SUMMARY
The Antarctic notothenioid Trematomus bernacchii (rock cod) lives at a constant mean temperature of −1.9 °C. Gastric digestion under these conditions relies on the proteolytic activity of aspartic proteases such as pepsin. To understand the molecular mechanisms of Antarctic fish pepsins, T. bernacchii pepsins A1 and A2 were cloned, overexpressed in E. coli, purified and characterized with a number of biochemical and biophysical methods. The properties of these two Antarctic isoenzymes were compared to porcine pepsin and found to be unique in a number of ways. Fish pepsins were found to be more temperature sensitive, generally less active at lower pH and more sensitive to inhibition by pepstatin than the mesophilic counterpart. The specificity of Antarctic fish pepsins was similar but not identical to pig pepsin, likely owing to changes in the sequence of fish enzymes near the active site. Gene duplication of Antarctic rock cod pepsins is the likely mechanism for adaptation to the harsh temperature environment in which these enzymes must function.
Keywords: Rock cod, aspartic proteases, specificity, cold-adapted protein
INTRODUCTION
One of the peculiarities characterizing Antarctic notothenioids is their ability to live at a temperature of −1.9 °C with seasonal variations of about 0.2 °C [1]. To cope with such a harsh habitat, Antarctic fish evolved a number of physiological adaptations including the presence of antifreeze glycoproteins [2], the capability to keep tubulin polymerized at −2 °C [3], and a substantial reduction of the hematocrit, a process wherein Channichthyidae reaches its extreme with the complete loss of both erythrocyte and hemoglobin [4]. In addition to that, cold-adapted organisms have developed a number of adjustments at the molecular level to maintain metabolic function at low temperatures. Among other features, they can produce enzymes characterized by a high turnover number or high catalytic efficiency.
In our studies on aspartic proteinases in notothenioids, a fish group endemic to Antarctica, we have focused our attention on pepsins because of their important nutritional role. Pepsins are a family of aspartic proteinases accomplishing important digestive functions in both invertebrates and vertebrates [5]. Like other aspartic proteinases, pepsin is produced as a zymogen. The primary structure of the zymogen includes a signal peptide (or presequence) and the so-called propart, whose autocatalytic cleavage leads to the formation of the active enzyme [6]. The catalytic mechanism depends on the presence of two aspartic acid residues positioned roughly in the center of a deep cleft forming the active site and covered by a hairpin loop (flap) protruding from N-terminal lobe of molecule. The active site cleft can accommodate about 7 residues of a substrate. These residues are usually designated as P4-P3-P2-P1*P1'-P2'-P3' with the scissile peptide bond between P1 and P1' indicated by "*" and normally flanked by two hydrophobic residues. The corresponding subsites that constitute the topography of the active site cleft in each enzyme are designated accordingly as S4-S3-S2-S1-S1'-S2'-S3' [7]. The two major groups of immunogenetically and biochemically distinct pepsins are pepsin A and pepsin C, the latter also know as gastricsin. These enzymes are active at extremely low pH values [8, 9] in both stomach and in the duodenum and are quickly denatured when the pH exceeds 5.5 [10], thus preventing continuous proteolytic action that may damage these organs. Pepsin C has an optimal pH slightly higher than pepsin A, but substrate specificity is nearly the same for the two enzymes [7]. Most of our knowledge on pepsin derives from studies on human and mammalian enzymes [11–14], whilst much less data are available on enzymes from other vertebrates including fish [13, 15, 16].
We have previously [17] cloned and sequenced three forms of pepsin A (which were named A1, A2 and A3), and a single form of gastricsin (named pepsin C) from the gastric mucosa of the Antarctic notothenioid, Trematomus bernacchii (rock cod). Note that the naming convention used here is tentative because fish pepsin A is a close relative of mammalian pepsins A, F and chymosin while fish gastricsin is close to a common ancestor of mammalian gastricsin and pepsin B. Phylogenetic analysis has shown that rock cod pepsin C and A form two distinct clades, and the three fish pepsin A isotypes result from two rounds of gene duplication leading to the most ancestral pepsin A3 and to the most recent forms represented by pepsin A1 and pepsin A2 [17]. Molecular modeling studies have unraveled significant structural differences in these enzymes with respect to their mesophilic counterparts [17]. Rock cod pepsin A2 displayed local changes of the substrate binding cleft with a significant reduction of the hydropathic character and concomitant increase in the flexibility of this region with respect to the two isoforms A1 and A3, the gastricsin and the other fish pepsins [17]. In general, a reduced level of hydropathy and higher molecular flexibility characterize cold-adapted proteins [18]. Moreover, the central role of pepsin A in food digestion makes it an attractive candidate for genetic engineering and identification of amino acid residues modulating its activity or stability.
To better understand the molecular mechanisms responsible for adaptation of food digestion at temperatures below 0 °C, we have produced the two T. bernacchii fish pepsin variants A1 and A2 (hereafter referred to as fish pepsin A1 and A2) by heterologous expression in E. coli. The enzymes were purified, and their biochemical properties were studied in comparison to pepsin A from porcine stomach.
RESULTS
1) Expression and purification of recombinant fish pepsinogens
IPTG induction of E. coli BL21 (DE3) cells transformed with the plasmid pET22b-PepA1 or pET22b-PepA2 resulted in the overexpression of recombinant fish pepsinogens A1 and A2 as inclusion bodies. The inclusion bodies were purified by several buffer washes, solubilized with 6 M urea, and the refolding was initiated by rapid dilution at alkaline pH as described in “Materials and Methods” Refolded pepsinogens A1 and A2 were loaded onto a Sephacryl S-300 column and each collected fraction was monitored for protease activity and absorbance at 280 nm (See Supporting Information Figure S1,A). For both refolded fish pepsinogens, the peak of proteolytic activity eluted just before the end of the A280 peak. Fractions from 15 to 23 did not show proteolytic activity but contain most of the recombinant fish pepsinogen in association with several contaminants, as estimated by SDS-PAGE analysis (data not shown). Both recombinant fish pepsinogens eluted under the A280 peak were assumed to be the incorrectly refolded zymogen forms [19]. The collected gel filtration fractions containing the active pepsinogen form (fractions from 25 to 40) were pooled and further purified by anion-exchange chromatography on a Resource Q column. Fish pepsinogens A1 and A2 were both eluted with 0.1 M NaCl (Supporting Information Figure S1,B). The enzyme-containing fractions were pooled and dialyzed against 20 mM Tris-HCl, pH 8.0.
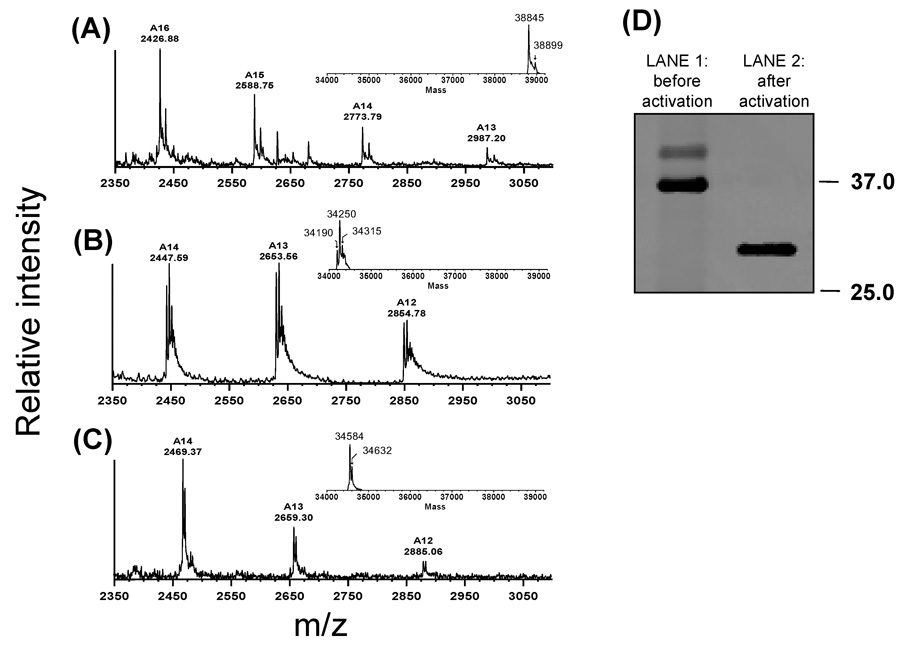
The dialyzed solution containing pepsinogen A1 was found to be homogeneous by SDS-PAGE and mass spectrometry (Fig. 1) and similar data were obtained for fish pepsinogen A2 (data not shown). To determine the purity and validate the correct mass of the purified fish pepsinogens, electrospray mass spectrometry (ESI) was used. Because pepsins traditionally do not ionize well in the positive ion mode, the negative ion mode was used. Figure 1 shows representative ESI spectra for fish pepsinogen A1 (panel A), fish pepsin A1 (panel B) and, for comparison, pig pepsin (panel C). The mass spectra confirmed that the proteins were relatively pure as the major peaks in the spectra were those of pepsinogen (Fig. 1, panel A). Although a second band of higher molecular weight was seen in the SDS-PAGE, mass spectrometry did not indicate that this was a major contaminant.
Figure 1.
Negative-ion ESI mass spectra of (A) fish pepsinogen A1 and (B) activated fish pepsin A1. The measured molecular weights were 38,845 Da for the pepsinogen form (theoretical mass: 38,850.6 Da) and 34,250 Da for the activated form (theoretical mass: 34,241.2 Da). The conversion of pepsinogen to pepsin leads to a mass shift of ~ 4600 Da due to the removal of the N-terminal residues 1–38. (C) Negative-ion ESI mass spectra of pig pepsin A from Sigma, as a control (theoretical mass: 34,583.8 Da; measured mass: 34,584 Da). (D). Commassie-stained SDS-PAGE gel of fish pepsin A1 before (lane 1) and after (lane 2) activation.
2) Characterization of recombinant fish pepsins
Fish pepsins were prepared by activating purified recombinant pepsinogens by acidification with 2 M glycine-HCl, pH 2.0. The solution was neutralized by dialysis against sodium acetate buffer, pH 5.3. The recovered solution containing the active form of fish pepsin A1 showed a single band on SDS-PAGE (Fig. 1, D, lane 2) and similar results were observed with activated fish pepsin A2 (data not shown). The conversion of pepsinogen to pepsin is expected to decrease the molecular mass by approximately 4,600 Da owing to the loss of the N-terminal enzyme propart (residues 1 to 38). The measured molecular masses of fish pepsinogen A1 and fish pepsin A1 were 38,845 and 34,250 Da, respectively (Fig 1, panels A and B). The observed experimental mass difference (4,595 Da) is very close to the theoretical difference indicating the removal of the N-terminal propart during the activation procedure. In addition, the amino terminal sequence determined by Edman degradation of the electroblotted recombinant fish pepsins was YQSGTESMTNDADLSYYGVI for both isoforms confirming that the mature enzymes were generated by autoactivation of the refolded zymogen. Taken together, the SDS-PAGE, Edman degradation and ESI mass spectrometry analyses all indicate that both fish pepsinogens were efficiently converted to the pepsin form at pH 2.0 by elimination of the N-terminal propart sequence.
The secondary structure of fish pepsin A1 and A2 was also investigated by far-UV CD spectroscopy (Supporting Information Fig. S2). The CD spectra for the two fish pepsin isoforms were overlapping indicating no substantial secondary-structure differences. Secondary structure calculations from the far-UV CD spectral data showed that both Antarctic enzymes contained a high proportion of β-sheets (56.1 % for A1 and 52.2 % for A2), as reported for mammalian pepsins [20]. This result indicates that the secondary structure of the recombinant proteins was not altered during protein processing.
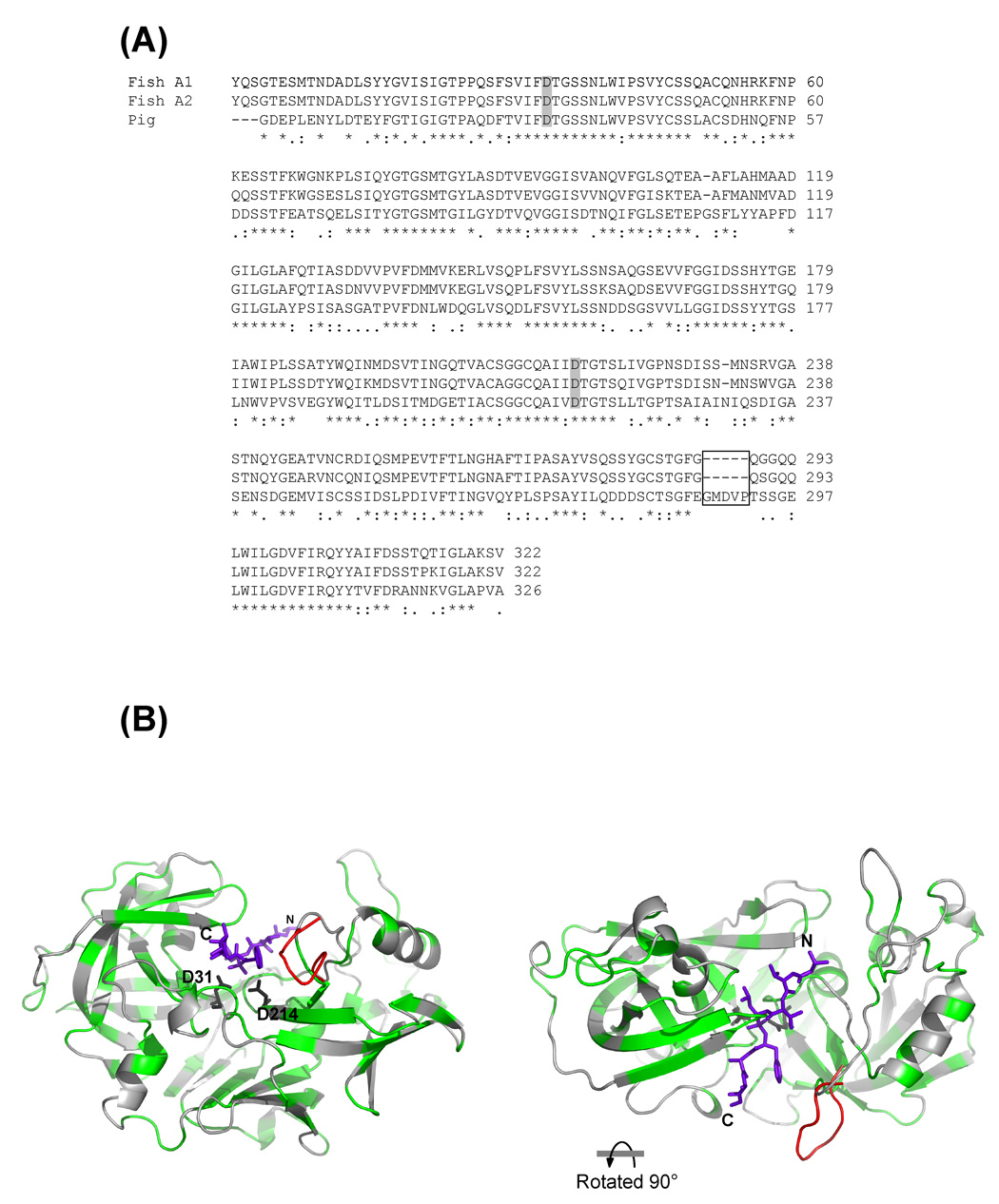
Alignment of Antarctic fish versus pig pepsins also reveals a strong sequence identity (52.7 %) between the three enzymes (Fig. 2). Compared to pig pepsin, both Antarctic fish enzymes lack the C-terminal sequence GMDVP (Fig. 2A and B). Interestingly some of these residues belong to the subsites that constitute the topography of the active site cleft. Notably, the four amino acid residues E287, M289, V291 and T293 forming the loop EGMDVPT are residues of the S4 through S1 subsites (E287 and M289), and S1’ through S3’ subsites (V291 and T293) of pig pepsin that make contact with the residues of an ideal heptapeptide substrate [21].
Figure 2.
(A) Alignment of amino acid sequences of the Antarctic recombinant pepsins with pig pepsin. Multialignment was achieved using the program Clustal W version 1.83 [44]. The two aspartic acid residues present in the catalytic site are marked in gray. The missing residues in fish pepsin isoforms are boxed. (B) Model of fish pepsin A1 based on the crystal structure of pig pepsin (PDB code 5PEP, [21]), prepared as described previously [17]. The rendering on the right has been turned 90 degrees towards the observer, as shown, to look down onto the top of the active site. The conserved residues between pig and fish pepsin, as determined by the alignment, are colored green. The extra residues in pig pepsin that are boxed in panel A are shown in the position they occupy in the crystal structure of pig pepsin (5PEP) and are colored red. The remained of the pig pepsin residues are virtually superimposable on the fish pepsin A1 model. The two conserved aspartic acid residues present in the catalytic site are drwan as sticks and labeled. The synthetic phosphonate inhibitor IVA-Val-Val-LeuP-(O)Phe-Ala-Ala-OMe (IVA = isovaleryl; LeuP = phosphinic acid analogue of leucine; (O)Phe = L-3-phenyllactic acid) in the active site in this PDB file is shown in stick form with the N- and C-termini labeled. Modeling of the inhibitor binding to the fish pepsin model was obtained with overlaying the crystal structure of the model with human pepsin 3A in complex with the synthetic phosphonate inhibitor (PDP code 1QRP, [24]).
2.a) pH and temperature studies
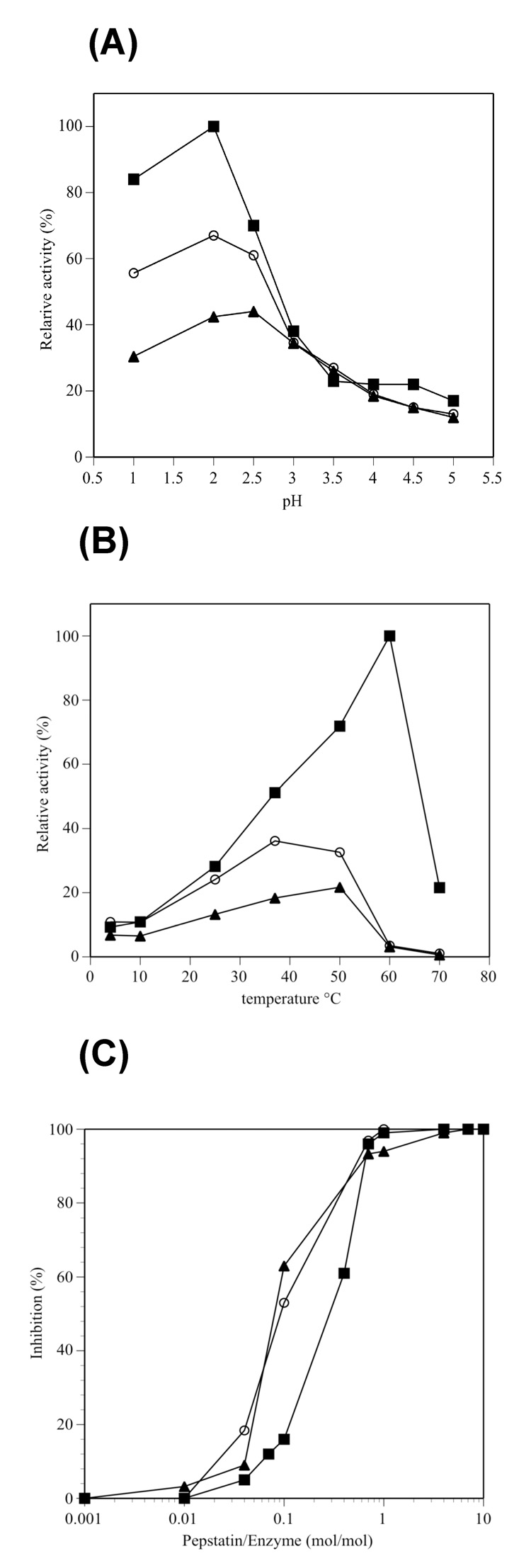
The enzymatic activity of fish pepsins A1 and A2 was investigated at different pH’s and temperatures and compared with pig pepsin. A broad pH range was tested using hemoglobin as substrate (Fig. 3A). The mesophilic porcine enzyme exhibited 100 % relative activity at pH 2.0, but a rapid reduction in activity at pH 2.5. In contrast, both Antarctic isoenzymes were less active at their optimum pH (2.5 for fish pepsin A1 and 2.0 for fish pepsin A2), and showed a slow decline of their relative activity at pH values above their optimum. The effect of the temperature on the enzymes activity was also examined for fish and pig pepsins (Fig. 3B). The optimum temperature (the temperature at which there was the most activity) for Antarctic fish pepsins A1 and A2 was found to be 50 and 37 °C, respectively. At lower temperatures (4, 10 and 25 °C), both pig and fish pepsin A2 showed similar relative activities while fish pepsin A1 was slightly less active. Pig pepsin exhibited 100 % relative enzyme activity at 60 °C and its activity dropped sharply to 20 % at 70 °C. At the temperatures at which they were optimal active, fish isoforms were less active than pig enzyme and rapidly lost activity above 50 °C (Fig. 3B). This prompted an examination of the stability of all three enzymes at 50 °C and at pH 5.3 (at this pH all three pepsins show low activity): aliquots were removed at appropriate times and the residual activity was measured at pH 2.0 using hemoglobin as substrate (Table I). Under these conditions, a half-life of 310 min was observed for the mammalian enzyme. By comparison, the half-life of Antarctic fish pepsins A1 and A2 under the same conditions was 270 and 72 min, respectively. Compared to pig pepsin and fish pepsin A1, fish isoform A2 appeared to be more labile at their maximally active temperature.
Figure 3.
(A) Effect of pH on the activity of commercial pig pepsin and recombinant fish pepsin isoforms A1 and A2. Active fish isoenzymes were obtained as described in “Materials and Methods” (see section “Conversion of pepsinogen in the active form”). The relative activity has been expressed as a percentage of the highest activity over the pH range examined. Each data point represents the mean of three determinations. (B) Influence of temperature on enzymatic activity. Protease activity was determined using denatured hemoglobin as a substrate. Pepsin obtained from pepsinogen activation was added to the reaction mixture containing the buffer sodium citrate, pH 2.0. The assay was performed at various temperatures as described in “Material and Methods” The results are representative of three independent sets of measurements. Relative activity has been expressed as a percentage of the highest activity over the temperature range examined for the enzymes considered. (C) Effect of pepstatin A on fish and pig pepsins. Enzymes were assayed in the presence of increasing concentrations of pepstatin A using a 2.5 % hemoglobin as substrate. Pepsin activity was determined at pH 2.0 and 37 °C The fish and pig pepsin concentration in each assay was 0.021 µM. Legend: activity of commercial pepsin (■); activity of fish pepsin A1 (▲); activity of fish pepsin A2 (○).
Table I.
Stability of pig and fish pepsins at 50 °C and pH 5.3. Aliquots were removed at appropriate times and the residual activity was measured at pH 2.0 and 37 °C using hemoglobin as substrate.
| Enzyme | Half-life time (min) |
|---|---|
| Pig pepsin | 310±1.55 |
| Fish pepsin A1 | 270±1.35 |
| Fish pepsin A2 | 72±0.46 |
2.b) Effect of pepstatin A
Fish and pig pepsin activity was assayed in the presence of increasing concentrations of pepstatin A using hemoglobin as substrate. Pepstatin A is a well known acid protease inhibitor which binds in the active site cleft of the enzyme [22, 23]. Fig 2B shows a ribbon representation of pig pepsin in complex with the phosphonate inhibitor IVA-Val-Val-LeuP-(O)Phe-Ala-Ala-OMe. The occupancy as well as the hydrogen bond network between the enzyme and this inhibitor are essentially the same as those observed with pepstatin A [24]. Pepsin activity was determined at pH 2.0 and 37 °C for all three enzymes (Fig. 3C). Fish and porcine pepsins are inhibited completely by the equivalent molar amount of pepstatin, but fish isoforms were found to have a different behavior in pepstatin A inhibition. Commercial pig pepsin required an inhibitor:enzyme ratio of 0.3:1 to produce 50% inhibition, whereas fish pepsin A1 and A2 required an inhibitor:enzyme ratio of 0.1:1 to reach about 50 and 60% inhibition, respectively.
2.c) Kinetic analysis
Kinetic parameters were determined for the purified Antarctic enzymes and pig pepsin using the synthetic chromogenic substrate Pro-Thr-Glu-Phe-(p-nitro-Phe)-Arg-Leu-OH (peptide #1) and are shown in Table II. The Km values were lower for fish enzymes, as might be expected for an enzyme from an Antarctic specie [25]. The measured kcat values for fish pepsins A1 and A2 were 22 and 130 times smaller than that of pig pepsin, respectively. Although the Km values of fish pepsins were lower, the resultant values for the specificity constant (kcat/Km) of fish pepsin A1 and A2 were 6 and 12 times smaller than that of pig pepsin, respectively. These data suggest that the differences in the 3D-structure of fish and pig pepsins may well affect the kinetic behavior of the porcine and the fish enzymes. In porcine pepsin, there is a loop formed by the following residues: EGMDVPT (Fig. 2A and B). It contains four amino acids that are residues of the S4 through S3’ subsites. This loop is missing in the Antarctic fish enzymes, as shown in the model of fish pepsin A1 in Figure 2B. As a consequence, fish pepsins might accommodate the synthetic substrate in the catalytic pocket better than pig pepsin due to a reduction is steric hinderance in fish pepsin. This hypothesis is corroborated by the fact that fish pepsins were more sensitive to pepstatin A than pig pepsin (Fig. 3C). On the other hand, the specificity constants (kcat/Km) of fish pepsins were lower than mammalian enzyme. That happens perhaps because the differences in subsite residues decrease the velocity of hydrolysis. To explore the possibility that differences between fish and pig pepsins result from sequence changes that effect the substrate recognition region of the active site, we analyzed the digestion of two different substrates, as shown below.
Table II.
Kinetic parameters for the hydrolysis of a chromogenic substrate #1 [sequence Pro-Thr-Glu-Phe-(p-nitro-Phe)-Arg-Leu; see also Figure 4A] by Antarctic and pig pepsins.
| Enzyme | Km (mM) | Kcat (s−1) | Kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Pig pepsin | 0.26±0.023 | 72.02±3.67 | 277±30.47 |
| Fish pepsin A1 | 0.074±0.006 | 3.32±0.29 | 45±6.75 |
| Fish pepsin A2 | 0.025±0.002 | 0.54±0.043 | 22±3.52 |
3. Specificity analysis
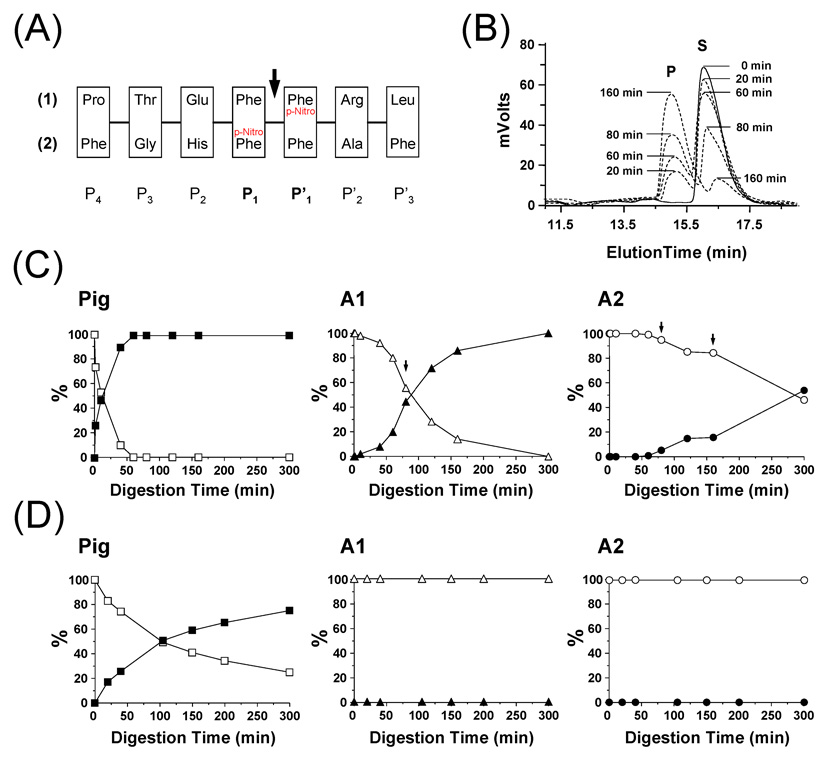
To determine if changing the residues in proximity of the scissile peptide bond Phe-Phe, would cause a difference in the activity of Antarctic pepsins, we used an HPLC assay involving two substrates that have different sequences on either side of the Phe-Phe scissile bond residues (Fig 4A). In this assay, the p-Nitro Phe group acts as a reporter and when the substrate is cleaved, the resulting smaller peptide containing the reporter has a different elution time in the HPLC (Fig 4B). Digestion of peptide #1 is shown in Fig 4C and digestion of peptide #2 is shown in Fig 4D. Both fish A1 and A2 were less efficient at cleaving peptide substrate #1 than was pig pepsin, a result consistent with the previous data described in Figure 3 (A and B) and Table II. However, both fish pepsins were unable to cleave peptide substrate #2 and pig pepsin activity towards this substrate was reduced over its digestion of substrate #1. These results are consistent with the hypothesis made above where the missing loop in Antarctic pepsins (formed by residues EGMDVPT in pig pepsin) alters the substrate specificity of the fish pepsins by removing some of the residues near the scissile peptide bond.
Figure 4.
Analysis of pepsin digestion of synthetic test peptides. (A) Amino-acid sequence of the two synthetic peptides used in this study. The P4 to P’3 nomenclature refers to the position of each residue. The arrow indicates the position of the scissile peptide bond between P1 and P’1. (B) Example of the HPLC elution profile observed at 280 nm for A1 digestion of synthetic peptide #1 (Legend: S = Substrate; P = Product). The digestion was performed in 100 mM sodium acetate buffer, pH 2.5, 0 °C using a 1 to 10 protease/substrate ratio (w/w). At defined time points, 600 pmoles were loaded onto a Jupiter 4u Proteo 90A column and peptides were eluted using a 5 to 70 % ACN gradient in 20 min. (C) Digestion of synthetic peptide #1 by pig pepsin, A1 and A2. Solid and empty symbols correspond to the substrate and the product, respectively. Arrows indicate time points at which additional amounts of fish pepsins were added [60 min: 4 µg (A1 and A2); 160 min: 4 µg (A2)]. (D) Digestion of synthetic peptide #2 by pig pepsin, A1 and A2. No digestion was observed with A1 and A2. Solid and empty symbols correspond to the substrate and the product, respectively.
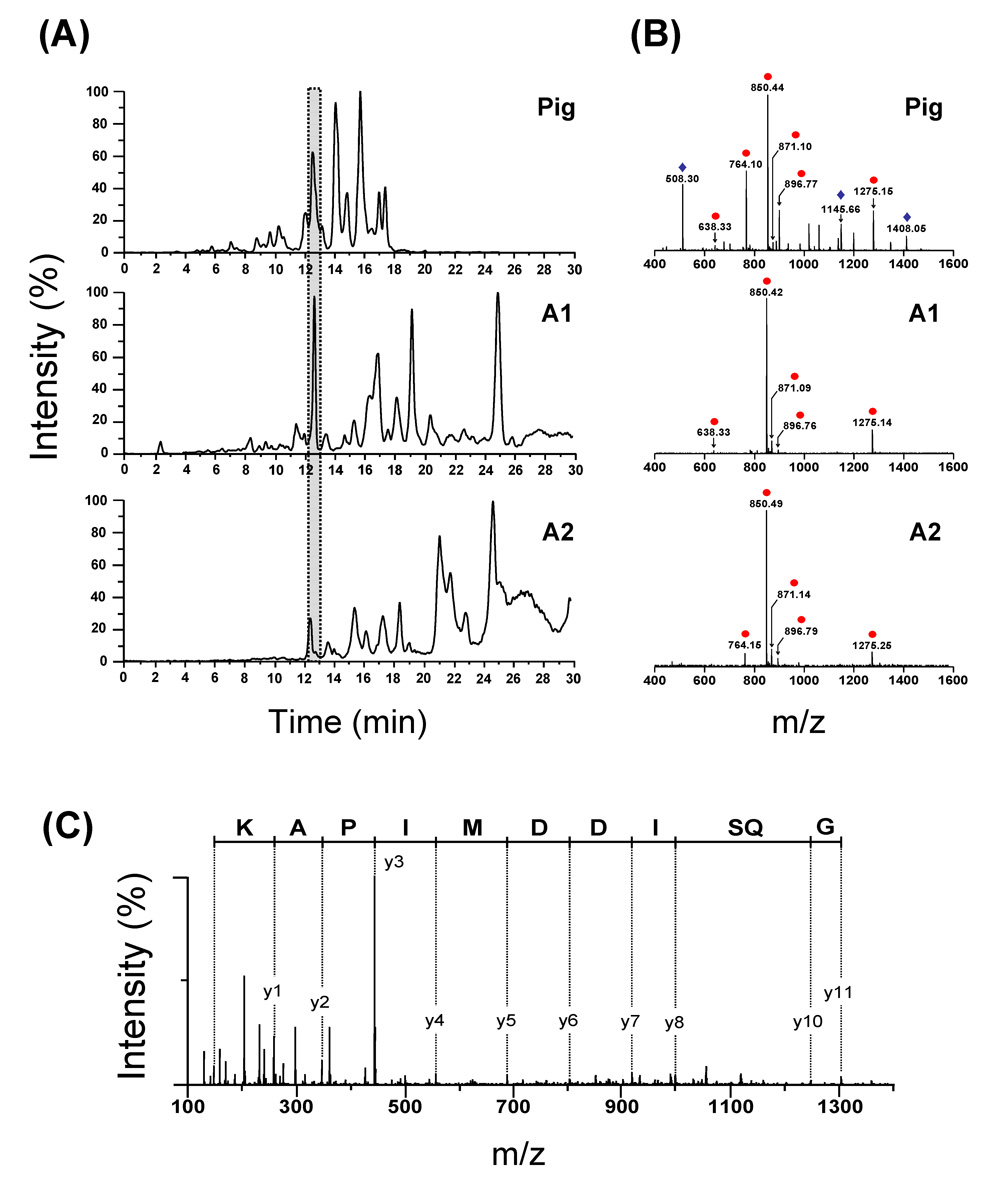
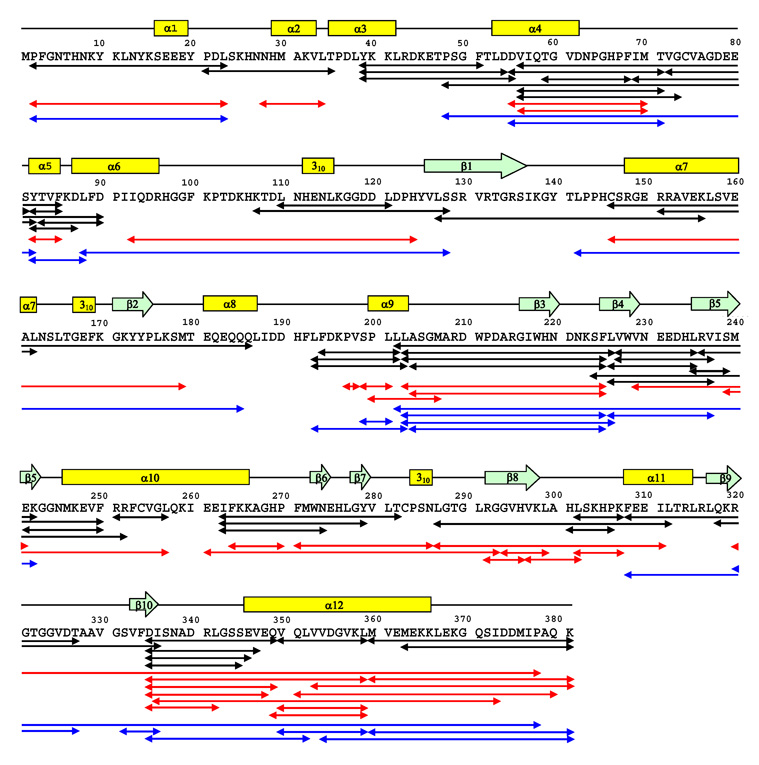
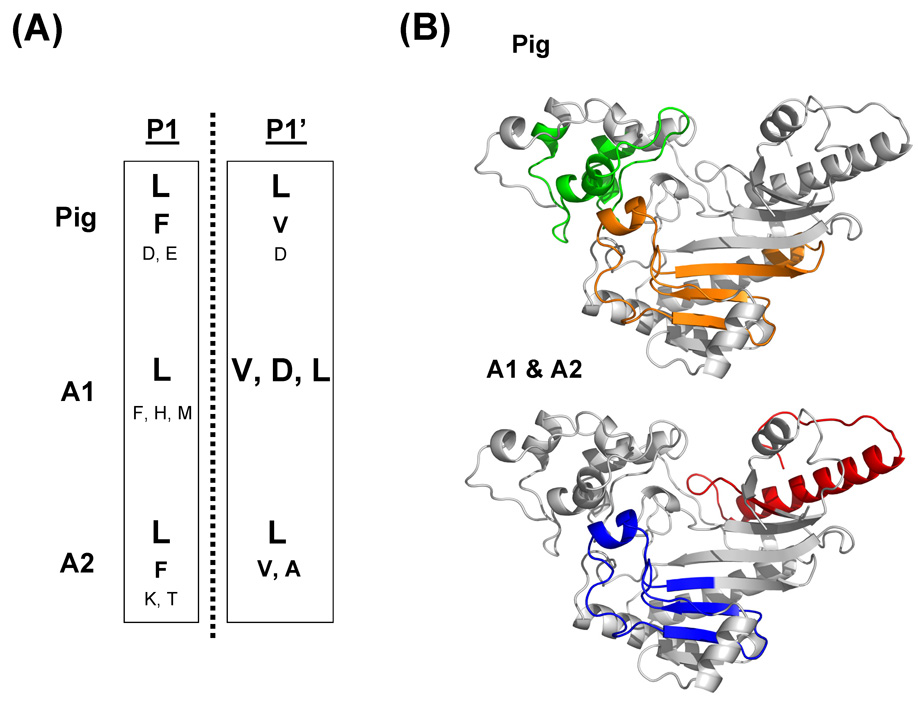
The specificity of the Antarctic fish pepsins A1 and A2 towards residues at the P1 and P1’ positions that flank the scissile peptide bond was determined by digesting a test protein with each enzyme (also with pig pepsin) and sequencing the resulting peptic peptide with mass spectrometry. The digestion pattern of the test protein, rabbit-muscle creatine phosphokinase (CK-MM), was different for digestion with pig versus fish pepsins (Fig 5A). In general, the fish pepsins produced much larger pieces than the pig pepsin during the 3.5 minute digestion time, again indicating the difference in the overall activity of the mesophilic versus the Antarctic enzymes. The peptides in each part of the HPLC separation were visualized, compared (Fig 5B) and then sequenced with tandem mass spectrometry (MS/MS). An example MS/MS spectrum is shown in Fig 5C and is representative of the quality of the spectra obtained for the majority of the peptic peptides. High quality MS/MS allowed the preparation of a digestion map of CK-MM, as shown in Fig 6. The location of each peptic cleavage was tallied (See Supporting Information Figure S3 and Tables S1–S3) and the P1-P1’ amino acid preference for CK-MM was summarized (Fig 7). The results of this analysis indicate the both pig and fish pepsin A1 and A2 prefer hydrophobic residues in the P1-P1’ positions and that their overall specificity was similar among the three enzymes. However, important differences were apparent. Fish pepsin A1 could tolerate histidine or methionine in the P1 position while fish pepsin A2 tolerated lysine and threonine in the P1 position. Pig pepsin was able to accommodate aspartic and glutamic acids in the P1 position while neither of the fish pepsins produced any peptides that had these charged amino acids in the P1 position. As a result, the overall digestion profile of the three pepsin on CK-MM was different. Fig 7B shows that pig pepsin produced many fragments in one portion of the substrate protein while the fish pepsins preferred other regions.
Figure 5.
(A) Comparison of the total ion chromatograms (TIC) obtained after digestion of CK-MM by pig pepsin, A1 and A2. The gray box corresponds to the region that was selected to compare the CK-MM peptide fragments generated after 3.5 min digestion on ice (see below). (B) Representative mass spectra comparing the different peptide fragments eluted after 12 min elution time. Identical ions are noted with a circle; non identical ions with a diamond. (C) MS-MS of the [M + 3H]3+ ion (m/z 850.49) of the peptide Met360-Lys381, generated after A2 digestion.
Figure 6.
CK-MM peptide map generated after 3.5 min digestion on ice (protease/substrate ratio: 1/1 (w/w)) by the pig pepsin (black arrows), A1 (red arrows) and A2 (blue arrows). Secondary structural elements of CK-MM determined by X-ray crystallography [45] are shown above the sequence.
Figure 7.
(A) Cleavage preference of pig, A1 and A2 pepsins at the P1, P1’ positions. The size of letters indicates the probability of cleavage. (B) Three dimensional structures of the monomeric form of CK-MM showing the regions that were extensively covered by the pig pepsin [Tyr39-Asp90 (green); Leu193-Phe250 (orange)], A1 [Asp335-Lys381 (red)] and A2 [Leu193-Val237 (blue)].
DISCUSSION
Antarctic fish are highly cold adapted and remarkably stenothermal [26] as a consequence of evolution of antifreeze glycoproteins and the higher catalytic efficiency of many enzymes at low temperatures [27]. For this reason, the study of enzymes in these poikilothermic species is of interest, especially in relation to the strategies adopted by these organisms to achieve a normal level of proteolysis at temperatures well below that of homeotherm species. Organisms can follow two strategies to achieve cold adaptation at metabolic level: one way is to increase enzyme production in order to compensate for their reduced kinetic efficiency; the other is via the expression of enzymes provided with relatively higher substrate turnover and capacity to maintain ligand-binding properties at low temperature [28]. Multiple forms of type-A pepsinogens are known to occur in various animals [29] and most of them are known to be products of different genes as has been shown in human [30] and cow [31]. The presence of multiple forms of pepsin A in some species may be correlated to the type of food or to the feeding habitat [32].
The present paper describes the characteristics of the enzymatic properties of two recombinant pepsinogen isoforms, A1 and A2 from the Antarctic rock cod. The two Antarctic isoenzymes are, to our knowledge, the first Antarctic fish pepsinogens obtained using the strategy of expression in E. coli followed by refolding and purification. The effectiveness of this strategy was evident as demonstrated by the fact that the purified fish pepsinogens were efficiently converted to the active form upon incubation in acidic conditions. The high percentage of the β-sheet structures, estimated by CD spectroscopy conducted on fish pepsins, was consistent with the literature data for properly folded forms of pepsin, including pig pepsins [20], suggesting that the Antarctic recombinant fish pepsins were correctly folded. The enzymatic assays further demonstrated that the recombinant proteins were active and therefore properly folded.
All reported cold-adapted enzymes display modifications in their kinetic parameters that allow catalytic reactions to take place at low temperatures [33]. These changes may be a high specific activity at temperatures ranging from 0 to 30 °C [34]. Fish pepsins from Antarctic rock cod seem to constitute an exception, since their specific activities at low temperatures (4, 10 and 25 °C) were similar or lower than that of pig pepsin. On the other hand, compared to pig pepsin and fish pepsin A1, fish isoform A2 appeared to be more temperature sensitive showing a half-life at 50 °C of 72 min (pig pepsin, 310 min; fish pepsin A1, 270 min). The full inactivation observed in the isoform A2 might be a consequence of the increased structural flexibility of the protein. Carginale and coworkers [17] have described that apparently, fish pepsin A2 underwent local changes such as reduced hydropathy and increased flexibility at the level of the substrate cleft.
The most striking feature of the primary structure of Antarctic pepsin isoenzymes with respect to the mammalian counterparts is the absence of a loop near the active site. In the 3-dimensional structure of pig pepsin, the sequence EGMDVPT forms a loop near the active site and is missing in the fish isoforms (see red segment on the model of fish pepsin A1, Figure 2). The loop is positioned close to the enzyme pocket and contains four amino acids (E, M, V and T) that are residues of the S4 through S3’ subsites. Because these residues are missing in fish pepsins, they might accommodate substrates in the catalytic pocket better than pig pepsin. This hypothesis was corroborated by kinetic analysis carried out on fish and pig pepsins using the synthetic substrate #1 (Figure 4A). Fish pepsins showed lower values of Km than pig pepsin for this substrate. In addition, both fish isoenzymes required an inhibitor:enzyme ratio lower that pig pepsin to reach 50 % of inhibition. The specificity constants (kcat/Km) of fish pepsins were lower than mammalian enzyme. That happens perhaps because the differences in subsite residues decrease the velocity of hydrolysis.
In comparison experiments with two different substrate peptides (Figure 4A), there was a significant difference between the activity of fish isoenzymes and pig pepsin. Fish pepsins were able to cleave the substrate peptide #1 with sequence PTEFF*RL but unable to cleave the #2 peptide with the sequence FGHF*FAF (where the p-nitro group is indicated by the *). Pig pepsin was active towards both substrates, with diminished activity towards substrate #2. These data suggest that the flanking residues on either side of the P1-P1’ position play a role in the specificity of the fish enzymes. To further investigate which residues fish pepsins preferred in the P1-P1’ positions, in comparison to pig pepsin, test digestion with CK-MM was performed. The results (Figure 7A) suggest that the general specificity of all three enzymes at the P1-P1’ positions are highly similar with some notable exceptions. Fish pepsin A1 could tolerate His or Met in the P1 position while fish pepsin A2 tolerated Lys and Thr in the P1 position. Pig pepsin was able to accommodate aspartic and glutamic acids in the P1 position while neither of the fish pepsins produced any peptides that had these charged amino acids in the P1 position. In the P1’ position, pig pepsin and both fish pepsins were similar except for the ability of fish pepsin A1 to accommodate Asp. The size of the peptide population in this study was too small to make any definitive conclusions about the contribution to specificity from amino acids in positions outside P1 and P1’ [35]. Taken together, our data indicate that the specificity of fish pepsins A1 and A2 are similar but not identical to that of pig pepsin and we attribute the differences to the key changes in the active site of the fish enzymes.
In terms of the cold adaptation of pepsins from Antarctic fish, our study suggests that fish pepsin A2 meets the criterion for a cold-adapted enzyme showing limited stability at moderate temperature (i.e. 50 °C). However, both isoforms do not meet the criterion that cold-adapted enzymes have a specific activity higher than that of their mesophilic counterparts over a temperature range from 0 to 30 °C. We now believe that Antarctic rock cod to accomplish efficient food digestion at low temperature have adopted primarily gene amplification. Gene amplification increases enzyme production to compensate for the reduced kinetic efficiency. Antarctic rock cod have three forms of pepsin A [17] resulting from two events of gene duplication: the first event consisted in the duplication of the pepsin A3 ancestor gene; the second duplication brought to the appearance of pepsin A1 and A2 genes that contributed to further increase enzyme concentration‥ Through the production of isoenzymes that perform nearly the same type of digestion with similar specificity, Antarctic fish may accomplish a similar overall rate of digestion as their mesophilic counterparts.
MATERIALS AND METHODS
Chemicals
Hemoglobin, pepstatin A, porcine pepsinogen and rabbit-muscle creatine phosphokinase (CK-MM), were from Sigma. Sephacryl S-300 was from Pharmacia Biotech. Isopropyl β-D-thiogalactoside (IPTG) was from BIO-RAD. Immobilon-P membranes were from Perkin-Elmer. All other reagents were of analytical grade. Chromophoric peptides Pro-Thr-Glu-Phe-(p-nitro-Phe)-Arg-Leu-OH and Phe-Gly-His-(p-nitro-Phe)-Phe-Ala-Phe-OMe were from Bachem.
Amplification, cloning and sequence analysis of the recombinant fish pepsinogen isoforms
The cDNAs encoding the propart and the mature regions of the two Antarctic fish isoforms (Pepsin A1, Genbank Accession No: AJ550949; Pepsin A2, Genbank Accession No: AJ550950) previously cloned in the pCR4-TOPO vector [17] were amplified by PCR using the forward primer (5’-GGAATTCCATATGTTCCACAAGATTCCCCT-3’) for fish pepsinogen A1 and A2; and the reverse primers (5’-ACCGGTCGACTTAGACAGACTTGGCCAA-3’) and (5’-ACCGGTCGACTTACACAGACTTGGCCAG-3’) for fish pepsinogen A1 and A2, respectively. The upstream primer contained an NdeI restriction site just before the start of the coding region. In the downstream primers, an adjacent SalI restriction site followed the stop codon. Amplification was performed with 5 units of Taq DNA polymerase (Perkin-Elmer), 50 pmol of each of the above primers and 0.2 mM dNTPs (final concentration). The mix was buffered with Perkin-Elmer PCR storage buffer (100 mM Tris-HCl at pH 8.3, 500 mM KCl, 15 mM MgCl2, 0.01 % gelatin). The reaction was carried out in a DNA Thermocycler Express (Hybaid), with an initial denaturation step at 95 °C for 3 min, 30 cycles of 95 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min, and a final step at 72 °C for 15 min. The PCR fragments were gel-purified using a QIAGEN gel-extraction kit (QIAGEN, Germany) and sub-cloned in a pCR4-TOPO vector using a Topo TA Cloning kit (Invitrogen, The Netherlands). Pepsinogen cDNAs were sequenced with an automatic sequencer available at PRIMM (DNA Sequencing Service Naples, Italy). The pCR4-TOPO vectors, containing fish pepsinogen A1 or fish pepsinogen A2, were digested with NdeI and SalI restriction enzymes, and the resulting fragments were ligated into the corresponding sites of expression vector pET22b(+) (Novagen). The resultant recombinant plasmids can produce the recombinant pepsinogen. The sequence of the fish pepsinogens cloned in pET22b(+) was again verified by sequencing in both directions.
Expression of recombinant pepsinogens
Competent BL21(DE3) E. coli were transformed with recombinant plasmid DNA containing either pepsinogen A1 or pepsinogen A2 cDNA to produce pET22b-PepA1 and pET22b-PepA2 cells. Transformed E. coli were grown overnight at 37 °C in Luria-Bertani (LB) broth supplemented with 100 µg/mL ampicillin. The overnight culture was diluted 100-fold using 1 L of fresh LB broth containing ampicillin. The cells were then incubated at 37 °C until mid-exponential growth phase (D600, 0.6). At this point isopropyl β-D-thiogalactoside (IPTG) was added to a final concentration of 1.0 mM. After 10 h induction at 37 °C, cells were collected by centrifugation at 7,410×g for 20 min and washed twice in PBS buffer (140 mM NaCl/2.7 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4, pH 7.3).
Isolation and solubilization of inclusion bodies
The bacterial pellet was frozen and thawed twice, resuspended in 50 mM Tris-HCl, pH 8.0, containing 150 mM NaCl and subjected to sonication. Lysozyme (1 mg), RNase (1 mg), DNase (1 mg) and 30 mM MgCl2 were added to the suspension and incubated for 30 min at 37 °C. The suspension was then diluted in 1 L of 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and gently shaken for 2 h at 4 °C After 30 min centrifugation at 7,000×g, the pellet containing the inclusion bodies was resuspended in 1 L of 100 mM Tris-HCl, pH 8.0, and 50 mM β-mercaptoethanol. The suspension was gently shaken for 2 h at 4 °C and then centrifuged at 7,000×g for 30 min. The pellet was solubilized in 6 M urea, 100 mM Tris-HCl, pH 8.0, 1 mM glycine, 1 mM EDTA, 50 mM β - mercaptoethanol at 4 °C with a gently shaking for 12 h.
Refolding and purification
The solubilized material was centrifuged at 44,380×g for 2 h using a 60Ti rotor. Renaturation of the supernatant containing pepsinogen A1 or A2 was achieved by a 200-fold dilution of the denaturant in 100 mM Tris-HCl buffer, pH 8.0, and the resultant solution was re-concentrated using a Prep/Scale-TFF cartridge, 10 kDa cut-off, tangential flow concentrator (Millipore Corporation, Billerica, Ma, USA) and then further concentrated to about 50 mL in an Amicon ultrafiltration cell (Millipore). After removing residual insoluble material by centrifugation at 20,000×g for 1 h, the clear supernatant was subjected to chromatographic purification using Sephacryl S-300 (1.5 × 45 cm) gel filtration. About 40 mg of total protein was loaded onto the Sephacryl S-300 column which was equilibrated and eluted with 20 mM Tris-HCl, pH 8.0, at a flow rate of 0.8 mL/min. Fractions containing active pepsin were pooled and loaded onto a Resource Q column (30 × 6.4 mm, Pharmacia) equilibrated with 20 mM Tris-HCl, pH 8.0. Proteins were eluted with a linear gradient of 0 to 500 mM NaCl in 20 min at a flow rate of 1 mL/min.
Conversion of pepsinogen in the active form
Acidification was carried out by adding five milliliters of 2 M glycine-HCl, pH 2.0, to 40 ml of the pepsinogen preparation and incubating the mixture at 25 °C for 2 h [36]. The solution was neutralized by dialyzing against 20 mM sodium acetate, pH 5.3 and at 4 °C, using dialysis membranes with 12–14 KDa cut-off. The recovered solution containing the active form of pepsinogen was centrifuged at 20,000×g for 30 min, at 4 °C and used as purified Antarctic fish pepsin A1 or A2.
CD spectra of fish pepsins
CD spectroscopy was performed on homogenous samples of recombinant fish pepsin A1 and A2 (0.1 mg/mL in 2.5 mM sodium acetate buffer, pH 5.3), at 25 °C, using a J-710 spectropolarimeter (Jasco, Tokyo, Japan) calibrated with a standard solution of (+)-10-camphosulphonic acid. A cuvette with 0.1 cm path length was used to record spectra in the far-UV region (240-200 nm) and each spectrum was averaged 6 times. Photomultiplier high voltage did not exceed 600 V in the spectral region measured. The results are expressed in terms of residue molar ellipticity. The percentages of secondary structures were estimated according to the method of Yang et al. [37].
Negative ion electrospray mass spectra of fish pepsinogens and pepsins
Negative-ion mass spectra were acquired on a Waters LCT-Premier mass spectrometer equipped with a standard electrospray source, a capillary voltage of 2.8 kV and a cone voltage of 50 V. The m/z scale was calibrated in negative mode with a 10 ng/mL sodium formate solution in 90/10 isopropanol/H2O. Pepsin samples were loaded onto a self-packed POROS 20R2 trap (POROS media from Applied Biosystems, Framingham, MA) equilibrated with H2O, 0.3 % NH4OH (pH 8.0), desalted for 10 min and eluted in the mass spectrometer with a 5 to 50 % isopropanol gradient in 20 min at 50 µL/min.
SDS-PAGE, Western blotting and sequencing
In addition to mass spectrometry, the purity of the pepsinogen and pepsin was examined by SDS-PAGE carried out according to Laemmli [38]. Samples were dissolved in buffer containing 5 % β-mercaptoethanol. Gels were stained with Coomassie blue. Blotting from gels onto Immobilon-P membrane was performed as described by Matsudaira [39]. N-terminal sequencing was performed on the blotted protein by automated Edman degradation. Protein concentration was determined using two methods: a) BioRad Protein assay, based on the Bradford method [40]; b) absorbance at 280 nm using an absorption coefficient of 56,840 M−1cm−1 and 62,340 M−1cm−1 for pepsin A1 and A2, respectively. Extinction coefficients were determined using ProtParam program on the ExPASy server [41].
Activity assay with hemoglobin
All enzyme assays were performed using a Cary 50 UV-Visible spectrophotometer (Varian). The proteolytic assay was measured using a 2.5 % denatured bovine hemoglobin as substrate, as described previously [42]. Pepsin activity was determined at acidic pH and 37 °C. After 60 min incubation, the reaction was stopped by the addition of 5 % TCA. The samples were centrifuged twice for 10 min at 16,000×g, and the resulting supernatant containing the small peptides produced by proteolytic digestion was analyzed at 280 nm. One enzyme unit was defined as the amount capable of producing a ΔOD280 (OD280 [sample] − OD280 [blank]) = 0.1 in 60 min.
Determination of pH optimum
In order to determine the optimum pH values, 0.35 µg of fish pepsins (A1 or A2) were assayed in 100 mM HCl/KCl buffer at pH 1.0 and 150 mM sodium citrate buffer at pH 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0 using hemoglobin as substrate. Incubation was carried out at 37 °C for 60 min. Pig pepsin was also tested using the same experimental conditions.
Temperature studies
The optimum temperature for pig and fish pepsin activities was determined by incubating pepsins in 0.15 M sodium citrate buffer, pH 2.0, at 4, 10, 25, 37, 50, 60 and 70 °C with 2.5 % hemoglobin. After 60 min, the reaction was terminated by the addition of 5 % TCA. All the determinations were done in triplicate. The stability of pepsins at 50 °C in 20 mM sodium acetate, pH 5.3, was also investigated. Aliquots were removed at appropriate times and the residual activity was measured at 37 °C, pH 2.0, using hemoglobin as substrate.
Inhibitory effect of pepstatin A
Fish and pig pepsins were assayed in the presence of increasing concentrations of pepstatin A in the range from 2.1×10−5 to 2.1×10−1 µM, using a 2.5 % hemoglobin solution as substrate (the final hemoglobin concentration in the assay was 1 %). Pepsin activity was determined at pH 3.0 and 37 °C After 60 min incubation, the reaction was stopped by the addition of 5 % TCA. The samples were centrifuged twice for 10 min at 16,000×g and the resulting supernatant was analyzed at 280 nm. Fish and pig pepsin concentrations used in each assay were 0.021 µM.
Cleavage of chromogenic substrates following the rate of decrease in absorbance at 300 nm
The enzymatic cleavage of the Phe-(p-nitro-Phe) bond in two chromogenic substrates Pro-Thr-Glu-Phe-(p-nitro-Phe)-Arg-Leu-OH (peptide #1) and Phe-Gly-His-Phe-(p-nitro-Phe)-Ala-Phe-OMe (peptide #2) was followed spectrophotometrically at 300 nm, as described by Dunn [43]. Stock solution (2.0 mM) of peptide substrate was prepared in 0.1 M acetic acid. The incubation mixture contained 0.2 mM of chromogenic substrate and 0.35 µg of fish or pig pepsins in 0.15 M sodium citrate buffer, pH 2.0. The assay was performed by monitoring the decrease in absorbance at 300 nm (ΔOD300/min) at 25 °C for at least 10 min.
Determination of kinetic parameters
The chromogenic peptide, Pro-Thr-Glu-Phe-(p-nitro-Phe)-Arg-Leu-OH (peptide #1), was used as the substrate for kinetic analyses. The assay with the synthetic substrate was performed as described in the section “Cleavage of chromogenic substrates following the rate of decrease in absorbance at 300 nm” The kinetic constants, Km and Vmax, were determined from the plot of the reciprocal of the initial velocity versus the reciprocal of the concentrations of the synthetic substrate (up to 0.3 mM). kcat values were obtained from the equation: Vmax = kcat[E] where [E] is the enzyme concentration. The reported kinetic values correspond to the average of three independent determinations.
Analysis of chromogenic substrates cleavage by HPLC
The chromogenic substrates Pro-Thr-Glu-Phe-(p-nitro-Phe)-Arg-Leu-OH and Phe-Gly-His-Phe-(p-nitro-Phe)-Ala-Phe-OMe were reconstituted in 30 % acetic acid, pH 1.8 to a final concentration of 1 mg/mL. Digestions were performed at 0 °C and 37 °C in 100 mM sodium acetate buffer, pH 2.5, with a 1 to 10 protease / substrate ratio (w/w). Fish pepsins and pig pepsin were diluted to a final concentration of 0.05 mg/mL. At defined time points, 25 µL of the digestion mixture was removed and immediately loaded onto a Jupiter 4µ Proteo 90A column (50 × 1.00 mm, Phenomenex Corp) equilibrated with 5 % acetonitrile (ACN). Peptides were eluted in 20 min with a linear gradient of 5 to 70 % ACN at 50 µL/min with a Shimadzu HPLC (LC-10ATvp). The elution profiles were recorded at 214 and 280 nm with a Shimadzu UV-Vis detector (SPD-10Avp).
Protein digestions - Identification of the generated peptide fragments
Creatine phosphokinase b from rabbit muscle was reconstituted in ddH2O to a final concentration of 0.5 mg/mL. Digestions were performed at 0 °C in 20 mM Tris-HCl buffer, pH 2.5 using a 1 to 1 protease/substrate ratio (w/w). After 3.5 min digestion, samples were loaded onto a Magic C18 5µ 200Å column (1.0 × 50 mm, Michrom Bioresources) equilibrated with 5 % ACN, 0.05 % TFA. Peptide fragments were eluted into the mass spectrometer using a 5 to 50 % ACN gradient in 30 min (for MS analysis) or in 50 min (for MS-MS analysis) at 50 µL/min with a Shimadzu HPLC (LC-10ADvp). Mass spectral analyses were performed on a Waters Q-TOF Ultima mass spectrometer equipped with a standard electrospray source and a lockspray interface. The capillary and the cone voltages were set to 3.00 kV and 35 V, respectively. For each acquisition, a 250 fmol/µL GFP solution in 50:50 ACN/H2O, 0.2 % formic acid was continuously infused through the reference probe at 5 µL/min. Data from the reference spray were used to calculate a mass correction factor in order to provide exact mass information. Peptide mass assignments were confirmed by exact mass measurement and/or MS-MS experiments.
Supplementary Material
A pdf file containing three Supplementary Figures (Figure S1, S2, S3) and three Supplementary Tables (Table S1, S2, S3).
ACKNOWLEDGEMENTS
We are pleased to acknowledge funding from the Italian ”National Program for Antarctic Research (PNRA)” and the National Institutes of Health (GM070590, J.R.E.). This work is contribution number 909 from the Barnett Institute.
REFERENCES
- 1.Brodeur JC, Calvo J, Clarke A, Johnston IA. Myogenic cell cycle duration in Harpagifer species with sub-Antarctic and Antarctic distributions: evidence for cold compensation. J Exp Biol. 2003;206:1011–1016. doi: 10.1242/jeb.00204. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, DeVries AL, Cheng CH. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc Natl Acad Sci U S A. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Detrich HW., 3rd Microtubule assembly in cold-adapted organisms: functional properties and structural adaptations of tubulins from antarctic fishes. Comp Biochem Physiol A Physiol. 1997;118:501–513. doi: 10.1016/s0300-9629(97)00012-1. [DOI] [PubMed] [Google Scholar]
- 4.D'Avino R, Caruso C, Camardella L, Schninà ME, Rutigliano B, Romano M, Carratore V, Barra D, di Prisco G. An overwiew of the molecular structure and funtional properties of the hemoglobins of a cold-adapted Antarctic teleost. Berlin: Springer-Verlag; 1991. [Google Scholar]
- 5.Kageyama T. Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell Mol Life Sci. 2002;59:288–306. doi: 10.1007/s00018-002-8423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kageyama T, Ichinose M, Miki K, Athauda SB, Tanji M, Takahashi K. Difference of activation processes and structure of activation peptides in human pepsinogens A and progastricsin. J Biochem (Tokyo) 1989;105:15–22. doi: 10.1093/oxfordjournals.jbchem.a122610. [DOI] [PubMed] [Google Scholar]
- 7.Fusek M, Vetvicka V. Aspartic proteinases. Physiology and Pathology. Boca Raton: CRC press; 1995. [Google Scholar]
- 8.Dunn BM, Valler MJ, Rolph CE, Foundling SI, Jimenez M, Kay J. The pH dependence of the hydrolysis of chromogenic substrates of the type, Lys-Pro-Xaa-Yaa-Phe-(NO2)Phe-Arg-Leu, by selected aspartic proteinases: evidence for specific interactions in subsites S3 and S2. Biochim Biophys Acta. 1987;913:122–130. doi: 10.1016/0167-4838(87)90320-7. [DOI] [PubMed] [Google Scholar]
- 9.Fruton JS. The specificity and mechanism of pepsin action. Adv Enzymol Relat Areas Mol Biol. 1970;33:401–443. doi: 10.1002/9780470122785.ch9. [DOI] [PubMed] [Google Scholar]
- 10.Foltmann B. Gastric proteinases: structure, function, evolution and mechanism of action. Essays Biochemestry. 1981;17:52–84. [PubMed] [Google Scholar]
- 11.Abad-Zapatero C, Rydel TJ, Erickson J. Revised 2,3, a structure of porcine pepsin: evidence for a flexible subdomain. Proteins. 1990;8:62–81. doi: 10.1002/prot.340080109. [DOI] [PubMed] [Google Scholar]
- 12.Fujinaga M, Chernaia MM, Tarasova NI, Mosimann SC, James MN. Crystal structure of human pepsin and its complex with pepstatin. Protein Sci. 1995;4:960–972. doi: 10.1002/pro.5560040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gildberg A, Olsen RL, Bjarnason JB. Catalytic properties and chemical composition of pepsins from Atlantic cod (Gadus morhua) Comp. Biochem. Physiol. Part B. 1990;96:323–330. doi: 10.1016/0305-0491(90)90382-4. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez LM, Freije JP, Merino AM, Vizoso F, Foltmann B, Lopez-Otin C. Isolation and characterization of a pepsin C zymogen produced by human breast tissues. J. Biol.Chem. 1992;267:24725–24731. [PubMed] [Google Scholar]
- 15.Gildberg A. Aspartic proteinases in fishes and acquatic invertebrates. Comp Biochem Physiol B. 1988;91:425–435. doi: 10.1016/0305-0491(88)90002-8. [DOI] [PubMed] [Google Scholar]
- 16.Tanji M, Kageyama T, Takahashi K. Tuna pepsinogens and pepsins. Purification, characterization and amino-terminal sequences. Eur. J. Biochem. 1988;177:251–259. doi: 10.1111/j.1432-1033.1988.tb14369.x. [DOI] [PubMed] [Google Scholar]
- 17.Carginale V, Trinchella F, Capasso C, Scudiero R, Riggio M, Parisi E. Adaptive evolution and functional divergence of pepsin gene family. Gene. 2004;333:81–90. doi: 10.1016/j.gene.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Marshall CJ. Cold-adapted enzymes. Trends Biotechnol. 1997;15:359–364. doi: 10.1016/S0167-7799(97)01086-X. [DOI] [PubMed] [Google Scholar]
- 19.Lin XL, Wong RN, Tang J. Synthesis, purification, and active site mutagenesis of recombinant porcine pepsinogen. J Biol Chem. 1989;264:4482–4489. [PubMed] [Google Scholar]
- 20.Cooper JB, Khan G, Taylor G, Tickle IJ, Blundell TL. X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution. J Mol Biol. 1990;214:199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- 21.Sielecki AR, Fedorov AA, Boodhoo A, Andreeva NS, James MN. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 A resolution. J Mol Biol. 1990;214:143–170. doi: 10.1016/0022-2836(90)90153-D. [DOI] [PubMed] [Google Scholar]
- 22.Fujinaga M, Chernaia MM, Tarasova NI, Mosimann SC, James MN. Crystal structure of human pepsin and its complex with pepstatin. Protein Sci. 1995;4:960–972. doi: 10.1002/pro.5560040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marciniszyn J, Jr, Hartsuck JA, Tang J. Mode of inhibition of acid proteases by pepstatin. J Biol Chem. 1976;251:7088–7094. [PubMed] [Google Scholar]
- 24.Fujinaga M, Cherney MM, Tarasova NI, Bartlett PA, Hanson JE, James MN. Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog. Acta Crystallogr D Biol Crystallogr. 2000;56:272–279. doi: 10.1107/s0907444999016376. [DOI] [PubMed] [Google Scholar]
- 25.Genicot S, Rentier-Delrue F, Edwards D, VanBeeumen J, Gerday C. Trypsin and trypsinogen from an Antarctic fish: molecular basis of cold adaptation. Biochim Biophys Acta. 1996;1298:45–57. doi: 10.1016/s0167-4838(96)00095-7. [DOI] [PubMed] [Google Scholar]
- 26.Cheng CH, Detrich WH. Molecular ecophysiology of Antarctic notothenioid fishes. Philos Trans R Soc Lond B Biol Sci. 2007 doi: 10.1098/rstb.2006.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somero GN. Biochemical mechanisms of cold adaptation and stenothermality in Antarctic fish. Berlin: Springer-Verlag; 1991. [Google Scholar]
- 28.D'Amico S, Claverie P, Collins T, Georlette D, Gratia E, Hoyoux A, Meuwis MA, Feller G, Gerday C. Molecular basis of cold adaptation. Philos Trans R Soc Lond B Biol Sci. 2002;357:917–925. doi: 10.1098/rstb.2002.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foltmann B. Gastric proteinases--structure, function, evolution and mechanism of action. Essays Biochem. 1981;17:52–84. [PubMed] [Google Scholar]
- 30.Zelle B, Evers MP, Groot PC, Bebelman JP, Mager WH, Planta RJ, Pronk JC, Meuwissen SG, Hofker MH, Eriksson AW, et al. Genomic structure and evolution of the human pepsinogen A multigene family. Hum Genet. 1988;78:79–82. doi: 10.1007/BF00291240. [DOI] [PubMed] [Google Scholar]
- 31.Lu Q, Wolfe KH, McConnell DJ. Molecular cloning of multiple bovine aspartyl protease genes. Gene. 1988;71:135–146. doi: 10.1016/0378-1119(88)90085-6. [DOI] [PubMed] [Google Scholar]
- 32.Kageyama T. New world monkey pepsinogens A and C, and prochymosins. Purification, characterization of enzymatic properties, cDNA cloning, and molecular evolution. J Biochem (Tokyo) 2000;127:761–770. doi: 10.1093/oxfordjournals.jbchem.a022668. [DOI] [PubMed] [Google Scholar]
- 33.Feller G, Narinx E, Arpigny JL, Aiattaleb M, Daise E, Genicot S, Gerday C. Enzymes from psycrophilic organisms. FEMS Microbiol. Rev. 1996;18:189–202. [Google Scholar]
- 34.Feller G, Payan F, Theys F, Qian M, Haser R, Gerday C. Stability and structural analysis of alpha-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. Eur J Biochem. 1994;222:441–447. doi: 10.1111/j.1432-1033.1994.tb18883.x. [DOI] [PubMed] [Google Scholar]
- 35.Keil B. Specificity of proteolysis. Berlin ; New York: Springer-Verlag; 1992. [Google Scholar]
- 36.Shintani T, Nomura K, Ichishima E. Engineering of porcine pepsin. Alteration of S1 substrate specificity of pepsin to those of fungal aspartic proteinases by site-directed mutagenesis. J Biol Chem. 1997;272:18855–18861. doi: 10.1074/jbc.272.30.18855. [DOI] [PubMed] [Google Scholar]
- 37.Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dame JB, Reddy GR, Yowell CA, Dunn BM, Kay J, Berry C. Sequence, expression and modeled structure of an aspartic proteinase from the human malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 1994;61:177–190. doi: 10.1016/0166-6851(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 43.Dunn BM, Jimenez M, Parten BF, Valler MJ, Rolph CE, Kay J. A systematic series of synthetic chromophoric substrates for aspartic proteinases. Biochem J. 1986;237:899–906. doi: 10.1042/bj2370899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 45.Rao JK, Bujacz G, Wlodawer A. Crystal structure of rabbit muscle creatine kinase. FEBS Lett. 1998;439:133–137. doi: 10.1016/s0014-5793(98)01355-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A pdf file containing three Supplementary Figures (Figure S1, S2, S3) and three Supplementary Tables (Table S1, S2, S3).