Abstract
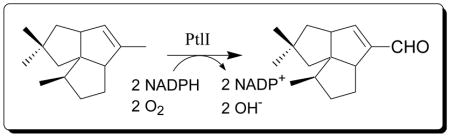
A gene cluster encoding all of the enzymes for the biosynthesis of the antibiotic pentalenolactone (1) has recently been identified inStreptomyces avermitilis. The biosynthetic gene cluster contains theptlI (SAV2999) gene which encodes a cytochrome P450 (CYP183A1). PtlI was cloned by PCR and expressed inEscherichia coli as a C-terminal-His6-tag protein. Recombinant PtlI bound pentalenene (3) with high affinity (KD 1.44 ± 0.14 μM). Incubation of recombinant PtlI with (±)-3 in the presence of NADPH,E. coliflavodoxin and flavodoxin reductase, and O2 resulted in conversion to a single enantiomer of pentalen-13-al (7), by stepwise allylic oxidation via pentalen-13-ol (6). The steady state kinetic parameters for the oxidation of pentalenene (3) to pentalen-13-ol (6) werekcat 0.503 ± 0.006 min−1 andKm3.33 ± 0.62 μM for3.
Streptomyces are a rich source of bioactive secondary metabolites. The 9.03 Mb linear chromosome of S. avermitilis, the producer of the widely used antiparasitic avermectins, harbors 7,575 open reading frames (ORFs)1 of which 33 encode cytochrome P450 enzymes.2 One of these CYP genes, ptlI (SAV2999, CYP183A1) is found within the gene cluster for the biosynthesis of the sesquiterpene antibiotic pentalenolactone (1). This cluster lies in a 13.4-kb segment, centered at 3.75 Mb in the S. avermitilis genome, that contains 13 unidirectionally-transcribed ORFs (Figure 1).3 Among these ORFs, the 1011-bp ptlA, encodes pentalenene synthase (PtlA), which catalyzes the cyclization of farnesyl diphosphate (FPP) (2) to pentalenene (3), the established parent hydrocarbon of the pentalenolactone family of antibiotics (Scheme 1).3,4 Besides the heme-dependent monooxygenase CYP183A1 (ptlI),2 seven of the remaining ORFs correspond to redox enzymes, including the non-heme iron dioxygenase encoded by ptlH,5 and six additional monooxygenases and dioxygenases.
Figure 1.
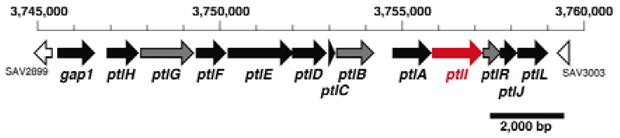
Pentalenolactone biosynthetic gene cluster from S. avermitilis. (See http://avermitilis.ls.kitasato-u.ac.jp/.)
Scheme 1.
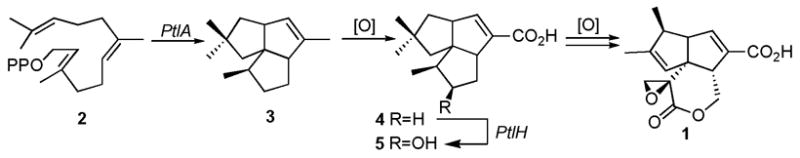
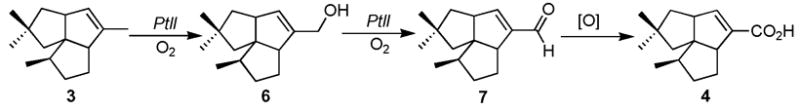
We recently showed that PtlH, an Fe2+/α-ketoglutarate-dependent hydroxylase, catalyzes the conversion of 1-deoxypentalenic acid (4) to a new biosynthetic intermediate, 11β-hydroxy-1-deoxypentalenic acid (5).5 Although several presumptive intermediates of pentalenolactone biosynthesis have been isolated from a wide variety of Streptomyces species,3,4 PtlH is the only enzyme linking pentalenene (3) to pentalenolactone (1) that has been characterized to date. Here we describe the biochemical characterization of PtlI, the cytochrome P450 that is shown to catalyze the conversion of pentalenene (3) to pentalen-13-al (7) by stepwise oxidation via pentalen-13-ol (6).
According to the proposed biosynthetic pathway (Scheme 1), the enzymes responsible for the conversion of pentalenene (3) to pentalenolactone (1) must first oxidize pentalenene to the corresponding unsaturated carboxylic acid 4. Cytochrome P450s are known to catalyze numerous oxygenation reactions of nonactivated hydrocarbons.6 Amongst these reactions is the three-step oxidation of a methyl group to a carboxylic acid.7–9 We therefore speculated that PtlI might be responsible for all or part of the allylic oxidation of pentalenene (3) to 1-deoxypentalenic acid (4) (Scheme 2).
Scheme 2.
PtlI was amplified by polymerase chain reaction (PCR) from DNA of S. avermitilis cosmid CL_216_D07 and cloned between the NdeI and XhoI sites of the vector pET31b. The resulting construct pET31b-PtlI was transformed into Escherichia coli BL21(DE3). After induction with IPTG, the expressed PtlI protein, carrying a C-terminal His6-tag, was purified to homogeneity by Ni-NTA chromatography.10 MALDI-TOF MS of purified protein showed subunit MD m/z 51667±50 (calc. 51723 for apo-protein) and m/z 52078±50 (calc. 52339 for holo-protein). Treatment of the sodium dithionite-reduced protein with carbon monoxide gave the characteristic P450 UV difference spectrum.11
Titration of PtlI with pentalenene (3)12 resulted in the typical blue-shift from 420 nm to 390 nm (type I binding).13 The dissociation constant for 3 was determined by non-linear fitting of the UV-difference spectra to give KD=1.44±0.14 μM. By contrast, the control sesquiterpene (−)-trans-caryophyllene showed no type I binding when added to PtlI.
A mixture of recombinant PtlI (0.57 μM), E. coli flavodoxin (Fld, 3.9 μM),14,15a E. coli flavodoxin reductase (Fdr, 6.3 μM),14,15b NADPH (0.45 mM), and a NADPH-regeneration system [glucose-6-phosphate (3.1 mM) and glucose-6-phosphate dehydrogenase (10 u)] in 3.0 mL of 50 mM phosphate buffer, 10% glycerol (v/v), pH 7.4, was incubated with (±)-(3) (1.1 mM) plus 0.8% DMSO for 16 h at room temperature. GC-MS analysis of the pentane extract revealed exclusively two new peaks with m/z 218 (retention time 10.96 min) and 220 (retention time 11.03 min), identical to authentic pentalen-13-al (7) and pentalen-13-ol (6), respectively (Figure 2 and Supporting Information).12 The 1H NMR spectrum of the crude neutral extract also showed the characteristic aldehydic and olefinic signals at δ 9.71 and 6.704 (d, J=0.8 Hz), respectively for 7 (Figure S8). Chiral GC-MS analysis, under conditions in which individual enantiomers of (±)-pentalen-13-ol (6) and (±)-pentalen-13-al (7) were well resolved, confirmed that enzymatically-produced 6 and 7 were each single enantiomers. Preparative-scale incubation with (±)-pentalenene (3) gave a mixture containing 6 and 7, which was dissolved in methanol and treated with sodium borohydride to give alcohol 6, identical by 1H NMR to chemically synthesized pentalen-13-ol (6). Incubation using alcohol 6 as substrate confirmed that PtlI catalyzes the oxidation of 6 to aldehyde 7.16 By contrast, only trace amounts of 1-deoxypentalenic acid 4 could be detected under a wide variety of incubation conditions.
Figure 2.
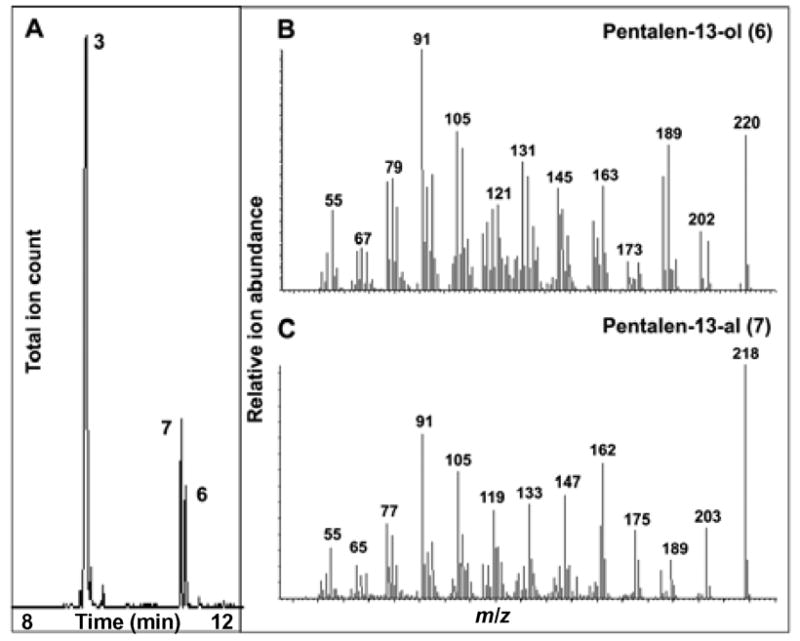
GC-MS analysis of incubation of PtlI with (±)-pentalenene (3). A, GC trace of pentane extract. B MS of 6 from PtlI-catalyzed oxidation of 3. C MS of 7 from PtlI-catalyzed oxidation of 3.
PtlI showed a pH optimum of 8.0 for the oxidation of pentalenene to pentalen-13-ol. The apparent steady-state kinetic parameters for the first oxidation step were determined by carrying out a series of 10-min incubations with 4–40 μM of (±)-pentalenene (3) and quantitation of the product pentalen-13-ol (6) by GC-MS. Under these conditions, further oxidation of 6 was negligible.8b Fitting of the initial velocities to the Michaelis-Menten equation gave kcat 0.503±0.006 min−1 and a Km of 3.33±0.62 μM for the active enantiomer of 3.
These results establish that the ptlI gene product can catalyze the two-step oxidation of pentalenene (3) to pentalen-13-al (7) (Scheme 2). At this point, it remains an open question how aldehyde 7 gets converted to 1-deoxypentalenic acid (4). Although it remains possible that PtlI might support the latter oxidation under the appropriate conditions,17 by analogy to other P450s,7–9 it is also conceivable that another redox enzyme from within the biosynthetic gene cluster could be responsible for this conversion. The work reported here sheds new light on the biosynthetic gap between pentalenene (3), generated by PtlA-catalyzed cyclization of FPP, and 11β-hydroxy-1-deoxypentalenic acid (5), the product of PtlH-catalyzed hydroxylation of 1-deoxypentalenic acid. Biochemical characterization of the remaining ORFs of the pentalenolactone biosynthetic gene cluster is in progress.
Supplementary Material
Expression of recombinant PtlI, binding spectra of 3 to PtlI, CO difference spectrum, mass spectra of products, chiral GC traces, NMR data, kinetic assays, and full citation for Ref 14b. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Dr. Tun-Li Shen for determining the mass spectra and Dr. Michael Waterman, Vanderbilt University, for providing us with plasmids harboring fld/fdr. Plasmid pPtlI-camAB encoding putidaredoxin and putidaredoxin reductase was generously provided by Mercian Corp, Japan. This work was supported by NIH grant GM30301 to DEC, by Grant of the 21st Century COE Program, Ministry of Education, Culture, Sports, Science and Technology, Japan to H.I and S.O, by Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science No. 17510168 to H.I and by a postdoctoral fellowship by the Swiss National Funds to R.Q.
References and Notes
- 1.(a) Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Proc Natl Acad Sci U S A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Nature Biotech. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 2.Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL. Biochem Biophys Res Commun. 2003;307:610–619. doi: 10.1016/s0006-291x(03)01231-2. [DOI] [PubMed] [Google Scholar]
- 3.Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H. Biochemistry. 2006;45:6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Cane DE, Sohng JK, Lamberson CR, Rudnicki SM, Wu Z, Lloyd MD, Oliver JS, Hubbard BR. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]; (b) Cane DE, Oliver JS, Harrison PHM, Abell C, Hubbard BR, Kane CT, Lattman R. J Am Chem Soc. 1990;112:4513–4524. [Google Scholar]
- 5.You Z, Omura S, Ikeda H, Cane DE. J Am Chem Soc. 2006;128:6566–6567. doi: 10.1021/ja061469i.(b) The 1-deoxypentalenic acid (4) has been isolated as the glucuronate ester: Takahashi S, Takeuchi M, Arai M, Seto H, Otake N. J Antibiot. 1983;36:226–228. doi: 10.7164/antibiotics.36.226.
- 6.Sono M, Roach MP, Coulter ED, Dawson JH. Chem Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 7.Gibberellin biosynthesis: Helliwell CA, Poole A, Peacock WJ, Dennis ES. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507.Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. Proc Natl Acad Sci U S A. 2001;98:2065–2070. doi: 10.1073/pnas.041588998.
- 8.Bile acid biosynthesis: Cali JJ, Russell DW. J Biol Chem. 1991;266:7774–7778.Pikuleva IA, Babiker A, Waterman MR, Bjorkhem I. J Biol Chem. 1998;273:18153–18160. doi: 10.1074/jbc.273.29.18153.
- 9.Artemisinic acid biosynthesis: Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 2006;440:940–943. doi: 10.1038/nature04640.Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065.
- 10.The nonionic detergent Triton X-100 (0.1%) was a required component of the lysis buffer in order for PtlI to bind to the Ni-NTA resin, suggesting that PtlI may be membrane-associated.
- 11.Omura T, Sato R. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 12.Compounds (±)-(3), (±)-(6), (±)-(7), and (±)-(4) were synthesized as previously described: Ohfune Y, Shirahama H, Matsumoto T. Tetrahedron Lett. 1976;17:2869–2872.Ohtsuka T, Shirahama H, Matsumoto T. Tetrahedron Lett. 1983;24:3851–3854.Cane DE, Tillman AM. J Am Chem Soc. 1983;105:122–124.Tillman AM. PhD Thesis. Brown Univ; Providence, RI: 1984. pp. 115–163.
- 13.Jefcoate CR. Methods Enzymol. 1978;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- 14.The natural electron transport pair for PtlI is unknown. S. avermitilis harbors six putative ferredoxin reductases and nine ferredoxins (cf. Ref 2). Numerous attempts to observe PtlI-catalyzed oxidation of pentalenene (3) using typical redox pairs such as spinach ferredoxin and NADPH:ferredoxin oxidoreductase or putidaredoxin and putidaredoxin reductase were uniformly unsuccessful. By contrast, incubations of 3 and NADPH with unpurified cell-free extracts of E. coli PtlI expression cultures indicated that the endogenous E. coli flavodoxin (Fld) and flavodoxin reductase (Fdr) supported activity of PtlI. Although S. avermitilis does not harbor a native flavodoxin, similar observations have been previously reported for other P450s. Jenkins CM, Waterman MR. J Biol Chem. 1994;269:27401–27408.Zhao B, et al. J Biol Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200.
- 15.(a) FldA, Swissprot P61949: Jenkins CM, Pikuleva I, Kagawa N, Waterman MR. Arch Biochem Biophys. 1997;347:93–102. doi: 10.1006/abbi.1997.0318.(b) Fdr, Swissprot P28861: Jenkins CM, Waterman MR. Biochemistry. 1998;37:6106–6113. doi: 10.1021/bi973076p.
- 16.Some background oxidation of (±)-(6) to pentalen-13-al (7) could also be detected. Under typical incubation conditions, the ratio of P450-catalyzed oxidation to oxidation in the absence of PtlI was ~3:1, as judged by GC-MS (PtlI (1.4 μM), Fld (6.0 μM) Fdr, (3.4 μM), NADPH (0.53 mM), glucose-6-phosphate (0.53 mM) and glucose-6-phosphate dehydrogenase (5 u) in 2.9 mL of 50 mM Tris-HCl buffer, 10% glycerol (v/v), pH 8, was incubated with (±)-(6) (0.1 mM) for 2 h at 25 °C). This finding was consistent with chiral GC-MS analysis of an incubation of (±)-(6) with PtlI, which revealed formation of both enantiomers of aldehyde 7 in a ratio of 2.6:1 natural 7 to enantio-7.
- 17.Incubations carried out with PtlI, ferredoxin FdxD (SAV3129) and ferredoxin reductase FprD (SAV5675), the most abundant electron carriers in S. avermitilis, showed no enhancement in pentalenene oxidation and no detectable formation of 1-deoxypentalenic acid (4).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of recombinant PtlI, binding spectra of 3 to PtlI, CO difference spectrum, mass spectra of products, chiral GC traces, NMR data, kinetic assays, and full citation for Ref 14b. This material is available free of charge via the Internet at http://pubs.acs.org.


