bauer U, Hollinshead M, Ovitt C, Lobel P, Hoflack B. Differential sorting of lysosomal enzymes in mannose 6-phosphate receptor-deficient fibroblasts. EMBO J. 1994;13:3430–3437. doi: 10.1002/j.1460-2075.1994.tb06648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciewicz, R.A., Wardale, R.J., Wotton, S.F., Duance, V.C., and Etherington, D.J. (1990). Mode of activation of the precursor to cathepsin L: implication for matrix degradation in arthritis. Biol. Chem. Hoppe-Seyler 371 (suppl), 223–228. [PubMed]
- McIntyre GF, Erickson AH. Procathepsins L and D are membrane-bound in acidic microsomal vesicles. J Biol Chem. 1991;266:15438–15445. [PubMed] [Google Scholar]
- McIntyre GF, Erickson AH. The lysosomal proenzyme receptor that binds procathepsin L to microsomal membranes at pH 5 is a 43-kDa integral membrane protein. Proc Natl Acad Sci USA. 1993;90:10588–10592. doi: 10.1073/pnas.90.22.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre GF, Godbold GD, Erickson AH. The pH-dependent membrane association of procathepsin L is mediated by a 9-residue sequence within the propeptide. J Biol Chem. 1994;269:567–572. [PubMed] [Google Scholar]
- Nielsen-Hamilton M, Hamilton RT, Adams G. Rapid selective stimulation by growth factors of the incorporation by Balb/c 3T3 cells of [35S]methionine into a glycoprotein and five superinducible proteins. Biochem Biophys Res Commun. 1982;108:158–166. doi: 10.1016/0006-291x(82)91845-9. [DOI] [PubMed] [Google Scholar]
- Owada M, Neufeld EF. Is there a mechanism for introducing acid hydrolases into liver lysosomes that is independent of mannose 6-phosphate recognition? Evidence from I-cell disease. Biochem Biophys Res Commun. 1982;105:814–820. doi: 10.1016/0006-291x(82)91042-7. [DOI] [PubMed] [Google Scholar]
- Pagano M, Dalet-Fumeron V, Engler R. The glycosylation state of the precursors of the cathepsin B-like proteinase from human malignant ascitic fluid: possible implication in the secretory pathway of these proenzymes. Cancer Lett. 1989;45:13–19. doi: 10.1016/0304-3835(89)90030-x. [DOI] [PubMed] [Google Scholar]
- Pohlmann R, Wendland M, Boeker C, von Figura K. The two mannose 6-phosphate receptors transport distinct complement of lysosomal proteins. J Biol Chem. 1995;270:27311–27318. doi: 10.1074/jbc.270.45.27311. [DOI] [PubMed] [Google Scholar]
- Prence EM, Dong J, Sahagian GG. Modulation of the transport of a lysosomal enzyme by PDGF. J Cell Biol. 1990;110:319–326. doi: 10.1083/jcb.110.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VY, Zhang Z-Y, Weiss SJ. Pericellular mobilization of the tissue-destructive cysteine proteinases, cathepsins B, L, and S, by human monocyte-derived macrophages. Proc Natl Acad Sci USA. 1995;92:3849–3853. doi: 10.1073/pnas.92.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnboutt S, Aerts HMFG, Geuze HJ, Tager JM, Strous GJ. Mannose 6-phosphate-independent membrane association of cathepsin D, glucocerebrosidase, and sphingolipid-activating protein in HepG2 cells. J Biol Chem. 1991a;266:4862–4868. [PubMed] [Google Scholar]
- Rijnboutt S, Kal AJ, Geuze HJ, Aerts H, Strous GJ. Mannose 6-phosphate-independent targeting of cathepsin D to lysosomes in HepG2 cells. J Biol Chem. 1991b;266:23586–23592. [PubMed] [Google Scholar]
- Sahagian GG, Gottesman MM. The predominant secreted protein of transformed murine fibroblasts carries the lysosomal mannose 6-phosphate recognition marker. J Biol Chem. 1982;257:11145–11150. [PubMed] [Google Scholar]
- Sloane BF, Honn KV, Sadler JG, Turner WA, Kimpson JJ, Taylor JD. Cathepsin B activity in B16 melanoma cells: a possible marker for metastatic potential. Cancer Res. 1982;42:980–986. [PubMed] [Google Scholar]
- Smith SM, Kane SE, Gal S, Mason RW, Gottesman MM. Glycosylation of procathepsin L does not account for species molecular-mass differences and is not required for proteolytic activity. Biochem J. 1989;262:931–938. doi: 10.1042/bj2620931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A, Omura K, Suzuki Y. I-cell disease: evidence for a mannose 6-phosphate independent pathway for translocation of lysosomal enzymes in lymphoblastoid cells. Clin Chim Acta. 1988;176:115–122. doi: 10.1016/0009-8981(88)90181-7. [DOI] [PubMed] [Google Scholar]
- Ulbricht B, Hagmann W, Ebert W, Spiess E. Differential secretion of cathepsins B and L from normal and tumor human lung cells stimulated by 12(S)-hydroxy-eicosatetraenoic acid. Exp Cell Res. 1996;226:255–263. doi: 10.1006/excr.1996.0226. [DOI] [PubMed] [Google Scholar]
- von Figura K, Hasilik A. Lysosomal enzymes and their receptors. Annu Rev Biochem. 1986;55:167–193. doi: 10.1146/annurev.bi.55.070186.001123. [DOI] [PubMed] [Google Scholar]
- Waheed A, Pohlmann R, Hasilik A, von Figura K, van Elsen A, Leroy JG. Deficiency of UDP-N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase in organs of I-cell patients. Biochem Biophys Res Commun. 1982;105:1052–1058. doi: 10.1016/0006-291x(82)91076-2. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Conner GE. Intermolecular association of lysosomal protein precursors during biosynthesis. J Biol Chem. 1994;269:3846–3851. [PubMed] [Google Scholar]