Abstract
Background and Aims
CRABS CLAW (CRC) encodes a transcription factor of the YABBY family that plays important roles in carpel and nectary development in Arabidopsis thaliana. Combined evolutionary and developmental studies suggest an ancestor of the CRC gene to have controlled carpel development in the last common ancestor of the angiosperms. Roles for CRC orthologues in leaf development and carpel specification in rice, and in nectary development in core eudicots, have accordingly been interpreted as derived. The aim of this study was to assess the capacity of CRC orthologues from a basal angiosperm and from rice to complement CRC mutants of arabidopsis. These experiments were designed to test the hypothesized ancestral role of CRC in the angiosperms, and to indicate whether putatively novel roles of various CRC orthologues resulted from changes to their encoded proteins, or from other molecular evolutionary events.
Methods
The crc-1 mutant of arabidopsis was genetically transformed with the coding sequences of various CRC orthologues, and with paralogous YABBY coding sequences, under the control of the arabidopsis CRC promoter. The phenotypes of transformed plants were assessed to determine the degree of complementation of the crc-1 mutant phenotype in carpel fusion, carpel form and nectary development.
Key Results
The CRC orthologue from the basal angiosperm Amborella trichopoda partially complemented the crc-1 mutant phenotype in carpels, but not in nectaries. The CRC orthologue from rice partially complemented all aspects of the crc-1 mutant phenotype. Though most non-CRC YABBY coding sequences did not complement crc-1 mutant phenotypes, YABBY2 (YAB2) proved to be an exception.
Conclusions
The data support a hypothesized ancestral role for CRC in carpel development and suggest that novel roles for CRC orthologues in monocots and in core eudicots resulted principally from molecular changes other than those affecting their coding sequences.
Key words: Carpel, gynoecium, nectary, Amborella trichopoda, Arabidopsis thaliana, Oryza sativa, CRABS CLAW, DROOPING LEAF, YABBY
INTRODUCTION
CRABS CLAW (CRC) encodes one of six putative transcription factors of the YABBY family in Arabidopsis thaliana. This gene plays an important role in carpel development, and is also necessary for the development of nectaries (Alvarez and Smyth, 1999). Its precise expression pattern in carpel tissues (Bowman and Smyth, 1999) reflects the general role of YABBY genes in specifying the abaxial (facing away from the axis) side of plant lateral organs (Bowman, 2000; Bowman et al., 2002). In the case of CRABS CLAW, this role is apparent in double mutant plants in which the strong crc-1 mutation is combined with mutations in any one of the genes KANADI, GYMNOS and AKETHE (Eshed et al., 1999), none of which belong to the YABBY family. Such double-mutants show a breakdown of abaxial–adaxial (adaxial = facing towards the axis) polarity at the margins of the two fused carpels in the arabidopsis gynoecium. Loss of developmental polarity in this case leads to the formation of placentas and ovules both inside and outside of the ovary. However, in single crc-1 mutants, polarity defects are not apparent (Alvarez and Smyth, 1999). Instead, these mutants show defects in carpel fusion and in the overall size and shape of the gynoecium. In addition, nectaries are absent from crc-1 mutants.
Strong evidence exists to suggest that the three ‘ANA’ (formerly ‘ANITA’) orders, Amborellales, Nymphaeales and Austrobaileyales, diverged from a remaining common lineage near the base of the phylogenetic tree of the extant angiosperms (Stevens, 2001 onwards). The remaining common lineage would later have diverged to form the two major angiosperm groups of the eudicots and monocots, in addition to several less species-rich clades. The expression pattern in carpel tissues of the putative orthologue of CRC from the ANA angiosperm Amborella trichopoda (Fourquin et al., 2005) is remarkably similar to that of CRC in arabidopsis. This gene, AmbCRC, is expressed in an abaxial-specific manner in the ovary wall, and is not generally expressed in leaves and other non-reproductive organs. As Amborella trichopoda is the only member of what is generally regarded as the most basal angiosperm group (Amborellales), the correspondence in expression patterns between CRC and AmbCRC strongly suggests these genes to have conserved their expression patterns since the common ancestor of the extant angiosperms. The conservation of such a distinctive expression pattern is highly suggestive of a conserved role, and so it has been argued that CRC and AmbCRC have conserved a common role in the control of carpel development since the last common ancestor of the extant flowering plants (Fourquin et al., 2005).
Yamaguchi et al. (2004) have identified a putative orthologue of CRC from rice (Oryza sativa), termed DROOPING LEAF (DL). In dl mutants, carpels are replaced by stamens in the fourth floral whorl, and leaf defects are also present, in contrast to the crc mutant phenotype of arabidopsis. The conservation of expression patterns of CRC orthologues between basal angiosperms such as Amborella, and eudicots such as arabidopsis, suggests the expression pattern and functions of DL in rice to be derived characters that originated after the divergence of the monocot and eudicot lineages (Fourquin et al., 2005).
Similarly, the nectary development function associated with CRC in arabidopsis, and with CRC orthologues in other species of core eudicots (i.e. the crown group of the eudicots, excluding the most basal lineages of this clade), is not thought to be ancestral to the angiosperms (Lee et al., 2005b). Angiosperm species that are external to the core eudicots do not show expression of CRC orthologues in nectary tissues, where these exist. Thus, CRC is hypothesized to have acquired a role in nectary specification at the base of the core eudicot clade. Floral nectaries are not present in most ANA grade angiosperms, including Amborella (Endress, 2001), or in rice.
Novel functions associated with CRC orthologues may have resulted from changes to their expression patterns, to their coding regions, or from changes to other genes acting together with, or downstream of, CRC orthologues in transcriptional control networks. In the present study, the aim is to assess to what extent the coding sequences of CRC orthologues from various species, and those of paralogous YABBY genes from arabidopsis, are able to functionally replace the wild-type CRC coding sequence in arabidopsis. Results from these analyses provide functional evidence for evolutionary hypotheses relating to the ancestral role of the CRC gene, and also indicate to what extent coding sequence changes, or other factors, may have been responsible for the hypothesized novel functions of CRC orthologues in monocots and in core eudicots.
MATERIALS AND METHODS
Transgene constructions
The plant transformation vector pCAMBIA3300 was modified by the addition of the nopaline synthase gene terminator (nos), ligated in a 5′–3′ orientation between unique Sac1 and EcoR1 restriction sites. The resulting plasmid was further modified by the ligation of a ‘GATEWAY’ (Invitrogen) ccdB selection cassette, bordered by attR1 and attR2 recombination sites, into a unique Sma1 restriction site immediately upstream of the nos terminator. A 3608-bp fragment from immediately upstream of the arabidopsis CRC start codon, taken to represent the CRC promoter (proCRC), was obtained by PCR amplification from genomic DNA of the Columbia ecotype of Arabidopsis thaliana using a proof-reading thermo-stable DNA polymerase. The resulting PCR product was cloned into a plasmid vector and fully sequenced. This CRC promoter fragment was subsequently excised from its cloning vector and ligated into the unique Xba1 restriction site of the modified pCAMBIA plasmid (above), adjacent to the ccdB cassette. Coding sequences of various YABBY genes from Arabidopsis thaliana (Columba ecotype), Amborella and Orzya sativa ‘Nipponbare’, were amplified from the appropriate full-length cDNAs by PCR to incorporate attB1 and attB2 GATEWAY recombination sites situated 2 bp upstream and 1 bp downstream, respectively, of their start and stop codons. The resulting coding sequences were inserted into pDONR207 (Invitrogen) by GATEWAY ‘BP’ recombination reactions. The exact nucleotide sequences of the coding sequences were confirmed, and these were then transferred by GATEWAY ‘LR’ recombination reactions to the proCRC-containing plant transformation vector, thereby replacing its ccdB cassette. The YABBY coding sequences were thus finally situated in the plant transformation vector, downstream of the CRC promoter and upstream of the nos terminator, and immediately flanked by attB1 and attB2 recombination sites. GATEWAY-type plasmids containing β-glucuronidase (GUS) and improved green fluorescent protein (Heim et al., 1994) reporter gene coding regions, flanked by attL1 and attL2 recombination sites, were obtained from Dr Vanessa Vernoud in our laboratory. These reporter genes were also transferred to the proCRC-containing plant transformation vector by GATEWAY ‘LR’ recombination reactions. Completed plant transformation vectors were transferred by electroporation to Agrobacterium tumefaciens strain C58pmp90 for plant transformation. Full nucleotide sequences of the constructions used for plant transformation will be made available on request.
Plant material and genetic transformation
Seeds of the Columbia ecotype of Arabidopsis thaliana (L.) Heynh., and of the crc-1 mutant in the Landsberg erecta genetic background, were obtained from the Nottingham Arabidopsis Stock Centre (UK). Agrobacterium tumefaciens-mediated genetic transformation of arabidopsis plants was performed by the floral dip method (Clough and Bent, 1998), to generate transformants carrying YABBY or reporter gene coding sequences under the control of the CRC promoter. Seeds harvested from dipped plants (T1 seeds) were sown initially onto sand and selected by watering with ammonium glufosinate (BASTA) herbicide (7·5 mg dm−3). BASTA-resistant seedlings were transferred to potting compost. These plants were then grown under 8-h day-length conditions in a growth chamber for 4 weeks and then transferred to 18-h day-length conditions to induce flowering.
Northern blot hybridization
Northern blot hybridization was performed as previously described (Fourquin et al., 2005).
Microscopy
Plant tissues were photographed at low magnification using a binocular dissecting microscope (Leica MZFIII) fitted with a digital camera Leica (DC 300F). GUS assays were performed as described by Nakayama et al. (2005). Excitation of GFP was performed using a UV light source fitted with a band-pass filter of 450–490 nm. The presence or absence of nectaries in arabidopsis flowers was recorded in fresh plant material using an environmental scanning electron microscope (Hitachi S800).
RESULTS
A 3·6-kb DNA fragment immediately upstream of the CRC coding region shows CRC promoter activity
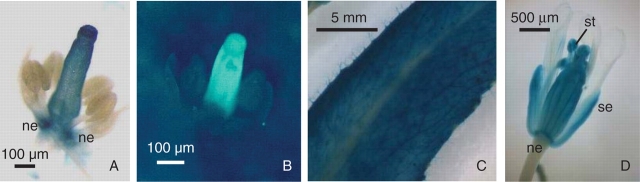
A genomic DNA fragment of 3608 bp, from immediately upstream of the arabidopsis CRC start codon, was taken to represent the CRC promoter in these studies. The promoter activity of this fragment was initially assayed using GUS and GFP reporter genes in populations of 40 transgenic Arabidopsis thaliana plants of the Columbia ecotype. Sixty per cent of plants transformed with GUS, and 5 % of those transformed with GFP reporter genes, showed moderate reporter gene activities that were largely restricted to carpels and nectaries, similar to the wild-type expression pattern of CRC (Fig. 1A, B). However, very strong GUS expression in leaves (Fig. 1C) and floral organs (Fig. 1D) was noted in 25 % of T1 plants containing the proCRC::GUS transgene (Fig. 1C). The presence of high levels of reporter gene mRNAs in leaves and inflorescences in 37·5 % and 25 % of two further T1 populations of 16 GUS and 16 GFP transformants, respectively, was subsequently demonstrated by northern blotting (results not presented).
Fig. 1.
GUS and GFP reporter gene expression under the control of the CRC gene promoter. (A) GUS staining in the gynoecium and nectaries (ne) of a flower from a proCRC::GUS transformant at developmental stage 11–12 (Smyth et al., 1990), showing the expected CRC-like pattern of reporter gene expression. (B) GFP localization in the gynoecium of a flower from a proCRC::GFP transformant at developmental stage 10–11 (Smyth et al., 1990), showing the expected CRC-like pattern of reporter gene expression. (C) GUS staining in leaf tissue of a proCRC::GUS transformant showing reporter gene activity outside of the normal zone of CRC expression. (D) GUS staining in mature flower tissues (se = sepal, st = stamen) in a proCRC::GUS transformant showing reporter gene activity outside of the normal zone of CRC expression.
Subsequent to the start of this study, a detailed analysis of sequences upstream of the CRC coding region, performed by comparison between three species of Brassicaceae, has indicated the presence of five conserved ‘modules’ of functional significance within the CRC promoter (Lee et al., 2005a). A synthetic promoter containing only these five modules was found to be capable of correctly directing reporter gene expression, as did an intact CRC promoter fragment of 3·8 kb. The 3·6-kb CRC promoter fragment used in the experiments described here contained all five of the conserved modules identified by Lee et al. (2005a), terminating some 548 bp upstream of the most distal (to the CRC coding sequence) of these modules. Therefore, the expression of reporter genes outside of the normal CRC expression domain, noted in a proportion of transformed plants in the present study, probably does not reflect the absence of important cis-acting regulatory sequences in the 3·6-kb promoter fragment used. As a proportion of T1 transformants in each population examined showed reporter gene expression that was largely limited to the domain of CRC expression in wild-type plants (Fig. 1A, B), the 3·6-kb promoter fragment used in these experiments was considered to have generated the expected CRC expression profile in at least a proportion of transgenic plants. The ectopic reporter gene expression noted here, in the leaves and other organs of a further group of T1 transformants (Fig. 1C, D), was concluded to have been produced by some position-dependent mechanism associated with the sites of transgene insertion.
The CRC orthologues from Amborella and rice show both quantitative and qualitative differences in their functional conservation with CRC from arabidopsis
Compared with wild-type plants (Fig. 2A–C), crc-1 mutants produce carpels that are only partially fused together (Fig. 2D) to form gynoecia, and later siliques, that are considerably shorter and wider than wild-type (Fig. 2E). These mutants also lack floral nectaries (Fig. 2F). In this study, a proCRC::CRC transgene was found to almost completely restore the wild-type phenotype to 65 % of transformed crc-1 mutants in a T1 population of 40 plants. Carpels and siliques of these tranformants were completely closed (Fig. 2G), and were close to wild-type length and width (Fig. 2H). The flowers of these transformants possessed nectaries (Fig. 2I). The remainder of the transformed plants from a T1 population containing the proCRC::CRC transgene showed lesser degrees of complementation, or no difference from the crc-1 mutant phenotype, possibly reflecting lesser levels of protein expression due to position-specific effects associated with the insertion of transgenes. These results form a positive control for the present study, providing a near-perfect complementation of the crc-1 mutation in a high proportion of transformants, against which the effects of other YABBY coding sequences can be compared.
Fig. 2.
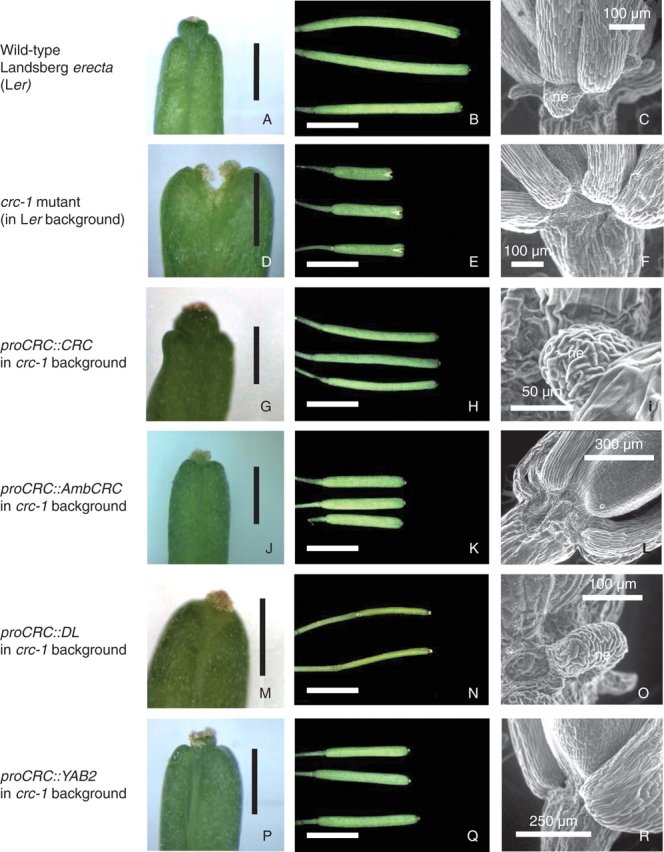
Complementation of the crc-1 mutation by transformation with various YABBY coding sequences under the control of the CRC promoter. (A, D, G, J, M, P) Silique apices showing degrees of carpel fusion in wild-type plants (A), crc-1 mutants (D) and crc-1 mutants transformed with the YABBY coding sequences CRC (G), AmbCRC (J), DL (M) and YAB2 (P). Scale bars = 1 mm. (B, E, H, K, N, Q) Fully elongated siliques in wild-type plants (B), crc-1 mutants (E) and crc-1 mutants transformed with the YABBY coding sequences CRC (H), AmbCRC (K), DL (N) and YAB2 (Q). Scale bars = 5 mm. (C, F, I, L, O, R) The base of the third floral whorl, showing the presence or absence of nectaries (ne), in mature flowers of wild-type plants (C), crc-1 mutants (F) and crc-1 mutants transformed with the YABBY coding sequences CRC (I), AmbCRC (L), DL (O) and YAB2 (R).
To test the conservation of CRC function between distantly related angiosperms, the degree of complementation of the crc-1 mutation was assessed in arabidopsis plants transformed with constructions containing the probable orthologues of CRC from the basal angiosperm Amborella, AmbCRC, and from rice, DL. Transformation of the crc-1 mutant with a proCRC::AmbCRC construction resulted in a partial restoration of the wild-type phenotype in 55 % of transformants from a T1 population of 40 plants. These plants showed complete carpel closure (Fig. 2J), and slightly increased silique length, compared with that of crc-1 mutants (Fig. 2K). No floral nectaries were present in any of the proCRC::AmbCRC transformants examined (Fig. 2L). These results indicate the AmbCRC coding sequence to be partially capable of substituting for CRC in the control of carpel development, but incapable of such a substitution in the control of nectary development.
Transformation of arabidopsis crc-1 mutants with a proCRC::DL transgene produced a range of distinct phenotypes. In 15 % of a T1 population of 40 plants, the gynoecium apex was completely closed (Fig. 2M) and gynoecium and silique dimensions were intermediate between those of crc-1 mutants and wild-type plants (Fig. 2N). These transformants possessed floral nectaries (Fig. 2O), indicating the DL coding sequence to be capable of replacing CRC functions in nectary development, in addition to carpel development, when correctly expressed under the control of the arabidopsis CRC promoter. A proportion of plants from the T1 population transformed with proCRC::DL exhibited a range of partially complemented phenotypes (25 %), or were apparently not complemented (20 %), closely resembling crc-1 mutants.
The proCRC::DL transformants described above presented no striking differences to wild-type plants in leaf development or in general plant architecture (Fig. 3A). However, a further proportion (40 %) of proCRC::DL transformants from the same T1 population exhibited aberrant development in all above-ground organs (Figs 3B–D), suggesting the generalized overexpression of the DL transgene. Similar phenotypes were noted in plants in which the CRC coding sequence was overexpressed under the control of the cauliflower mosaic virus 35S promoter (Fig. 3E). The leaves in proCRC::DL transformants that showed leaf development phenotypes, and in all pro35S::CRC transformants examined, were somewhat narrowed, and contorted out of their normal plane of growth by irregular blade expansion (Fig. 3B–F). Approximately half of the highly aberrant plants transformed with a proCRC::DL transgene eventually produced flowers (e.g. Fig. 3C), though these were also developmentally abnormal (data not presented) and almost completely sterile. ProCRC::DL transformants showing aberrant leaf development also showed a lack of internode extension in their reproductive phase (Fig. 3C), unlike p35S::CRC transformants (Fig. 3E). The presence of high levels of expression of DL in the leaves and inflorescences (where produced) of proCRC::DL transformants that showed altered leaf development was confirmed by northern blotting. High levels of DL transcripts in leaf tissue correlated with abnormal leaf phenotypes in a sample of six proCRC::DL transformants, three of which had abnormal leaves (Fig. 3G). The generalized transgene expression observed in a proportion of proCRC::DL transformants seems, therefore, to be similar to that observed in a proportion of proCRC::GUS (Figs 1C and D) and proCRC::GFP (data not presented) transformants.
Fig. 3.
Phenotypic effects and expression of DL in leaves and other organs in a proportion of proCRC::DL transformants. (A) A mature plant transformed with proCRC::DL (corresponding to plant 1 in Fig. 3G), showing normal development of leaves and inflorescence architecture. (B) A seedling transformed with proCRC::DL, showing aberrant leaf development. (C) A mature plant transformed with proCRC::DL (corresponding to plant 9 in Fig. 3G), showing aberrant leaf development and plant architecture. (D) The underside of a leaf from the proCRC::DL transformant shown in (C). (E) A pro35S::CRC transformant showing a leaf phenotype similar to that of some proCRC::DL transformants. (F) Mature rosette leaves, showing the effect of overexpression of CRC and DL transgenes in leaf tissue. (G) Northern hybridization of a DL cDNA probe to RNA from leaves and inflorescences of three proCRC::DL transformants (plants 1, 3 and 5) that showed normal leaf development, and to RNA from leaves of three proCRC::DL transformants (plants 9, 13 and 33) that showed aberrant leaf development. High levels of DL expression in leaves correlate with aberrant leaf phenotypes.
The YABBY2 coding sequence shows partially conserved functions with that of CRC
The capacities of two further YABBY coding sequences from arabidopsis, FILAMENTOUS FLOWER (FIL) and YABBY2 (YAB2), to replace the functions of CRC were tested. A proCRC::FIL transgene showed no capacity to complement the crc-1 mutation. All crc-1 transformants examined from a T1 population containing a proCRC::FIL transgene showed a phenotype identical to that of crc-1 mutants (data not presented). However, 70 % of crc-1 plants from a T1 population transformed with a proCRC::YAB2 construction showed a partial restoration of the wild-type phenotype. The most highly complemented individuals from this population showed complete carpel closure (Fig. 2P) and silique dimensions that were intermediate between those of crc-1 mutants and wild-type plants (Fig. 2Q). Silique length was more completely restored to wild-type dimensions by transformation with this construction than with a proCRC::AmbCRC construction (Fig. 2K). No floral nectaries were present in proCRC::YAB2 transformants (Fig. 2R), indicating the YAB2 coding sequence to be incapable of replacing the nectary development function of CRC.
DISCUSSION
Studies of functional conservation between orthologous coding sequences support a role for CRC in carpel development in the last common ancestor of the flowering plants
A principal aim of this study was to obtain functional data concerning the ancestral role of CRC in the angiosperms. Previous comparative studies, performed between the putatively most-basal angiosperm, Amborella, and the model angiosperm, arabidopsis, had already shown the likely orthologous genes from these two species, AmbCRC and CRC, to share a common expression pattern in carpel tissues. This result was highly suggestive of a conserved role in carpel development, implying that the ancestral CRC gene would have already played this role in the last common ancestor of the flowering plants. The present study aimed to determine the extent to which the AmbCRC coding sequence could compensate for the lack of a functional CRC gene in arabidopsis mutants, and thereby provide some functional evidence for the hypothesized conserved role of the CRC gene.
AmbCRC has been shown to be capable of partially restoring a wild-type phenotype to the strong crc-1 mutant of arabidopsis. AmbCRC was fully able to restore carpel fusion, and slightly increased carpel and silique lengths, though not completely to wild-type dimensions. AmbCRC was not, however, able to restore nectary development to crc-1 mutants. These results lend some support to the hypothesis that the carpel development role of CRC, as determined by genetic studies in arabidopsis (Alvarez and Smyth, 1999), has been conserved since the common ancestor of the living flowering plants, some 160 Mya (Davies et al., 2004).
Novel functions of DL in rice may principally have resulted from changes in gene expression, rather than from changes to the DL coding sequence
The phenotype associated with the dl mutation in rice (Yamaguchi et al., 2004) is strikingly different to the crc mutant phenotype in arabidopsis (Alvarez and Smyth, 1999), despite the probable orthology between DL and CRC. dl mutants show a homeotic replacement of carpels by stamens and their leaves lack a mid-rib, contrasting with the effects on carpel form and nectary specification of crc mutants. As the role played by CRC in arabidopsis carpel development has been interpreted as the ancestral role of this gene in the angiosperms (Fourquin et al., 2005), the roles of DL in carpel specification and leaf development may be considered as derived.
The results of the present study, showing that the DL coding sequence is able to largely replace that of CRC in the control of carpel development, suggest the differences between the phenotypes of dl and crc mutants to be principally due to factors other than physical differences between the DL and CRC proteins. The complementary experiment of transforming rice dl mutants with a proDL::CRC construction would be useful to provide a fuller answer to this question. At least one of the factors contributing to the novel roles of DL in rice must be the control of its expression, as DL and CRC expression patterns differ markedly (Bowman and Smyth, 1999; Yamaguchi et al., 2004). The novel functions of DL may thus have arisen in part by evolutionary changes to its cis-acting control regions, or to trans-acting factors controlling its expression.
The acquisition by CRC of a role in nectary development in the core eudicots may have arisen independently of changes to the CRC coding sequence
The DL coding sequence from rice was able to restore nectary development to crc-1 mutants of arabidopsis, even though the role of CRC in nectaries is thought to have evolved within the eudicot clade, after the last common ancestor shared between this group and the monocots (Lee et al., 2005b). This result suggests that the acquisition of a role in the control of nectary development by CRC did not occur as a direct result of evolutionary changes to the CRC coding sequence. The novel role of CRC in nectary development may instead have resulted from one or more changes affecting, for example, the expression of CRC in nectaries, its repertoire of target genes, or aspects of its potential partner proteins.
Seven crc mutants are known, which form an allelic series showing a range of carpel phenotypes, though a uniformly complete absence of nectaries (Bowman and Smyth, 1999). In the present study, only the CRC and DL coding sequences were able to restore both wild-type carpel and nectary development to crc-1 mutants, whereas the more distantly related coding sequences AmbCRC and YAB2 only restored carpel phenotypes. Taken together, these data suggest nectary development to be more sensitive than carpel development in arabidopsis to changes to the CRC coding sequence.
YAB2 shows a level of functional conservation with CRC that cannot be explained by its phylogenetic position within the YABBY family
It was shown that YAB2 from arabidopsis was able to replace CRC functions in transgenic plants to a somewhat greater extent than was AmbCRC from Amborella. Thus, a proCRC::YAB2 construction completely restored carpel fusion and showed a marked effect on gynoecium and silique length, but failed to restore nectary development to crc-1 mutants. Of the six YABBY coding sequences from arabidopsis, only YAB5 has yet to be tested for its capacity to compensate for a lack of CRC activity in transgenic plants. Y. Eshed and J. L. Bowman (unpubl. res., cited in Meister et al., 2005) found that neither FIL, nor its paralogue YABBY3 (YAB3), was capable of restoring wild-type phenotypes to crc-1 mutants. In the case of FIL, these findings have been confirmed in the present study. Meister et al. (2005) found that INNER NO OUTER (INO) was unable to replace CRC when expressed under the control of the CRC promoter. CRC is only distantly related to the other YABBY genes present in arabidopsis, having arisen as a distinct member of the YABBY family by a gene duplication event that occurred before the radiation of the flowering plants (Fourquin et al., 2005; Lee et al., 2005b). Globally, the results of the present and previous studies indicate the different members of the YABBY family to exhibit distinct protein activities, and the partial CRC activity of which YAB2 is capable represents an exception to this rule. Meister et al. (2005) found, by constructing chimeric CRC/INO proteins, that the three domains of CRC: the zinc-finger, central, and YABBY (DNA binding) domains, contributed equally to CRC activity. Further experiments will be required to determine which domains of the YAB2 coding sequence confer its capacity to partially replace CRC in carpel development.
A 3·6-kb fragment of the CRC promoter shows erratic position effects in transgenic plants
A currently unexplained phenomenon, noted in the present study, led to the very high level of expression of DL, GUS and GFP coding sequences under the control of a 3·6-kb promoter fragment from the CRC gene in leaves and other organs in a proportion of transgenic plants. As this effect was only present in some transformants from each T1 population, with other plants showing the expected CRC-like pattern of transgene expression, it would seem to be a position-related effect. Further experiments are in progress to investigate the erratic expression of reporter genes noted in the present study with the use of the 3·6-kb CRC promoter fragment. However, the main conclusions of the present work were not affected by this gene expression phenomenon as they were based on transformed plants in which developmental changes were limited to tissues in which CRC is expressed in wild-type plants.
CONCLUSIONS
It is concluded from the present study that putative orthologues of CRC from very widely diverged angiosperm groups show a conserved activity in carpel tissues. These data support the hypothesis of an ancestral role for CRC in the control of carpel development. The present data suggest the putatively derived roles of CRC orthologues in the control of nectary development in the core eudicots, and in carpel determination and leaf development in monocots, to have been acquired by molecular evolution events that did not principally affect the coding sequences of the CRC orthologues present in these plant groups. These conclusions have been reached by studying the coding regions of genes taken from a few widely diverged taxa (Amborella, arabidopsis and rice). Studies of intermediate taxa would be useful to confirm our conclusions.
ACKNOWLEDGEMENTS
We thank Miss Geraldine Brunoud for contributing pro35S::CRC transformants, Dr Vanessa Vernoud for contributing ‘Gateway’ plasmids, Dr Sophie Jasinski for help with figure preparation, and the technical and secretarial staff of the RDP Laboratory for their general assistance. Our laboratory is financially supported by the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Recherche Agronomique (INRA), the Ecole Normale Supérieure de Lyon, and the Université Claude-Bernard (Lyon). It is also a member of the Institut Fédératif de Recherche-128 (Biosciences, Lyon-Gerland).
LITERATURE CITED
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999;126:2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- Bowman JL. The YABBY gene family and abaxial cell fate. Current Opinion in Plant Biology. 2000;3:17–22. doi: 10.1016/s1369-5266(99)00035-7. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix–loop–helix domains. Development. 1999;126:2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF. Establishment of polarity in angiosperm lateral organs. Trends in Genetics. 2002;18:134–141. doi: 10.1016/s0168-9525(01)02601-4. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences of the USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. The flowers in extant basal angiosperms and inferences on ancestral flowers. International Journal of Plant Sciences. 2001;162:1111–1140. [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- Fourquin C, Vinauger-Douard M, Fogliani B, Dumas C, Scutt CP. Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proceedings of the National Academy of Sciences of the USA. 2005;102:4649–4654. doi: 10.1073/pnas.0409577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proceedings of the National Academy of Sciences of the USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Baum SF, Alvarez J, Patel A, Chitwood DH, Bowman JL. Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. The Plant Cell. 2005a;17:25–36. doi: 10.1105/tpc.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Baum SF, Oh SH, Jiang CZ, Chen JC, Bowman JL. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005b;132:5021–5032. [Google Scholar]
- Meister RJ, Oldenhof H, Bowman JL, Gasser CS. Multiple protein regions contribute to differential activities of YABBY proteins in reproductive development. Plant Physiology. 2005;137:651–662. doi: 10.1104/pp.104.055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Arroyo JM, Simorowski J, May B, Martienssen R, Irish VF. Gene trap lines define domains of gene regulation in Arabidopsis petals and stamens. The Plant Cell. 2005;17:2486–2506. doi: 10.1105/tpc.105.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. The Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens PF. Angiosperm Phylogeny Website. 2001 onwards. Version 7, May 2006 (http://www.mobot.org/MOBOT/research/APweb/ [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. The Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]