Abstract
Background and Aims
This study examines the pattern of genetic variability and genetic relationships of wild olive (Olea europaea subsp. europaea var. sylvestris) populations in the north-western Mediterranean. Recent bottleneck events are also assessed and an investigation is made of the underlying population structure of the wild olive populations.
Methods
The genetic variation within and between 11 wild olive populations (171 individuals) was analysed with eight microsatellite markers. Conventional and Bayesian-based analyses were applied to infer genetic structure and define the number of gene pools in wild olive populations.
Key Results
Bayesian model-based clustering identified four gene pools, which was in overall concordance with the Factorial Correspondence Analysis and Fitch–Margoliash tree. Two gene pools were predominantly found in southern Spain and Italian islands, respectively, in samples gathered from undisturbed forests of the typical Mediterranean climate. The other two gene pools were mostly detected in the north-eastern regions of Spain and in continental Italy and belong to the transition region between the temperate and Mediterranean climate zones.
Conclusions
On the basis of these results, it can be assumed that the population structure of wild olives from the north-western Mediterranean partially reflects the evolutionary history of these populations, although hybridization between true oleasters and cultivated varieties in areas of close contact between the two forms must be assumed as well. The study indicates a degree of admixture in all the populations, and suggests some caution regarding genetic differentiation at the population level, making it difficult to identify clear-cut genetic boundaries between candidate areas containing either genuinely wild or feral germplasm.
Key words: Olea europaea, genetic variability, gene pools, microsatellites, oleasters, population structure
INTRODUCTION
Olive, the emblematic tree of the Mediterranean Basin, is found in two forms, namely wild (Olea europaea subsp. europaea var. sylvestris) and cultivated (Olea europaea subsp. europaea var. europaea) (Green, 2002). Cultivated and wild olives have the same chromosome number (2n = 2x = 46) (Contento et al., 2002), are interfertile (Zohary and Spiegel-Roy, 1975; Besnard and Bervillé, 2000) and show very good grafting affinity (Mulas and Deidda, 1998). Wild olives reproduce sexually, are pollinated by wind and their seeds are mainly dispersed by birds (Alcantara and Rey, 2003). The presence of wild olives is considered the best bioindicator of the Mediterranean Floristic Region (Rubio et al., 2002).
Wild olives have colonized many Mediterranean environments, characterized by semi-arid climatic conditions with different altitudes, plant communities and soils, including those with extreme levels of salinity (Zohary and Spiegel-Roy, 1975). The distribution of genetic diversity of oleaster populations has been reconstructed on the basis of recolonization of the Mediterranean basin from Ice Age refugia located in western and eastern regions (Breton et al., 2006). From an ecological point of view, wild olive populations fulfil a role in protecting soils against desertification, due to their great resistance to wind and drought, their ability to resprout after fire or frost and, particularly, their very long life span, which allows plants to survive up to many thousands of years (Mulas and Deidda, 1998). Despite all these characteristics, wild olives have been considered of low value in sustainable forestry, and only in a few recent cases have they been used for reforestation purposes with funding from the European Union. Thus far, little selection work on wild olives has been carried out for their potential in silviculture or as a source of valuable traits to introduce into cultivars. The long coexistence of the two forms of olive in the Mediterranean has been inferred from archaeological and paleobotanical findings (Terral et al., 2004).
Wild olives include true oleasters (wild forms present in natural areas) and feral forms, which may be seedlings of the cultivated clones or products of hybridization between the true oleasters and cultivars (Zohary and Hopf, 1994; Angiolillo et al., 1999; Lumaret et al., 2004). Morphological distinction between these two forms is difficult owing to their similar phenotypes (Bronzini de Caraffa et al., 2002a). Attempts have been made to establish criteria to distinguish oleasters from cultivated forms clearly taking into account suitable climatic conditions for wild olive growth, isolation from areas of cultivation and sufficiently large areas in order to minimize the influence of occasional pollen and seed flow from cultivated areas (Lumaret et al., 2004), but the interrelationships between the two forms of olive appear quite complex.
Genetic variation of wild olives and particularly their relationships with cultivars have been evaluated by allozymes, randomly amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP) and cytoplasmic (chloroplast and mitochondrial) DNA markers, both in restricted areas (Angiolillo et al., 1999; Vargas and Kadereit, 2001; Bronzini de Caraffa et al., 2002a,b) and over the whole Mediterranean Basin (Besnard et al., 2002a,b; Lumaret et al., 2004). Evidence of the survival of indigenous oleaster populations in the western Mediterranean including Spain and Italy has been provided by allozymes (Lumaret and Ouazzani, 2001; Lumaret et al., 2004), inter-simple sequence repeats (ISSRs; Vargas and Kadereit, 2001) and simple sequence repeats (SSRs) (Breton et al., 2006). These markers, together with cytoplasmic variation, have shown a clear distinction between eastern and western Mediterranean oleasters (Besnard et al., 2002b; Lumaret et al., 2004). Only a few local western Mediterranean cultivars presented an allelic distribution similar to that observed in the oleaster populations growing in the same areas (Lumaret et al., 2004). Detailed analyses at a lower scale may produce new insights and give a better understanding of the distribution of genetic diversity at the regional level (Baldoni et al., 2006).
The present study examines both the level of genetic diversity in wild olive and its distribution in the north-western Mediterranean, an area of intense olive cultivation. Samples were collected from 11 wild olive populations in different regions of Spain and Italy, an area where wild and cultivated forms have long coexisted (Terral et al., 2004), and their genetic diversity was assessed with eight microsatellite markers. The specific objectives were: (1) to study the genetic diversity within and among populations, (2) to investigate the genetic relationships among populations, (3) to estimate the level of inbreeding and assess recent bottleneck events, and (4) to detect underlying population structure in wild olive populations.
MATERIAL AND METHODS
Sampling of wild olive populations
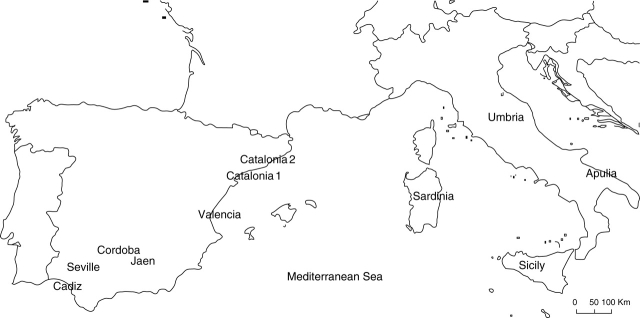
Eleven wild olive populations from Italy and Spain were chosen for the analysis by means of eight SSR markers (Fig. 1, Table 1); seven populations were from southern and north-eastern Spain, while the remaining four populations came from the Italian islands and mainland. A total of 171 individuals were sampled with at least 14 individuals per population (Table 2). Some collection sites are representatives of the typical Mediterranean climate while others are located at the border with the Eurosiberian region (Fig. 1).
Fig. 1.
Locations of the wild olive populations sampled.
Table 1.
Source, repeat motifs, size ranges, number of alleles (Na), observed (Ho) and expected heterozygosity (He) and polymorphic information content (PIC) for eight microsatellite loci analysed in 11 wild olive populations
| Source | Locus | Repeat motif | Size range | Na | Ho | He | PIC |
|---|---|---|---|---|---|---|---|
| Sefc et al. (2000) | ssrOeUA-DCA03 | (GA)19 | 227–255 | 13 | 0·679 | 0·722 | 0·696 |
| ssrOeUA-DCA09 | (GA)23 | 160–210 | 23 | 0·909 | 0·920 | 0·914 | |
| ssrOeUA-DCA16 | (GT)13(GA)29 | 122–222 | 37 | 0·914 | 0·929 | 0·925 | |
| ssrOeUA-DCA18 | (CA)4CT(CA)3(GA)19 | 158–182 | 15 | 0·888 | 0·902 | 0·893 | |
| de la Rosa et al. (2002) | EMO03 | A15(CA)7 | 203–218 | 10 | 0·710 | 0·822 | 0·801 |
| Cipriani et al. (2002) | UDO99-019 | (GT)20(AT)5 | 98–166 | 10 | 0·432 | 0·635 | 0·598 |
| UDO99-039 | (AT)5(GT)11 | 139–190 | 18 | 0·503 | 0·894 | 0·885 | |
| UDO99-043 | (GT)12 | 165–190 | 22 | 0·704 | 0·917 | 0·911 | |
| Mean | 18·5 | 0·717 | 0·843 | 0·828 | |||
| Min. | 10 | 0·432 | 0·635 | 0·598 | |||
| Max. | 37 | 0·914 | 0·929 | 0·925 |
Table 2.
Population, sample size and genetic variability estimates based on data from eight microsatellite loci in 11 wild olive populations
| No. | Population | n | Nat | Nr | Nal | Nar | Npr | Ho | He |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Cadiz | 16 | 78 | 34 | 9·75 | 8·14 | 4 | 0·727 | 0·732 |
| 2 | Seville | 15 | 73 | 25 | 9·13 | 7·93 | 3 | 0·616 | 0·731 |
| 3 | Cordoba | 14 | 72 | 27 | 9·00 | 8·14 | 0 | 0·753 | 0·779 |
| 4 | Jaen | 15 | 78 | 26 | 9·75 | 8·38 | 2 | 0·713 | 0·766 |
| 5 | Valencia | 16 | 72 | 24 | 9·00 | 7·64 | 3 | 0·734 | 0·749 |
| 6 | Catalonia 1 | 16 | 74 | 36 | 9·25 | 7·43 | 2 | 0·792 | 0·731 |
| 7 | Catalonia 2 | 15 | 54 | 20 | 6·75 | 5·86 | 1 | 0·619 | 0·654 |
| 8 | Sardinia | 18 | 76 | 32 | 9·50 | 7·64 | 3 | 0·757 | 0·770 |
| 9 | Sicily | 16 | 74 | 34 | 9·25 | 7·81 | 7 | 0·703 | 0·746 |
| 10 | Apulia | 15 | 75 | 23 | 9·38 | 8·29 | 4 | 0·768 | 0·782 |
| 11 | Umbria | 15 | 68 | 25 | 8·50 | 7·34 | 1 | 0·689 | 0·737 |
n, number of individuals per population; Nat, total number of alleles per population; Nr, number of rare alleles (alleles present in fewer than 5 % of the individuals) per population; Nal, number of alleles per locus; Nar, number of alleles per locus independent of sample size (allelic richness); Npr, number of private alleles per population; observed (Ho) and expected heterozygosity (He).
Although Andalusia (southern Spain) represents the most intensive area of olive cultivation in the world, the samples from this region (Cadiz, Cordoba, Jaen and Seville) were wild individuals taken from large undisturbed forest areas. In contrast, the north-eastern Spanish regions of Catalonia and Valencia lack extensive wild olive forests, and the majority of wild individuals from here were sampled near olive groves. Catalonia is represented by two populations (Catalonia 1 and Catalonia 2) collected in the southern part of the region (Tarragona), the area where most wild olives are concentrated (J. Tous, IRTA, Spain, pers. comm.). Catalonia 1 includes individuals sampled in Baix-Ebre Montsia (southern Tarragona) and Catalonia 2 samples collected in Priorat (northern Tarragona).
In Italy, wild olive populations were collected from two mainland regions (Apulia and Umbria) and two islands (Sicily and Sardinia). Apulia is represented by samples collected in the northern (Gargano-Foggia), central (Bari) and southern (Lecce) parts of the region, where some trees were found close to cultivated fields. In Umbria, plants were sampled from an isolated, woody area in the southern part of the region (Terni) as well as from a site near to cultivated olive fields in the central part (Perugia). The islands were chosen because of their location in the centre of the Mediterranean area and as representatives of the most favourable conditions for wild olive expansion. The wild population of Sardinia is composed of individuals collected from different sites, all of which were located in undisturbed areas. In Sicily, the wild individuals were sampled in the western, eastern and south-eastern area of the island, mostly from sites quite close to cultivated fields.
DNA extraction and microsatellite genotyping
Total genomic DNA was extracted from mature leaf tissue collected from 1-year-old shoots in the upper part of the trees. DNA from Spanish wild populations was extracted following the protocol described by de la Rosa et al. (2002) while the method described by Angiolillo et al. (1999) was used for the DNA extraction of samples from Italy. The PCR conditions described by de la Rosa et al. (2002) were used for the amplifications of the eight SSR (Table 1) primer pairs (Sefc et al., 2000; Cipriani et al., 2002; de la Rosa et al., 2002).
The detection of amplification products was carried out with an automated sequencer (ABI PRISM 3100 DNA sequencer, Applied Biosystems, Foster City, CA, USA) at the Unit of Genomics of the Central Service for Research Support of the University of Córdoba (Spain). Sizing was performed using the program genescan 3·7 and genotyper 3·7 from Applied Biosystems. Three reference samples were used in all runs.
Data analysis
The average number of alleles per locus (Na), the observed heterozygosity (Ho), the expected heterozygosity or gene diversity (He) and the Polymorphism Information Content (PIC) for each microsatellite locus as well as the average number of alleles Nal, Ho, He and inbreeding coefficient (f) in each population across loci were calculated using PowerMarker v3·23 software (Liu, 2002). The allelic richness (Nar) was calculated using fstat v2·9·3·2 program package (Goudet, 1995). The number of ‘private’ alleles (Npr) was assessed by microsat (Minch et al., 1997).
genepop v3·4 (Raymond and Rousset, 1995) was used to test genotypic frequencies for conformance to Hardy–Weinberg (HW) expectations, to test the loci for linkage disequilibrium and to estimate the significance of genotypic differentiation between population pairs. All probability tests were based on the Markov chain method (Guo and Thompson, 1992; Rousset and Raymond, 1995) using 10 000 de-memorization steps, 100 batches and 5000 iterations per batch. The sequential Bonferroni adjustment (Holm, 1979; Rice, 1989) was applied to correct for the effect of multiple tests using SAS Release 8·02 (SAS Institute, 1999).
The program bottleneck v1·2·02 (Cornuet and Luikart, 1996; Piry et al., 1999) was used to test for evidence of recent bottleneck events on the basis of theoretical expectations under the Two-Phase mutation Model (TPM), assuming 5 % multistep changes as suggested by di Rienzo et al. (1994).
A factorial correspondence analysis (FCA) was carried out using Genetix 4·05 (Belkhir et al., 2004) in order to represent genetic relationships among individual olive trees graphically.
The proportion-of-shared-alleles distance (Bowcock et al., 1994) between pairs of individuals was calculated using microsat (Minch et al., 1997) and the distance matrix was subjected to the analysis of molecular variance (AMOVA) approach using the winamova v1·55 program (Excoffier et al., 1992). The AMOVA was also performed to examine hierarchical structure considering among- and within-population diversity. The significance of the ϕ statistics was tested non-parametrically by 1000 permutations.
Pairwise standard genetic distances (Nei, 1972) were calculated and an unrooted tree was constructed using a least-squares algorithm (Fitch and Margoliash, 1967) with 10 000 bootstraps over microsatellite loci as implemented in the phylip v3·6b software package (Felsenstein, 1993).
A model-based clustering method was applied to multilocus microsatellite data to infer genetic structure and define the number of clusters (gene pools) in the dataset using the software structure (Pritchard et al., 2000). Six runs of structure were done by setting the number of clusters (K) from 1 to 11 (number of sampled populations). Each run consisted of a burn-in period of 200 000 steps followed by 106 MCMC (Monte Carlo Markov Chain) replicates, assuming an admixture model and correlated allele frequencies. No prior information was used to define the clusters. The choice of the most likely number of clusters (K) was carried out comparing log probabilities of data [Pr(X|K)] for each value of K (Pritchard et al., 2000), as well as by calculating an ad hoc statistic ΔK based on the rate of change in the log probability of data between successive K values, as described by Evanno et al. (2005). The runs with the highest ln Pr(X|K) values were chosen and the proportion of ancestry of each population in each of the gene pools was calculated by averaging the estimated membership coefficients of the individuals i.e. ancestry estimates. On the individual level, the admixture level was analysed by assigning the individual olive trees sampled to a gene pool if an arbitrary value of 75 % of their genome was estimated to belong to that gene pool (Matsuoka et al., 2002), while those individuals with membership probabilities of < 75 % for all gene pools were considered as of ‘mixed origin’ (see Supplementary Information, available online).
RESULTS
Overall microsatellite diversity
A total of 148 alleles were observed across the eight markers, the number of alleles per locus ranging from ten (EMO03; UDO99-019) to 37 (ssrOeUA-DCA16) with a mean value of 18·5 alleles per locus (Table 1). For all loci, most alleles varied in steps of two nucleotides and only loci UDO99-019 and ssrOeUA-DCA16 showed alleles that differed from the others by 10 and 12 nucleotides, respectively. Some odd-sized alleles were clearly identified in UDO99-039, and a few alleles differing by one nucleotide were found for loci UDO99-043 and EMO03. The repeat motif of the latter locus may explain the presence of alleles differing by only one nucleotide. The allelic frequencies for each locus were generally low with many alleles detected in only one or a few individuals (data not shown). Only three loci (ssrOeUA-DCA03, EMO03 and UDO99-019) displayed alleles with (overall) frequency values greater than 0·30. Allelic frequency values ranged from 0·003 (alleles present in single individual) to 0·55.
Overall observed heterozygosity values (per marker) ranged from 0·432 to 0·914, with a mean value of 0·717. The expected heterozygosities showed slightly higher values than the observed heterozygosities, with values ranging from 0·635 to 0·929 and a mean value of 0·843. All microsatellite loci scored in this study were highly polymorphic, displaying high values of PIC (from 0·598 to 0·925).
Intrapopulation diversity and HW equilibrium
The total number of alleles per population ranged from 54 (Catalonia 2) to 78 (Cadiz). Low values of allelic frequencies were observed and, as a consequence, a high number of rare alleles (alleles present in fewer than 5 % of the individuals in a population) was observed for each population (Table 2). The number of rare alleles ranged from 20 (Catalonia 2) to 36 (Catalonia 1). The mean number of alleles per locus and per population ranged from 6·75 (Catalonia 2) to 9·75 (Cadiz and Jaen). The number of alleles per locus independent of sample size (the allelic richness) ranged from 5·86 (Catalonia 2) to 8·38 (Jaen). The populations with most private alleles were Sicily (seven) together with Apulia and Cadiz (four), while no such alleles were found in the population of Cordoba. The observed heterozygosity per population ranged from 0·616 (Seville) to 0·792 (Catalonia 1) and the expected heterozygosity per population ranged from 0·654 (Catalonia 2) to 0·782 (Apulia). The population Catalonia 2 had the lowest values among all the populations for all the genetic variability estimates.
No significant departures from HW equilibrium were observed at most loci or in most of the populations (Table 3). No consistent patterns across loci and populations were observed. The Wahlund effect cannot be completely ignored because in some cases the sampled trees within a region were geographically distant. The presence of null alleles would be a likely explanation for significant deviations from HW equilibrium in the case of locus UDO-39 in several populations. When the recent bottleneck history of olive populations from Spain and Italy was assessed using the TPM, a recent reduction in effective population size (genetic bottleneck) was found only for Catalonia 1.
Table 3.
Inbreeding coefficients f (Weir and Cockerham, 1984) and expected heterozygosities (He) across eight microsatellite loci in 11 olive populations
| No. | Population | DCA-03 | DCA-09 | DCA-16 | DCA-18 | EMO-03 | UDO-19 | UDO-39 | UDO-43 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Cadiz | f | −0·096 | −0·103 | 0·007 | −0·006 | 0·028 | 0·390 | 0·158 | 0·311 |
| He | 0·548 | 0·837 | 0·944 | 0·862 | 0·900 | 0·513 | 0·871 | 0·830 | ||
| 2 | Seville | f | 0·080 | 0·154 | 0·055 | 0·022 | 0·117 | 0·661 | 0·640** | 0·255 |
| He | 0·507 | 0·945 | 0·917 | 0·886 | 0·906 | 0·590 | 0·794 | 0·863 | ||
| 3 | Cordoba | f | −0·055 | −0·117 | 0·027 | −0·099 | 0·055 | 0·378 | 0·505 | 0·254 |
| He | 0·745 | 0·896 | 0·949 | 0·910 | 0·814 | 0·690 | 0·918 | 0·894 | ||
| 4 | Jaen | f | −0·194 | −0·057 | −0·042 | 0·165 | 0·184 | 0·616* | 0·247 | 0·224 |
| He | 0·644 | 0·883 | 0·960 | 0·879 | 0·736 | 0·695 | 0·886 | 0·945 | ||
| 5 | Valencia | f | −0·152 | 0·060 | 0·012 | 0·003 | 0·138 | 0·391 | 0·437 | −0·079 |
| He | 0·752 | 0·931 | 0·885 | 0·869 | 0·798 | 0·438 | 0·888 | 0·869 | ||
| 6 | Catalonia 1 | f | −0·341 | −0·174 | −0·157 | −0·151 | −0·184 | 0·143 | 0·652** | −0·013 |
| He | 0·574 | 0·852 | 0·865 | 0·869 | 0·792 | 0·583 | 0·898 | 0·802 | ||
| 7 | Catalonia 2 | f | 0·219 | −0·098 | −0·098 | −0·180 | 0·274 | −0·167 | 0·686** | 0·394 |
| He | 0·640 | 0·850 | 0·850 | 0·848 | 0·643 | 0·400 | 0·795 | 0·660 | ||
| 8 | Sardinia | f | 0·026 | −0·151 | 0·027 | −0·034 | −0·195 | 0·166 | 0·418 | 0·292 |
| He | 0·742 | 0·869 | 0·890 | 0·859 | 0·640 | 0·799 | 0·859 | 0·914 | ||
| 9 | Sicily | f | 0·167 | 0·124 | −0·063 | −0·037 | 0·231 | 0·400 | 0·082 | 0·170 |
| He | 0·640 | 0·856 | 0·869 | 0·771 | 0·827 | 0·729 | 0·885 | 0·883 | ||
| 10 | Apulia | f | 0·064 | 0·074 | 0·154** | −0·006 | 0·282 | −0·157 | 0·222 | 0·040 |
| He | 0·855 | 0·936 | 0·929 | 0·894 | 0·836 | 0·576 | 0·857 | 0·893 | ||
| 11 | Umbria | f | 0·201 | −0·040 | −0·117 | 0·025 | 0·079 | −0·022 | 0·683** | 0·217 |
| He | 0·805 | 0·898 | 0·836 | 0·879 | 0·724 | 0·489 | 0·840 | 0·912 |
Significant deviations from Hardy–Weinberg equilibrium after sequential Bonferroni corrections: **, significance at the 1 % nominal level; *, significance at the 5 % nominal level; no marking depicts non-significant values.
Among a total of 308 tests for linkage disequilibrium between pairs of loci, 20 (approx. 6 %) were significant (P < 0·05). Only one individual test was significant after sequential Bonferroni corrections for multiple testing (Holm, 1979; Rice, 1989).
Interpopulation differentiation and genetic relationships
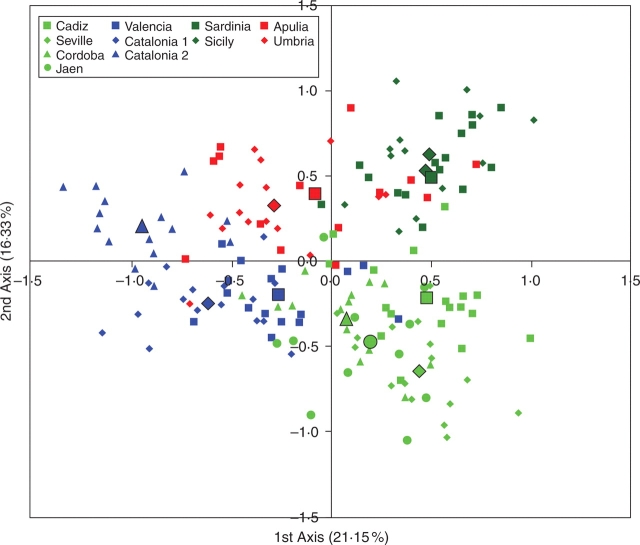
Figure 2 represents the projection of the individuals and population barycentres on the plane defined by the first two axes of the FCA. The first two axes accounted for 21·15 and 16·33 % of the total inertia, respectively. Along the first axis, most wild individuals from southern Spain (Cadiz, Seville, Cordoba, Jaen) and the Italian island populations (Sardinia, Sicily) plot separately from the majority of those collected from north-eastern Spain (Valencia, Catalonia 1, Catalonia 2) and continental Italy (Apulia, Umbria). Slight differentiation between wild individuals from southern Spain and Italian islands was observed along the second FCA axis. At the same time most individuals from Valencia and Catalonia 1 clustered separately from those sampled in Catalonia 2, Umbria and Apulia along this axis. However, some individuals plotted distant from the barycentre of their group of origin, suggesting that they could be first-generation migrants or admixed trees.
Fig. 2.
Factorial correspondence analysis (FCA) of 171 olive trees belonging to 11 populations. Each individual genotype is indicated by a small symbol, while the population barycentres are represented by larger ones.
The majority of tests for pairwise genetic differentiation among populations were significant (Table 4) except in the case of six pairwise comparisons between Spanish olive populations. Interestingly, the greatest FST values were observed between Catalonia 2 and the populations of Seville, Sardinia and Cadiz (0·129, 0·125 and 0·124, respectively). Lower but still significant values of FST were observed between the pairs of populations Sardinia–Sicily (0·022) and Apulia–Umbria (0·020). A similar pattern of differentiation among populations was seen using Nei's genetic distance.
Table 4.
Nei's standard genetic distance (upper diagonal) and pairwise FST values (lower diagonal) between 11 wild olive populations
| No. | Population | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Cadiz | 0·18 | 0·28 | 0·29 | 0·35 | 0·55 | 0·66 | 0·39 | 0·31 | 0·48 | 0·43 | |
| 2 | Seville | 0·001 | 0·24 | 0·24 | 0·32 | 0·46 | 0·72 | 0·39 | 0·38 | 0·51 | 0·50 | |
| 3 | Cordoba | 0·026* | 0·012 | 0·19 | 0·22 | 0·44 | 0·55 | 0·40 | 0·38 | 0·36 | 0·37 | |
| 4 | Jaen | 0·032* | 0·015 | 0·000 | 0·24 | 0·48 | 0·57 | 0·43 | 0·46 | 0·52 | 0·50 | |
| 5 | Valencia | 0·048*** | 0·039** | 0·011 | 0·020 | 0·32 | 0·31 | 0·43 | 0·41 | 0·26 | 0·23 | |
| 6 | Catalonia 1 | 0·088*** | 0·072*** | 0·055*** | 0·068*** | 0·046** | 0·41 | 0·71 | 0·55 | 0·42 | 0·47 | |
| 7 | Catalonia 2 | 0·124*** | 0·129*** | 0·091*** | 0·101*** | 0·057*** | 0·085*** | 0·71 | 0·63 | 0·41 | 0·29 | |
| 8 | Sardinia | 0·052*** | 0·048*** | 0·037*** | 0·049*** | 0·055*** | 0·100*** | 0·125*** | 0·25 | 0·37 | 0·43 | |
| 9 | Sicily | 0·038** | 0·050*** | 0·035*** | 0·052*** | 0·054** | 0·083*** | 0·115*** | 0·022* | 0·38 | 0·40 | |
| 10 | Apulia | 0·057*** | 0·060*** | 0·025** | 0·053*** | 0·021** | 0·053*** | 0·070*** | 0·037*** | 0·038*** | 0·27 | |
| 11 | Umbria | 0·062** | 0·070*** | 0·037** | 0·063*** | 0·021** | 0·071*** | 0·051*** | 0·057*** | 0·053*** | 0·020* |
Pairwise significance after sequential Bonferroni corrections: ***, significance at the 0·1 % nominal level; **, significance at the 1 % nominal level; *, significance at the 5 % nominal level; no marking depicts non-significant values.
The AMOVA analysis showed that although most of the genetic diversity was attributable to differences among individuals within populations (91·8 %), ϕ values among populations were significant (ϕst = 0·082; P < 0·001), suggesting the existence of weak population differentiation.
The unrooted Fitch–Margoliash tree illustrates that southern Spanish (Cadiz, Seville, Cordoba, Jaen) along with insular Italian (Sardinia, Sicily) populations exhibit a clear genetic differentiation from northern Spanish (Valencia, Catalonia 1, Catalonia 2) and continental Italian (Apulia, Umbria) populations. This result was supported by the bootstrap value of 80 % (Fig. 3). However, a clear separation is visible between the populations of Andalusia and the populations of Sardinia and Sicily, where Italian populations grouped together, with bootstrap support of 88 %. In contrast, the rest of the populations did not follow the geographical pattern. Thus, the group formed by the populations from Catalonia 2 and Umbria was the most distant from the rest.
Fig. 3.
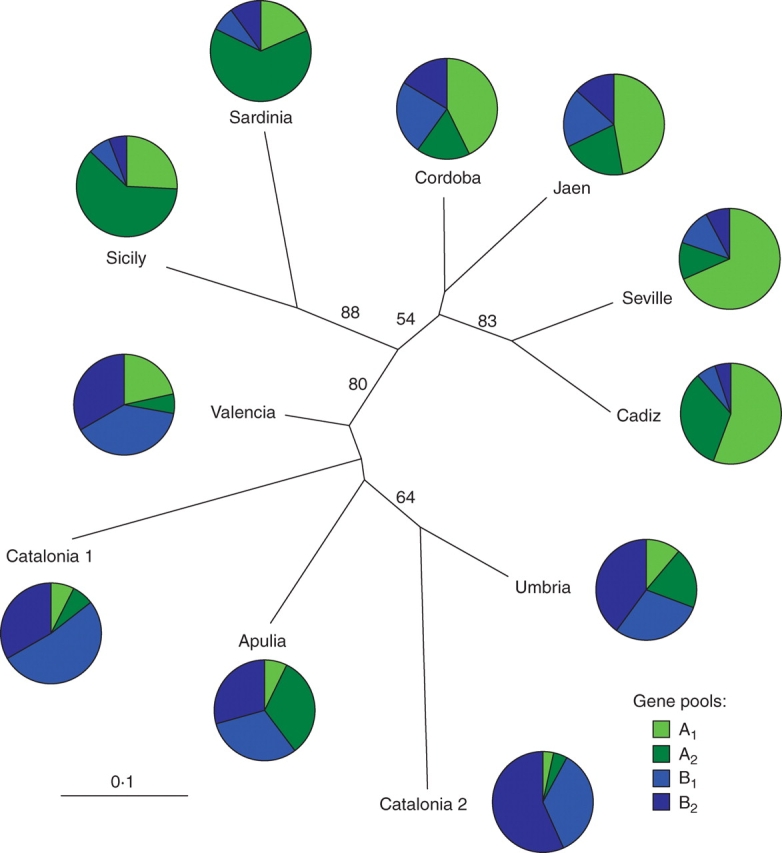
Unrooted Fitch–Margoliash tree based on Nei's standard genetic distance between 11 olive populations and the proportions of ancestry of each population in each of the four gene pools defined with the model-based clustering method from Pritchard et al., (2000). Numbers above branches indicate bootstrap support percentage over 50 % in 10 000 pseudoreplicates. Inferred gene pools: A1, southern Spanish genepool; A2, insular Italian genepool; B1, north-eastern Spanish genepool; B2, gene pool detected predominantly in Catalonia 2 and Umbria.
The results of structure analysis are reported in Table 5. At K = 2 the southern Spanish populations (Cadiz, Seville, Cordoba, Jaen) and Italian island populations (Sardinia, Sicily) are predominantly composed of a single gene pool (gene pool A), whereas the north-eastern Spanish populations (Valencia, Catalonia 1, Catalonia 2) and continental Italian populations (Apulia, Umbria) come from a different gene pool (gene pool B). At K = 3 the gene pool A defined for K = 2 splits into the gene pools (A1) and (A2), the former predominating in southern Spain and the latter in insular Italy, suggesting the existence of genetic differences between Spanish and Italian wild olive populations.
Table 5.
Proportion of ancestry of each population in each of two, three and four olive gene pools defined with the model-based clustering method from Pritchard et al., (2000)
| Population |
K = 2 |
K = 3 |
K = 4 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | A1 | A2 | B | A1 | A2 | B1 | B2 | |
| Cadiz | 0·911 | 0·089 | 0·568 | 0·367 | 0·065 | 0·556 | 0·330 | 0·060 | 0·054 |
| Seville | 0·819 | 0·181 | 0·717 | 0·147 | 0·136 | 0·682 | 0·120 | 0·118 | 0·079 |
| Cordoba | 0·601 | 0·399 | 0·474 | 0·231 | 0·295 | 0·427 | 0·169 | 0·242 | 0·163 |
| Jaen | 0·663 | 0·337 | 0·533 | 0·236 | 0·230 | 0·476 | 0·202 | 0·187 | 0·135 |
| Valencia | 0·282 | 0·718 | 0·282 | 0·104 | 0·614 | 0·214 | 0·063 | 0·386 | 0·336 |
| Catalonia 1 | 0·194 | 0·806 | 0·150 | 0·136 | 0·713 | 0·075 | 0·073 | 0·515 | 0·336 |
| Catalonia 2 | 0·067 | 0·933 | 0·048 | 0·089 | 0·862 | 0·032 | 0·049 | 0·356 | 0·563 |
| Sardinia | 0·800 | 0·200 | 0·211 | 0·650 | 0·139 | 0·187 | 0·635 | 0·078 | 0·100 |
| Sicily | 0·896 | 0·104 | 0·281 | 0·638 | 0·080 | 0·262 | 0·608 | 0·071 | 0·059 |
| Apulia | 0·425 | 0·575 | 0·122 | 0·399 | 0·479 | 0·074 | 0·323 | 0·311 | 0·292 |
| Umbria | 0·272 | 0·728 | 0·134 | 0·236 | 0·629 | 0·114 | 0·191 | 0·295 | 0·400 |
Proportion of ancestry from a given sample assigned to the best-scoring gene pool, PMAX, is given in bold type.
The most probable division is K = 4, which received the strongest support in terms of log-likelihood values for the data conditional of K, ln Pr(X|K), as suggested by Pritchard et al. (2000), and on ΔK values (data not shown). Under these conditions, gene pool B defined for K = 2 splits into two different gene pools, B1 and B2, revealing an apparently odd separation that does not follow the geographical origin of the populations. Gene pool B1 is found mostly in populations of Valencia, and Catalonia 1, while gene pool B2 was detected predominantly in Catalonia 2 and Umbria. The average admixture of gene pools within sampled populations, expressed as the proportion of genome from a given sample assigned to the best-scoring gene pool, PMAX, in the sample, was 50·6 % and ranged from 32·2 % (Apulia in gene pool A2) to 68·2 % (Seville in gene pool A1). The population from Apulia was clearly the most admixed, deriving almost equal proportions of ancestry (approx. 30 %) from three gene pools (A2, B1, B2).
Levels of admixture
In individual-level admixture analyses, 27 individual trees (25 from Spanish oleaster populations and two from Italian oleaster populations) were assigned to gene pool A1. Twenty-one individual trees, 19 originating from Italian oleaster populations and two from the Spanish oleaster populations, belong to gene pool A2. No individuals from Valencia, Catalonia 1 or Catalonia 2 belong to gene pool A1 and the same is true in the case of Umbria and gene pool A2. However, no single individual was found with more than 75 % of its genome estimated as belonging to either gene pool B1 or B2, although these gene pools were the best-scoring (QMAX) in the case of 39 and 37 individuals, respectively. When the estimates of the proportions of ancestry in gene pools B1 and B2 are pooled together, 62 individual trees belong to that gene pool; among these, eight were from the southern Spanish oleaster populations and two from the Italian oleaster populations. The remaining trees, 61, were considered as of ‘mixed origin’. It is worth noting that the majority of individuals sampled in Sicily and Cadiz, and classified as of ‘mixed origin’, represent putative hybrids between Spanish and Italian oleasters (six out of nine in Sicily and seven out of eight in Cadiz).
DISCUSSION
Microsatellite variation in wild olive populations
The present study uncovered abundant allelic variation over eight loci and high overall genetic diversity. These results are consistent with studies of morphological variation in wild olive populations (Mulas et al., 2004) as well as with studies of allozymes (Lumaret and Ouazzani, 2001; Lumaret et al., 2004), cytoplasmic DNA markers (Besnard et al., 2002a,b), RAPDs, AFLPs and SSRs (Angiolillo et al., 1999; Bronzini de Caraffa et al., 2002a, b; Baldoni et al., 2006; Breton et al., 2006). The higher intrinsic diversity of microsatellites may explain the higher allelic diversity reported here than that found with allozymes in wild olive populations from around the Mediterranean Basin (Lumaret et al., 2004). The values of observed and expected heterozygosity detected by SSRs in this study were relatively high (Ho = 0·717 and He = 0·843). These values are higher than those reported for cultivated olives (de la Rosa et al., 2000; Sefc et al., 2000; Cipriani et al., 2002), and those revealed by SSRs in oleaster populations (Breton et al., 2006) were lower (Ho = 0·67 and He = 0·72). The relatively large number of alleles observed in the present study in comparison with the studies mentioned above can be attributed to the large number and diversity of individuals surveyed by us.
The positive values of the inbreeding coefficient f for most populations may be due to the presence of null alleles (Bruford et al., 1998) at loci UDO99-039, UDO99-019 and UDO99-043, which show a high deficit of heterozygosity, perhaps due to allele dropout. Furthermore, observed deviations from HW equilibrium at some loci may also derive from limited sample sizes. SSRs are highly multi-allelic and the number of individuals genotyped per population (around 15) is relatively limited for sampling all possible genotypes at a locus.
Genetic differentiation and underlying populations structure in wild olive populations
The AMOVA results show a very high level of intrapopulation diversity and low but significant genetic differentiation among olive populations as in most woody perennial outbreeding species maintaining most of their variation within populations (Hamrick and Godt, 1989; Hamrick et al., 1992).
The Bayesian-based analysis with structure allowed the detection of four different gene pools. Two gene pools, predominantly found in southern Spain and the Italian islands, respectively, represent populations situated in regions of typical Mediterranean climate. The other two gene pools were mostly detected in the north-eastern regions of Spain and in continental Italy and belong to the transition region between the temperate and Mediterranean climate zones. These results are in overall concordance with both the FCA at the individual level and Fitch–Margoliash tree at the population level. The results obtained by FCA, the dendrogram and the structure analysis leave room for alternative explanations. The pluviothermal regimes could have imposed complex selective pressures on populations that resulted in genome-wide responses, although recombination would quickly erase associations between neutral markers and adaptive traits in this outcrossing species. For instance, when analysing a pluviothermal parameter such as the De Martonne index (data not shown) the regions of Cadiz, Seville, Cordoba, Jaen, Valencia, Catalonia 1, Sardinia and Sicily were clearly separated from those of Catalonia 2, Umbria and Apulia. Similar impacts on population structure have already been observed for divergent selection of adaptive traits in maize (Remington et al., 2001). However, the distribution of genetic diversity observed among wild olive populations and their relationships show also striking coincidence with their proximity to areas of intensive olive cultivation. The gene flow from varieties could have contributed to the observed distribution of diversity within wild olive populations. Thus, the populations sampled in Andalusia and Italian islands may not have hybridized with cultivated plants and may thus represent genuine wild populations, while those sampled in Apulia, Umbria, Catalonia and Valencia could be contaminated with the cultivated gene pool and would then be classified as feral.
Hybridization with cultivars, and the long history of exchange of cultivated genetic resources throughout the Mediterranean countries (Besnard et al., 2001) could have contributed to the low degree of differentiation of the relatively distant Italian populations of Umbria and Apulia, and would explain their clustering with the populations of Catalonia and Valencia, as well as why geographically distant populations such as those of Apulia and Catalonia are so little differentiated.
However, the presence of genuine wild individuals within the populations of Apulia, Umbria, Catalonia and Valencia cannot be excluded. AFLP analysis suggested that at least some of the Umbrian oleasters may be considered as genuine wild olives (Baldoni et al., 2006). The wild status of the olive in the Eurosiberian region, north of Iberia has been strongly indicated by Vargas and Kadereit, (2001) and confirmed by Rubio de Casas et al. (2002, 2006), who have also demonstrated that colonization of such northern latitudes may have occurred no earlier than the warm Holocene period, around 5000–8000 years BP. The traditional use of wild olives from mountain areas as rootstocks to graft suitable cultivars (J. Tous, IRTA, Spain, pers. comm.) could have contributed to the reduction of wild olive forest areas in the north-eastern regions of Spain. Finally, to be fully verified, the above-mentioned classification of the wild olive populations studied here should be compared with the distribution of diversity of cultivated material. This may not be a straightforward task given that intensive exchange of cultivars and various supplanting campaigns may have continuously reshuffled the distribution of cultivars, making it difficult to document past hybridization.
The first two gene pools detected by this study, Cadiz–Seville–Cordoba–Jaen (A1) and Sardinia–Sicily (A2), may indicate the presence of local oleaster germplasm of Spanish and Italian origin, respectively. The assignment of individual trees to a gene pool has allowed the identification of potential genuine oleaster trees from both of these gene pools. The presence of hybrids between Spanish and Italian oleasters and the fact that some wild Spanish individuals belong to the Italian gene pool, and vice versa, may indicate a common origin of these wild olive populations. Describing the patterns of genetic structure at a smaller scale than in previous studies (Lumaret and Ouazzani, 2001; Lumaret et al., 2004), the present study identifies the survival of oleasters in various areas where many cultivars are present.
The low level of differentiation between the Sicilian and Sardinian populations may be explained by a common genetic pool, lack of differential selective pressure and longevity of the trees, as well as gene flow between the two islands. There is evidence that olive pollen may travel over 200 km (Ribeiro et al., 2005), although a strong genetic isolation has been observed among olive populations of the Canary islands analysed by RAPD and ISSR markers, suggesting that inter-island dispersal and establishment were very rare (Hess et al., 2000). The present results did not reveal any genetic uniqueness in Sicilian wild olive populations, in contrast to results obtained with chloroplast DNA markers (Besnard et al., 2002a).
FCA results and the average genome proportions derived from each gene pool at the population level indicate a certain degree of admixture in all the populations, Apulia being the clearest case. The findings suggest some caution regarding the differentiation at the population level; it is likely that, in nature, the difference is gradual, making it difficult to identify clear-cut genetic boundaries between the candidate areas containing either genuinely wild or feral germplasm. The current results and available estimates of gene flow in olives across the Mediterranean (Rubio de Casas et al., 2006) indicate that reproductive isolation is highly improbable and that genetic material seems to be exchanged frequently among populations. However, some caution should be paid in evaluating the genetic exchange with cultivated forms. Due to continuous reshuffling and supplanting of cultivars it would be rather difficult to identify a reference gene pool of cultivars.
The present study suggests that there is no immediate reason for concern about demographic bottlenecks in wild olive populations, and the presence of a high number of rare alleles is clear evidence of this fact. However, a strong and continuous gene flow between wild and cultivated germplasm may lead to slow but steady genetic erosion of the genuine oleaster gene pool. Future prospecting, diversity analyses and monitoring in native Mediterranean forests should help to identify additional genuine individuals and/or oleaster populations as well as to establish suitable conservation strategies.
SUPPLEMENTARY INFORMATION
Data for the proportion of ancestry of each individual tree in each of two, three and four gene pools is available online at http://aob.oxfordjournals.org/.
ACKNOWLEDGMENTS
We thank the Agentes de Medio Ambiente of Junta de Andalucia (Spain) for their help with collecting plant material in Andalusia, J. Tous, J. F. Hermoso, V. Cabus, F. Prats, J. Franch and L. Sanchez for sampling plant material in Catalonia and Valencia, and L. Rallo, R. de la Rosa and B. Sutherland for their contribution to the improvement of the manuscript. We are particularly grateful to K. R. Tobutt for helpful discussion and advice.
LITERATURE CITED
- Alcantara JM, Rey PJ. Conflicting selection pressures on seed size: evolutionary ecology of fruit size in a bird-dispersed tree. Olea europaea. Journal of Evolutionary Biology. 2003;16:1168–1176. doi: 10.1046/j.1420-9101.2003.00618.x. [DOI] [PubMed] [Google Scholar]
- Angiolillo A, Mencuccini M, Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoretical and Applied Genetics. 1999;98:411–421. [Google Scholar]
- Baldoni L, Tosti N, Ricciolini C, Belaj A, Arcioni S, Pannelli G, et al. Genetic structure of wild and cultivated olives in the Central Mediterranean Basin. Annals of Botany. 2006;98:935–942. doi: 10.1093/aob/mcl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4·05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Université de Montpellier II; 2004. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. [Google Scholar]
- Besnard G, Bervillé A. Multiple origins for Mediterranean olive (Olea europaea L-ssp europaea) based upon mitochondrial DNA polymorphisms. Comptes Rendus de l'Academie des Sciences, Sciences de la Vie – Life Sciences. 2000;323:173–181. doi: 10.1016/s0764-4469(00)00118-9. [DOI] [PubMed] [Google Scholar]
- Besnard G, Baradat P, Berville A. Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theoretical and Applied Genetics. 2001;102:251–258. [Google Scholar]
- Besnard G, Khadari B, Baradat P, Berville A. Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theoretical and Applied Genetics. 2002a;104:1353–1361. doi: 10.1007/s00122-001-0832-x. [DOI] [PubMed] [Google Scholar]
- Besnard G, Khadari B, Baradat P, Berville A. Combination of chloroplast and mitochondrial DNA polymorphisms to study cytoplasm genetic differentiation in the olive complex (Olea europaea L.) Theoretical and Applied Genetics. 2002b;105:139–144. doi: 10.1007/s00122-002-0868-6. [DOI] [PubMed] [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Breton C, Tersac M, Bervillé A. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. Journal of Biogeography. 2006;33:1916–1928. [Google Scholar]
- Bronzini de Caraffa V, Maury J, Gambotti C, Breton C, Bervillé A, Giannettini J. Mitochondrial DNA variation and RAPD mark oleasters, olive and feral olive from Western and Eastern Mediterranean. Theoretical and Applied Genetics. 2002a;104:1209–1216. doi: 10.1007/s00122-002-0883-7. [DOI] [PubMed] [Google Scholar]
- Bronzini de Caraffa V, Giannettini J, Gambotti C, Maury J. Genetic relationships between cultivated and wild olives of Corsica and Sardinia using RAPD markers. Euphytica. 2002b;123:263–271. [Google Scholar]
- Bruford MW, Hanotte O, Brookfield JFY, Burke T. Multilocus and single-locus DNA fingerprinting. In: Hoelzel AR, editor. Molecular genetic analysis of population. New York: Oxford University Press; 1998. pp. 287–336. [Google Scholar]
- Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theoretical and Applied Genetics. 2002;104:223–228. doi: 10.1007/s001220100685. [DOI] [PubMed] [Google Scholar]
- Contento A, Ceccarelli M, Gelati MT, Maggini F, Baldoni L, Cionini PG. Diversity of Olea genotypes and the origin of cultivated olives. Theoretical and Applied Genetics. 2002;104:1229–1238. doi: 10.1007/s00122-001-0799-7. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:1119–1127. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) version 3·5c. Seattle, WA: Department of Genetics, University of Washington; 1993. [Google Scholar]
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Green PS. A revision of Olea L. (Oleaceae) Kew Bulletin. 2002;57:91–140. [Google Scholar]
- Goudet J. FSTAT (vers. 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Guo SW, Thompson EA. Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer Associates; 1989. pp. 43–63. [Google Scholar]
- Hamrick JL, Godt MJW, Sherman-Broyles S. Factors influencing levels of genetic diversity in woody plant species. New Forests. 1992;6:95–124. [Google Scholar]
- Hess J, Kadereit JW, Vargas P. The colonization history of Olea europaea L. in Macaronesia based on internal transcribed spacer 1 (ITS-1) sequences, randomly amplified polymorphic DNAs (RAPD), and intersimple sequence repeats (ISSR) Molecular Ecology. 2000;9:857–868. doi: 10.1046/j.1365-294x.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- Liu J. POWERMARKER – A powerful software for marker data analysis. Raleigh, NC: North Carolina State University, Bioinformatics Research Center; 2002. (http://www.powermarker.net. ) [Google Scholar]
- Lumaret R, Ouazzani N. Ancient wild olives in Mediterranean forests. Nature. 2001;413:700. doi: 10.1038/35099680. [DOI] [PubMed] [Google Scholar]
- Lumaret R, Ouazzani N, Michaud H, Vivier G, Deguilloux MF, Di Giusto F. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity. 2004;92:343–351. doi: 10.1038/sj.hdy.6800430. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proceedings of the National Academy of Science of the USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minch E, Ruiz-Linares A, Goldstein D, Feldman M, Cavalli-Sforza LL. MICROSAT: a computer program for calculating various statistics on microsatellite allele data, ver. 1·5d. Stanford, CA: Stanford University; 1997. available at: http://hpgl.stanford.edu/projects/microsat . [Google Scholar]
- Mulas M, Deidda P. Domestication of woody plants from Mediterranean maquis to promote crops for mountain lands. Acta Horticulturae. 1998;457:295–301. [Google Scholar]
- Mulas M, Fadda A, Cauli E. Prime osservazioni su cloni di oleastro (Olea europaea var. sylvestris Hoff. E Link) selezionati per l'utilizzo forestale. Italus Hortus. 2004;4:214–217. [Google Scholar]
- Nei M. Genetic distance between populations. American Naturalist. 1972;106:283–292. [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer programme for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;89:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Science of the USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro H, Abreu I, Cunha M, Mota T, Castro R. Aeropalynological study of Vitis vinifera in the Braga region (1999–2003) Aerobiologia. 2005;21:131–138. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB. Mutational processes of simple-sequence repeat loci in human populations. Proceedings of the National Academy of Science of the USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa R, James C, Tobutt KR. Isolation and characterization of polymorphic microsatellite in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Molecular Ecology Notes. 2002;2:265–267. [Google Scholar]
- Rousset F, Raymond M. Testing heterozygote excess and deficiency. Genetics. 1995;140:1413–1419. doi: 10.1093/genetics/140.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio de Casas R, Balaguer L, Manrique E, Pérez-Corona ME, Vargas P. On the historical presence of the wild olive Olea europaea L. var. sylvestris (Miller) Leh. in the Eurosiberian North of the Iberian Peninsula. Anales de Jardín Botánico. 2002;59:342–344. [Google Scholar]
- Rubio de Casas R, Besnard G, Schönswetter P, Balaguer L, Vargas P. Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L. Theoretical and Applied Genetics. 2006;113:575–583. doi: 10.1007/s00122-006-0306-2. [DOI] [PubMed] [Google Scholar]
- SAS Institute. SAS Procedures Guide. Cary, NC: SAS Institute Inc; 1999. Version 8. [Google Scholar]
- Sefc KM, Lopes S, Mendonca D, Dos Santos MR, Machado MLD, Machado AD. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Molecular Ecology. 2000;9:1171–1173. doi: 10.1046/j.1365-294x.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- Terral JF, Alonso N, Capdevila RBI, Chatti N, Fabre L, Fiorentino G, et al. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. Journal of Biogeography. 2004;31:63–77. [Google Scholar]
- Vargas P, Kadereit JW. Molecular fingerprinting evidence (ISSR, inter-simple sequence repeats) for a wild status of Olea europaea L. (Oleaceae) in the Eurosiberian North of the Iberian Peninsula. Flora. 2001;196:142–152. [Google Scholar]
- Zohary D, Hopf M. Domestication of plants in the Old World. 2nd edn. Oxford, UK: Clarendon Press; 1994. [Google Scholar]
- Zohary D, Spiegel-Roy P. Beginning of fruit growing in the old world. Science. 1975;187:319–327. doi: 10.1126/science.187.4174.319. [DOI] [PubMed] [Google Scholar]