Abstract
tRNA-guanine transglycosylase (TGT) is a key enzyme involved in the post-transcriptional modification of certain tRNAs in their anticodon wobble positions with queuine. In order to maintain the correct Watson-Crick base pairing properties of the wobble base (and hence proper translation of the genetic code), TGT must recognize its heterocyclic substrate with high specificity. The X-ray crystal structure of a eubacterial TGT bound to preQ1 (Romier et al., EMBO J. (1996) 15, 2850–2857) suggested that aspartate 143 (E. coli TGT numbering) was involved in heterocyclic substrate recognition. Subsequent mutagenic and computational modeling studies from our lab (Todorov et al., Biophys. J. (2005), 89 (3), 1965–1977) provided experimental evidence supporting this hypothesis. Herein, we report further studies probing the differential heterocyclic substrate recognition properties of the aspartate 143 mutant TGTs. Our results are consistent with one of the mutants exhibiting an inversion of substrate recognition preference (xanthine vs. guanine) relative to the wild-type, as evidenced by Km values. This confirms the key role of aspartate 143 in maintaining the anticodon identities of the queuine-containing tRNAs and suggests that TGT mutants could be developed that would alter the tRNA wobble base base-pairing properties.
Keywords: queuine, transglycosylase, TGT, RNA modification
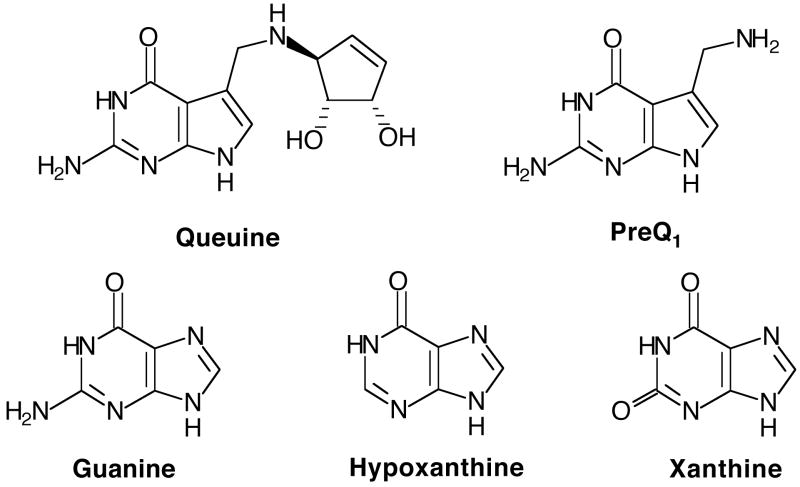
There are approximately 100 modified nucleosides that occur in RNA, with the vast majority of them occurring in tRNAs (1). The tRNA anticodon is one “hot spot” for hypermodification (e.g., the incorporation of elaborately modified nucleosides). Queuine (Figure 1) is one example of a hypermodified base that occurs in the wobble position of the anticodon of tRNAs Asp, Asn, His and Tyr. Queuine is incorporated into tRNA via a base exchange reaction (replacing guanine) catalyzed by tRNA-guanine transglycosylase (TGT). In eukaryotes, queuine is directly exchanged into tRNA while in eubacteria, a queuine precursor (preQ1, Figure 1) is incorporated and ultimately modified to queuine (2). Queuine, guanine and preQ1 all share the same 2-amino-pyrimidin-4-one moiety that is involved in the Watson-Crick type base pairing between the tRNA anticodon and the mRNA codon during translation. In order to accurately maintain this interaction, and hence the identity of the tRNAs and the fidelity of translation, TGT must exercise precise and specific recognition of its heterocyclic substrate.
Fig. 1.
Physiological and Potential Alternative Heterocyclic Substrates for TGT.
The X-ray crystal structure of the Zymomonas mobilis TGT bound to preQ1 revealed that aspartate 143 (D143, E. coli TGT numbering) appears to make two hydrogen bonds to the amino pyrimidone portion of preQ1 (3). In order to experimentally probe the role of D143 in heterocyclic substrate recognition, we carried out a thorough biochemical and computational characterization of wild-type and D143 mutant TGTs. Their interactions with guanine confirmed that D143 does play a vital role in heterocyclic substrate recognition (4). Computational simulations of guanine binding to wild-type and D143 mutant TGTs provided insight as to which interactions assisted in binding guanine (4). Since D143 was ascertained to be the determinant for guanine specificity, it follows that mutating this residue may allow for alternate substrate recognition. Not only is there precedence for altering substrate specificity with guanine binding proteins using hypoxanthine and xanthine (Figure 1) (5–7), but these purines are readily available in vivo and therefore provide an interesting and physiologically-relevant study of substrate specificity. We herein report biochemical studies to probe the recognition between wild-type and D143 mutant TGTs and the alternate purine heterocycles, hypoxanthine and xanthine.
MATERIALS AND METHODS
Reagents
Reagents were purchased from Sigma-Aldrich unless otherwise noted. Dithiothreitol (DTT) was obtained form Gibco BRL. HEPES (1M solution, pH 7.3) was from Amersham Pharmacia. [8-3H]-Guanine (1–10 Ci/mmol), [8-3H]-xanthine (9 Ci/mmol) and [2,8-3H]-hypoxanthine (24.2 Ci/mmol) were purchased from Moravek Biochemicals, Inc. Whatman GF/C 24mm glass microfibre filters were obtained from Fisher Scientific. Biodegradable liquid scintillation counting cocktail Bio-Safe II was from Research Products. Wild-type and mutant TGTs were expressed and purified with amino terminal histidine tags as described previously (4). Based on SDS-PAGE analysis (not shown), the mutants were free of any detectable, contaminating wild-type. E. coli tRNATyr (ECY) was prepared via in vitro transcription as previously described (8) and purified under native conditions by anion exchange chromatography.
Determination of the Concentrations and Solubilities of Hypoxanthine and Xanthine
Due to the high hypoxanthine and xanthine concentrations necessary for the TGT kinetics, the solubility of these compounds was tested at pH 7.3. Excess hypoxanthine or xanthine was added to a mixture containing 100 mM HEPES, 20 mM MgCl2, and 5 mM DTT. This mixture was incubated for 5 hours; at one-hour intervals, the solution was mixed and a 100 μL aliquot was removed. These aliquots were clarified via centrifugation (5 min at 13000 RPM in a benchtop microfuge) and the concentrations of the supernatant solutions were determined spectrophotometrically (see below). These determinations were repeated in triplicate. The concentration limits of hypoxanthine and xanthine were evaluated over the maximum time used for the assays (Table 1).
Table 1.
Solubility Determination of Hypoxanthine and Xanthine.
| Compound | Wavelength | Absorption Coefficient (ε) | Limit of Solubility |
|---|---|---|---|
| Hypoxanthine | 250 nm | 10.55 (0.02) × 10−3 cm−1μM−1 | ~ 2 mM |
| Xanthine | 225 nm | 4.71 (0.04) × 10−3 cm−1μM−1 | ~ 1.5 mM |
Solubilities were determined spectrophotometrically in assay buffer (100 mM HEPES (pH 7.3), 20 mM MgCl2 and 5 mM DTT) as described in Methods. Standard errors are shown in parentheses.
Concentrations were determined spectrophotometrically using a Cary Bio-100 spectrophotometer. Values of the absorption coefficient for each compound varied widely in the literature and were pH dependent. For accurate concentration determination, an absorption coefficient was experimentally determined in the buffer solution used for the TGT assays. A stock solution of 500 mg of each compound was dissolved in dilute NaOH and the pH of the solution was adjusted to 7.3. Serial dilutions were made and a standard curve was generated from a plot of absorption versus concentration. The absorption coefficient was calculated from the slope of the line (e.g., slope divided by path length). Obtained values of absorption coefficients were similar to those previously reported.
Determination of Kinetic Parameters for Hypoxanthine and Xanthine
The TGT-catalyzed incorporation of xanthine and hypoxanthine into tRNA was monitored by following the incorporation of radioactivity from tritium-labeled xanthine and hypoxanthine into tRNA as previously described for labeled guanine (9). KM, kcat, and kcat/KM for hypoxanthine and xanthine were determined for htTGT(wt), htTGT(D143A), htTGT(D143N), htTGT(D143S), and htTGT(D143T). Therefore, tRNA was kept at saturating (20 μM) concentration (see Results) for the KM and kcat determinations for hypoxanthine and xanthine.
htTGT(wt) (100 nM) was incubated with tRNA (20 μM) in the presence of radiolabeled 3H-hypoxanthine or 3H-xanthine (varied concentrations), MgCl2 (20 mM), DTT (5 mM) and HEPES, pH 7.3 (100 mM) in a total reaction volume of 400 μL. At varying intervals over an appropriate time course (10–180 minutes, longer time course for less active enzymes), 70 μL aliquots were withdrawn and quenched in 2 mL of 5% TCA. This reaction was allowed to precipitate for 1 hour. The precipitated tRNA was then collected on Whatman GF/C glass microfibre filters. The filters were dried and the radioactivity was counted via liquid scintillation to quantitate incorporation of 3H-hypoxanthine or 3H-xanthine into tRNA. The assays were performed, at least, in triplicate.
In order to determine the kinetic parameters for xanthine and hypoxanthine for wild-type and the D143 mutant TGTs, various concentrations of xanthine and hypoxanthine and differing time courses, dependent upon the enzyme were used (Table 2). Note that the high concentrations of hypoxanthine were found to adsorb to the glass filters and cause inaccurately high and unstable readings of radioactivity. To prevent this, all filters used for hypoxanthine kinetics were pre-soaked in unlabeled hypoxanthine (2.5mM) dissolved in 5% TCA and dried. All substrates were tested to their maximal level of solubility. Reaction time courses were followed to a maximum of 10% turnover and the enzyme concentration was never greater than 0.1 × substrate concentration. Initial velocities were determined via linear fits of radioactivity (converted to pmoles xanthine or hypoxanthine using the appropriate specific activity) versus time. The initial velocities were then plotted against the concentration of the varied substrate. KM and kcat were calculated (averages of three independent determinations) from non-linear fits of the initial velocity data to the Michaelis-Menten equation. All relative values were determined relative to htTGT(wt). The efficiency constants, defined as kcat/KM, were also calculated. All errors are the standard errors of the fits with the errors in kcat/KM normally propagated from the standard errors in KM and kcat.
Table 2.
Experimental Conditions for Hypoxanthine and Xanthine Kinetics Determinations.
| enzyme | [Hypoxanthine] μM | [Xanthine] μM | Time Courses (both substrates) |
|---|---|---|---|
| htTGT(wt) | 25–3000 | 10–1500 | 120 min |
| htTGT(D143A) | 50–2000 | 1–750 | 120 min |
| htTGT(D143N) | 50–2000 | 0.5–750 | 120 min |
| htTGT(D143S) | 1–3000 | 1–3000 | 4 hr |
| htTGT(D143T) | 1–3000 | 1–3000 | 4 hr |
[tRNA] = 20 μM for all enzymes and [TGT] = 100 nM for all except htTGT(D143S) & htTGT(D143T) = 250 nM. Assays were conducted in 100 mM HEPES (pH 7.3), 20 mM MgCl2 and 5 mM DTT at 37 °C as described in Methods.
Determination of Ki Values for Xanthine
htTGT(D143S or D143T) (200 nM) was incubated with tRNA (20 μM) in the presence of MgCl2 (20 mM), DTT (5 mM), HEPES, pH 7.3 (100 mM), 3H-guanine (20- 100 μM), and xanthine (25–500 μM) in a total reaction volume of 400 μL. Aliquots (70 μL) were taken at 15 min intervals over a 75 min time course. The aliquots were withdrawn, filtered, and quantified as described above. Initial velocities were calculated from linear fits of plots of guanine incorporated versus time. The initial velocities were re-plotted, and fit by three dimensional, non-linear regression to a competitive (with respect to guanine) inhibition equation (eq. 1) using the preprogrammed equations in GraFit (10). The results were then plotted using Kaleidagraph to graphically display both the data and the fitted lines.
| (eq. 1) |
| (eq. 2) |
| (eq. 3) |
For graphical analysis of the data, equation 1 can be rearranged in the form of 1/vi vs. [I] (a Dixon plot, eq. 2) (11) or [S]/vi vs. [I] (the Cornish-Bowden method, eq. 3) (12). In fact, both plots are complementary when analyzing data such as these. Therefore, the data were plotted by both methods based on the methodology described below. In the case of competitive inhibition plots of 1/vi vs. [I] (eq. 2) are intersecting. Plots of [S]/vi vs. [I] (eq. 3) are parallel (the [S] term in the slope falls out of the equation and the slope is a constant independent of [S]).
RESULTS
Kinetic Characterization of Wild-type and Mutant TGTs with Hypoxanthine and Xanthine
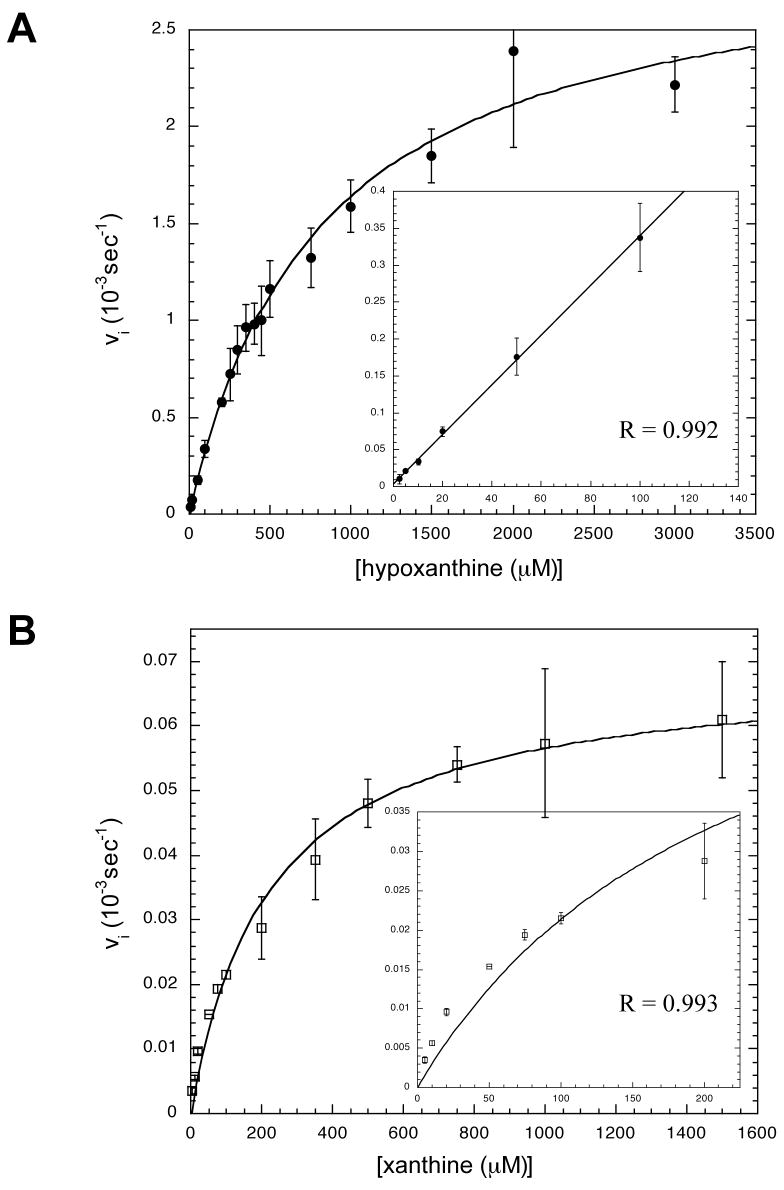
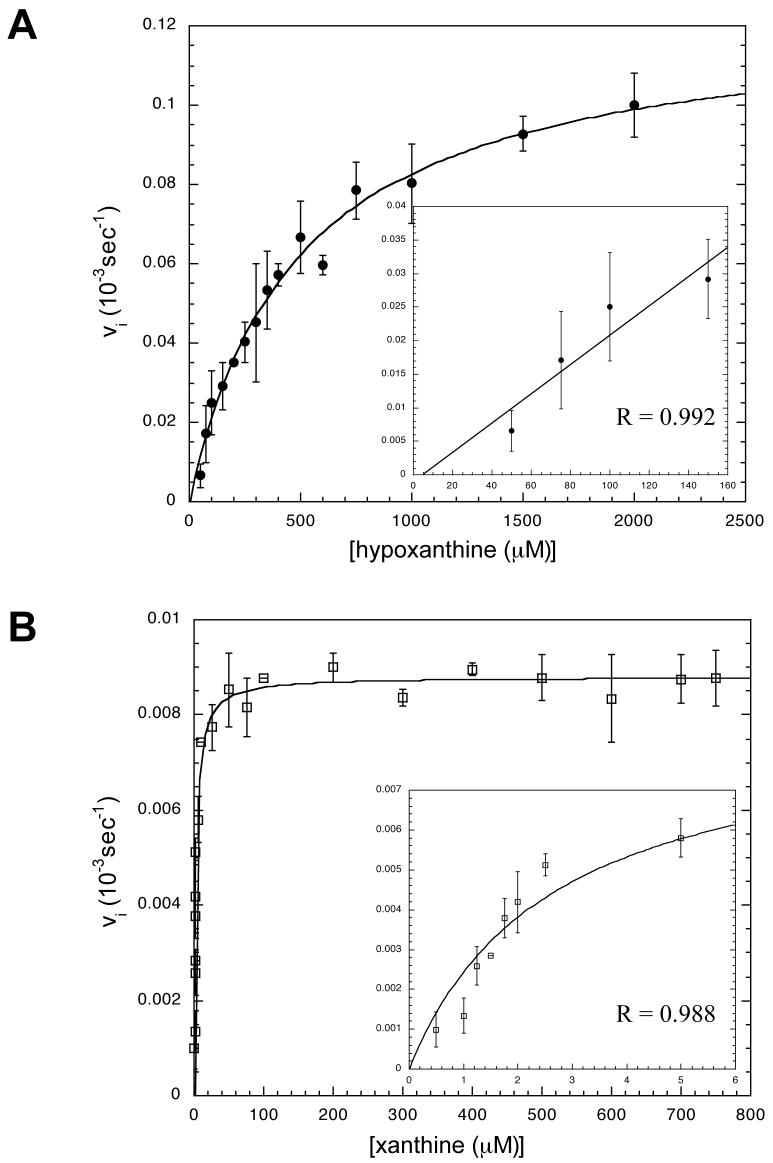
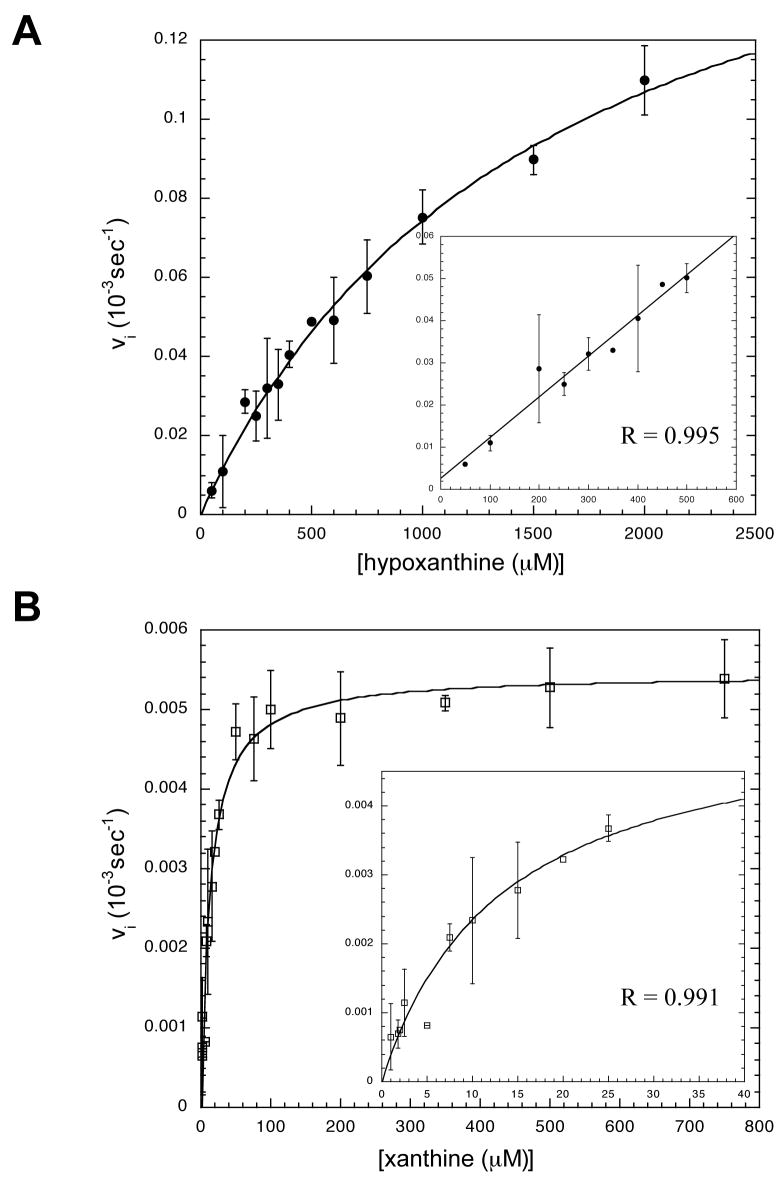
The kinetic parameters (KM and kcat) for htTGT(wt), htTGT(D143A), and htTGT(D143N) were determined for hypoxanthine (Figures 2–4, Table 3) and for xanthine (Figures 2–4, Table 4). The incorporation of the alternate substrate was followed by monitoring the increase in 3H radioactivity in the tRNA caused by the exchange of an unlabelled guanine in position 34 of tRNA with a radiolabeled hypoxanthine or xanthine. It is reasonable to assume that the KM values of tRNA would not be affected by the nature of the heterocyclic substrate based on the ping-pong kinetic mechanism for TGT (13). The tRNA binds to TGT, guanine-34 is then removed forming a covalent tRNA-TGT intermediate. The incoming base then binds to the tRNA-TGT complex and is inserted into the tRNA in position 34 and the tRNA dissociates from the TGT. We have previously shown that the KM values for tRNA for each of these mutants are not significantly different from those for the wild-type TGT when studied with guanine (4). To confirm this, we have determined that at 10 μM, 20 μM (saturating, 10x KM), and 40 μM tRNA, the rate of the reaction for both xanthine and hypoxanthine with each mutant TGT did not change (data not shown). For both the serine and threonine mutants, neither xanthine nor hypoxanthine exhibited any detectable substrate activity at concentrations up to the limit of their solubility.
Fig. 2.
Michaelis-Menten Kinetics Plots for htTGT(wt). A: Kinetic Fit for Hypoxanthine, B: Kinetic Fit for Xanthine. Curves were obtained from a non-linear fit of the average of three independent determinations of initial velocity data. Error bars are generated from the standard deviation in each point. The inset is an expansion of the low concentration. The fit of the curve is represented by R. Note that the scales of the axes change from plot to plot.
Fig. 4.
Michaelis-Menten Kinetics Plots for htTGT(D143N). A: Kinetic Fit for Hypoxanthine, B: Kinetic Fit for Xanthine. See Fig. 4 legend.
Table 3.
Table of Hypoxanthine Kinetic Parameters for TGTs.
| Enzyme | kcata,b,d (10−6sec−1) | KMa,b,d (μM) | kcat/KMa,b,d (10−6sec−1μM−1) | Relative KMa,c,d | Relative kcat/KMa,c,d |
|---|---|---|---|---|---|
| htTGT(wt) | 2970 (140) | 811 (86) | 3.7 (0.4) | 1.0 | 1.0 |
| htTGT(D143A) | 190 (13) | 1550 (170) | 0. 1 (0.02) | 1.9 | 0.03 |
| htTGT(D143N) | 120 (05) | 490 (45) | 0. 2 (0.02) | 0.6 | 0.07 |
| htTGT(D143S) | NAe | NA | - | - | - |
| htTGT(D143T) | NA | NA | - | - | - |
Standard errors are shown in parentheses.
Kinetic parameters are calculated from the average of three replicate determinations of initial velocity data.
Relative values were determined relative to that of His-tagged wild-type.
NA is no activity within the limits of solubility (2 mM hypoxanthine).
Table 4.
Table of Xanthine Kinetic Parameters for TGTs.
| Enzyme | kcata,b,d (10−6sec−1) | KMa,b,d (μM) | kcat/KMa,b,d (10−6sec−1μM−1) | Relative KMa,c,d | Relative kcat/KMa,c,d |
|---|---|---|---|---|---|
| htTGT(wt) | 69 (3) | 224 (28) | 0. 31 (0.04) | 1.0 | 1.0 |
| htTGT(D143A) | 5.5 (0. 1) | 13 (1) | 0. 42 (0.04) | 0.059 | 1.35 |
| htTGT(D143N) | 8.8 (0.1) | 2.6 (0.2) | 3.4 (0. 3) | 0.012 | 10.9 |
| htTGT(D143S) | NAe | - | - | - | - |
| htTGT(D143T) | NAe | - | - | - | - |
Standard errors are shown in parentheses.
Kinetic parameters are calculated from the average of three replicate determinations of initial velocity data.
Relative values were determined relative to that of His-tagged wild-type.
NA is no activity within the limits of solubility (1.5 mM xanthine).
The insets on each plot of the kinetics with hypoxanthine (Figures 2–4) show an independent linear fit of the low concentration range to determine kcat/KM. This additional validation of the kinetic parameters determined from the Michaelis-Menten equation was necessary because saturating concentrations (10x KM) could not be reached due to the limit of solubility (ca. 2 mM, as discussed above). The determination of kcat/KM using the linear fit of the data matched the kcat/KM calculated from the kinetic parameters derived from the fits to the Michaelis-Menten equation (data not shown). On each plot of xanthine kinetics is an inset of the low concentration range to better depict how well the data fits to the Michaelis-Menten equation.
Inhibition of the D143S and D143T Mutants by Xanthine
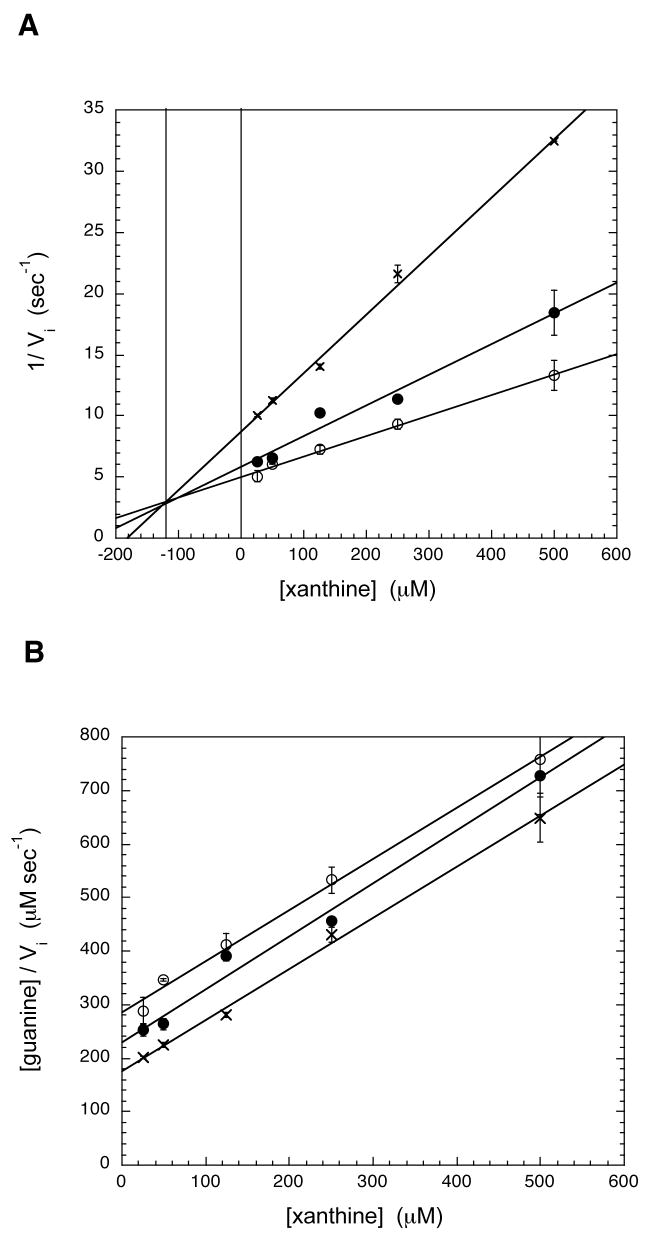
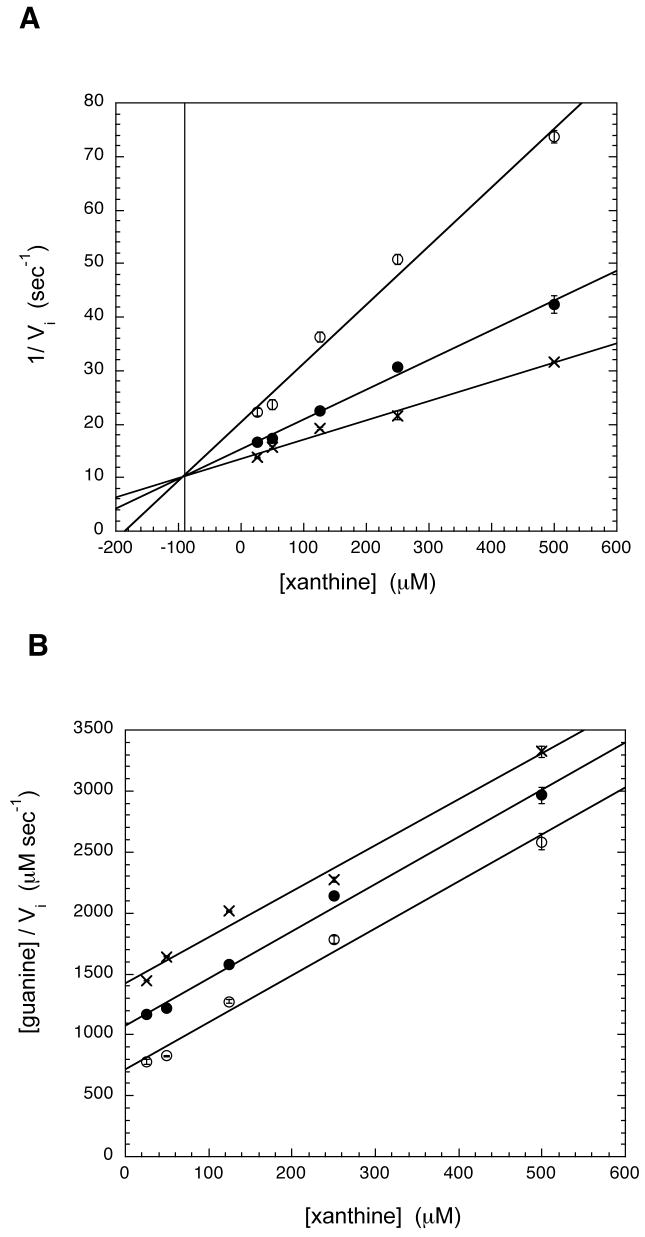
The inhibition constants for xanthine with htTGT(D143S) and htTGT(D143T) have been determined (Figures 5 & 6, Table 4) by monitoring their ability to inhibit the incorporation of radiolabeled guanine into tRNA. For both htTGT(D143S) and htTGT(D143T), xanthine was found to be competitive, with respect to guanine, based on intersecting lines in a Dixon plot (Figures 5 (A) & 6 (A)) and parallel lines when plotted according to the method of Cornish-Bowden (Figures 5 (B) & 6 (B)). The competitive inhibition constant (Ki) for htTGT(D143S) was 120 μM and for htTGT(D143T) was 90 μM; the data were calculated from the point of intersection in the Dixon plot. There was no inhibition seen with hypoxanthine.
Fig. 5.
Inhibition Plots of htTGT(D143S) by Xanthine. A: Dixon Plot, B: Cornish-Bowden Plot. The mode of inhibition of the guanine incorporation by xanthine was determined by varying the concentrations of xanthine (25–500 μM), while holding tRNA constant at a saturating concentration (20 μM) at three different concentrations of guanine (open circle: 20 μM; solid circle: 40 μM; X: 57 μM). The data were analyzed graphically by plotting either (A) 1/vi vs. [xanthine] (eq. 2) or (B) [guanine]/vi vs. [xanthine] (eq. 3). Ki (competitive with respect to guanine) corresponds to the point of intersection in the Dixon plot (A). The data points arise from an average of two independent determinations of initial velocities under these conditions.
Fig. 6.
Inhibition Plots of htTGT(D143T) by Xanthine. A: Dixon Plot, B: Cornish-Bowden Plot. The mode of inhibition of the guanine kinetics by xanthine was determined by varying the concentrations of xanthine (25–500 μM), while holding tRNA constant at a saturating concentration (20 μM) at three different concentrations of guanine (open circle: 35 μM; solid circle: 70 μM; X: 105 μM). The data were analyzed as described in Fig. 7.
DISCUSSION
Previously, X-ray structural (3), biochemical (4) and computational (4) studies have all indicated that aspartate 143 is critical to the recognition of the 2-amino-pyrimidin-4-one portion of the heterocyclic substrates for tRNA-guanine transglycosylase (TGT). This recognition is absolutely requisite to maintain the proper Watson-Crick base pairing in the queuine-containing tRNAs’ anticodons, and hence the fidelity of translation of the corresponding codons. Given the critical role of aspartate 143, it seemed likely that mutants of aspartate 143 may exhibit altered heterocyclic substrate recognition properties. Therefore, we have examined the recognition of xanthine and hypoxanthine by wild-type and D143 mutant TGTs.
We have previously reported the construction and biochemical and computational characterization of these mutants and their interactions with guanine (4). Consistent with the postulate that heterocyclic substrate specificity is critical for the proper functioning of the corresponding tRNAs, we observed that the D143 mutants could only be expressed in the presence of chromosomally encoded wild-type TGT (which presumably maintained the integrity of the tRNAs). The mutants were expressed in histidine-tagged form to allow for purification away from contaminating wild-type. The mutants exhibited an increasing trend in KM for guanine. Given that TGT follows the ping-pong kinetics with guanine binding second (13) and assuming that a step after guanine binding is rate limiting (for which we have evidence (14)), then the KM for guanine is very likely to be equivalent to its KD and can be treated as a measure of binding affinity/recognition. These results clearly demonstrated that asparate 143 is intimately involved in recognizing guanine.
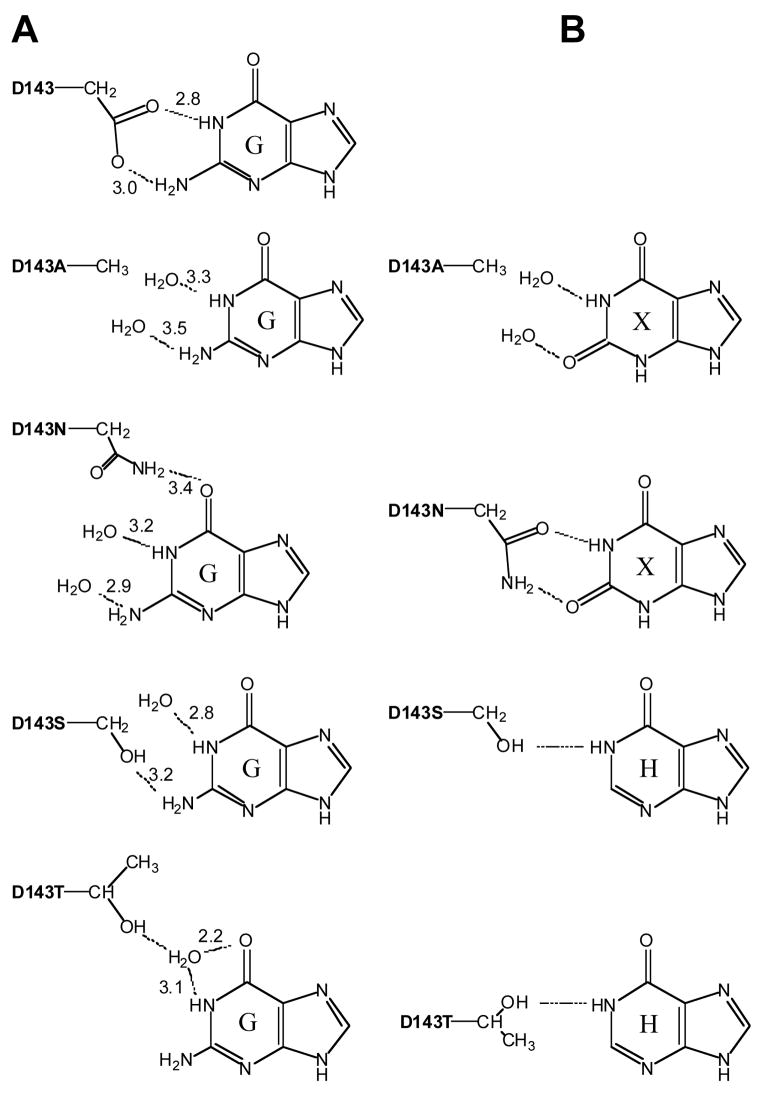
Hypoxanthine and xanthine were selected because they are close analogues of guanine, a well-characterized substrate for TGT and because they are naturally occurring, physiologically relevant molecules. In vivo, TGT must be able to discriminate against hypoxanthine and xanthine in favor of its natural substrate queuine (or preQ1). Perhaps more importantly, hypoxanthine and xanthine differ from guanine in the 2-amino-pyrimidin-4-one portion of the molecule, the exact portion that is recognized by aspartate 143. Therefore they are the logical choices to use to probe the differential substrate recognition properties of aspartate 143 mutants of TGT. Figure 7 shows the interactions between guanine and wild-type and D143 mutant TGTs as previously determined by biochemical and computational studies (4). Based on a manual hydrogen-bonding analysis, we have selected those TGT mutants which seem likely to have the highest affinity for hypoxanthine and for xanthine. The expected interactions between these mutants and hypoxanthine and xanthine are also displayed in Figure 7. Most interestingly, the D143N mutant is predicted to restore the hydrogen bond that is lost between the wild-type TGT and xanthine relative to guanine.
Fig. 7.
Recognition of Guanine (A) by the D143 Mutant TGTs and the Alternate Purine Substrates (B) Paired With Their Presumptively Most Appropriate Mutant (G denotes guanine, X denotes xanthine, H denotes hypoxanthine).
In-depth biochemical analyses for xanthine and hypoxanthine were carried out to explore the recognition of these alternate bases to wild-type and D143 mutant TGTs. Some unexpected results were obtained. Hypoxanthine was not recognized, as initially hypothesized, by htTGT(D143S) or htTGT(D143T). Xanthine was found to act as a substrate with wild-type, htTGT(D143A), and htTGT(D143N), but unexpectedly as an inhibitor of htTGT(D143S) and htTGT(D143T). Initially, it was expected that the overall affinity of htTGT(D143N) for xanthine would be comparable to the affinity of the wild-type for guanine. However, the overall catalytic efficiency (kcat/KM) of htTGT(D143N) with xanthine was much lower than that for the wild-type with guanine. This was due to a substantial and unexpected decrease in kcat.
Recognition of Hypoxanthine
The recognition of hypoxanthine, relative to that for guanine and xanthine, by wild-type and all D143 mutant TGTs was poor (Tables 3–5). It is interesting to note that the catalytic rates of the reactions, kcat, observed for wild-type, D143A, and D143N with hypoxanthine were similar to those observed with guanine (2300, 380, and 580 × 10−6 sec−1 respectively (4)). The serine (D143S) and threonine (D143T) TGT mutants were initially hypothesized to exhibit a strong binding preference for hypoxanthine, but, within the limits of detection and at concentrations up to its solubility limit, hypoxanthine was not a substrate for TGT(D143S) or TGT(D143T). Given the similarity of the kcat values for hypoxanthine and guanine with wild-type and the alanine and asparagine mutants, it is clear that, once bound, hypoxanthine is a good substrate for the chemical steps in the TGT reaction. Therefore, it is likely that the inactivity of hypoxanthine with the serine and threonine mutants is due to very poor binding/recognition.
Table 5.
Comparison of Experimental KM and Ki Values of Guanine, Hypoxanthine, and Xanthine with TGTs.
| Enzyme | Guanine KM (μM) | Hypoxanthine KM (μM) | Xanthine KM or Ki (μM) |
|---|---|---|---|
| htTGT(wt) | 0.15a | 811 | 224 |
| htTGT(D143A) | 11.4 | 1550 | 13.2 |
| htTGT(D143N) | 21.8 | 490 | 2.6 |
| htTGT(D143S) | 57 | NAb | 109 (17)c |
| htTGT(D143T) | 105 | NAb | 75 (11)c |
Data from Todorov et al. (Biophys. J. (2005) 89, 1965–1977).
NA indicates that no activity was determined given the limits of substrate solubility (2 mM for hypoxanthine, and 1.5 mM for xanthine).
For the serine and threonine mutants, xanthine was found to competitively inhibit the incorporation of guanine. Values were determined via a non-linear regression fit to a competitive inhibition model (eq. 2). Note that the errors in the KM values (which are found in Tables 3 and 4) have been omitted here for clarity.
Recognition of Xanthine As a Substrate
Biochemical characterization of the interaction between TGT (wild-type and D143 mutants) and xanthine demonstrates that xanthine is better recognized than hypoxanthine by all TGTs (Tables 3–5). The alanine mutant (D143A), which was initially expected to have similar recognition for guanine and xanthine, does indeed exhibit comparable values of KM (Table 5) for both substrates.
The asparagine mutant (D143N) was hypothesized to have an affinity for xanthine comparable to that of wild-type for guanine, suggesting a potential inversion of substrate specificity. Indeed, the experimental data clearly indicate that an inversion of specificity (in terms of KM) was seen with the mutant D143N recognizing xanthine in preference to guanine (Table 5). The expected hydrogen-bonding pattern of xanthine bound to D143N closely mimics that of guanine bound to wild-type, suggesting that the positioning of, and the interactions with the substrate are similar (Figure 7). An investigation by Kang et al. (6), involving E. coli adenylosuccinate synthetase (AMPSase), yielded a similar inversion of specificity. They established that the determinant of AMPSase for guanosine 5′-triphosphate specificity is D333, and mutating this residue to asparagine alters substrate specificity from GTP to XTP. They proposed that the aspargine mutant most likely makes complementary hydrogen bonds to XTP; accepting a hydrogen to the 2-oxo group and donating a hydrogen from the N1 position of the base of XTP (6).
Although the hydrogen bonding patterns for wild-type with guanine and D143N with xanthine are almost identical, their KM values are significantly different. It is likely that this is, in part, due to differences in the strengths of the hydrogen bonds in which they take part. The wild-type D143 side chain has two oxygens that share a negative charge. This carboxylate provides two hydrogen bond acceptors with a formal negative charge. Guanine has two nitrogens (one amine and one amide), which donate hydrogen bonds. The mutant D143N side chain has a neutral amide group that provides one hydrogen bond donor (nitrogen) and one acceptor (oxygen). Xanthine also has an amide group, within the structure of the ring that provides the complementary hydrogen bond acceptor (amide oxygen) and donor (amide nitrogen).
In contrast, Munagala and Wang discovered that a mutation from aspartate 163 to asparagine resulted in a loss of specificity for xanthine in HGXPRTase (hypoxanthine-guanine-xanthine phosphoribosyltransferase), whereas, the recognition of guanine and hypoxanthine remained unaltered (15). Based on a crystal structure of xanthine-bound to T. foetus HGXPRTase, aspartate 163 recognizes the O2 of xanthine via a water-mediated hydrogen bond (15). The amide of asparagine would not be expected to participate in such a water-mediated hydrogen bond, accounting for the loss of xanthine recognition in this case.
Recognition of Xanthine As an Inhibitor
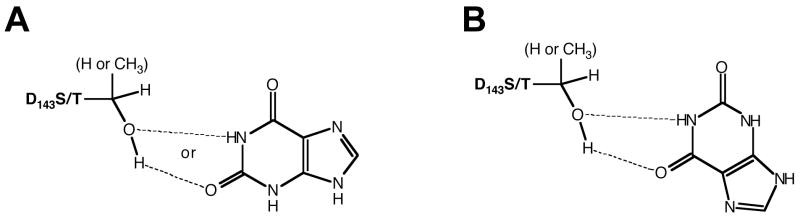
Interestingly, xanthine was found to act as an inhibitor, competitive with respect to guanine, for TGT(D143S) and TGT(D143T) (Figures 5 and 6, Table 4). It is very interesting that xanthine inhibits D143S and D143T more efficiently than xanthine acts as a substrate for wild-type TGT (Table 5). Xanthine exhibits a kind of pseudo-symmetry around its 2,4-dioxo-pyrimidine moiety. It is possible that xanthine may bind in an alternative, non-productive mode to these two TGT mutants (Figure 8). If this is true, it may also explain the reduced kcat values for xanthine with wild-type and other D143 mutant TGTs (see below).
Fig. 8.
Potential Pseudosymmetric Binding Modes Between TGT(D143S/T) and Xanthine. A: “Normal” binding mode. B: “Non-productive” binding mode.
Reduction in kcat for Xanthine
The biochemical studies with xanthine reveal that, although the kcat values for hypoxanthine are similar to those for guanine, the values for xanthine decrease 30-fold (wild-type) to 70-fold (D143 mutants) relative to those for guanine. The first half of the TGT reaction is identical for all heterocyclic substrates: the tRNA binds and G34 of tRNA is displaced and dissociates, resulting in a covalent TGT-tRNA complex (16). The second half of the reaction involves heterocyclic substrate binding, deprotonation of the N9 position of the base, and lastly, attack of the ribose displacing TGT by the base. The rate-limiting step of this reaction is unknown, but evidence suggests that it occurs after the formation of the covalent intermediate (14). It is possible that the rate-limiting step for the reaction may change upon changing the heterocyclic substrate. Therefore, it is difficult to definitively explain the decrease in kcat for xanthine with wild-type and D143 mutant TGTs. However, there are four possibilities which may be considered: 1) the altered chemistry of the purine ring, 2) a potential slow release of xanthine-modified tRNA, 3) the binding of xanthine to cause a change in the active site affecting the position of catalytic residues, 4) or the potential for xanthine to act as both a substrate and an inhibitor.
The first possibility is that since the chemistry of the ring is altered by substituting a carbonyl oxygen (xanthine) for an amine (guanine) on C2, this may affect the reactivity of xanthine in the chemical steps of the TGT reaction. If the pKa of the N9 position is higher, it may be more difficult to deprotonate the incoming base, thereby slowing down the reaction. However, the first pKa (i.e., neutral species to anionic) of xanthine is within one pKa unit of that of guanine (9.9 versus 9.26 respectively) (17), inconsistent with a 30- to 70-fold decrease in kcat. For HGXPRTase, Munagala and Wang (15) found that the kcat values of the conversion from the bases (hypoxanthine, guanine, and xanthine) to the nucleotide monophosphates (IMP, GMP, and XMP) are very similar (8.9, 2.5 and 4.8 sec−1 respectively). This reaction involves deprotonation of the base and formation of a glycosidic bond. These two steps are mimicked in the second half of the TGT reaction (16) in which the incoming base (guanine, hypoxanthine, or xanthine) is deprotonated. This base then attacks the TGT-tRNA complex, displacing TGT and forming a glycosidic bond. In the case of HGXPRTase, the kcat value of hypoxanthine is ca. 3-fold decreased relative to that for guanine. This result is mirrored by the minor differences we have observed in the kcat values of hypoxanthine relative to guanine for TGTs (wild-type and D143 mutants) (Tables 3 and 4). In HGXPRTase, the kcat value of xanthine is decreased ca. 2-fold relative to guanine, suggesting that it is unlikely that differences in the heterocyclic ring chemistry are responsible for the significant reductions in kcat that we have observed for xanthine relative to guanine.
The second possibility is that if product release is the rate-limiting step for the xanthine reaction, then a slow dissociation of the xanthine(34)tRNA·TGT complex would result in a lower kcat. This is unlikely since the tRNA has multiple interactions and one small binding interaction would most likely result in little contribution to the binding. Additionally, Goodenough-Lashua investigated the binding of guanine(34)-tRNA and preQ1(34)-tRNA (18). It was determined that preQ1(34)-tRNA binds 2-fold more tightly than guanine(34)-tRNA; however, the rate of the reactions of TGT with guanine versus preQ1 were previously determined to be comparable (19).
The third possibility is that a reorganization of the active site residues occurs upon xanthine binding. It was previously discussed (4) that D143 is involved in a key hydrogen bond with S90; this hydrogen bond orients S90, which, in turn, orients both the substrate and a catalytic residue, D89. In computational simulations of the D143 mutants with guanine, S90 moves 2–3 Å away from guanine in comparison to wild-type (4). This was proposed to be the reason that the mutants exhibited a 10-fold decrease in activity relative to wild-type TGT. It is certainly possible that the binding of xanthine causes a reorganization of the active site residues S90 and D89, thereby causing a decrease in kcat values for the incorporation of xanthine.
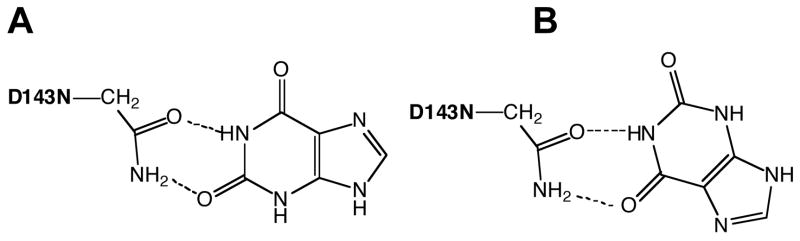
The fourth and perhaps most likely possibility is that xanthine may act as both a substrate and an inhibitor. Inhibition may occur via xanthine binding in a 180°-flipped conformation owing to the pseudo-symmetry of the molecule (Figure 9). This binding orientation would position the N9 such that it could not be deprotonated in the normal reaction and would therefore result in a nonproductive binding mode. Above, we postulated that this type of binding mode may be responsible for the xanthine inhibition of TGT(D143S) and TGT(D143T) that we have observed. For these mutants, the changes in the active site may have enhanced the affinity of xanthine for this nonproductive binding mode to the extent that no normal reaction is seen. For the wild-type and other D143 mutants, both binding modes may have similar affinities, resulting in a lower kcat due to the presumed nonproductive binding. Further studies are necessary to clarify this issue.
Fig. 9.
Potential Binding Modes Between TGT(D143N) and Xanthine. A: “Normal” binding mode. B: “Non-productive” binding mode.
One final note: we have previously developed and used a computational model system to probe the molecular details of the interaction between guanine and wild-type and D143 mutant TGTs (4). However, attempts to computationally model the interaction of xanthine with wild-type and D143 mutant TGTs (Todorov, Tan, Garcia & Carlson, unpublished) were unsuccessful in correlating the predicted free energies of binding with the observed values. It should be noted that the modeling was carried out for a time frame (1 ns) that is not sufficient to allow the xanthine to flip into the alternate, nonproductive binding mode that we postulate. Given that, it is not surprising that the calculations do not correlate with experiment in the xanthine case. In fact, this is consistent with our nonproductive binding mode hypothesis.
Our results have provided further confirmation of the important role that aspartate 143 plays in heterocyclic substrate recognition in the TGT reaction. The inversion of recognition of xanthine over guanine that we have observed for the asparagine mutant indicates that it is possible that TGT could be engineered to incorporate a base with different Watson-Crick type base pairing properties into tRNA. If this were done in conjunction with altered tRNA recognition, then a novel tRNA with a novel anticodon wobble base could theoretically be generated in vivo and could be used to study genetic code evolution and perhaps in unnatural amino acid mutagenesis.
Fig. 3.
Michaelis-Menten Kinetics Plots for htTGT(D143A). A: Kinetic Fit for Hypoxanthine, B: Kinetic Fit for Xanthine. See Fig. 4 legend.
Acknowledgments
We would like to thank Dr. Jeff Kittendorf for critical review of the experimental procedures and results reported in this manuscript. This work was supported in part by the National Institutes of Health (GM065489, GAG) and the University of Michigan, College of Pharmacy, Vahlteich Research Fund (GAG). KAT would like to acknowledge the support of the Chemistry-Biology Interface Training Program (GM08597).
Footnotes
The abbreviations used are: TGT, tRNA-guanine transglycosylase; PCR, polymerase chain reaction; DTT, dithiothreitol; HEPES, hydroxyethylpiperazine-ethylsulfonate; Tris·HCl, tris(hydroxymethyl)aminomethane hydrochloride; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; TCA, trichloroacetic acid; ECY, E. coli tRNAtyr transcript; htTGT(wt), histidine-tagged wild-type TGT. Histidine-tagged TGT mutant enzymes are referred to according to the following pattern: htTGT(D143A) (i.e., aspartate 143 mutated to alanine).
References
- 1.Grosjean H, Benne R. Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press; Washington, D. C: 1998. [Google Scholar]
- 2.Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorganic Chemistry. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- 3.Romier C, Reuter K, Suck D, Ficner R. Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. EMBO Journal. 1996;15:2850–2857. [PMC free article] [PubMed] [Google Scholar]
- 4.Todorov K, Tan XJ, Nonekowski ST, Garcia GA, Carlson HA. The Role of Aspartic Acid 143 in E. coli tRNA-Guanine Transglycosylase: Insights from Mutagenesis Studies and Computational Modeling. Biophysical Journal. 2005;89:1965–1977. doi: 10.1529/biophysj.105.059576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes CM, Kollman PA. Investigating the Binding Specificity of U1A-RNA by Computational Mutagenesis. J Mol Biol. 2000;295:1–6. doi: 10.1006/jmbi.1999.3319. [DOI] [PubMed] [Google Scholar]
- 6.Kang C, Sun N, Honzatko RB, Fromm HJ. Replacement of Asp333 with Asn by Site-directed Mutatgenesis Changes teh Substrate Specificity of Escherichia coli Adenylosuccinate Synthetase from Guanosine 5′-Triphosphate to Xanthosine 5′-Triphosphate. J Biol Chem. 1994;269:24046–24049. [PubMed] [Google Scholar]
- 7.Zhong JM, Chen-Hwang MC, Hwang YW. Switching Nucleotide Specificity of Ha-Ras p21 by a Single Amino Acid Substitution at Aspartate 119. J Biol Chem. 1995;270:10002–10007. doi: 10.1074/jbc.270.17.10002. [DOI] [PubMed] [Google Scholar]
- 8.Curnow AW, Kung FL, Koch KA, Garcia GA. tRNA-Guanine Transglycosylase from Escherichia coli: Gross tRNA Structural Requirements for Recognition. Biochemistry. 1993;32:5239–5246. doi: 10.1021/bi00070a036. [DOI] [PubMed] [Google Scholar]
- 9.Okada N, Nishimura S. Isolation and Characterization of a Guanine Insertion Enzyme, a Specific tRNA Transglycosylase, from Escherichia coli. Journal of Biological Chemistry. 1979;254:3061–3066. [PubMed] [Google Scholar]
- 10.Leatherbarrow R. GraFit Users Manual. 2.0. Erithacus Software Ltd; Staines, U.K: 1990. [Google Scholar]
- 11.Dixon M. The Determination of Enzyme Inhibitor Constants. Biochemistry Journal. 1953;56:170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornish-Bowden A. A Simple Graphical Method for Determining the Inhibition Constants of Mixed, Uncompetitive, and Non-Competitive Inhibitors. Biochemistry Journal. 1974;137:143–144. doi: 10.1042/bj1370143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodenough-Lashua DM, Garcia GA. tRNA-Guanine Transglycosylase from Escherichia coli: a Ping-Pong Kinetic Mechanism is Consistent with Nucleophilic Catalysis. Bioorganic Chemistry. 2003;31:331–344. doi: 10.1016/S0045-2068(03)00069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kittendorf JD. Medicinal Chemistry. University of Michigan; Ann Arbor: 2004. [Google Scholar]
- 15.Munagala NR, Wang CC. Altering Purine Specificity of Hypoxanthin-Guanine-Xanthine Phosphoribosyltransferase from Trichomonas foetus by Structure-Based Point Mutations in the Enzyme Protein. Biochemistry. 1998;37:16612–16619. doi: 10.1021/bi9818974. [DOI] [PubMed] [Google Scholar]
- 16.Garcia GA, Kittendorf JD. Transglycosylation: A mechanism for RNA modification (and editing?) Bioorganic Chemistry. 2005;33:229–251. doi: 10.1016/j.bioorg.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Windholz M, Budavari S, Blumetti RF, Otterbein ES. The Merck Index. 10. Merck & Co., Inc; Rahway, NJ: 1983. [Google Scholar]
- 18.Goodenough-Lashua DM. Medicinal Chemistry. University of Michigan; Ann Arbor: 2002. [Google Scholar]
- 19.Hoops GC, Townsend LB, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: Structure-activity studies investigating the role of the aminomethyl substituent of the heterocyclic substrate preQ1. Biochemistry. 1995;34:15381–15387. doi: 10.1021/bi00046a047. [DOI] [PubMed] [Google Scholar]
- 20.Harris DC. Quantitative Chemical Analysis. 3. W. H. Freeman and Company; New York: 1991. [Google Scholar]
- 21.Gordon AJ, Ford RA. The Chemist’s Companion: A Handbook of Practical Data, Techniques, and References. Wiley-Interscience; New York: 1972. [Google Scholar]