Abstract
A series of c- and z•-type product ions formed via gas-phase electron transfer ion/ion reactions between protonated polypeptides with azobenzene radical anions are subjected to ion trap collision activation in a linear ion trap. Fragment ions including a-, b-, y-type and ammonia-loss ions are typically observed in collision induced dissociation (CID) of c ions, showing almost identical CID patterns as those of the C-terminal amidated peptides consisting of the same sequences. Collisional activation of z• species mainly gives rise to side-chain losses and peptide backbone cleavages resulting in a-, b-, c-, x-, y-and z-type ions. Most of the fragmentation pathways of z• species upon ion trap CID can be accounted for by radical driven processes. The side-chain losses from z• species are different from the small losses observed from the charge-reduced peptide molecular species in electron transfer dissociation (ETD), which indicates rearrangement of the radical species. Characteristic side-chain losses are observed for several amino acid residues, which are useful to predict their presence in peptide/protein ions. Furthermore, the unique side-chain losses from leucine and isoleucine residues allow facile distinction of these two isomeric residues.
Keywords: Ion/ion reactions, electron transfer dissociation, c and z• ions, MS3, linear ion trap/triple quadrupole
Introduction
Gas-phase ion dissociation techniques have been investigated extensively in mass spectrometry to meet expanding requirements in the analysis and characterization of biomolecules.1 Collision induced dissociation (CID) is the most commonly used approach to derive structural information from peptide and protein ions through energetic collisions with a neutral bath gas,2 which cleaves the amide bonds along the backbone and gives rise to a-, b- and y-type product ions. However, during CID processes labile post-translational modifications, such as phosphorylation, are often more readily cleaved than the amide bonds along the peptide/protein backbone, thereby complicating both the identification of the post-translationally modified peptide/protein and location of the site of modification. In the past decade, electron capture by gaseous multiply charged proteins and peptides has drawn significant attention due to the rich structural information that can be derived as well as the ability to characterize post-translational modifications by this means.3–5 This technique is termed electron capture dissociation (ECD) and its implementation has been mostly restricted to Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometers.6 An analogue of ECD is the dissociation induced by electron transfer between oppositely charged ions, referred to as electron transfer dissociation (ETD). ETD has been implemented on electrodynamic ion traps and it has been noted that the dissociation due to electron transfer is very similar to that observed in ECD.7,8 In both ECD and ETD processes, the major fragmentation process is the cleavage of the N-Cα bonds along the peptide/protein backbone to give rise to complementary c- and z-type fragment ions. In most cases, the c-product is observed as an even-electron species leaving the complementary z-type fragments as an odd-electron species, denoted as z•. The proposed mechanisms accounting for ECD and ETD processes suggest that the radical site on z• ions is located on the alpha-carbon of the cleaved bond when the z• ions are initially formed.6,8 In addition to the peptide backbone fragmentation, neutral losses from the charge-reduced molecular radical species are commonly observed in ECD/ETD as well.9–11 Cooper et al. characterized small neutral losses and loss of amino acid side chains from several amino acid residues and found most of the losses were from basic residues, e.g. arginine, histidine and lysine.9 Haselmann et al. showed that not only protonation but charge solvation is responsible for the small molecule losses in ECD.10 However, the information regarding amino acid composition derived from the neutral losses is limited. For example, a 59 Da loss can indicate the presence of arginine, asparagine, and/or glutamine.
The odd-electron species formed from ECD or ETD can give rise to unimolecular dissociation behavior quite distinct from that of the corresponding even-electron species. In an ECD study of cyclic peptides, multiple backbone cleavages and side-chain losses are caused by multiple free radical rearrangement initiated by the radical propagation along the peptide backbone.12 Radical site migration in the surviving ions from ETD or ECD has also been demonstrated with deuterium labeled linear peptides.13 So far, there are only few studies of the fragmentation patterns of the resultant z• fragments formed from either ECD or ETD process. Kjeldsen et al. utilized the so-called hot ECD approach to induce secondary fragmentation of the z• fragments. It was demonstrated that the resultant w ions allow for the distinction between leucine/isoleucine isomers.14,15 Chan’s group has recently reported the loss of amino acid side chains due to secondary dissociation of the z• fragments from linear peptides under standard ECD conditions. Characteristic side-chain losses from many amino acids have been identified and corresponding mechanisms have been proposed.16,17 Although the reports showed that useful structural information can be acquired upon secondary fragmentation of z• fragments, it is desirable to conduct MS3 studies on the z• fragments so that precursor/product ion relationships can be determined with certainty.
In this work, we report a systematic study of the collisional activation of a series of c and z• ions formed via ETD of protonated polypeptides in a linear ion trap. The fragmentation patterns of c ions were investigated and compared to that of the C-amidated peptide ions of the same sequences. Rich backbone fragmentation of z• ions was noted and the possible mechanisms giving rise to the fragments are discussed. Unique side-chain losses are observed from the CID of z• ions, which show promise as a method for deriving confident amino acid residue compositions. An example of distinguishing isomeric residues such as leucine and isoleucine by performing CID on z• ions is also demonstrated.
Experimental Procedures
Materials
Methanol, glacial acetic acid and ammonium hydroxide were purchased from Mallinckrodt (Phillipsburg, NJ). The model peptides VDPVNFK, GAILKGAILR, GNRDADA, KGAILKGAILR, MRFA, KGAGGKGAGGKL, RGAGGRGAGGKL, RAAGFSPFR and GLSDGEWQQVLNVWGK were synthesized by SynPep (Dublin, CA). The model peptides PHCKRM, MQMKKVLDS and SAPRATISHYLMGG are obtained from AnaSpec Inc. (San Jose, CA). Peptide IFVQK is obtained from tryptic digestion of bovine cytochrome c. Peptides LFTGHPETLEK and HPGDFGADAQGAMTK are obtained from tryptic digestion of horse myoglobin. Peptides GAILK and GNR were obtained from tryptic digestion of model peptides GAILKGAILR and GNRDADA, respectively. Angiotensin II (sequence: DRVYIHPF), TPCK-treated trypsin, ammonium bicarbonate, acetyl chloride and azobenzene were obtained from Sigma-Aldrich (St. Louis, MO). Trifluoroacetic acid (TFA) was purchased from Pierce (Rockford, IL). All materials were used without further purification. Model peptides were diluted to 20μM in 50/50/1 (v/v/v) methanol/water/acetic acid for positive nano-electrospray ionization (nano-ESI).
Tryptic Digestion
Peptides (1 mg) were dissolved in 0.5 mL of aqueous 200 mM ammonium bicarbonate. TPCK-treated trypsin (20 μL of a 1 mg/mL aqueous solution) was added to the peptide solutions to effect the digestion. The peptide solutions were incubated at 38°C for ~15 min followed by separation on a reverse-phase HPLC (Agilent 1100, Palo Alto, CA) using an Aquapore RP-300 (7 μm pore size, 100 × 4.6 mm i.d.) column (Perkin-Elmer, Wellesley, MA) operated at 1 mL/min. A linear 20 min gradient from 0 to 100% buffer B was used, where buffer A was 0.1% (v) TFA aqueous solution and buffer B was 60/40/0.09 (v/v/v) acetonitrile/water/TFA. The fractions were dried in vacuum. Similar procedure is used for tryptic digestion of proteins except two differences. The two differences are: (1) the digestion time used for protein was ~12 h; (2) A linear 60 min gradient from 0 to 100% buffer B was used to separate protein digest.
Methyl Esterification
A solution of 2N HCl in methanol was prepared by adding 40 μL acetyl chloride drop wise to 250 μL chilled methanol.18 Methyl esterification was performed by adding 100 μL the prepared solution to the dried peptides, e.g. GAILK and GNR. The reaction was allowed to proceed for 2 h at room temperature and then the samples were dried in vacuum.
C-Terminal Amidation
C-terminal amidation of peptides was performed by adding 0.5 mL ammonium hydroxide to the dried methyl esterified peptide samples prepared above for ~20 h.19 The amidated samples were dried and dissolved in 50/50/1 (v/v/v) methanol/water/acetic acid to 0.5 mL for positive nano-ESI.
Mass Spectrometry
Most experiments were performed using a prototype version of Q TRAP mass spectrometer (Applied Biosystems/MDS SCIEX, Concord, Ontario, Canada).20 The Q TRAP electronics were modified to superpose auxiliary RF signals on IQ2 and IQ3, the containment lenses of the Q2 quadrupole array, allowing mutual storage of oppositely charged ions in the Q2 cell. The frequency and amplitude of the auxiliary RF signals applied to IQ2 and IQ3 lenses were optimized for the electron transfer ion/ion reaction experiments. A home-built pulsed dual nano-ESFatmosphere pressure chemical ionization (APCI) source21 was coupled directly to the interface of the Q TRAP mass spectrometer to generate multiply charged peptide cations and azobenzene radical anions, respectively. Sequential pulsing and accumulation of the oppositely charged ions were controlled by the Daetalyst 3.10 software, provided by MDS SCIEX. The high mass accuracy experiments were done using a QqTOF tandem mass spectrometer22 (QSTAR XL, Applied Biosystems/MDS SCIEX, Concord, ON, Canada), modified for ion/ion reactions.23
The experimental procedure for the electron transfer ion/ion reactions used here has been reported in detail elsewhere.21 In brief, a typical experiment employed in this work comprised sequences as follows: (1) pulsing the high voltage (−3 kV) applied on the APCI needle and injection of azobenzene radical anions into the Q2 linear ion trap (LIT), selected by Ql in the mass-resolving mode; (2) switching off the high voltage on the APCI needle while the anions were cooled in Q2 for 50 ms; (3) switching on the high voltage (+1.0–1.5 kV) on the nano-ESI emitter and injection of positive ions selected by Ql in the mass-resolving mode into the Q2 LIT with relatively low kinetic energies; (4) mutual storage of oppositely charged ions in Q2 LIT with nitrogen as bath gas at ~6 mTorr; (5) ejecting the remaining anions from Q2 by applying attractive DC potentials to the Q2 containment lenses; (6) transfer of the ETD product ions from Q2 to Q3; (7) cooling the transferred ions for 50 ms in Q3; (8) isolating product ions of interest under RF/DC mode in Q3; (9) cooling the isolated ions for 50 ms in Q3; (10) performing ion trap CID of the isolated ions; (11) cooling the product ions for 50 ms and (12) mass analysis of ions in Q3 via mass selective axial ejection (MSAE)24 using a supplementary RF signal at a frequency of 380 kHz. The spectra shown here were typically the averages of 50–200 individual scans.
Results and Discussion
MS3 of c Ions
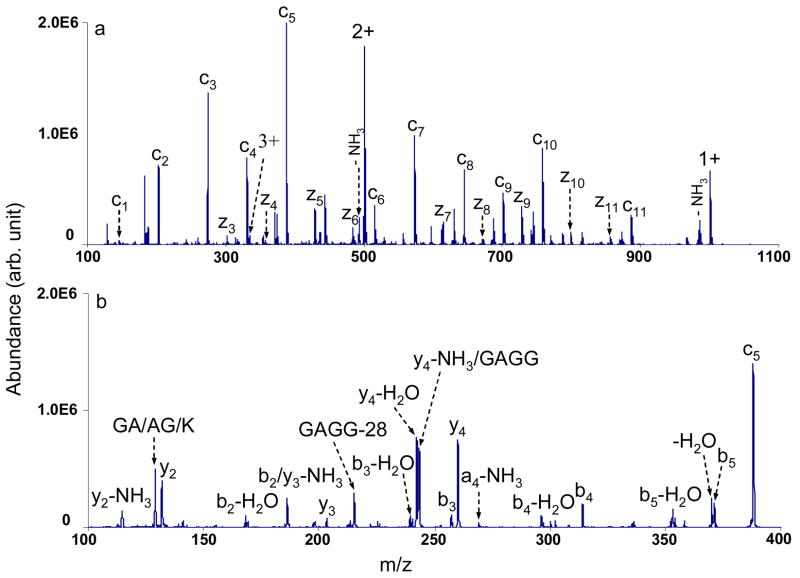
A series of c- and z•-type ions were formed via ETD by reacting different protonated model peptides with azobenzene radical anions in the Q2 linear ion trap (LIT) buffered with nitrogen at ~6 mTorr. The resulting c- and z•-type ions were isolated and the fragmentation patterns of those ions were investigated by collision induced dissociation (CID) in the low pressure Q3 LIT (~3×10−5 Torr). Figure 1a, for example, shows the c and z• ions formed from reacting triply protonated KGAGGKGAGGKL cations with azobenzene radical anions. The c5 ions were isolated and fragmented by applying a dipolar radio frequency (RF) in resonance with the secular frequency of the ions. The resulting MS3 spectrum is shown in Figure 1b, where b and y ions as well as their ammonia or water losses are predominant. In addition, peaks corresponding to a-type ions and internal fragments are observed. The major CID fragmentation channels of all the c ions studied in this work (thirty c ions were examined) are similar to that of the c5 ions shown in Figure 1b, including the loss of NH3 from cn ions to generate bn ions and the cleavages of backbone amide bonds resulting in the formation of other a-, b- and y-type ions.
Figure 1.
a) Electron transfer ion/ion reaction of triply protonated KGAGGKGAGGKL cations with azobenzene radical anions in the Q2 LIT; b) CID mass spectrum of c5 species from KGAGGKGAGGKL (77.50 kHz, 540 mVpp).
The c ions, if no rearrangements occur after their initial formation, can be regarded as a tautomer of C-terminal amidated peptides with the same sequences. Comparing the fragmentation patterns of c ions formed from ETD with those of the ions of amidated peptides with the same sequence formed directly from ESI, therefore, may shed light on the structure of the c ions. A direct comparison of the CID of the c5 species from GAILKGAILR, the [M+H]+ ions of the synthesized peptide GAILK-NH2 and the [M+H]+ ions of GAILK-OH is shown in Figure 2. Figures 2a and 2b show the CID spectra of the c5 ions and the singly protonated GAILK-NH2 respectively, upon applying the same activation frequency and amplitude. It is quite evident that these two CID spectra are essentially identical, with similar relative abundances of all the product ions.
Figure 2.
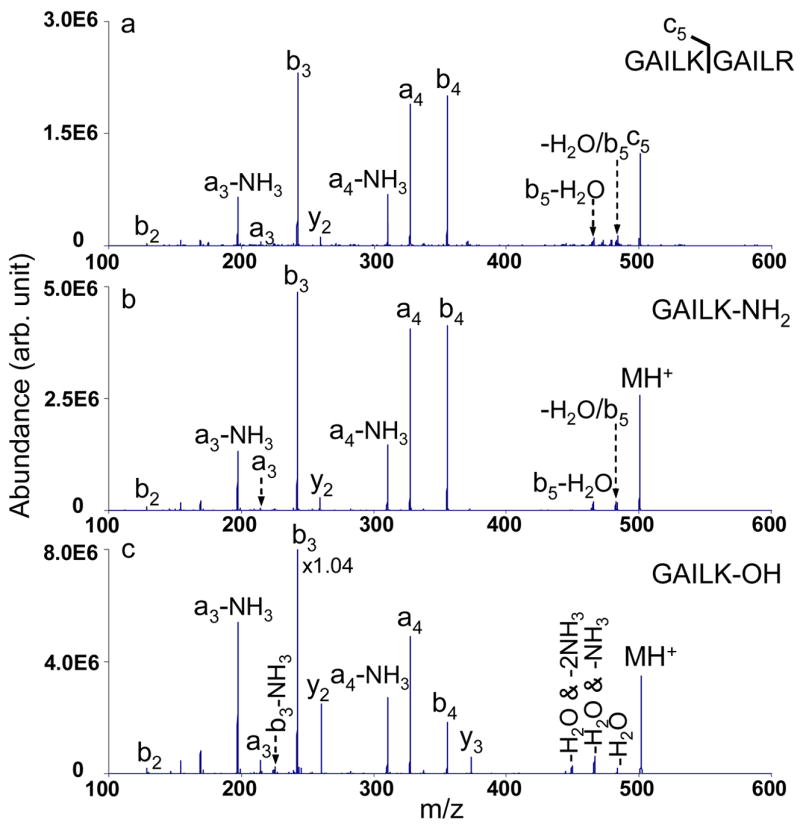
CID mass spectra of singly charged species: a) c5 ions from GAILKGAILR (90.59 kHz, 480 mVpp); b) GAILK-NH2 (90.59 kHz, 480 mVpp); c) GAILK-OH (90.44 kHz, 480 mVpp).
The CID spectrum of the singly protonated peptide GAILK-OH is provided in Figure 2c as well, showing a few differences in the fragment channels, e.g. the presence of the y3 species and different neutral-loss species compared with spectra shown in Figures 2a and 2b, which is consistent with the literature.25 Collisional activation results for several other c ions were compared with those from their C-terminal amidated peptides and the same CID spectra, within experimental reproducibility, were observed for each pair. The similarity in the dissociation behaviors for the c ions and the amidated peptides does not necessarily prove that they have identical structures, since ion trap collisional activation is a slow heating method.26 However, these results certainly show that the same structural information can be retrieved from c ions generated from ETD and the corresponding amidated peptide directly formed by ESI when ion trap CID method is employed. These results, therefore, provide supporting evidence that c ions formed via ETD effectively yield the corresponding amidated peptide.
MS3 of z• Ions
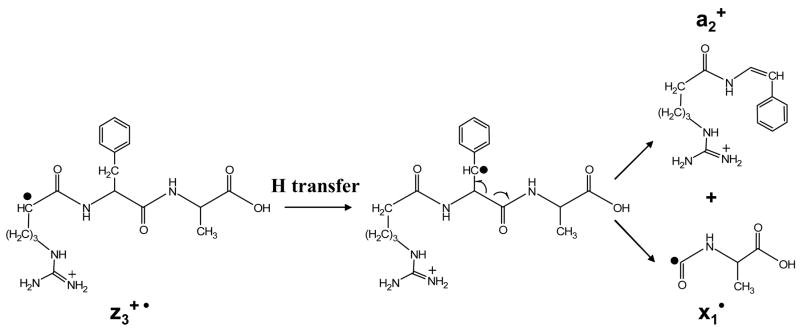
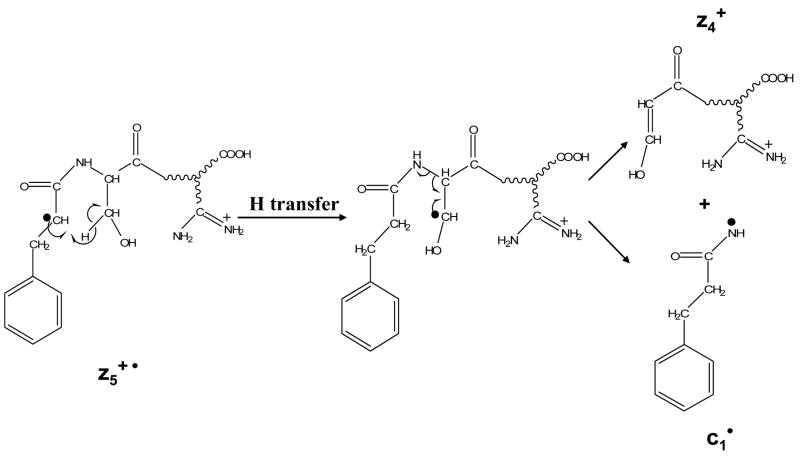
Ion trap collisional activation was applied to twenty-eight zn• ions derived from ETD of various protonated peptides. Compared to the even-electron c-ion species, the fragmentation patterns of the z• ions were mostly dominated by radical driven processes, although charge directed fragmentations were also noted as minor processes. It is worth pointing out that under ion trap collisional activation conditions, the z• ions may comprise a number of tautomers with the radical site scrambled to more favorable sites than the initial N-Cα cleavage site.13 Therefore, the fragmentation pattern of the z• ions is highly dependent on the partitioning of various tautomers. For example, Figure 3 a shows the CID spectrum of z1• species derived from ETD of the triply protonated peptide KGAILKGAILR. The radical site initially formed via ETD should be on the a-carbon of arginine residue in the z1• species. Losses of 72 Da and 73 Da, corresponding to the cleavage of the β-γ carbon-carbon bond and the McLafferty rearrangement, respectively, support such a structure.27 (The z1•-l species shown in Figure 3a is from the isolation of z1• ions.) However, in the CID spectrum of the z3• species from MRFA, as shown in Figure 3b, the most abundant fragment is assigned to be the a2 species from the z3• ions. The possible mechanism associated with forming the a2 ion is shown in Scheme 1, which involves a hydrogen transfer to generate the more stable radical on the benzylic group of the phenylalanine residue side chain and the subsequent cleavage of the Cα -C=O bond. Note that no detectable signals corresponding to the losses from the arginine side chain are observed in Figure 3b, although both of the two z• species should have the radical side initially on arginine residues after ETD.
Figure 3.
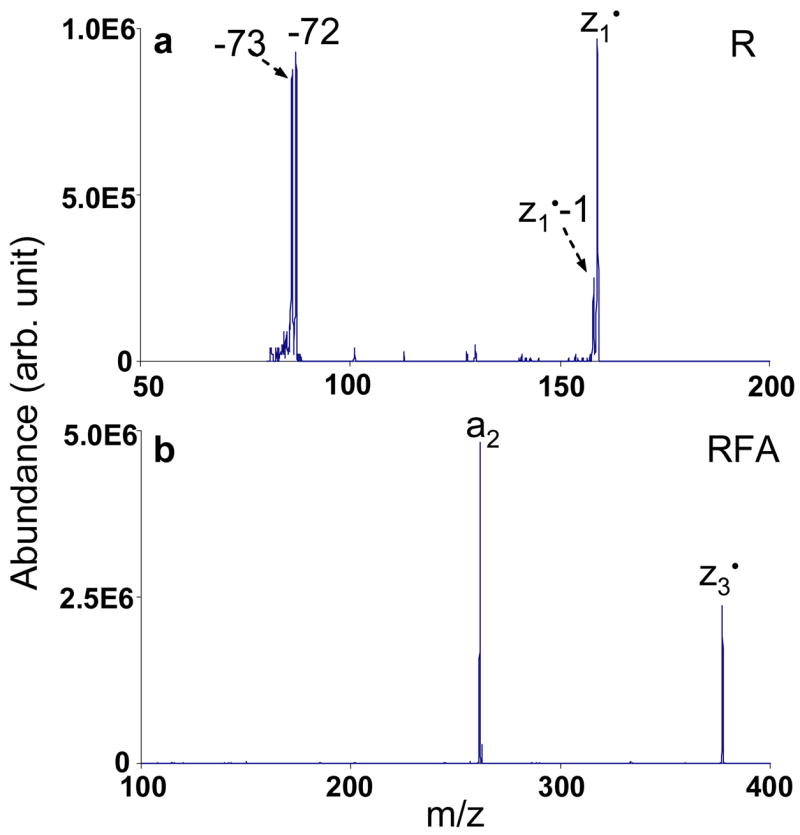
CID mass spectra of singly charged species: a) z1•ions from KGAILKGAILR (155.73 kHz, 340 mVpp); b) z3•ions from MRFA (79.77 kHz, 510 mVpp).
Scheme 1.
Fragmentation pathway of z3• ions of MRFA under ion trap CID to form a2 and x1• species.
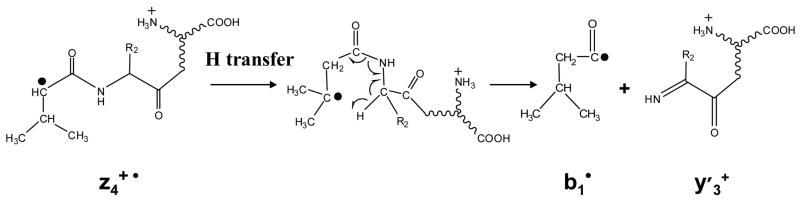
Figure 4a shows the CID spectrum of the z 4•ions formed from doubly protonated VDPVNFK. Sequential radical ions z2• and z3• are observed upon the CID of the z4• species. The formation of smaller zn• ions has also been noted in the free radical cascade process of cyclic pep tide ions.12 Note that although z2• ions are the second most abundant species among the CID products of the z4• ions, they are absent in the ETD spectrum of the VDPVNFK (see supporting material). The x2• ions are also observed, which can be explained by invoking a mechanism to form a and x•species similar to that shown in Scheme 1. A peak corresponding to the formation of y3′ions is also present. The possible mechanism for forming y′ ions and the related b• species involves homolytic cleavage of the amide bond due to a McLafferty rearrangement, as shown in Scheme 2. The y′ ions formed through this radical driven fragmentation process have different structures from the y ions typically observed in CID of protonated peptides and are 2 Da lower in mass. The reason that y3′ ions are observed might also be related to the favorable structure with radical site on the tertiary carbon of the valine side chain, which then can subsequently undergo McLafferty rearrangement to form the b• and y′ species.
Figure 4.
CID mass spectra of singly charged species: a) z4• ions from VDPVNFK (92.17 kHz, 540 mVpp); b) z4• ions from methyl esterified VDPVNFK (72.40 kHz, 390 mVpp).
Scheme 2.
Fragmentation pathway of z4• ions of VDPVNFK under ion trap CID to form b1• and y3′ species.
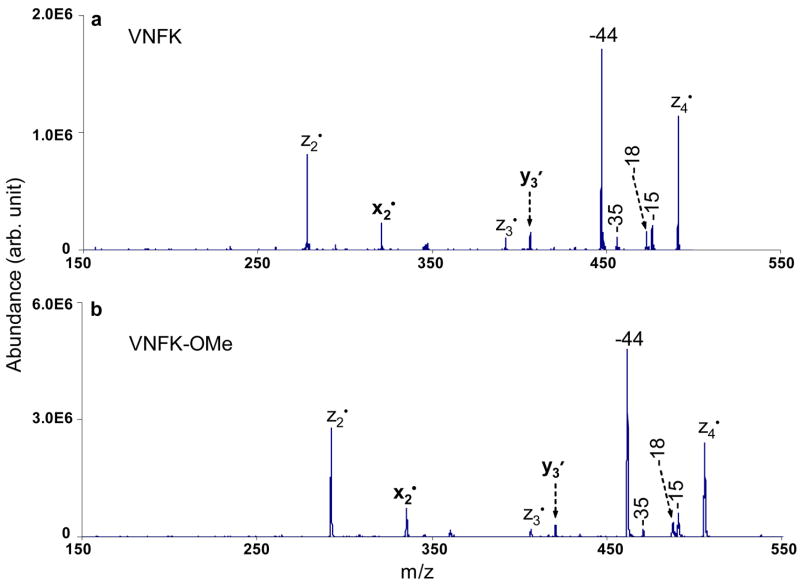
In addition to the radical driven backbone cleavages, side-chain losses from z4• ions are also observed. The loss of 15 Da from the z4• species is most likely to be the side-chain loss from valine (•CH3). The 18 Da and 35 Da losses correspond to a water loss and a combined loss of H2O and NH3, respectively. The loss of 44 Da is the most abundant peak among the CID products of z4• ions, which can have two possible origins, the loss of •CONH2 (theoretical mass: 44.0136 Da) from the asparagine side chain and the loss of CO2 (theoretical mass: 43.9898) from C-terminal carboxylic acid. To clarify the origin of the loss of 44 Da, the peptide VDPVNFK was methyl esterified to block the potential CO2 loss. The CID spectrum of the z4• species formed from ETD of doubly protonated methyl esterified VDPVNFK with azobenzene radical anions is shown in Figure 4b, which is very similar to the CID spectrum of the unmodified version shown in Figure 4a, in terms of the fragmentation channels and the relative product abundances. In Figure 4b, the 44 Da loss gives rise to the most abundant CID product of methyl esterified z4• ions, which indicates that the side-chain loss from asparagine (•CONH2) is the major contributor to the loss of 44 Da. Accurate mass measurements (measured loss = 44.0145 Da) taken on a quadrupole/time-of-flight instrument confirms the asparagine side-chain loss (•CONH2) upon the CID of the z4• ions as well (data not shown).
Figure 5 compares the fragmentation patterns of the z6• ions from VDPVNFK and methyl esterified VDPVNFK upon CID. The loss of 44 Da is the dominant process among the few noted in the CID of the z6• species from VDPVNFK, as shown in Figure 5a. The loss of 44 Da could arise from three sources: the side chain of aspartic acid (CO2), the side chain of asparagine (•CONH2) and the C-terminus (CO2). CID of z6• ions from methyl esterified VDPVNFK (Figure 5b) shows that the 44 Da loss remains the dominant process, which is due to the side-chain loss from the asparagine residue. Contributions from CO2 loss from the aspartic acid residue and the C-terminus cannot be precluded in the unmodified z6• species based on these results, however. The greatest contrast between Figures 5a and 5b is that much more extensive fragmentation is observed after methyl esterification, as exemplified by the appearance of fragment ions due to backbone cleavages: zn• ions, an ions, bn/bn• ions, xn/xn• ions and yn′ ions (2<n<6). The reason why more structural information is apparent from CID of the methyl esterified Zn• ions is not clear. However, one possibility is that CO2 loss does contribute somewhat to the fragmentation of the unmodified species and the preclusion of CO2 loss channels resulting from methyl esterification allows other dissociation channels to make greater relative contributions.
Figure 5.
CID mass spectra of singly charged species: a) z6• ions from VDPVNFK (85.68 kHz, 370 mVpp); b) z6• ions from methyl esterified VDPVNFK (73.96 kHz, 330 mVpp).
Distinguishing Leucine and Isoleucine Residues
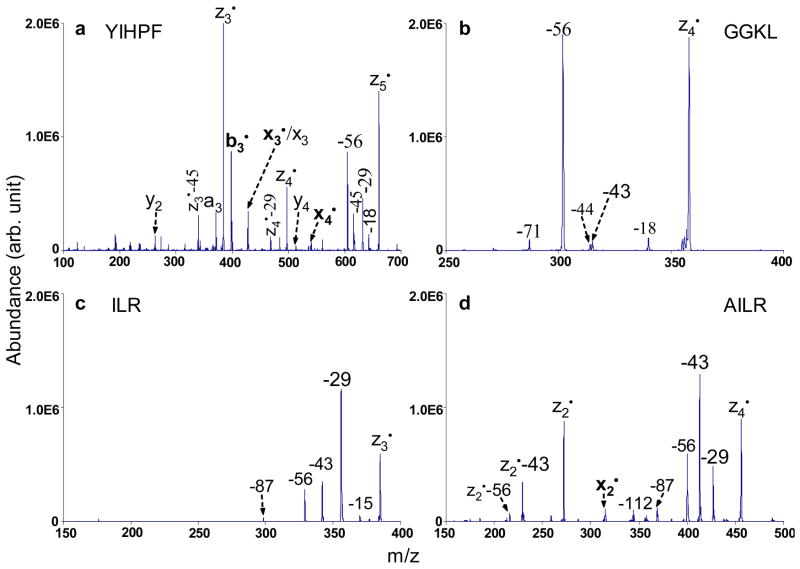
The isomeric amino acids leucine and isoleucine in polypeptides have been successfully determined from specific side-chain losses via high energy CID28 and hot BCD14,15. In our study of the collisional activation of a series of zn• ions, we also observed characteristic side-chain losses from leucine and isoleucine residues, which should enable facile determination of these two isomeric residues in polypeptides. Figure 6 compares the CID spectra derived from four different z• ions containing either isoleucine, leucine or both residues.
Figure 6.
CID mass spectra of singly charged species: a)z5• ions from DRVYIHPF (68.23 kHz, 300 mVpp), b) z4• ions from KGAGGKGAGGKL (84.13 kHz, 340 mVpp), c) z3• ions from KGAILKGAILR (118.64 kHz, 380 mVpp), and d) z4• ions from KGAILKGAILR (99.51 kHz, 410 mVpp).
Figure 6a summarizes the CID results for the z5• ions derived from doubly protonated DRVYIHPF. The characteristic side-chain losses from isoleucine are present as the loss of 29 Da (•CH2CH3) and the loss of 56 Da (CH3CH=CHCH3) together with backbone cleavage products such as an, xn, yn, bn•, xn• and zn• ions. Note that b3• and y2 ions are formed due to the amide bond cleavage N-terminal to the proline residue, which is driven by charge directed fragmentation. However, if only leucine is contained in a z• species, an odd-electron side-chain loss of 43 Da (•CH(CH3)2) or an even-electron side-chain loss of 56 Da (CH2=C(CH3)2) or both is observed in the ion trap CID spectra of the zn• ions. One example can be found in Figure 6b, which shows the CID spectrum of z4• ions generated from the peptide KGAGGKGAGGKL. In Figure 6b, the most abundant product peak corresponds to the loss of 56 Da, while the product due to loss of 43 Da is less abundant. Other neutral losses such as H2O (18 Da) loss, CO2 (44 Da) loss from the C-terminus and side-chain loss from lysine (71 Da) are also present. Figure 6c shows the CID spectrum of the z3• ions generated from the triply protonated peptide KGAILKGAILR. The peaks corresponding to the losses of 29 Da, 43 Da and 56 Da indicate that leucine and isoleucine residues are both present in the z3• species. The loss of 29 Da, which is unique to the isoleucine residue, has the greatest abundance in Figure 6c as compared to that in Figure 6a. A possible explanation might be that when the isoleucine residue is present at the cleavage site from initial ETD, more of the z3• ions (from KGAILKAILR) might have the radical site on isoleucine to induce its side-chain loss as compared to z5• ions (from DRVYIHPF) where a hydrogen abstraction step is needed before the isoleucine side-chain loss. It is interesting to note that when leucine and isoleucine are both present in the z• ions but neither of them is at the ETD cleavage site, the loss of 43 Da from leucine usually has higher abundance than the loss of 29 Da from isoleucine under ion trap CID conditions. This is probably due to the fact that forming •CH(CH3)2 is thermodynamically more favorable than forming •CH2CH3. One example is shown in Figure 6d. The losses of 29 Da, 43 Da and 56 Da from z4• ions (from KGAILKGAILR) in Figure 6d indicate the presence of leucine and isoleucine residues, with the loss of 43 Da as the most abundant peak. The z2• species are also observed from CID of z4• ions, together with the characteristic 43 Da and 56 Da leucine side-chain losses from the z2• species. It is quite clear that only leucine is contained in the z2• species. Since the 29 Da loss from z4• is less abundant than the 43 loss, isoleucine is less likely at the cleavage site, which places it in the third position from C-terminus. The loss of 112 Da arises from a combination of two even-electron 56 Da species from both leucine and isoleucine side chains. The loss of 87 Da has been attributed by Fung et al. to the loss of CH3CH2NHC(NH2)=NH from the side chain of the arginine residue.16 Therefore, leucine and isoleucine residues can be identified and differentiated by characteristic side-chain losses upon CID of z• species formed via ion/ion electron transfer.
Characteristic Cleavages of z• Ions Containing Proline or Serine Residues
The N-Cα cleavage at the proline residue is not usually observed under low-energy ECD or ETD conditions because two bonds need to be cleaved since proline is a cyclic amino acid. The characteristic cleavage of proline-containing z• ions is the C-N linkage at the N-terminal side of the proline, producing y′ or y species containing proline residue. For example, the y3′ species were observed in both the ion trap CID spectra of z4• and z5• ions of RAAGFSPFR. And z6• ions of both unmodified and methyl esterified VDPVNFK produced y5′ species upon ion trap CID. The formation of y′ species is radical-directed andthe mechanism is similar as the one shown in scheme 2. The formation of y species is charge-directed and less common in this study, only observed in the CID spectrum of z5• ions of DRVYIHPF (the presence of y2 species in Fig. 6a).
Besides the characteristic side-chain losses of 17 Da (•OH) and 30 Da (H2C=O) from serine residue when serine-containing z• species were subjected to ion trap CID, the N-Ca linkage at the N-terminal side of the serine was also cleaved, which produced even-electron z•-l species (denoted as z). For example, z5• ions of RAAGFSPFR and z8• ions of SAPRATISHYLMGG produced z4 species and z7 species under ion trap CID, respectively (data not shown). The proposed mechanism for the formation of z4 species under ion trap CID of z5• ions of RAAGFSPFR is shown in scheme 3.
Scheme 3.
Fragmentation pathway of z5• ions of RAAGFSPFR under ion trap CID to form c1• and z4 species.
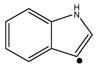
The fragmentation patterns of the forty-four z• ions under ion trap collisional activation are summarized in Table 1 and the characteristic side-chain losses are listed in Table 2. Valine, isoleucine, leucine, serine, threonine, cysteine, methionine, glutamic acid, asparagine, glutamine, tryptophan and lysine residues were found to lose odd-electron fragments, i.e., 15 Da (•CH3), 29 Da (•CH2CH3), 43 Da (•CH(CH3)2), 17 Da (•OH), 15 Da (•CH3) and 17 Da (•OH), 33 Da (•SH), 47 Da (•SCH3) and 61 Da (•CH2SCH3), 59 Da (•CH2COOH), 44 Da (•CONH2), 58 Da (•CH2CONH2), 116 Da (
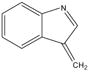
 ) and 58 Da (•CH2CH2CH2NH2) from z• species, respectively. The elimination of an odd-electron side chain from z• species is associated with the α-carbon radical center bearing the side chain and the β-γ carbon-carbon bond cleavage, resulting in the formation of an α-β carbon-carbon double bond. In addition to the loss of odd-electron species, the loss of even-electron species from labile z• fragments are also observed upon CID of z• species. Isoleucine, leucine, serine, methionine, aspartic acid, glutamine, tryptophan and lysine residues were found to lose even-electron fragments, i.e., 56 Da (CH3CH=CHCH3), 56 Da (CH2=CH(CH3)2), 30 Da (H2C=O), 74 Da (CH2=CHSCH3), 44 Da (CO2), 71 Da (CH2=CHCONH2), 129 Da (
) and 58 Da (•CH2CH2CH2NH2) from z• species, respectively. The elimination of an odd-electron side chain from z• species is associated with the α-carbon radical center bearing the side chain and the β-γ carbon-carbon bond cleavage, resulting in the formation of an α-β carbon-carbon double bond. In addition to the loss of odd-electron species, the loss of even-electron species from labile z• fragments are also observed upon CID of z• species. Isoleucine, leucine, serine, methionine, aspartic acid, glutamine, tryptophan and lysine residues were found to lose even-electron fragments, i.e., 56 Da (CH3CH=CHCH3), 56 Da (CH2=CH(CH3)2), 30 Da (H2C=O), 74 Da (CH2=CHSCH3), 44 Da (CO2), 71 Da (CH2=CHCONH2), 129 Da (
 ) and 71 Da (CH2=CHCH2CH2NH2) from z• species, respectively. The loss of 87 Da (CH3CH2NHC(NH)NH2) or 99 Da (CH2=CHCH2NHC(NH)NH2) or both is observed from z• species containing arginine residue upon CID. The possible mechanism for loss of even-electron species has two steps: (1) abstraction of a γ-hydrogen from the side chain by its a-carbon radical center; (2) the cleavage of the α-β carbon-carbon single bond and formation of a β-γ carbon-carbon double bond. After the loss of even-electron species, the radical center migrates back to the α-carbon. When z• species are subjected to CID, the characteristic odd- and even-electronside-chain losses from certain amino acid residues are still observed even when these residues are not at the initial cleavage sites from ETD because of the migration of radical center through hydrogen abstraction along the peptide backbone of z• species. Both odd-electron and even-electron side-chain losses appear to be due to radical-directed fragmentation pathways.
) and 71 Da (CH2=CHCH2CH2NH2) from z• species, respectively. The loss of 87 Da (CH3CH2NHC(NH)NH2) or 99 Da (CH2=CHCH2NHC(NH)NH2) or both is observed from z• species containing arginine residue upon CID. The possible mechanism for loss of even-electron species has two steps: (1) abstraction of a γ-hydrogen from the side chain by its a-carbon radical center; (2) the cleavage of the α-β carbon-carbon single bond and formation of a β-γ carbon-carbon double bond. After the loss of even-electron species, the radical center migrates back to the α-carbon. When z• species are subjected to CID, the characteristic odd- and even-electronside-chain losses from certain amino acid residues are still observed even when these residues are not at the initial cleavage sites from ETD because of the migration of radical center through hydrogen abstraction along the peptide backbone of z• species. Both odd-electron and even-electron side-chain losses appear to be due to radical-directed fragmentation pathways.
Table 1.
Summary of fragmentation patterns of z• ions. The amino acid residues with side-chain losses are indicated with ▲.

|
Table 2.
Losses of odd-electron and even-electron species from amino acid side chains upon CID of z• ions.
| Ammo acids | Odd-electron losses (Da) | Assignment | Even-electron losses (Da) | Assignment |
|---|---|---|---|---|
| Valine (V) | 15 | •CH3 | - | - |
| Isoleucine (I) | 29 | •CH2CH3 | 56 | CH3CH=CHCH3 |
| Leucine (L) | 43 | •CH(CH3)2 | 56 | CH2=CH(CH3)2 |
| Serine (S) | 17 | •OH | 30 | H2C=0 |
| Threonine (T) | 15,17 | •CH3, OH | - | - |
| Cysteine (C) | 33 | •SH | - | - |
| Methionine (M) | 47,61 | •SCH3, •CH2SCH3 | 74 | CH2=CHSCH3 |
| Aspartic acid (D) | - | - | 44 | O=C=O |
| Glutamic acid (E) | 59 | •CH2COOH | - | - |
| Asparagine (N) | 44 | •CONH2 | - | - |
| Glutamine (Q) | 58 | •CH2CONH2 | 71 | CH2=CHCONH2 |
| Tryptophan (W) | 116 |
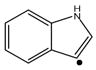
|
129 |
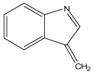
|
| Lysine (K) | 58 | •CH2CH2CH2NH2 | 71 | H2C=CHCH2CH2NH2 |
| Arginine (R) | - | - | 99 | H2C=CHCH2NHC(NH)NH2 |
| 87 | CH3CH2NHC(NH)NH2 |
The mass of the side-chain loss is rounded to its nominal mass.
It is noteworthy that the neutral losses from zn• ions observed via ion trap collisional activation in this study are quite different from the neutral losses in the (M•-X) region reported in ETD11 or BCD10 processes. In an ETD or ECD spectrum, the neutral losses from parent ions are most frequently associated with the charged residues, where the electron is initially transferred to or captured, e.g. N-terminus, arginine side chain, lysine side chain, histidine side chain. However, for the CID of zn• ions, the radical site is expected to be present at a different location initially. Furthermore, the ion trap collisional activation experiment takes place over a much longer time-frame, in the tens of ms time scale, allowing structural rearrangements of the initially formed z• ions. The radical site therefore, can be scrambled to the sites with more favorable structures. For the CID of zn• ions containing arginine, e.g. zn• from KGAILKGAILR, no 44 Da ((NH2)HC=NH), 59 Da ((NH2)2C=NH) or 101 Da losses are observed, while all are present in the ETD spectrum of triply protonated KGAILKGAILR.11 On the other hand, the side-chain losses from amino acid residues with alkyl chains upon CID of zn• ions are quite abundant, as shown in Figures 6c and 6d, while they are absent from the parent ion losses in ETD or ECD.
Conclusions
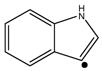
In this study, the fragmentation patterns of c and z• ions formed via ETD are systematically investigated using ion trap collisional activation. Ion trap CID of c ions gives rise to a-, b-, y-type fragment ions and ammonia losses from these ions, showing similar charge-directed fragmentation pathways as singly protonated C-terminal amidated peptides. Side-chain losses and amino acid backbone cleavages are commonly observed upon CID of z• ions. The fragmentation of the z• ions is mostly dominated by radical-driven processes, although charge-directed fragmentation is also noted. Certain amino acids generate their characteristic side-chain losses, including valine, leucine, isoleucine, serine, threonine, cysteine, methionine, aspartic acid, glutamic acid, asparagine, glutamine, tryptophan, lysine and arginine residues. A strong signal of 44 Da loss (•CONH2) from the z• species of methyl esterified peptides indicates the presence of asparagine residue, although the presence of aspartic acid residue cannot be excluded from unmodified z• species. Leucine/isoleucine isomers can be successfully identified and distinguished by their diagnostic side-chain losses, 43 Da (•CH(CH3)2) and 29 Da (•CH2CH3), respectively, while the loss of 56 Da can predict the presence of either leucine or isoleucine residue. Theloss of 58 Da or 71 Da or both demonstrates that glutamine or lysine or both can be present. The loss of 30 Da (H2C=O) is diagnostic to indicate the presence of serine. The 33 Da loss (•SH) is characteristic to identify the presence of cysteine. Ion trap CID of methionine-containing z• ions usually gives rise to abundant peaks corresponding to the loss of 61 Da (•CH2SCH3) and 74 Da (CH2=CHSCH3). In some cases, the loss of 47 Da (•SCH3) is also observed. Therefore, these characteristic side-chain losses from methionine residue can be used to confirm its presence. The loss of 116 Da (
 ) or 129 Da (
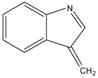
) or 129 Da (
 ) or both is used to identify the presence of tryptophan residue. The observation of the 87 Da loss (CH3CH2NHC(NH)NH2) or 99 Da loss (H2C=CHCH2NHC(NH)NH2) or both losses indicates the presence of arginine residue. Peptide backbone cleavages observed in the collisional activation of z• ions may provide more structure information for peptide characterization. For example, the characteristic cleavages of z• ions containing proline or serine residue could be used to further confirm the presence of these amino acid residues in the peptide/protein sequence.
) or both is used to identify the presence of tryptophan residue. The observation of the 87 Da loss (CH3CH2NHC(NH)NH2) or 99 Da loss (H2C=CHCH2NHC(NH)NH2) or both losses indicates the presence of arginine residue. Peptide backbone cleavages observed in the collisional activation of z• ions may provide more structure information for peptide characterization. For example, the characteristic cleavages of z• ions containing proline or serine residue could be used to further confirm the presence of these amino acid residues in the peptide/protein sequence.
Supplementary Material
Acknowledgments
This research was sponsored by the National Institute of General Medical Sciences, under Grant GM 45372.
References
- 1.Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 2.Wells JM, McLuckey SA. Collision–induced dissociation (CID) of peptides and proteins. Methods Enzymol. 2005;402:148–85. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 3.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptide to yield complementary sequence information. Anal Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 4.Mirgorodskaya E, Roepstorff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a fourier transform mass spectrometer. Anal Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 5.Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh L, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 6.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 7.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4:628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Nat Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper HJ, Hudgins RR, Hakansson K, Marshall AG. Characterization of amino acid side chain losses in electron capture dissociation. J Am Soc Mass Spectrom. 2002;13:241–249. doi: 10.1016/S1044-0305(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 10.Haselmann KF, Budnik BA, Kjeldsen F, Polfer NC, Zubarev RA. Can the (M•-X) region in electron capture dissociation provide reliable information on amino acid composition of polypeptides? Eur J Mass Spectrom. 2002;8:461–469. doi: 10.1255/ejms.555. [DOI] [PubMed] [Google Scholar]
- 11.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Electron transfer ion/ion reactions in a three-dimensional quadrupole ion trap: Reactions of doubly and triply protonated peptides with SO2•−. Anal Chem. 2005;77:1831–1839. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leymarie N, Costello CE, O’Connor PB. Electron capture dissociation initiates a free radical reaction cascade. J Am Chem Soc. 2003;125:8949–8958. doi: 10.1021/ja028831n. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor PB, Lin C, Cournoyer JJ, Pittman JL, Belyayev M, Budnik BA. Long-lived electron capture dissociation product ions experience radical migration via hydrogen abstraction. J Am Soc Mass Spectrom. 2006;17:576–585. doi: 10.1016/j.jasms.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Kjeldsen F, Haselmann KF, Budnik BA, Jensen F, Zubarev RA. Dissociative capture of hot (3–13 eV) electrons by polypeptide polycations: an efficient process accompanied by secondary fragmentation. Chem Phys Lett. 2002;356:201–206. [Google Scholar]
- 15.Kjeldsen F, Haselmann KF, Sorensen ES, Zubarev RA. Distinguishing of Ile/Leu amino acid residues in the PP3 protein by (hot) electron capture dissociationin Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2003;75:1267–1274. doi: 10.1021/ac020422m. [DOI] [PubMed] [Google Scholar]
- 16.Fung YME, Chan TWD. Experimental and theoretical investigations of the loss of amino acid side chains in electron capture dissociation of model peptides. J Am Soc Mass Spectrom. 2005;16:1523–1535. doi: 10.1016/j.jasms.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Savitski MM, Nielsen ML, Zubarev RA. Side-chain losses in electron capture dissociation to improve peptide identification. Anal Chem. 2007;79:2296–2302. doi: 10.1021/ac0619332. [DOI] [PubMed] [Google Scholar]
- 18.Reid GE, Simpson RJ, O’Hair RAJ. A mass spectrometric and ab initio study of the pathways for dehydration of simple glycine and cysteine-containing peptide [M+H]+ ions. J Am Soc Mass Spectrom. 1998;9:945–956. [Google Scholar]
- 19.Bibbs JA, Lehman De Gaeta LS, Jones H. U. S. Patent 5503989. Production of peptide amides. April 2, 1996
- 20.Hager JW. A new linear ion trap mass spectrometer. Rapid Commun Mass Spectrom. 2002;16:512–526. doi: 10.1002/rcm.1020. [DOI] [PubMed] [Google Scholar]
- 21.Liang X, Xia Y, McLuckey SA. Alternately pulsed nanoelectrospray ionization/atmospheric pressure chemical ionization for ion/ion reactions in an electrodynamic ion trap. Anal Chem. 2006;78:3208–12. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shevchenko A, Chernushevich L, Ens W, Standing KG, Thomson B, Wilm M, Mann M. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1997;11:1015–1024. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Xia Y, Chrisman PA, Erickson DE, Liu J, Liang X, Londry FA, Yang MJ, McLuckey SA. Implementation of ion/ion reactions in a quadrupole/time-of-flight tandem mass spectrometer. Anal Chem. 2006;78:4146–54. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Londry FA, Hager JW. Mass selective axial ion ejection from a linear quadrupole ion trap. J Am Soc Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 25.Mouls L, Subra G, Aubagnac JL, Martinez J, Enjalbal C. Tandem mass spectrometry of amidated peptides. J Mass Spectrom. 2006;41:1470–83. doi: 10.1002/jms.1118. [DOI] [PubMed] [Google Scholar]
- 26.McLuckey SA, Goeringer DE. Slow heating methods in tandem mass spectrometry. J Mass Spectrom. 1997;32:461–474. [Google Scholar]
- 27.Xia Y, Chrisman PA, Pitteri SJ, Erickson DE, McLuckey SA. Ion/molecule reactions of cation radicals formed from protonated polypeptides via gas-phase ion/ion electron transfer. J Am Chem Soc. 2006;128:11792–11798. doi: 10.1021/ja063248i. [DOI] [PubMed] [Google Scholar]
- 28.Johnson RS, Martin SA, Biemann K, Stults JT, Watson JT. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass-spectrometer - Differentiation of leucine and isoleucine. Anal Chem. 1987;59:2621–2625. doi: 10.1021/ac00148a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.