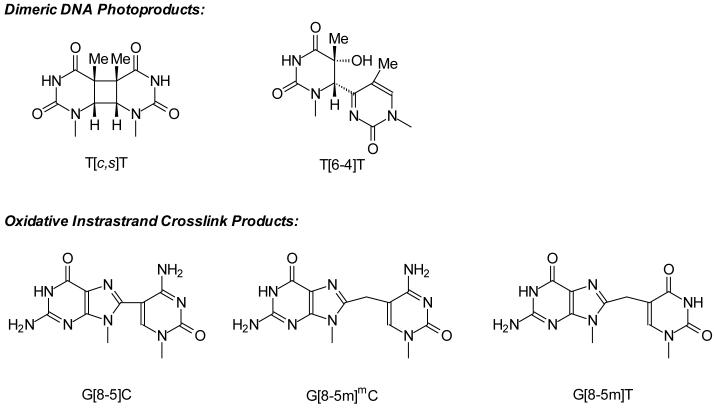
Abstract
Nucleotide excision repair (NER) is a repair pathway that removes a variety of bulky DNA lesions in both prokaryotic and eukaryotic cells. The perturbation of DNA helix structure caused by the oxidative intrastrand lesions could render them good substrates for the NER pathway. Here we employed Escherichia coli (E. coli) NER enzymes, i.e., UvrA, UvrB and UvrC, to examine the incision efficiency of duplex DNA carrying three different oxidative intrastrand cross-link lesions, that is, G[8-5]C, G[8-5m]mC, and G[8-5m]T, and two dithymine photoproducts, namely, the cis,syn-cyclobutane pyrimidine dimer (T[c,s]T) and pyrimidine(6-4)pyrimidone product (T[6-4]T). Our results showed that T[6-4]T was the best substrate for UvrA binding, followed by G[8-5]C, G[8-5m]mC and G[8-5m]T, and then by T[c,s]T. The efficiencies of the UvrABC incisions of these lesions were consistent with their UvrA binding affinities: The stronger the binding to UvrA, the higher the incision rate. In addition, flanking DNA sequences appeared to have little effect on the binding affinity of UvrA toward G[8-5]C as AG[8-5]CA was only slightly preferred over CG[8-5]CG. Consistently, these two sequences exhibited almost no difference in incision rates. Furthermore, we investigated the thermal stability of dodecameric duplexes containing a G[8-5m]mC or G[8-5m]T and our results revealed that these two lesions destabilized the duplex, due to an increase in free energy for duplex formation at 37°C, by approximately 5.4 kcal/mol and 3.6 kcal/mol, respectively. The destabilizations to DNA helix caused by those lesions, for the most part, are correlated with the binding affinities of UvrA and incision rates of UvrABC. Taken together, the results from this study suggest that oxidative intrastrand lesions might be substrates for NER enzymes in vivo.
Reactive oxygen species (ROS) can induce damages to various cellular components including DNA and proteins (1,2). Other than single-nucleobase lesions, a number of intrastrand crosslink lesions can form from ROS attack (3-15). In this respect, it was found that the intrastrand crosslink lesions G[8-5]C, G[8-5m]T and G[8-5m]mC (structures shown in Scheme 1) can be induced in duplex DNA upon exposure to γ rays (8,13,15). In addition, our recent study revealed that G[8-5m]T could also be generated from calf thymus DNA upon treatment with Fenton reagent under aerobic conditions (14). In mammalian cells, the methylated CpG sites are mutational hotspots and the most frequently observed mutations at these sites are C→T transitions (16). In addition, Lee et al. (17) reported that, upon treatment with Cu(II)/H2O2/ascorbate, a substantial frequency of mCG→TT tandem mutations was observed in nucleotide excision repair (NER)-deficient XP-A cells, but not in repair-proficient cells.
In both prokaryotic and eukaryotic cells, NER is an important pathway for removing bulky base damages or helix-distorting lesions from DNA thereby maintaining genomic integrity (18). Deficiency in NER has been shown to be associated with cancer-prone patients with different complementation groups of xeroderma pigmentosum (XP), Trichothiodystrophy (TTD), and Cockayne’s syndrome (18,19). The E. coli UvrABC nuclease system has been employed as a model system for investigating the repair of a variety of bulky DNA lesions. The repair process begins with the DNA damage recognition by UvrA2/UvrB complex, which binds the damaged sites through at least two steps (20), i.e., the recognition of the global duplex distortion by UvrA2 and the subsequent recognition of the specific type of DNA base modifications by UvrB. After the dissociation of UvrA2, UvrB-DNA complex recruits UvrC to the damaged site, which triggers the first cleavage at the fourth or fifth phosphodiester bond 3′ to the damage, followed immediately by the 5′ incision at the eighth phosphodiester bond. UvrD helicase then unwinds and removes this 12-13 mer damaged DNA strand, and DNA polymerase and ligase seal the gap in the DNA duplex. Although there is no structural homology between the NER proteins in prokaryotes and eukaryotes, they share similar functions of nucleases and overall sequences of the repair process (18).
Pyrimidine(6-4)pyrimidone product and its Dewar valence isomer, i.e., T[6-4]T (Scheme 1) and T[Dewar]T, are known to be good substrates for the UvrABC nucleases, which are attributed, at least in part, to the large structural distortion to duplex DNA induced by the two lesions (21,22). In contrast, another major UV photoproduct formed at TT site, the cis,syn-cyclobutane pyrimidine dimer (T[c,s]T, Scheme 1), cannot be repaired efficiently by NER (22). This latter lesion, however, can be bypassed in an error-free manner by translesion synthesis with polymerase η (23,24).
Only a few studies have been carried out for examining the biological effects of oxidative intrastrand lesions. In this context, we reported recently that the intrastrand crosslink lesion formed between guanine and cytosine, G[8-5]C (Scheme 1), could stall replicative DNA polymerases, i.e., the exonuclease-free (exo-) Klenow fragment of E. coli DNA polymerase I, exo- T7 DNA polymerase, and HIV reverse transcriptase (15,25). The lesion, however, can be partially bypassed by human (unpublished results) and yeast polymerase η, but incorrect nucleotides were preferentially inserted opposite the guanine portion of the lesion (15). In addition, G[8-5m]T has been recently shown to be a poorer substrate for UvrABC enzymes than T[6-4]T and AAF-dG adduct (26) .
Herein, we systematically examined the action of E. coli UvrABC enzymes on three oxidative intrastrand cross-link lesions, G[8-5]C, G[8-5m]mC, and G[8-5m]T. For comparison, we also investigated the corresponding recognition and incision of the structurally related T[6-4]T and T[c,s]T in the same sequence context as the G[8-5]C lesion. Furthermore, we examined the thermodynamic properties of G[8-5m]mC and G[8-5m]T, which, in combination with previous thermodynamic studies of G[8-5]C (25), allowed us to correlate the UvrABC incision efficiency with the extent of the duplex destabilization caused by different lesions.
Materials and Methods
Chemicals and Enzymes
All unmodified oligodeoxyribonucleotides (ODNs) used in this study were purchased from Integrated DNA Technologies (Coraville, IA). [γ-32P]ATP was obtained from Amersham Biosciences Co. (Piscataway, NJ). All other chemicals unless otherwise noted were obtained from Sigma-Aldrich (St. Louis, MO). UvrA, UvrB, and UvrC proteins were purified as reported previously (26). The purities of these enzymes were estimated to be better than 95%.
Preparation of ODN Substrates Containing Intrastrand Cross-link Lesions for in-vitro Repair Studies
The preparation of ODNs containing dimeric thymine photoproducts was described previously (27). Briefly, d(GTATTAT) was dissolved in H2O with an AU260 of 0.3 in a quartz tube, degassed by argon bubbling for 30 min, and irradiated with 254-nm light, which was generated by a TLC lamp (UVP Inc., Upland, CA), at room temperature for 30 min. The solution was continuously bubbled with argon during irradiation and, after irradiation, the solution was dried with a Speed-Vac.
The photoproduct-containing ODNs were separated by HPLC on a 4.6 × 250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Alltech Associates Inc., Deerfield, IL). The composition of buffer A was 50 mM TEAA (pH 6.5), and buffer B contained 50 mM TEAA and acetonitrile (70/30, v/v). The gradient program for the mobile phase was: 0 min, 0% B; 5 min, 20% B; 45 min, 50% B; 50 min, 100% B. The flow rate was 0.8 mL/min, and a UV detector was set at 260 nm to monitor the effluents. The identities of the photoproducts were confirmed by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) measurements (27).
The lesion-containing substrate d(GTAG[8-5]CAT) was obtained from the Pyrex-filtered UV irradiation of d(GTAGBrCAT) (“BrC” designates a 5-bromo-2′-deoxycytidine) under the similar conditions as described previously (15,28). The irradiation mixture was separated by HPLC as mentioned above, and a 40-min gradient of 0-40% methanol in 50 mM phosphate buffer was used. The dodecameric lesion-bearing substrate d(ATGGCG[8-5]CGCTAT) was obtained from the Pyrex-filtered UV light irradiation of a 5-bromocytosine-containing duplex DNA (15,28).
To prepare ODNs with G[8-5m]mC and G[8-5m]T, we first synthesized ODNs containing a 5-phenythiomethyl-2′-deoxycytidine or 5-phenythiomethyl-2′-deoxyuridine (11,13). After irradiation with 254-nm light under anaerobic conditions, the mixtures were separated by HPLC. The ODN d(ATGGCG[8-5m]TGCTAT) was purified first with the TEAA buffer system and then by the phosphate buffer with the gradient programs described above. The ODN d(ATGGCG[8-5m]mCGCTAT) was purified with the TEAA buffer system.
The above lesion-bearing heptameric and dodecameric substrates were used to construct 49- and 54-mer substrates by enzymatic ligation. In this regard, the above lesion-carrying ODNs were ligated with a 17-mer ODN, d(AGC TAC CAT GCC TGC AC) (the 5′ piece), and a 5′-phosphorylated 25-mer ODN, d(GCA ATT CGT AAT CAT GGT CAT AGC T) (the 3′ piece), in the presence of a template (Schemes S1&S2). The resulting 49-mer or 54-mer lesion-bearing ODNs (sequences shown in Table 1) were separated from the ligation reaction mixtures by denaturing-PAGE and desalted by ethanol precipitation. The purities of the ligation products were confirmed by denaturing PAGE analysis, and the yields for the ligation reactions were shown in Schemes S1&S2.
Table 1.
TT photoproduct- and oxidative intrastrand cross-link lesion-containing ODNs used for UvrA binding and UvrABC incision assays. The lesion-bearing ODNs used for ligation are shown in bold
| Name | Sequence |
|---|---|
| T[c,s]T-49bp | 5′-AGC TAC CAT GCC TGC ACG TAT[c,s]TAT GCA ATT CGT AAT CAT GGT CAT AGC T-3′ |
| T[6-4]T-49bp | 5′-AGC TAC CAT GCC TGC ACG TAT[6-4]TAT GCA ATT CGT AAT CAT GGT CAT AGC T-3′ |
| G[8-5]C-49bp | 5′-AGC TAC CAT GCC TGC ACG TAG[8-5]CAT GCA ATT CGT AAT CAT GGT CAT AGC T-3′ |
| G[8-5]C-54bp | 5′-AGC TAC CAT GCC TGC ACA TGG CG[8-5]C GCT ATG CAA TTC GTA ATC ATG GTC ATA GCT-3′ |
| G[8-5m]mC-54bp | 5′-AGC TAC CAT GCC TGC ACA TGG CG[8-5m]mC GCT ATG CAA TTC GTA ATC ATG GTC ATA GCT-3′ |
| G[8-5m]T-54bp | 5′-AGC TAC CAT GCC TGC ACA TGG CG[8-5m]T GCT ATG CAA TTC GTA ATC ATG GTC ATA GCT-3′ |
Electrophoretic Mobility Shift Assays (EMSA)
Binding affinities of UvrA to the ODNs were determined by EMSA. The 49-mer or 54-mer lesion-bearing substrates were labeled at the 5′-end with [γ-32P]-ATP and annealed with the corresponding complementary strand. The duplex substrate (1 nM) was then incubated with varying concentrations of UvrA in a 20-μL UvrABC buffer, which contained 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 5 mM DTT and 1 mM ATP, at 37°C for 15 min. A solution of 80% glycerol (v/v, at 37°C) was added, and the products were resolved on a 3.5% (1:29) native polyacrylamide gel, which was run at 80 V at room temperature (26). Gel band intensities of duplex DNA and DNA-protein complex were quantified by using a Typhoon 9410 Variable Mode Imager (Amersham Biosciences Co.) and ImageQuant 5.2 software (Amersham Biosciences Co.).
From the EMSA results, the fractions of protein-bound duplex were quantified from the radioactivity in each band (29). The dissociation constants (Kd) were obtained by non-linear curve fitting using Origin 6.0 (Microcal Software, Northampton, MA) with the following equation:
where [E]Total is the total concentration of UvrA.
Incision Assays
To determine the incision efficiency of UvrABC on different lesion-bearing substrates, we incubated the duplex ODNs (2 nM) with UvrABC (UvrA, 15 nM; UvrB, 250 nM; UvrC, 100 nM) in the UvrABC buffer at 37 °C for the indicated periods of time. The reactions were terminated by adding 100 mM EDTA (2 μL) and formamide gel-loading buffer (80% formamide, 1 mg/mL xylene cyanol, and 1 mg/mL bromophenol blue, 12 μL). The mixture was loaded onto a 12%, 1:29 cross-linked denaturing polyacrylamide gel containing 8 M urea. The percentage of incision was determined from the radioactivities of gel bands corresponding to incision product and uncleaved DNA. The incision rates were derived from the slope of the plot where the amounts of incision products were monitored as a function of reaction time. The error limits for incision rate were derived from the fitted parameters as described previously (30).
Measurement of Melting Curves and Data Processing
The above dodecameric ODNs harboring a G[8-5m]mC or G[8-5m]T were annealed with another 12-mer ODN at 1:1 ratio to form duplexes, which were employed for melting temperature measurements. The specific nucleotide sequences were:
| Strand 1 | 5′- ATGGCXYGCTAT-3′ |
| Strand 2 | 3′- TACCGNMCGATA-5′ |
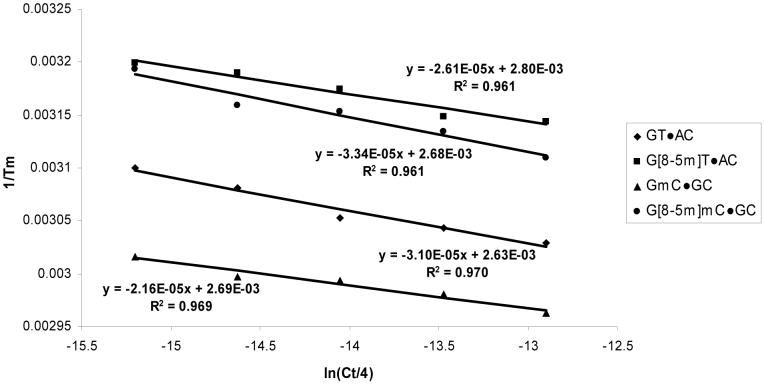
where XY was GmC, GT, G[8-5m]mC, or G[8-5m]T, and MN was GC or AC (Table 2). UV absorbance-versus-temperature profiles were recorded on a Cary 500 spectrophotometer (Varian Inc., Palo Alto, CA), and the ODNs were dispersed in a 1.2-mL solution containing 250 mM NaCl, 10 mM sodium cacodylate, and 0.1 mM EDTA (pH 7.0) at a total ODN concentration (Ct) of 1.0, 1.8, 3.2, 5.6, or 10 μM. The absorbance was recorded in the reverse and forward directions for a temperature range of 80°C to 10°C at a rate of 1°C/min, and the melting temperature (Tm) value was obtained by the derivative method. The thermodynamic parameters were obtained from the van’t Hoff plot (31), where the reciprocal of Tm was plotted against :
and
in which R is the ideal gas constant (= 1.987 cal·mol-1·K-1). The error limits for ΔG, ΔH and ΔS derived from fitted parameters were calculated by using previously described equations (32).
Table 2.
Thermodynamic parameters of duplex formation in a 250-mM NaCl solution
| Duplex | ΔH (kcal/mol) | ΔS (cal/mol K) | ΔG37°C (kcal/mol) | ΔΔG37°Ca (kcal/mol) |
|---|---|---|---|---|
| 5′-ATGGCGmCGCTAT-3′ 3′-TACCGC GCGATA-5′ |
-91.8 ± 4.7 | -247 ± 13 | -15.4 ± 0.3 | |
| 5′-ATGGCG[8-5m]mCGCTAT-3′ 3′-TACCGC GCGATA-5′ |
-59.6 ± 3.5 | -160 ± 10 | -9.9 ± 0.1 | 5.4 ± 0.3 |
| 5′-ATGGCGTGCTAT-3′ 3′-TACCGCACGATA-5′ |
-82.0 ± 2.0 | -221 ± 5 | -13.5 ± 0.1 | |
| 5′-ATGGCG[8-5m]TGCTAT-3′ 3′-TACCGC ACGATA-5′ |
-76.1 ± 4.6 | -213 ± 13 | -9.9 ± 0.1 | 3.6 ± 0.1 |
ΔΔG37°C = ΔG37°C (lesion-containing duplex) - ΔG37°C (undamaged duplex).
Results
Preparation of ODNs Harboring an Intrastrand Crosslink Lesion
After the UVC irradiation of d(GTATTAT), four major photoproduct-containing ODNs, d(GTAT[c,s]TAT), d(GTAT[6-4]TAT), d(GT[ ]ATTAT) and d(GTATT[ ]AT), can be clearly separated from each other as well as from the starting material by HPLC (Figure S2). The structure characterizations of these products have been reported previously (27) .
We recently found that the Pyrex-filtered UV light irradiation of a dodecameric duplex d(ATGGCGBrCGCTAT)/d(ATAGCGCGCCAT) can induce the formation of an intrastrand cross-link lesion where the C8 of guanine and the C5 of its adjacent 3′ cytosine are covalently bonded (15,28). Similarly, the Pyrex-filtered UV light irradiation of the single-stranded d(GTAGBrCAT) can result in the generation of the same lesion where the guanine at the fourth position is covalently bonded to the cytosine at the fifth position. The structure of this lesion was further confirmed by ESI-MS and MS/MS analyses of the lesion-bearing substrate [Figure S3, nomenclature for fragment ions follows that of McLuckey et al. (33)].
We prepared the G[8-5m]mC- and G[8-5m]T-containing ODN substrates according to previously published procedures (8,13). In this respect, we synthesized d(ATGGCGYGCTAT) (“Y” represents a 5-phenylthiomethyl-2′-deoxycytidine or 5-phenylthiomethyl-2′-deoxyuridine) by using phosphoramidite chemistry (13). The lesion-containing dodecamers d(ATGGCG[8-5m]mCGCTAT) and d(ATGGCG[8-5m]TGCTAT) were purified from the UVC irradiation mixtures of the mercaptonucleoside-bearing ODNs by HPLC. The molecular masses of the above two ODNs were measured by ESI-MS to be 3672.0 and 3673.2 Da, respectively, which are in accordance with the corresponding calculated average masses of 3672.6 and 3673.6 Da. Moreover, the site of the cross-link was confirmed by the product-ion spectra (MS/MS) of the [M - 3H]3- ions of these ODNs (spectra shown in Figures S4&S5).
With the above lesion-carrying ODNs, we constructed lesion-bearing 49- and 54-mer substrates by enzymatic ligation (see Materials and Methods).
Binding Affinities of UvrA to the Lesion-bearing ODNs
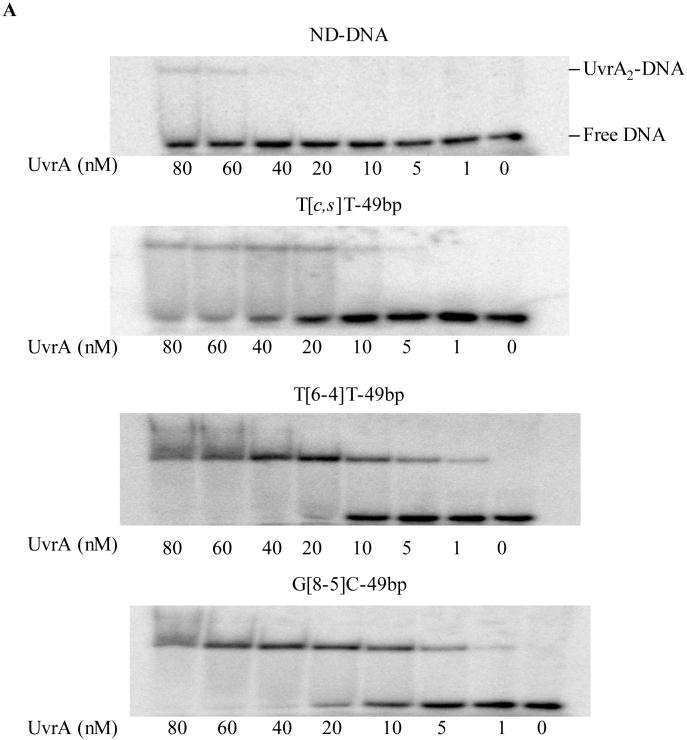
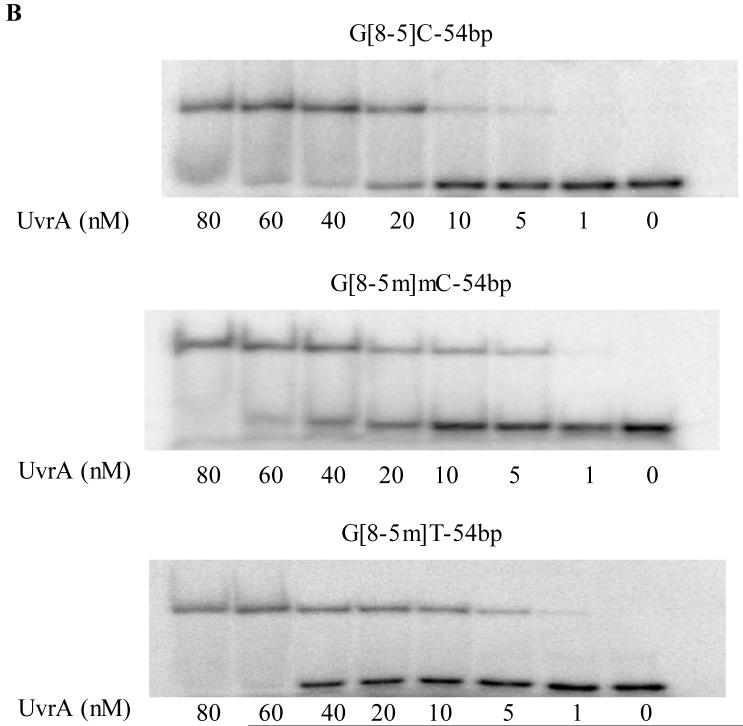
To improve our understanding on the recognition of these lesions by the E. coli NER enzymes, we next employed the electrophoretic mobility shift assays (EMSAs) and examined the binding affinities of these 49- and 54-mer substrates to UvrA. As depicted in Figure 2A, UvrA assumes a much stronger binding to the lesion-containing substrates than the control undamaged duplex (Figure 2). We next plotted the percentage of bound fraction against the concentration of UvrA and determined Kd by non-linear regression analysis (Figures 3&S6, Table 3). Consistent with previous findings (22), T[c,s]T was the poorest substrate for UvrA with the largest dissociation constant (91 nM), whereas the substrate exhibiting the strongest binding was T[6-4]T (Kd ∼ 11 nM). The Kd values for the two G[8-5]C-bearing substrates with different flanking bases, on the other hand, were 23 nM for AG[8-5]CA, and 29 nM for CG[8-5]CG. Thus, the sequences flanking the lesion had little effect on the binding of G[8-5]C-containing substrates to UvrA.
Figure 2.
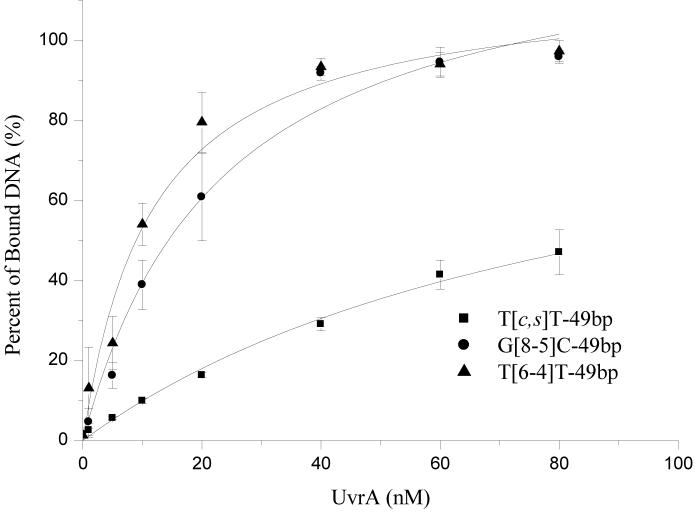
Binding of UvrA to (A) undamaged DNA, T[c,s]T-49bp, T[6-4]T-49bp and G[8-5]C-49bp, and (B) G[8-5]C-54bp, G[8-5m]mC-54bp and G[8-5m]T-54bp. ODN substrates (1 nM) were incubated with UvrA at the indicated concentrations in the UvrABC buffer at 37°C for 15 min.
Figure 3.
Non-linear regression analysis of the plot of average percentage of binding (determined from EMSA with three or more sets of titrations per duplex) versus the concentrations of UvrA for 49-mer ODNs containing T[c,s]T (■), T[6-4]T (▲) and G[8-5]C (●).
Table 3.
Equilibrium dissociation constants (Kd) for UvrA binding, initial incision rates of UvrABC, and free energy changes for duplex formation
Our results also revealed that the G[8-5m]mC-containing ODN had a lower binding affinity toward UvrA (Kd = 49 nM) than the two G[8-5]C-containing ODNs. On the other hand, the structurally-related G[8-5m]T-bearing 54-mer ODNs had a dissociation constant of 60 nM, which is the second poorest substrate for UvrA binding among all the lesion-carrying substrates that we investigated.
UvrABC Incisions of the Lesion-containing ODNs
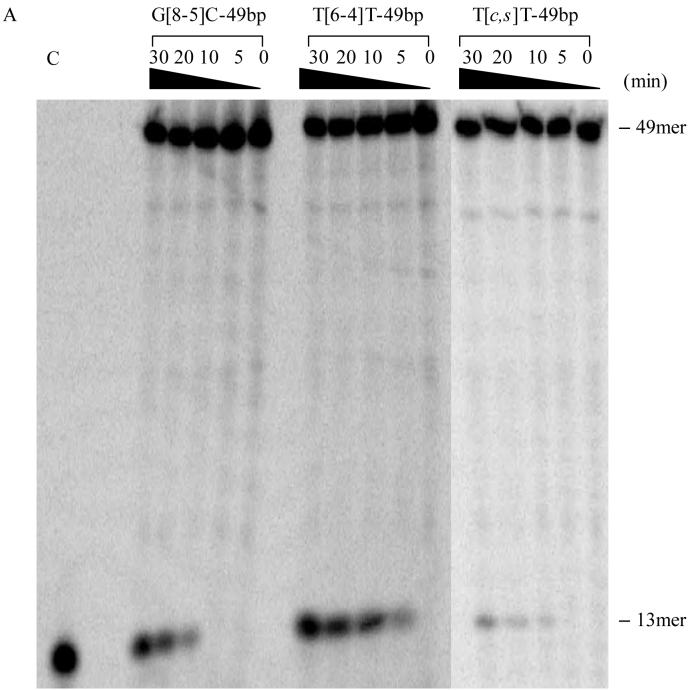
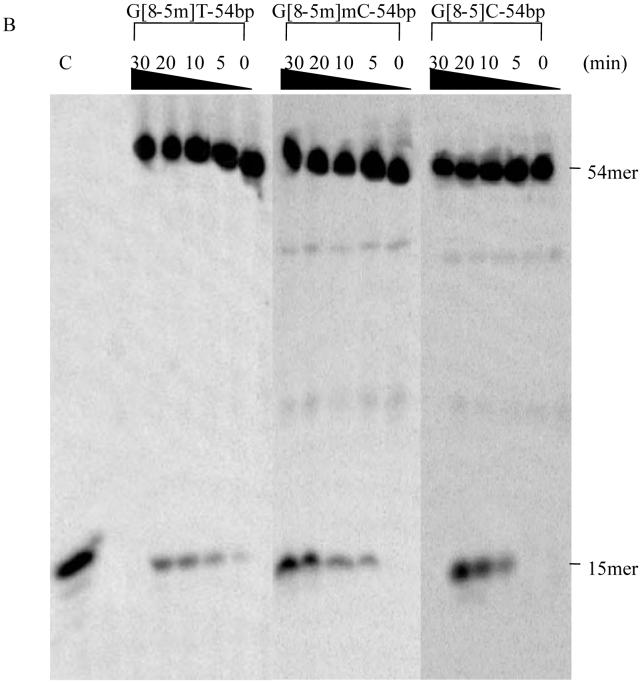
According to the currently accepted mechanism of UvrABC system, the first cleavage occurs at the 4th or 5th nucleoside 3′ to the damage, followed by incision at the 8th nucleoside 5′ to the lesion (34). We observed 13-mer and 15-mer incision products for 49- and 54-mer lesion-containing substrates, respectively (Figures 4A & 4B). Because all substrates were labeled at the 5′-end, the lengths of the labeled incision products were in keeping with the cleavage mechanism of UvrABC.
Figure 4.
UvrABC incision assays of ODNs containing (A) T[c,s]T-49bp, T[6-4]T-49bp and G[8-5]C-49bp; (B) G[8-5]C-54bp, G[8-5m]mC-54bp and G[8-5m]T-54bp. UvrABC was incubated with 2 nM substrates in the UvrABC buffer at 37°C for the indicated periods of time. “C” indicates control where the known length ODN is loaded. Lanes marked with a “C” correspond to authentic ODNs with the same sequences as the expected cleavage products: d(AGC TAC CAT GCC T) in panel A and d(AGC TAC CAT GCC TGC) in panel B. The bands observed between the full-length product and 13- or 15-mer cleavage product are due to contamination from short ODNs employed for the ligation reaction (integration shows that they constitute a few percent of the full-length product).
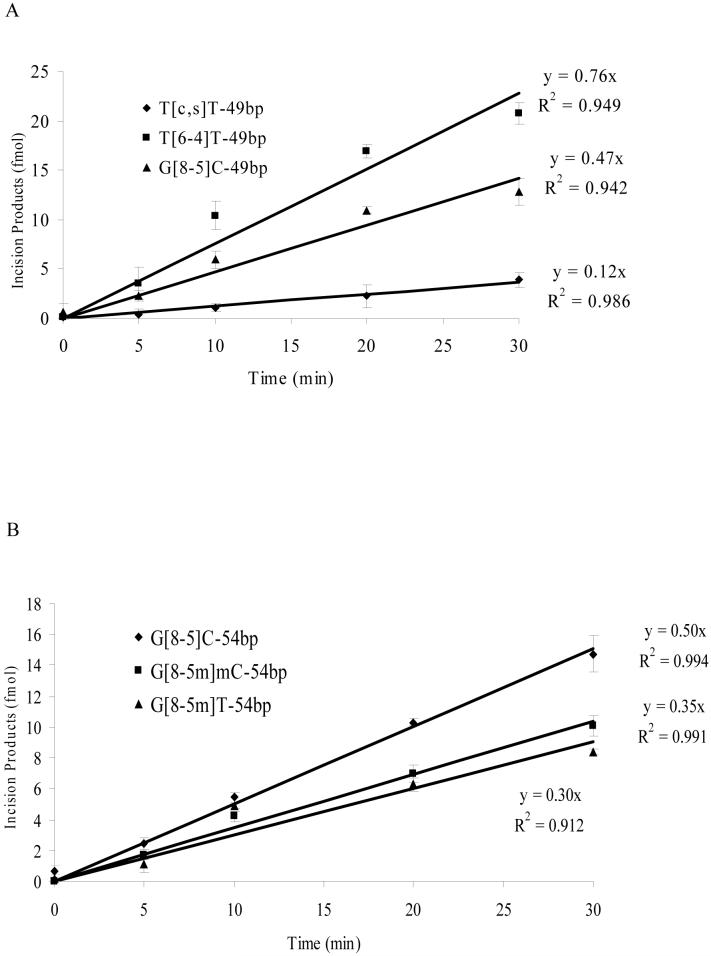
The initial incision rates of different substrates can provide insights into the repair efficiencies of UvrABC toward different substrates. To this end, we extracted the initial incision rates from the slope of the line where the amounts of incision products were plotted as a function of the reaction time (Figure 5). It turned out that the incision rates, for the most part, are correlated with UvrA binding affinity (Table 3). In line with its highest binding affinity toward UvrA, T[6-4]T-49bp was incised at the highest efficiency by UvrABC. Along this line, T[c,s]T-49bp exhibited the poorest binding to UvrA, and the initial incision rate was also the lowest, i.e., 0.12 fmol/min. G[8-5]C-containing ODNs had higher incision rates (0.50 fmol/min) than the corresponding ODNs harboring a G[8-5m]mC (0.35 fmol/min) or G[8-5m]T (0.30 fmol/min). In addition, the initial incision rates for AG[8-5]CA-49bp and CG[8-5]CG-54bp were comparable (i.e., 0.47 fmol/min and 0.50 fmol/min, respectively), which is consistent with what we found for the binding affinities of these two substrates to UvrA. The sequence context, therefore, had little effect on the incision rate of the G[8-5]C-bearing substrates.
Figure 5.
Kinetics of UvrABC incisions of DNA substrates. (A) 49-mer ODNs containing T[c,s]T (♦), T[6-4]T (■) and G[8-5]C (▲); (B) 54-mer ODNs containing G[8-5]C (♦), G[8-5m]mC (■) and G[8-5m]T (▲).
Thermodynamic Properties of Lesion-carrying DNA Duplexes
To understand the biochemical basis of the different binding affinities of UvrA and the different initial incision rates by UvrABC for the five lesions, we next examined the thermodynamic parameters for the G[8-5m]T- and G[8-5m]mC-containing duplexes by melting temperature measurements (Figure 6, details shown in Materials and Methods). In this respect, we used the dodecameric ODNs in the same sequence context as the repair substrates.
Figure 6.
Plots of 1/Tm versus ln(Ct/4) for the dodecameric ODN duplexes. The duplex sequence is d(ATGGCXYGCTAT)·d(ATAGCMNGCCAT), where XY·MN represents GmC·GC (▲),G[8-5m]mC·GC (●), GT·AC (♦), or G[8-5m]T·AC (■).
It turned out that the presence of G[8-5m]mC destabilized the duplex by 5.4 kcal/mol in free energy at 37°C, whereas the replacement of GT with G[8-5m]T caused a decrease in thermal stability of the duplex by 3.6 kcal/mol (Table 2). Our previous study showed that, in the same sequence context, the presence of G[8-5]C can destabilize the duplex by 4.0 kcal/mol (25).
Discussion
Recently, several oxidative intrastrand cross-link lesions of DNA have been identified (3-15). The three lesions employed in this study, that is, G[8-5]C, G[8-5m]mC and G[8-5m]T, could all be induced in duplex DNA upon exposure to γ-ray irradiation (8,13,15). In addition, G[8-5m]T could also form in calf thymus DNA upon treatment with Fenton reagents (14). In this context, it is worth noting that G[8-5]C, but not the corresponding C[5-8]G, can be induced in duplex DNA by γ irradiation (15). Likewise, intrastrand cross-link lesions formed between G and T or mC are produced in much higher yield at GT or GmC site than the corresponding lesions formed at TG or mCG site (8,13). Obtaining lesion-bearing ODN substrates constitutes a crucial step toward examining the biological implications of these lesions at the molecular level. As we reported previously, a photochemical approach enables us to obtain well-characterized intrastrand cross-link lesion-containing ODNs (13,28) .
To put our studies into the context of previous knowledge about the incision of the two extensively studied UV irradiation-induced thymine dimers, that is, T[c,s]T and T[6-4]T, by UvrABC enzymes, we also prepared ODN substrates containing these two lesions in the same sequence context as that for the G[8-5]C-bearing ODN. Our results with the ODN substrates carrying five different lesions showed that the initial incision rates of UvrABC correlated well with the relative binding affinities of UvrA to the lesion-carrying duplex ODNs. Both the efficiency of UvrABC incision and the affinity of UvrA binding follow the same order, that is, T[6-4]T > G[8-5]C > G[8-5m]mC > G[8-5m]T > T[c,s]T. This result suggests that these intrastrand cross-link lesions are discriminated primarily in the initial recognition step with UvrA, but recognized equally well in the second step by UvrB (20).
The thermal stability of DNA double helix parallels the repair efficiency as long as repair enzymes require a double-stranded substrate (35). Thus, the thermal stability measurement may provide some insights into the UvrA recognition mechanism and the following incision reaction by nucleases. Our results showed that this is indeed the case for most substrates examined in this study. T[c,s]T causes the least destabilization to DNA helix, which destabilizes duplex DNA by 1.5 kcal/mol of free energy at 37°C, and has the lowest binding affinity and incision rate among all the five substrates (Table 3). T[6-4]T, on the other hand, induces the greatest destabilization to duplex DNA, which is consistent with the observations that it has the strongest binding affinity to UvrA and can be incised at the highest efficiency (Table 3). These results are also in keeping with previous observations with T[6-4]T (22,26). When compared with these TT photoproducts, the three oxidative intrastrand cross-link lesions destabilize the duplex more than T[c,s]T, but less than T[6-4]T, which is in line with the fact that the binding affinities and incision rates of oxidative lesions fall between the two dithymine photoproducts. In this context, it is worth noting that different dodecameric sequences were employed for determining the thermodynamic parameters for the dithymine photoproducts and intrastrand crosslink lesions (21,25); different flanking sequences may also contribute partially to the difference in destabilization of DNA duplexes.
The only exception in this group is the G[8-5m]mC-bearing ODN. The presence of the lesion destabilizes the duplex by approximately 5.4 kcal/mol, which is more than that caused by G[8-5]C (4.0 kcal/mol) or G[8-5m]T (3.6 kcal/mol, Table 3). The binding affinity of UvrA and the incision rate for UvrABC towards G[8-5m]mC, however, were similar as those for G[8-5m]T, but lower than those for G[8-5]C. The results suggest that other factors may also affect the incision efficiency. The less efficient cleavage by the E. coli UvrABC nucleases toward G[8-5m]mC-bearing substrate prompts us to speculate that this lesion may accumulate more in cells than G[8-5]C. In addition, the AG[8-5]CA-49bp and CG[8-5]CG-54bp, which had different flanking sequences, showed a slight difference in UvrA binding and little difference in incision by UvrABC. The dissociation constant for the complex formed between UvrA and CG[8-5]CG-54bp is 29 nM, which is slightly higher than the corresponding complex for AG[8-5]CA-49bp (23 nM). This result is consistent with the observations of Zou et al. (36), which showed that the flanking bases of A/T cause more destabilization than the flanking bases of C/G, and hence can render the lesion to be recognized more readily by UvrA. However, since the difference of UvrA binding between the two sequences is relatively small, there could be little effect on the incision efficiency at the end.
It is worth noting that free radical can also induce the formation of 8,5′-cyclo-2′-deoxyadenosine (37), which is structurely related to oxidative intrastrand crosslink lesions discussed in this paper. Previous studies illustrated that this lesion can also be repaired by NER enzymes (38,39).
Deficiency in NER results in some severe genetic diseases, such as XP, TTD, and Cockayne’s syndrome (18,19,40). Surprisingly, XP patients often have neurological abnormalities due to premature neuronal death (41). These neuron losses occur without exposure to UV light, suggesting that these cells are deficient in repairing DNA damage formed from other mutagens (e.g., ROS). Given that neurons consume a large amount of oxygen, oxidative intrastrand cross-link lesions may also accumulate in neuron cells, which may confer neurotoxic effects in XP patients. The repair study of these oxidative intrastrand cross-link lesions by NER enzymes may set a stage for understanding further the pathophysiology of XP patients.
Supplementary Material
Figure 1.
Structures of DNA photoproducts and oxidative intrastrand cross-link lesions examined in this study.
Abbreviations
- NER
nucleotide excision repair
- XP
xeroderma pigmentosum
- TTD
Trichothiodystrophy
- T[c,s]T
cis,syn-cyclobutane pyrimidine dimer at TT site
- T[6-4]T
pyrimidine(6-4)pyrimidone product at TT site
- BrC
5-bromo-2′-deoxycytidine
- ODN
oligodeoxyribonucleotide
- T7-
exonuclease-deficient T7 DNA polymerase
- HIV-RT
human immunodeficiency virus reverse transcriptase
- PAGE
polyacrylamide gel electrophoresis
- EMSA
electrophoretic mobility shift assay
- Tm
melting temperature
- ESI
electrospray ionization
- (MS/MS)
tandem mass spectrometry
Footnotes
This work was supported by the National Institutes of Health (CA96906 and CA101864 to Y. W. and CA86927 to Y. Z.) and the University of California Toxic Substances Research & Teaching Program (to C. G.).
References
- 1.Lindahl T. DNA lesions generated in vivo by reactive oxygen species, their accumulation and repair. NATO ASI Ser., Ser. A. 1999;302:251–257. [Google Scholar]
- 2.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 3.Box HC, Budzinski EE, Dawidzik JD, Wallace JC, Evans MS, Gobey JS. Radiation-induced formation of a crosslink between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA) Radiat. Res. 1996;145:641–643. [PubMed] [Google Scholar]
- 4.Box HC, Budzinski EE, Dawidzik JB, Wallace JC, Iijima H. Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res. 1998;149:433–439. [PubMed] [Google Scholar]
- 5.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radic. Biol. Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 6.Budzinski EE, Dawidzik JB, Rajecki MJ, Wallace JC, Schroder EA, Box HC. Isolation and characterization of the products of anoxic irradiation of d(CpGpTpA) Int. J. Radiat. Biol. 1997;71:327–336. doi: 10.1080/095530097144210. [DOI] [PubMed] [Google Scholar]
- 7.Romieu A, Bellon S, Gasparutto D, Cadet J. Synthesis and UV photolysis of oligodeoxynucleotides that contain 5- (phenylthiomethyl)-2′-deoxyuridine: a specific photolabile precursor of 5-(2′-deoxyuridilyl)methyl radical. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 8.Bellon S, Ravanat JL, Gasparutto D, Cadet J. Cross-linked thymine-purine base tandem lesions: synthesis, characterization, and measurement in gamma-irradiated isolated DNA. Chem. Res. Toxicol. 2002;15:598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Gao Y, Wang Y. Identification and characterization of a novel crosslink lesion in d(CpC) upon 365 nm-irradiation in the presence of 2-methyl-1,4-naphthoquinone. Nucleic Acids Res. 2003;31:5413–5424. doi: 10.1093/nar/gkg736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Gao Y, Zeng Y, Fang F, Chi D, Wang Y. Isolation and characterization of a novel cross-link lesion in d(CpC) induced by one-electron photooxidation. Photochem. Photobiol. 2004;80:209–215. doi: 10.1562/2003-12-15-RA-029. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Wang Y. Independent generation of 5-(2′-deoxycytidinyl)methyl radical and the formation of a novel crosslink lesion between 5-methylcytosine and guanine. J. Am. Chem. Soc. 2003;125:12795–12802. doi: 10.1021/ja034866r. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Wang Y. Independent generation of the 5-hydroxy-5,6-dihydrothymidin-6-yl radical and its reactivity in dinucleoside monophosphates. J. Am. Chem. Soc. 2004;126:13287–13297. doi: 10.1021/ja048492t. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Wang Y. Generation of 5-(2′-deoxycytidyl)methyl radical and the formation of intrastrand cross-link lesions in oligodeoxyribonucleotides. Nucleic Acids Res. 2005;33:1593–1603. doi: 10.1093/nar/gki301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong H, Cao H, Wang Y, Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu C, Wang Y. LC-MS/MS identification and yeast polymerase eta bypass of a novel gamma-irradiation-induced intrastrand cross-link lesion G[8-5]C. Biochemistry. 2004;43:6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- 16.Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 17.Lee DH, O’Connor TR, Pfeifer GP. Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG-->TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. Nucleic Acids Res. 2002;30:3566–3573. doi: 10.1093/nar/gkf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 19.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 20.Zou Y, Luo C, Geacintov NE. Hierarchy of DNA damage recognition in Escherichia coli nucleotide excision repair. Biochemistry. 2001;40:2923–2931. doi: 10.1021/bi001504c. [DOI] [PubMed] [Google Scholar]
- 21.Jing Y, Kao JF, Taylor JS. Thermodynamic and base-pairing studies of matched and mismatched DNA dodecamer duplexes containing cis-syn, (6-4) and Dewar photoproducts of TT. Nucleic Acids Res. 1998;26:3845–3853. doi: 10.1093/nar/26.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svoboda DL, Smith CA, Taylor JS, Sancar A. Effect of sequence, adduct type, and opposing lesions on the binding and repair of ultraviolet photodamage by DNA photolyase and (A)BC excinuclease. J. Biol. Chem. 1993;268:10694–10700. [PubMed] [Google Scholar]
- 23.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 24.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu C, Wang Y. Thermodynamic and in-vitro replication studies of an intrastrand crosslink lesion G[8-5]C. Biochemistry. 2005;44:8883–8889. doi: 10.1021/bi050036+. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Colis LC, Basu AK, Zou Y. Recognition and incision of gamma-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chem Res Toxicol. 2005;18:1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Taylor JS, Gross ML. Differentiation of isomeric photomodified oligodeoxynucleotides by fragmentation of ions produced by matrix-assisted laser desorption ionization and electrospray ionization. J. Am. Soc. Mass Spectrom. 1999;10:329–338. doi: 10.1016/S1044-0305(98)00158-5. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, Wang Y. Facile formation of an intrastrand cross-link lesion between cytosine and guanine upon pyrex-filtered UV light irradiation of d(BrCG) and duplex DNA containing 5-bromocytosine. J. Am. Chem. Soc. 2004;126:6552–6553. doi: 10.1021/ja049760q. [DOI] [PubMed] [Google Scholar]
- 29.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer SL. Data analysis for scientists and engineers. Wiley; New York: 1975. [Google Scholar]
- 31.Breslauer KJ. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Methods Enzymol. 1995;259:221–242. doi: 10.1016/0076-6879(95)59046-3. [DOI] [PubMed] [Google Scholar]
- 32.Persmark M, Guengerich FP. Spectroscopic and thermodynamic characterization of the interaction of N7-guanyl thioether derivatives of d(TGCTG*CAAG) with potential complements. Biochemistry. 1994;33:8662–8672. doi: 10.1021/bi00195a006. [DOI] [PubMed] [Google Scholar]
- 33.McLuckey SA, Habibi-Goudarzi S. Ion trap tandem mass spectrometry applied to small multiply charged oligodeoxyribonucleotides with a modified base. J. Am. Soc. Mass Spectrom. 1994;5:740–747. doi: 10.1016/1044-0305(94)80006-5. [DOI] [PubMed] [Google Scholar]
- 34.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(-)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 35.Singer B, Hang B. Nucleic acid sequence and repair: role of adduct, neighbor bases and enzyme specificity. Carcinogenesis. 2000;21:1071–1078. [PubMed] [Google Scholar]
- 36.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dizdaroglu M. Free-radical-induced formation of an 8,5′-cyclo-2′-deoxyguanosine moiety in deoxyribonucleic acid. Biochem. J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 40.Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 41.Cleaver JE, Kraemer KH. In: The Metabolic Basis of Inherited Disease. Scriver C, Beaudet AL, Sly WS, Vale D, editors. Vol. 2. McGraw-Hill; New York: 1989. pp. 2949–2971. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.