Abstract
It has been sixty years since the Millers first described the covalent binding of carcinogens to tissue proteins. Protein covalent binding was gradually overshadowed by the emergence of DNA adduct formation as the dominant paradigm in chemical carcinogenesis, but re-emerged in the early 1970s as a critical mechanism of drug and chemical toxicity. Technology limitations hampered the characterization of protein adducts until the emergence of mass spectrometry-based proteomics in the late 1990s. The time since has seen rapid progress in the characterization of the protein targets of electrophiles and the consequences of protein damage. Recent integration of novel affinity chemistries for electrophile probes, shotgun proteomics methods and systems modeling tools has led to the identification of hundreds of protein targets of electrophiles in mammalian systems. The technology now exists to map the targets of damage to critical components of signaling pathways and metabolic networks and to understand mechanisms of damage at a systems level. The implementation of sensitive, specific analyses for protein adducts from both xenobiotic-derived and endogenous electrophiles offers a means to link protein damage to clinically relevant health effects of both chemical exposures and disease processes.
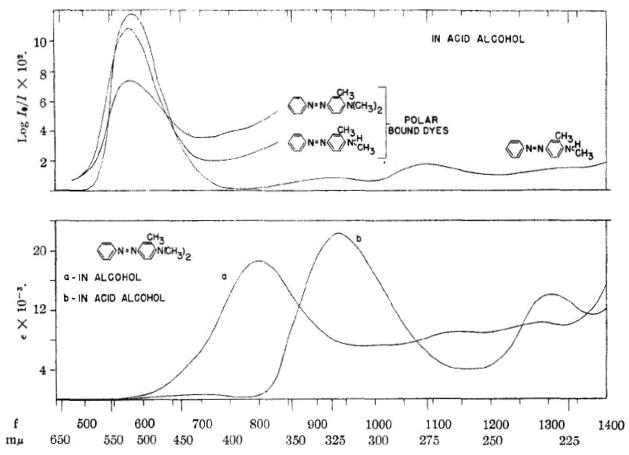
The covalent modification of proteins by xenobiotics was first reported sixty years ago by Elizabeth and James Miller, who observed yellow pigment irreversibly bound to liver proteins in rats treated with methylaminoazobenzene carcinogens (1). Figure 1 depicts visible spectra of the chromophores of the carcinogens 3-methyl-4-dimethylaminoazobenzene and its N-monomethyl derivative in solution and of the yellow pigments bound to protein following administration in vivo—the first covalent binding analysis (2). This initial observation was followed by several reports that characterized protein binding by polycyclic aromatic hydrocarbons and other azo dye carcinogens, as described in an early review by the Millers (3). Soon thereafter, characterization of the structure and function of DNA as genetic material and the discovery of carcinogen-DNA adducts in the early 1960s (4, 5) established a DNA damage/mutation/carcinogenesis paradigm that dominated the field for the next forty years. Interest in protein covalent binding by carcinogens gradually disappeared and did not resurface until twenty years later, when studies by Ehrenberg, Tannenbaum and Farmer employed adducts to hemoglobin as biomarkers of carcinogen exposure (6–9).
Figure 1.
Visible spectra of the carcinogens 3-methyl-4-dimethylaminoazobenzene, its N- monomethyl derivative and the protein-bound chromophores formed in liver following administration of the same compounds to rats in vivo. From reference (2).
The idea that protein covalent binding contributed to the toxicity of drugs and other chemicals emerged in the early 1970s from the laboratories of Brodie, Gillette and Reynolds who measured covalent binding of radiolabeled hepatic toxicants to liver proteins following treatment of rats and mice in vivo (10, 11). A classic series of papers published in 1973 by Jollow, Mitchell, Gillette and colleagues described the role of metabolism and covalent binding in acetaminophen hepatotoxicity (12–15). Key findings from this work were that microsomal enzymes metabolized acetaminophen to a reactive metabolite that covalently bound to microsomal proteins in vitro and that in vivo metabolism and covalent binding of the drug were tightly correlated. Moreover, covalent binding of the acetaminophen metabolite was blocked by reaction with hepatic glutathione, in what emerged as a critical detoxication process. These studies established the key elements of the covalent binding/toxicity paradigm, which has been extended to account for the toxicity of diverse xenobiotics.
The covalent binding hypothesis of chemical toxicity offered an appealing, broadly applicable mechanism to explain the toxicities of structurally diverse xenobiotics. However, the covalent binding hypothesis had two major problems. First, not all covalent binding necessarily leads to toxicity, as studies with nontoxic acetaminophen analogs were able to demonstrate covalent binding in the absence of toxicity (16, 17). The lack of a clear causal relationship between binding and toxicity complicates the interpretation of covalent binding data (18, 19). This does not necessarily mean that binding is unimportant, but simply that the relationship between binding and toxicity mechanisms is not simple. Second, the covalent binding hypothesis was untestable at the level of specific biochemical and chemical mechanisms, because it was impossible to identify protein targets of reactive electrophiles with the analytical technologies of the 1970s and early 1980s. Protein covalent binding was measured (and still is in most cases) as chemical-derived radiolabel bound to bulk tissue or cell protein (18). This analysis only allows overall trends in bioactivation and covalent binding to be estimated across all affected proteins, but indicates nothing about specific targets. In contrast, the technology for chemical characterization of specific nucleic acid adducts was much more advanced and contributed substantially to the validation of the DNA damage hypothesis of chemical carcinogenesis. This disparity in analytical methodology inevitably contributed to widespread ambivalence regarding the importance and relevance of protein adduction in toxicity and carcinogenesis. Nevertheless, analysis by relatively crude radiochemical assay of covalent binding to proteins in both in vitro and in vivo models has been widely used to identify compounds with significant potential to exert toxicity via bioactivation (18).
The above synopsis describes the understanding of protein covalent binding and its significance in chemical toxicity twenty years ago, when Chemical Research in Toxicology was founded. Since then, advances in analytical technology for immunochemistry, affinity labeling, MS1-based proteomics, definition of specific cell-signaling pathways associated with stress and the integrative tools of bioinformatics have revolutionized the field. The following sections describe the major developments in the analysis and significance of protein damage by reactive electrophiles.
The terms “protein covalent binding” and “protein adduction” have been widely used to denote protein covalent modifications generated by electrophiles, typically from xenobiotics. However, the scope of this phenomenon is broader than its original context of protein adducts formed by toxic drugs and carcinogens and is now understood to encompass modifications by endogenously generated electrophiles resulting from oxidative stress, glycation and related processes. Moreover, the changes induced may include formation of adducts with electrophiles, but also protein oxidations produced by reactive oxygen and nitrogen species. In addition, stress may perturb the distribution of endogenous regulatory modifications to proteins, including phosphorylation, acylation, methylation, ubiquitination and sumoylation (20–23). The modification of proteins by endogenously generated reactive intermediates is now thought to be a key element in the molecular pathology of diseases involving inflammation and oxidative damage, including atherosclerosis, neurodegenerative diseases, diabetes and associated metabolic diseases, asthma and related pulmonary diseases and cancer. Thus, damage by xenobiotic-derived electrophiles associated with toxicity and endogenous electrophiles and oxidants associated with disease represents essentially the same phenomenon. This mechanistic link between molecular toxicology and the molecular pathology of disease underscores the potential of fundamental research in toxicology to impact human health. In this article, I use the term “protein damage” to refer broadly to covalent protein adduction and oxidation and the more specific term “protein adduction” to refer to electrophile-derived covalent modifications.
Analysis of protein damage: an analytical challenge
The analysis of protein damage is complicated by two major problems. First, many different proteins are modified by reactive intermediates in cell and tissue proteomes. This was evident in the first reported electrophoretic studies of liver proteins modified by yellow pigment in azo-dyed treated rats and has been observed by similar approaches with many other protein modifications. Second, modified proteins apparently make up only a small fraction of proteins present in cell and tissue extracts. This reflects both the relatively low stoichiometry of protein modification (in most cases) and the diversity of protein targets. Often described as a “needle in a haystack” problem, analyzing protein damage is really more like looking for dozens of needles in thousands of haystacks. Thus, global analysis of protein covalent binding requires tools for affinity enrichment of specific adducted or modified forms and identification methods capable of resolving and detecting anywhere from dozens to thousands of different modified species. The evolution of approaches to detect and identify protein adducts is described in the following sections. The examples cited are illustrative of the approaches and their applications, but do not constitute a comprehensive literature review. Unfortunately, there is no comprehensive database resource for all data on protein modification by various reactive intermediates. However, Hanzlik and colleagues have implemented a useful database restricted to xenobiotics and their reported protein targets (see http://tpdb.medchem.ku.edu:8080/protein_database/) (24).
Immunochemical detection and analysis of protein adducts
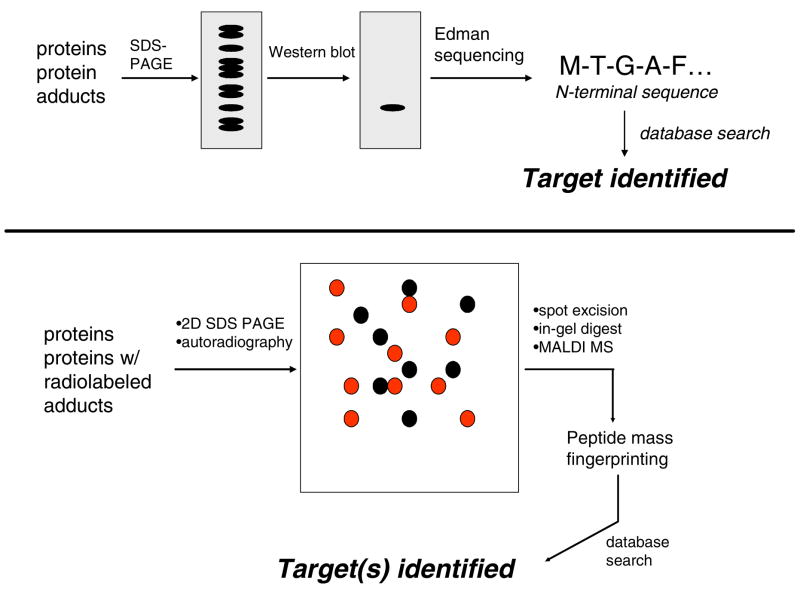
Initial progress in the molecular characterization of protein damage came from application of immunochemical methods in the 1980s and early 1990s. Satoh, Pohl and colleagues first reported that antibodies raised against Nε-trifluoroacetyllysine recognized hepatocytes and specific hepatocyte proteins from rats treated with the inhalation anesthetic halothane (25, 26). Trifluoroacetylation is a covalent modification produced by a reactive metabolite of halothane. Sera from humans with halothane-induced hepatitis recognized multiple ER proteins, including a trifluoroacetylated carboxylesterase (27). The antibodies against trifluoroacetylated proteins were used in Western blot analyses to identify numerous proteins modified by halothane or other trifluoroacetylating chemicals in vivo and in vitro (28–30). This approach was extended to analysis of other protein adducts by the laboratories of Hinson, Cohen, Pumford and several other groups to identify putative protein targets of covalent adduction by xenobiotic metabolites (Figure 2). Approximately 30 putative targets of about a half dozen electrophiles were identified in this manner (31–41). This body of work represented tremendous efforts by these groups in antibody development and target identification. Nevertheless, there were drawbacks to the approach. The antibodies used lacked the affinity for efficient immunoenrichment of adducts and were used primarily for detection by Western blotting. Proteins in immunoreactive bands were identified by Edman sequencing or peptide mass fingerprinting by MS. The problem with this approach is that these analyses did not actually detect adducts, but instead identified proteins that co-migrated with immunoreactivity. The use of 1D SDS-PAGE to resolve complex protein mixtures also meant that immunoreactive bands inevitably contained multiple proteins, thus adding further ambiguity to the identification process. Moreover, it is nearly impossible to distinguish detection of truly adducted proteins from nonspecific reactivity of the antibodies.
Figure 2.
Detection and identification of protein adducts using SDS-PAGE and western blotting with antibodies against adducts (top). Application of 2D-SDS-PAGE to resolution and detection of protein adducts (bottom). The example depicts use or radiolabel to locate adducts, although western blotting with antibodies against adducts is also used (see text for discussion).
Proteomics approaches to identify protein targets of electrophiles: initial studies and 2D gel-based proteomics approaches
A major advance in the analysis of protein adducts was the introduction of proteomics methods in the late 1990s. Qiu et al. employed 2D-SDS-PAGE to analyze liver proteins extracted from mice treated with [14C]-acetaminophen (42). Autoradiography of the gels revealed the locations of adducted proteins and these spots were excised, the proteins were digested with trypsin and the peptides were analyzed by MALDI-MS to identify the proteins (Figure 2). This study identified 23 putative targets of acetaminophen in a single analysis, several of which were also identified in earlier studies in which antibodies were used to detect acetaminophen-derived adducts. This approach was replicated to identify protein targets of reactive metabolites of naphthalene (43–46), monocrotaline pyrrole (47, 48), and bromobenzene (49, 50). These studies also provided some degree of cross-validation, as many of the proteins identified were targets of multiple electrophiles. Williams et al. (51) applied highly sensitive AMS to detect 14C enrichment in proteins from liver and bone marrow of mice after treatment with [14C]-benzene. The overall approach was analogous to that described above, except that AMS analysis detected attomolar levels of 14C. Detection of radiolabel associated with histones in bone marrow samples led them to analyze proteolytic digests by AMS and 14C label was found to be widely distributed across the protein sequences analyzed. The authors concluded that benzene metabolite adduction was “promiscuous” with respect to the protein nucleophile targets modified.
Strictly speaking, analyses based on identifying proteins that co-migrate with radioactivity do not detect adducts. This approach was nevertheless a significant advance over the immunochemical approach to adduct detection and analysis, as the increased resolution of 2D gel separations and MS-based analyses increased both the numbers and reliability of the identifications. However, the 2D gel-based approach using radiolabeled compounds is biased toward detection of more abundant protein targets of the electrophiles. Indeed, many of the proteins detected are expressed at high abudance in cells (e.g., cytoskeletal proteins, chaperones, enzymes of intermediary metabolism). The dynamic range of the approach depends on the specific activity of the radiochemical and only protein spots containing significant amounts of radiolabel are detected by autoradiography. Despite these limitations, the application of 2D gel-based proteomics together with radiolabeling provided the first method for global discovery of the protein targets of electrophiles.
A more recently applied variation of the 2D gel-based approach involves the use of antibodies directed against adducts to detect protein targets (i.e., 2D Western blotting). Petersen and colleagues reported adduction of the heat shock proteins Hsp72 and Hsp90 by the lipid peroxidation product HNE (52, 53). These studies were undertaken in the context of a rat model of alcoholic liver disease, in which HNE and other lipid oxidation products are generated in vivo. Adduction of both proteins by HNE in vivo was detected by 2D SDS-PAGE followed by immunoblotting with anti-HNE Michael adduct antibodies. HNE-adducted retinal proteins in aging rats were identified in a similar study (54). Immunochemical detection of adducts has been combined with 2D-SDS-PAGE and MS to identify adducts formed from 2,6-di-tert-butyl-4-methylphenol (55, 56).
Another strategy combines biotin-labeled electrophile probes with 2D-SDS-PAGE. The endogenous electrophile 15-deoxy-PGJ2 has garnered considerable interest as a contributor to the effects of oxidative stress and inflammation (57). Work by Uchida and colleagues employed a biotin-tagged analog of this electrophile to identify adducts with thioredoxin 1 (58), γ-actin (59), the electrophile sensor protein keap1 (60) and the DNA damage response mediator ATM (61). The biotinylated 15-deoxy-PGJ2 probe detectably adducted at least a dozen proteins in SH-SY5Y neuroblastoma cells (59) and nine proteins in cultured MC mesangial cells (62).
Although the 2D gel-based approach increased the numbers of targets identified, the absolute numbers of targets and the information contained in these lists still was quite limited. Discussion of the results typically included a rundown of the target list and speculation about the links, if any, between the identified targets and mechanisms of toxicity. Although only a small fraction of the proteome comprised the detected electrophile targets, greater numbers of protein targets presumably went unidentified due to the limitations of the technologies employed, especially the dynamic range for immunoblotting from 2D gels. Overcoming these limitations requires much more efficient affinity capture tools and the application of shotgun proteomics technology, as described in the next two sections.
Shotgun proteomics and affinity capture
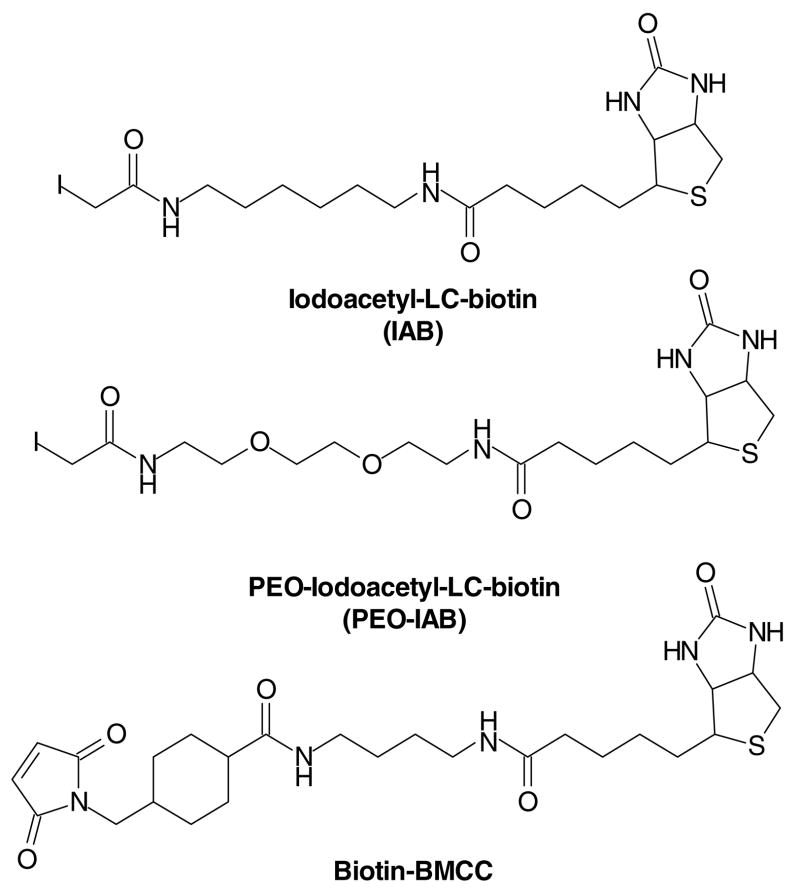
The most basic problem in analyzing protein targets of electrophiles is that the adducts represent a small fraction of all proteins present. If adducts can be selectively captured with minimal contamination by nonadducted proteins, then shotgun proteomics approaches can identify the peptides and proteins captured. It appeared to us from previous work (see above) that antibodies directed against adducts had significant limitations in both affinity and specificity. We decided instead to employ commercially available protein biotinylating agents as model electrophiles for studies with cultured cells. The three compounds we have studied, IAB, PEO-IAB and BMCC are shown in Figure 3. Our initial experiments indicated that both probes penetrate cells and react with proteins in multiple subcellular compartments. The biotin tag on alkylated proteins makes it possible to efficiently capture either alkylated proteins or peptides for shotgun LC-MS-MS analysis. This label and capture strategy is similar to the activity based proteome profiling approach developed by Cravatt and colleagues (63, 64). However, their probes contained functional groups recognized by specific enzyme classes and latent electrophiles that alkylated the bound enzymes, whereas our electrophile probes simply contain the electrophile, a linker and biotin. It is worth noting that the linker and biotin may determine the targeting of protein alkylation by these probes in some cases.
Figure 3.
Biotin electrophile probes used for shotgun LC-MS-MS analysis of cellular protein targets.
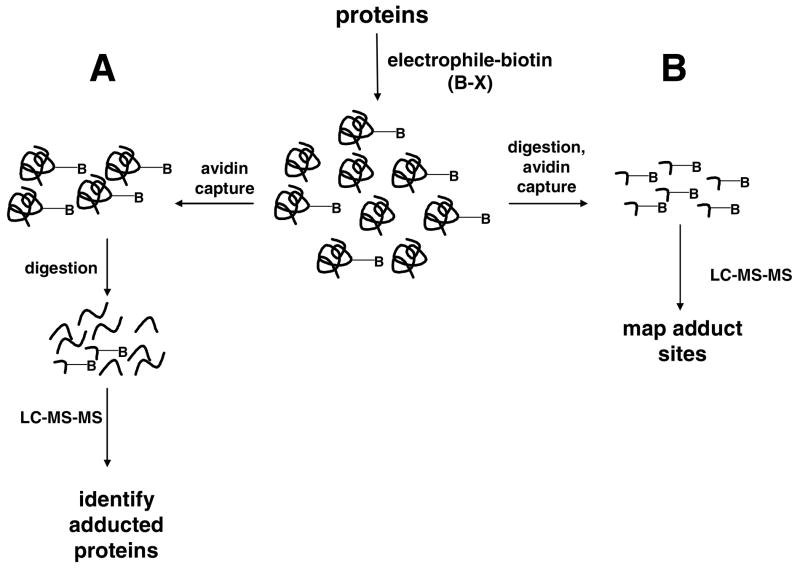
Shotgun proteomics is the analytical strategy in which complex protein mixtures are first digested to peptides and the peptides are then analyzed by LC-MS-MS to generate MS-MS spectra (65). Database search of the MS-MS spectra assigns them to peptides and reassembly of the information indicates the proteins present in the original mixture. If biotin-tagged proteins generated with the probes are analyzed, the result is an inventory of adducts—either peptides or proteins, depending on how the analysis is done (Figure 4). If a mixture of adducted and unadducted intact proteins is passed over an avidin column, adducted proteins are captured. Elution and digestion of this fraction yields mostly unadducted peptides and a few peptide adducts. LC-MS-MS analysis of this sample identifies peptides from the captured proteins, but few of these are adducts (Figure 4A). This indicates proteins that are most likely targets of the electrophile, but unless adducted peptides are detected, there is no absolute proof. The alternative approach is to digest the proteins first and then capture the adducted peptides with avidin. Analysis of the eluted peptides by LC-MS-MS primarily yields MS-MS spectra of adducted peptides. If the sequences of the peptides and the position of adduction can be established with confidence, this approach offers the benefit of unambiguously identifying both the protein target and the sequence position of the modification (Figure 4B). However, the matches of spectra to sequence must be of very high quality to establish the protein identifications, as the unmodified proteins are lost during avidin enrichment of the adducts and corroborating unadducted peptide sequences thus are not detected. Nevertheless, because even abundant protein targets are represented by only their adducted peptides, lower abundance peptide adducts are more likely to be detected and this approach is most likely to yield a broader range of high and low abundance targets.
Figure 4.
Strategies for analyses of proteins and peptide sequence targets modified by biotinylated electrophile probes. Capture of intact protein adducts (A) followed by digestion and LC-MS-MS analysis primarily yields unmodified peptides, which enables identification of protein targets, but results in few adduct identifications. Capture of adducted peptides following proteolytic digestion (B) results in an enriched adduct population for LC-MS-MS and identifies greater numbers of specific adduct sites.
We used the latter approach with biotin electrophile probes to label proteins in isolated subcellular fractions from human cells (66). This approach of labeling complex subproteomes under native conditions enabled adduction of a broad range of cell proteins that could be susceptible to damage in cells. Affinity capture of the adducted peptides following tryptic digestion yielded a sample highly enriched in peptide adducts for LC-MS-MS analysis (see strategy depicted in Figure 4B). Analysis of cytoplasmic and nuclear proteins alkylated by the probes indicated that 897 specific cysteine residues on 539 proteins were labeled by either PEO-IAB or BMCC. Surprisingly, only about 20% of the identified proteins were adducted by both PEO-IAB and BMCC, which indicates that two different thiol alkylation chemistries could target distinctly different populations of proteins.
This study represented the first large-scale characterization of proteins and protein sites targeted by electrophiles. The results provided several interesting observations. First, protein alkylation was selective and reproducible—replicate analyses indicated that the electrophile probes reproducibly targeted the same residues, thus indicating selective chemistry as opposed to random alkylation. Second, alkylation by the Michael acceptor BMCC, but not by the SN2 alkylator PEO-IAB was favored at Cys residues adjacent to Lys or Arg residues, which suggests that local structural features favor certain alkylation chemistries. Third, over 80% of all protein targets were modified at only a single Cys residue. Fourth, some protein domain and motif structures were highly susceptible to alkylation, including RNA recognition motifs and K-homology domains of RNA binding proteins. Finally, a core group of approximately 20% of proteins targeted by both electrophiles represented functions known to be highly susceptible to alkylation damage (e.g., regulation of cytoskeletal structures).
Similar studies with human liver microsomes identified 376 peptides in 263 proteins modified by the iodoacetamide IAB and the N-alkylmaleimide BMCC (67). As with cytoplasmic and nuclear protein targets, there was only a ~20% overlap in the protein and sequence targeting by the two electrophiles. An interesting feature of this study was that the two electrophile probes were shown to differ in their ability to trigger an ER stress response—only IAB induced the accumulation of the ER chaperone BiP. This observation indicated that differences in protein alkylation patterns produce different biological effects. The concomitant observation that Cys50 of microsomal glutathione-S-transferase is targeted by both probes and the quinoneimine metabolite of acetaminophen suggests that a subset of protein targets may be modified by multiple electrophile chemisties and that such alkylations may provide selective screens for reactive metabolites. Liu et al. developed a similar biotin-tagged probe containing a quinone methide structure derived from the selective estrogen receptor modifying drug raloxifene (68). This probe was used to label human microsomal proteins and the alkylated proteins, rather than peptides were captured and subjected to MS analysis. This study identified several targets, all of which were also targeted in microsomes by the IAB and BMCC probes.
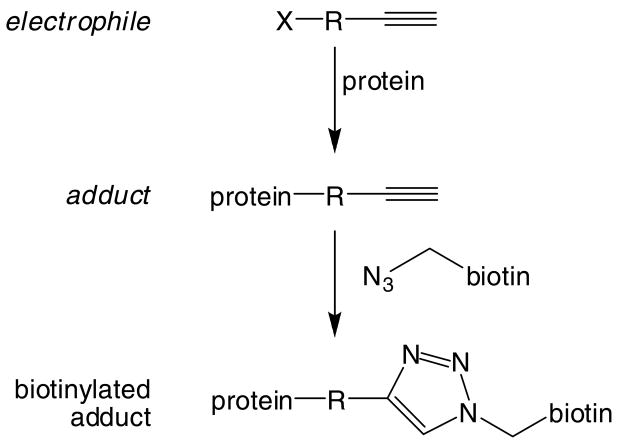
The ability of an affinity capture and shotgun proteomics strategy to identify hundreds to thousands of protein targets clearly indicates the ability of this approach to advance the global characterization of protein damage. Development of biotinylated analogs of other electrophiles could be envisioned. However, the influence of the linker and the biotin moiety, as opposed to electrophile chemistry per se on protein target selectivity is unknown. An alternative to bulky, linker/biotin probes is the application of new postlabeling strategies utilizing Click chemistry (69) (Figure 5) and the Staudinger ligation (70) to attach biotin probes to azido- or alkynyl-electrophiles. This approach has the advantage of adding the affinity tag during sample workup. The azido and alkynyl tags are not bulky and display little reactivity toward normal cellular constituents. This approach has been used successfully in an activity-based proteome profiling strategy with probes directed against serine hydrolase enzymes (71). Although this strategy requires the synthesis of azido or alkynyl electrophiles or their metabolic precursors and the complementary alkynyl- or azido-biotin tags, the power of the approach could nevertheless be realized with studies of a limited number of well-chosen electrophile probes.
Figure 5.
Click chemistry for post-reaction tagging of alkynyl-labeled adducts with biotin.
A final approach to adduct affinity capture is the use of a biotin-linked capture reagent that targets a specific feature of certain adducts for covalent capture. Maier et al. have reported the use of N′-aminooxymethylcarbonylhydrazino-d-biotin, which covalently captures the residual carbonyls on adducts formed by HNE and related lipid oxidation products (72, 73). The reaction forms a Schiff base that can be stabilized by borohydride reduction and captured for MS analysis. This interesting approach offers a means to capture broad classes of adducts (e.g., lipid oxidation electrophile-derived adducts) for analyses of the global impact of oxidative stress.
A final point worth emphasizing in global analysis of protein targets is that many adducts may be chemically unstable, which may complicate their analysis. For example, quinone adducts are often difficult to analyze because reoxidation of the initial addition products allows the formation of crosslinks and other unexpected structures (74, 75). Other adducts, such as those formed from acyl halides and some epoxides, may form esters that easily undergo hydrolysis, as was observed protein adducts formed from trichloroethylene oxide (76). Finally, secondary reactions of adducts can affect their detection, as was observed by Burcham et al., who reported that reaction of acrolein adducts with the amino group of Tris buffers prevented antibody-based adduct detection (77). Other work by these same authors actually showed that trapping of the aldehyde group in acrolein Michael adducts with the hydrazine-containing drug hydralazine prevented the formation of protein adduct oligomers, thus offering the novel prospect of adduct detoxication in situ (78).
Protein adduction selectivity and its determinants: studies with individual proteins
MS analyses have been used to characterize protein modifications by dozens of electrophiles. In most cases, incubation of the target protein with electrophiles was done to generate adducts, which were then analyzed by MS of either the intact protein or a proteolytic digest. Mapping of adducts to specific sequence locations was achieved by MS-MS or by peptide mass fingerprinting and chemical inference. Such studies have almost invariably revealed that protein modification is selective and specific—most proteins are reproducibly adducted at a subset of the sites that would appear to be available for reaction. Reactivity of solvent-exposed nucleophiles (e.g., cysteine, histidine, lysine) is expected, but solvent exposure per se is not necessarily the dominant factor in adduction site selectivity.
Several interesting studies have linked sequence-specific modifications to functional changes in proteins. In the following examples, the proteins of interest were found to be targets of the electrophiles in cell and tissue analyses by 2D-SDS-PAGE with biotinylated electrophiles or by immunoblotting for adducts. Follow-up studies in vitro were directed at identifying sequence targets and establishing effects on enzymatic activities or other functions. As discussed above, heat shock proteins were identified by the Petersen group as targets of HNE generated by oxidative stress in vivo (52, 53). Reaction of the purified Hsp72 and Hsp90 proteins with HNE, followed by LC-MS-MS analysis revealed adduction of Hsp72 at Cys267 and of Hsp90 at Cys572. However, the adduction at Cys267 of Hsp72 apparently accounted for inhibition of ATPase activity of the protein, whereas the Cys572 adduct on Hsp90 was not confirmed as responsible for inhibition of Hsp90 activity, although the activity was highly sensitive to the thiol modifier NEM. Adduction of thioredoxin 1 was detected using a biotinylated 15-deoxy-PGJ2 probe in cells and the adduct then was mapped to Cys35 and Cys69 by MS analysis when the protein was treated with 15-deoxy-PGJ2 in vitro (58). Cys35 is part of the functional thiol-disulfide redox couple of the enzyme. Similar studies in vitro identified the major actin target of 15-deoxy-PGJ2 and other α,β-unsaturated aldehydes as Cys374, which led to dysfunction in actin polymerization (59, 79).
Among the most interesting studies of protein adduction in vitro reveal features of protein structure that confer susceptibility to damage. Person et al. reported a novel adduct formed by benzoquinone and a benzoquinone-glutathione thioether conjugate with adjacent lysine residues on cytochrome c (74, 75). Both quinones formed crosslinking adducts as Lys25-Lys27 and Lys86-Lys87 on the protein. Initial formation of an addition product of one of the lysine ε-amino groups was followed by reoxidation of the quinone ring and then attack of the second lysine ε-amino nucleophile in a reaction favored at elevated pH. This case illustrates the potential importance of adjacent nucleophiles in protein targets generating crosslinks with electrophiles capable of sequential oxidation-adduction cycles (in the case of the quinones) or with bifunctional electrophiles, as in the case of the lipid oxidation products HNE and ONE (80, 81).
We previously characterized electrophile modifications of serine-threonine phosphatase PP2A, a three subunit protein that serves as a key regulator of signal transduction (82). Reaction with the biotinylated electrophile probes PEO-IAB and BMCC (see above and Fig. 3) labeled the structural (A), regulatory (B) and catalytic (C) subunits, as analyzed by affinity capture and Western blotting. However, only BMCC inhibited the phosphatase activity of the holoenzyme. LC-MS-MS-based adduct mapping studies revealed that the two electrophile probes differed only in the labeling pattern for the C subunit, in which BMCC labeled four cysteine residues not labeled by PEO-IAB. Two of these BMCC-specific adduct sites placed the electrophile side chain in direct contact with critical catalytic residues in the enzyme active site, which would account for the inhibition of activity.
One of the hazards of MS-based analysis of protein adducts is the determination of relative reactivities of different nucleophile targets. The inference of reactivity from MS spectral or signal intensities is often complicated by the different ionization efficiencies of different peptide sequences bearing the same adduct. It may seem that a particular sequence is highly reactive based on a strong MS signal or MS-MS spectrum of the peptide adduct, but this may reflect only highly favorable ionization properties in MS. The most rigorous way to assess competing reactivities is to measure the rates of reaction at the sites and compare the kinetic constants. However, the measurement of adduction kinetics presents a challenging problem, largely because of the difficulty of absolutely quantifying individual adducts. We recently reported an approach that employed differential stable isotope tagging of peptides in digests of adducted proteins (83). Derivatization of the N-termini of tryptic peptides with 12C6- or 13C6-PIC forms light and heavy derivatives that are easily distinguished by MS and MS-MS. To extract kinetic parameters from a time course study with electrophile-treated protein, sample from the last time point is digested and labeled with the heavy (13C6-PIC) and all other time points are digested and labeled with the light (12C6-PIC) (84, 85). Each light-labeled sample is spiked with an equal portion of the heavy-labeled sample as a reference standard and then the ratios of light to heavy signals extracted from MS-MS data are plotted as a function of reaction time. The plots are fitted to appropriate first order exponential curves to derive kobs values, which can be used directly to compare reactivities and different protein sites. Application of this approach to measure kobs for HNE adduction of several histidine and lysine residues in human serum albumin revealed a range of three orders of magnitude in the measured constants (84). The most reactive target of HNE was His242, which lies in a hydrophobic binding pocket in the IIa subdomain known to bind fatty acids and lipophilic xenobiotics. The reactivity of the His242 imidazole was further enhanced by electrostatic effects of nearby lysine residues, which reduce the estimated pKa of the imidazole nitrogen to less that 1.0.
Protein damage and signaling: mechanisms of toxicity and adaptation
The first studies linking covalent binding to toxicity in the early 1970s established the dominant paradigm in the field: xenobiotic metabolism yields reactive electrophiles, which form adducts with certain critical targets, which leads to toxicity and cell death. However, low dose exposures to electrophilic chemicals can protect against subsequent, larger doses that would induce toxic or carcinogenec responses (86, 87). These seemingly paradoxical consequences of exposure to electrophiles can be explained in part through the induction of protective responses due to low levels of protein damage. Two of the protective systems that appear to contribute to this effect are the ER stress response and the induction of genes regulated by the ARE/ERE. In both cases, stress leads to the induction of genes and proteins that express chaperone, antioxidant, xenobiotic detoxication, and protein degradation functions.
The ER stress response is the term given to the coordinated response triggered by binding of the ER resident chaperone BiP/GRP78 to unfolded or misfolded proteins in the ER lumen, thereby dissociating BiP from and activating the proteins PERK, ATF6 and IRE1. BiP is a member of the Hsp70 family of protein chaperones and increases in levels of unfolded proteins are thought to lead to BiP dissociation from these mediators, which then down regulate protein translation and up regulate transcription of ER stress response genes and activate complementary stress pathways (88). Studies by Stevens and colleagues in the mid 1990s established that activation of the ER stress response in LLC-PK1 cells block apoptosis in response to an otherwise lethal dose of iodoacetamide (89, 90).
Our recent studies with the electrophile probes IAB and BMCC revealed that ER stress induction differed with different patterns of protein alkylation in microsomes (67). The iodoacetamide probe IAB induced ER stress in HEK293 cells, whereas the N-alkylmaleimide BMCC did not. Interestingly, both probes alkylated BiP, as was also shown for the biotinylated raloxifene quinone methide probe used by Liu et al. (68). Direct alkylation of the critical BiP chaperone could provide a direct triggering mechanism for alkylation-induced ER stress. Alternatively, alkylation of other ER proteins could cause their misfolding and induce activation of ER stress through essentially the same mechanism as thiol reductants or modifiers of protein glycosylation (20, 88). At present, neither hypothesis has been adequately tested.
The induction of glutathione-S-transferase and antioxidant enzymes by phenolic antioxidants has now come to be understood as a broadly-based induction of over 200 genes by prooxidants, electrophiles, and cancer chemopreventive agents through the ARE/ERE (91, 92). Expression of these ARE/ERE-regulated genes is controlled by the transcription factor Nrf2 and its regulator, the thiol-rich sensor protein keap1 (93). Talalay and Yamamoto originally proposed that keap1 tethers Nrf2 in the cytoplasm until electrophiles and oxidants promote dissociation of the complex and translocation of Nrf2 to the nucleus (94, 95). Both the Hannink and Hayes groups subsequently demonstrated that keap1 is an adapter protein for the ubiquitination of Nrf2 by the E3 ubiquitin ligase Cul3 (96–98). Electrophile and oxidant inducers of the ARE/ERE disrupt the ubiquitination of Nrf2 by modifying the adapter protein (keap1), thus leading to stabilization of Nrf2 and activation of the ARE/ERE (95, 99–102).
Keap1 is perhaps the best example of a specific protein target of electrophiles whose covalent adduction leads to a specific biological effect—the stablization of Nrf2 and the induction of ARE/ERE-regulated genes. However, the keap1 sensor protein contains 27 cysteine residues and has not yet been structurally characterized (except for the Kelch domain (103)). A key question is which of the cysteines in keap1 are critical to its sensor function. Several reports have identified cysteine residues targeted by different electrophiles upon reaction with the bacterially expressed protein in vitro (95, 100–102, 104). However, these studies collectively reported that 25 of the 27 cysteines in human keap1 are adducted by at least one of the electrophiles studied to date. Among these, the most frequently reported targets include Cys151, Cys 257, Cys 273, Cys288, Cys297 and Cys613, although there are notable differences between laboratories studying mouse versus human keap1 or different electrophile probes, reaction conditions, sample workups and MS analysis methods. Studies with different electrophile probes under the same conditions and using the same analyses can be revealing, as the IAB and BMCC probes formed almost completely different patterns of keap1 adducts (101). This difference corresponded to a difference in ability of the electrophiles to activate ARE/ERE gene transcription in HepG2 cells (IAB activated, whereas BMCC was without effect).
Despite suggestions that three keap1 residues are particularly reactive (Cys151, Cys273 and Cys288), no rigorous kinetic analyses of the reactions of any electrophiles with keap1 cysteines have been reported. As noted above, assessments of the relative reactivities of different residues based on relative intensities of peptide adducts in MS analyses are unreliable.
Mutagenesis studies indicate that Cys273Ser and Cys288Ser mutations in the central linker domain blocked keap1-dependent ubiquitination of Nrf2 (96)and Cys273Ala and Cys288Ala mutations blocked keap1-dependent repression of an ARE/ERE reporter (100). Zhang et al. also reported that Cys151 in the keap1 BTB domain is required for inhibition of keap1-dependent degradation of Nrf2 by sulforaphane and oxidative stress, although this mutant does function as a constitutive repressor of Nrf2 (96, 97). Consideration of both the adduct data and mutagenesis data focuses attention on Cys151, Cys273 and Cys288 as potentially important sensor residues in keap1. Nevertheless, the issue of which keap1 cysteine is a critical target, or whether a single critical cysteine residue mediates activation by diverse electrophile chemistries remains to be resolved.
Other regulators of transcription factors and signaling networks are sensitive to oxidants or covalent modification, including several phosphotyrosine and phosphoserine/threonine phosphatases (82, 105–111), IkappaB kinase beta (112–117), the thiol-disulfide redox regulatory enzymes thioredoxin (58, 66, 118), protein disulfide isomerase (43, 46–49, 66, 68, 119–122), and the DNA damage response protein ATM (61). Many more proteins that directly or indirectly regulate signaling networks and pathways are targets of electrophiles (66, 67) and damage inventories using affinity capture and current shotgun proteomics methods most likely will range into the hundreds to thousands of protein targets. Interpretation of cellular outcomes of damage by different electrophiles thus will depend on 1) the combination of signaling-related targets modified, 2) the extent to which the adducts alter functions of the proteins and 3) the interplay between the affected pathways. Given our current ability to inventory targets of different electrophile chemistries and the degree of selectivity displayed by different electrophiles (e.g., 20% overlap in targets between the iodoacetamido- and N-alkylmaleimido electrophile probes), it should be possible to use systems “damage maps” to hypothesize effects on different pathways.
Studies with model organisms exposed to alkylating agents, oxidants and arsenic have integrated transcriptome profiling, genomic phenotyping and systems modeling to reveal agent-specific effects on signaling and metabolic networks (123–129). None of these studies have yet identified protein damage targets of the agents studied in these systems. Identification of protein targets in such well-characterized models could link the genomic and systems responses to initial damage events. Targeted assays of key intermediates and pathway outputs could enable tests of individual hypotheses regarding pathway effects of damage. This problem represents a fascinating challenge and opportunity that is certainly within the reach of presently available tools for proteomics, transcriptomics, metabolomics and systems modeling.
Conclusion
The most widely used assay of protein damage by chemicals still is the binding of radiolabel to bulk protein (18). Although this assay provides a useful basis for identifying molecules likely to cause toxicity via metabolic activation, the analytical toolkit for analyzing protein damage has advanced tremendously in the last 20 years. We are now on the brink of a comprehensive understanding of protein damage and its biological effects. Emerging evidence from studies with electrophile affinity probes and shotgun proteomics shows clearly that different binding patterns lead to distinct biological outcomes. Indeed, it seems reasonable to hypothesize that the binding pattern encodes the biology. The convergence of analytical technology, pathway analysis and computational systems biology tools will provide the means to interpret these codes.
The synthesis of probes representing diverse structures and chemistries is actually a fairly straightforward proposition and the application of shotgun proteomics analyses and bioinformatics analyses to identify the targets can now be standardized and implemented in reasonably high throughput. One of the most interesting challenges will be to define the molecular, cellular and tissue injury phenotypes that characterize different exposures. With these tools in hand, the most important task is to define the relationships between specific protein modifications and changes in function related to toxicity. This will constitute a significant challenge, but ultimately can offer the greatest insights into mechanisms.
It is important to emphasize that the problem of protein damage is not limited to the effects of exogenous chemical exposures. Endogenous electrophiles associated with oxidative stress and metabolic imbalances result in protein modifications that contribute to diverse diseases and understanding these mechanisms of injury represents an important opportunity for toxicology to impact human health (130).
The mapping of specific protein damage targets to pathway effects or injury phenotypes will lead to specific assays for these targets. The use of targeted analyses of specific adducts may offer clinically useful methods of characterizing adverse drug reactions or disease processes. Recent reports described the analysis of acetaminophen-cysteine adducts in serum and tissue protein hydrolysates to detect covalent acetaminophen adducts in both adults and children with liver failure (131, 132). Such studies take the analysis of protein damage from discovery science to the clinic and raise important issues about not only the analytical validity of the methods applied to measure adducts in clinical settings, but also about the application of such markers in a clinical context. It seems reasonable to expect that the next decade will bring increasing focus on the analysis of damaged proteins as markers for chemical toxicity and disease.
Acknowledgments
I thank past and present members of the Liebler laboratory for their contributions to the work discussed. I also thank Drs. F.P. Guengerich, M.A. Freeman, N.A. Porter, L.J. Marnett, S.S. Lau, T.J. Monks, A.J. Gandolfi, J.L. Bolton, L.M. Sayre, I.A. Blair, H. Ischiropoulos, D.R. Petersen, R.P. Hanzlik and S.R. Tannenbaum for helpful discussions of topics in this field. Many investigators have made important contributions to the topics discussed here and the citations of published work are meant to be representative, rather than comprehensive. I regret any omissions of relevant contributions by others. Work in the Liebler laboratory was supported by NIH Grants ES010056, ES000267, ES007028, ES001811, ES013125 and CA104590.
Footnotes
Abbreviations: 2D-SDS-PAGE; two dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis; 15-deoxy-PGJ2, 15-deoxy-Δ12,14-prostaglandin-J2; AMS, accelerator mass spectrometry; ARE/ERE, antioxidant response element/electrophile response element; ATF6, activating transcription factor 6; BiP/GRP78, immunoglobulin heavy chain-binding protein/glucose-regulated protein of molecular weight 78 kDa; BMCC, 1-biotinamido-4-(4′-[maleimidoethylcyclohexane]-carboxamido)butane; ER, endoplasmic reticulum; IAB, N-iodoacetyl-N-biotinylhexylenediamine; IRE1, inositol-requiring enzyme 1; LC-MS-MS, liquid chromatography-tandem mass spectrometry; HNE, 4-hydroxy-2-nonenal; keap1, Kelch-like ECH-associated protein 1; MALDI-MS, matrix assisted laser desorption-ionization mass spectrometry; MS, mass spectrometry; MS-MS, tandem mass spectrometry; Nrf2, nuclear factor-E2-related factor 2; PEO-IAB, (+)-biotinyl-iodoacetamidyl-3, 6-dioxaoctanediamine; PERK, PKR-like ER protein kinase; eIF2α (eukaryotic translation initiation factor 2, agr; subunit) kinase; PIC, phenylisocyanate; ONE, 4-oxo-2-nonenal; SDS-PAGE, sodium dodecyl sulfate- polyacrylamide gel electrophoresis.
References
- 1.Miller EC, Miller JA. The presence and significance of bound aminoazo dyes in the livers of rats fed p-dimethylaminoazobenzene. Cancer Res. 1947;7:468–480. [Google Scholar]
- 2.Miller JA, Sapp RW, Miller EC. The absorption spectra of certain carcinogenic aminoazo dyes and the protein-bound derivatives formed from these dyes in vivo. J Am Chem Soc. 1948;70:3458–3463. doi: 10.1021/ja01190a072. [DOI] [PubMed] [Google Scholar]
- 3.Miller EC, Miller JA. In vivo combinations between carcinogens and tissue constituents and their possible role in carcinogenesis. Cancer Res. 1952;12:547–556. [PubMed] [Google Scholar]
- 4.Lawley PD, Brooks P. The action of alkylating agents on deoxyribonucleic acid in relation to biological effects of the alkylating agents. Exp Cell Res. 1963;9:512–520. doi: 10.1016/0014-4827(63)90291-x. [DOI] [PubMed] [Google Scholar]
- 5.Magee PN, Farber E. Toxic liver injury and carcinogenesis. Methylation of rat-liver nucleic acids by dimethylnitrosamine in vivo. Biochem J. 1962;83:114–124. doi: 10.1042/bj0830114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green LC, Skipper PL, Turesky RJ, Bryant MS, Tannenbaum SR. In vivo dosimetry of 4-aminobiphenyl in rats via a cysteine adduct in hemoglobin. Cancer Res. 1984;44:4254–4259. [PubMed] [Google Scholar]
- 7.Farmer PB, Bailey E, Lamb JH, Connors TA. Approach to the quantitation of alkylated amino acids in haemoglobin by gas chromatography mass spectrometry. Biomed Mass Spectrom. 1980;7:41–46. doi: 10.1002/bms.1200070109. [DOI] [PubMed] [Google Scholar]
- 8.Ehrenberg L, Hiesche KD, Osterman-Golkar S, Wenneberg I. Evaluation of genetic risks of alkylating agents: tissue doses in the mouse from air contaminated with ethylene oxide. Mutat Res. 1974;24:83–103. doi: 10.1016/0027-5107(74)90123-7. [DOI] [PubMed] [Google Scholar]
- 9.Osterman-Golkar S, Ehrenberg L, Segerback D, Hallstrom I. Evaluation of genetic risks of alkylating agents. II. Haemoglobin as a dose monitor. Mutat Res. 1976;34:1–10. doi: 10.1016/0027-5107(76)90256-6. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds ES. Liver parenchymal cell injury. IV. Pattern of incorporation of carbon and chlorine from carbon tetrachloride into chemical constituents of liver in vivo. J Pharmacol Exp Ther. 1967;155:117–126. [PubMed] [Google Scholar]
- 11.Brodie BB, Reid WD, Cho AK, Sipes G, Krishna G, Gillette JR. Possible mechanism of liver necrosis caused by aromatic organic compounds. Proc Natl Acad Sci USA. 1971;68:160–164. doi: 10.1073/pnas.68.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jollow DJ, Mitchell JR, Potter WZ, Davis DC, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. II. Role of covalent binding in vivo. J Pharmacol Exp Ther. 1973;187:195–202. [PubMed] [Google Scholar]
- 13.Mitchell JR, Jollow DJ, Potter WZ, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. IV. Protective role of glutathione. J Pharmacol Exp Ther. 1973;187:211–217. [PubMed] [Google Scholar]
- 14.Mitchell JR, Jollow DJ, Potter WZ, Davis DC, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. I. Role of drug metabolism. J Pharmacol Exp Ther. 1973;187:185–194. [PubMed] [Google Scholar]
- 15.Potter WZ, Davis DC, Mitchell JR, Jollow DJ, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. 3. Cytochrome P-450-mediated covalent binding in vitro. J Pharmacol Exp Ther. 1973;187:203–210. [PubMed] [Google Scholar]
- 16.Roberts SA, Price VF, Jollow DJ. Acetaminophen structure-toxicity studies: in vivo covalent binding of a nonhepatotoxic analog, 3-hydroxyacetanilide. Toxicol Appl Pharmacol. 1990;105:195–208. doi: 10.1016/0041-008x(90)90181-s. [DOI] [PubMed] [Google Scholar]
- 17.Streeter AJ, Bjorge SM, Axworthy DB, Nelson SD, Baillie TA. The microsomal metabolism and site of covalent binding to protein of 3′-hydroxyacetanilide, a nonhepatotoxic positional isomer of acetaminophen. Drug Metab Dispos. 1984;12:565–576. [PubMed] [Google Scholar]
- 18.Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA. Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 19.Baillie TA. Future of toxicology-metabolic activation and drug design: challenges and opportunities in chemical toxicology. Chemical research in toxicology. 2006;19:889–893. doi: 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- 20.Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 21.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 22.Johnson AB, Barton MC. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat Res. 2007;618:149–162. doi: 10.1016/j.mrfmmm.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laine A, Ronai Z. Ubiquitin chains in the ladder of MAPK signaling. Sci STKE. 2005;2005:re5. doi: 10.1126/stke.2812005re5. [DOI] [PubMed] [Google Scholar]
- 24.Hanzlik RP, Koen YM, Theertham B, Dong Y, Fang J. The reactive metabolite target protein database (TPDB)--a web-accessible resource. BMC Bioinformatics. 2007;8:95. doi: 10.1186/1471-2105-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh H, Fukuda Y, Anderson DK, Ferrans VJ, Gillette JR, Pohl LR. Immunological studies on the mechanism of halothane-induced hepatotoxicity: Immunohistochemical evidence of trifluoroacetylated hepatocytes. J Pharmacol Exp Ther. 1985;233:857–862. [PubMed] [Google Scholar]
- 26.Satoh H, Gillette JR, Davies HW, Schulick RD, Pohl LR. Immunochemical evidence of trifluoroacetylated cytochrome P-450 in the liver of halothane-treated rats. Mol Pharmacol. 1985;28:468–474. [PubMed] [Google Scholar]
- 27.Satoh H, Martin BM, Schulick AH, Christ DD, Kenna JG, Pohl LR. Human anti-endoplasmic reticulum antibodies in sera of patients with halothane-induced hepatitis are directed against a trifluoroacetylated carboxylesterase. Proc Natl Acad Sci USA. 1989;86:322–326. doi: 10.1073/pnas.86.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden PJ, Ichimura T, McCann DJ, Pohl LR, Stevens JL. Detection of cysteine conjugate metabolite adduct formation with specific mitochondrial proteins using antibodies raised against halothane metabolite adducts. J Biol Chem. 1991;266:18415–18418. [PubMed] [Google Scholar]
- 29.Butler LE, Thomassen D, Martin JL, Martin BM, Kenna JG, Pohl LR. The calcium-binding protein calreticulin is covalently modified in rat liver by a reactive metabolite of the inhalation anesthetic halothane. Chemical research in toxicology. 1992;5:406–410. doi: 10.1021/tx00027a014. [DOI] [PubMed] [Google Scholar]
- 30.Pohl LR. An immunochemical approach of identifying and characterizing protein targets of toxic reactive metabolites. Chem Res Toxicol. 1993;6:786–793. doi: 10.1021/tx00036a006. [DOI] [PubMed] [Google Scholar]
- 31.Kleiner HE, Jones TW, Monks TJ, Lau SS. Immunochemical analysis of quinol-thioether-derived covalent protein adducts in rodent species sensitive and resistant to quinol-thioether-mediated nephrotoxicity. Chem Res Toxicol. 1998;11:1291–1300. doi: 10.1021/tx9801357. [DOI] [PubMed] [Google Scholar]
- 32.Kleiner HE, Rivera MI, Pumford NR, Monks TJ, Lau SS. Immunochemical detection of quinol--thioether-derived protein adducts. Chem Res Toxicol. 1998;11:1283–1290. doi: 10.1021/tx980134e. [DOI] [PubMed] [Google Scholar]
- 33.Bulera SJ, Birge RB, Cohen SD, Khairallah EA. Identification of the mouse liver 44-kDa acetaminophen-binding protein as a subunit of glutamine synthetase. Toxicol Appl Pharmacol. 1995;134:313–320. doi: 10.1006/taap.1995.1197. [DOI] [PubMed] [Google Scholar]
- 34.Hoivik DJ, Manautou JE, Tveit A, Mankowski DC, Khairallah EA, Cohen SD. Evidence suggesting the 58-kDa acetaminophen binding protein is a preferential target for acetaminophen electrophile. Fundam Appl Toxicol. 1996;32:79–86. doi: 10.1006/faat.1996.0109. [DOI] [PubMed] [Google Scholar]
- 35.Halmes NC, McMillan DC, Oatis JE, Jr, Pumford NR. Immunochemical detection of protein adducts in mice treated with trichloroethylene. Chem Res Toxicol. 1996;9:451–456. doi: 10.1021/tx950171v. [DOI] [PubMed] [Google Scholar]
- 36.Cohen SD, Pumford NR, Khairallah EA, Boekelheide K, Pohl LR, Amouzadeh HR, Hinson JA. Selective protein covalent binding and target organ toxicity. Toxicol Appl Pharmacol. 1997;143:1–12. doi: 10.1006/taap.1996.8074. [DOI] [PubMed] [Google Scholar]
- 37.Pumford NR, Halmes NC, Hinson JA. Covalent binding of xenobiotics to specific proteins in the liver. Drug Metab Rev. 1997;29:39–57. doi: 10.3109/03602539709037572. [DOI] [PubMed] [Google Scholar]
- 38.Hargus SJ, Amouzedeh HR, Pumford NR, Myers TG, McCoy SC, Pohl LR. Metabolic activation and immunochemical localization of liver protein adducts of the nonsteroidal anti-inflammatory drug diclofenac. Chem Res Toxicol. 1994;7:575–582. doi: 10.1021/tx00040a014. [DOI] [PubMed] [Google Scholar]
- 39.Kretz-Rommel A, Boelsterli UA. Selective protein adducts to membrane proteins in cultured rat hepatocytes exposed to diclofenac: radiochemical and immunochemical analysis. Mol Pharmacol. 1994;45:237–244. [PubMed] [Google Scholar]
- 40.Hargus SJ, Martin BM, George JW, Pohl LR. Covalent modification of rat liver dipeptidyl dipeptidase IV (CD 26) by the nonsteroidal anti-inflammatory drug diclofenac. Chem Res Toxicol. 1995;8:993–996. doi: 10.1021/tx00050a001. [DOI] [PubMed] [Google Scholar]
- 41.Wade LT, Kenna JG, Caldwell J. Immunochemical identification of mouse hepatic protein adducts derived from the nonsteroidal anti-inflammatory drugs diclofenac, sulindac, and ibuprofen. Chem Res Toxicol. 1997;10:546–555. doi: 10.1021/tx960153t. [DOI] [PubMed] [Google Scholar]
- 42.Qiu Y, Benet LZ, Burlingame AL. Identification of the hepatic protein targets of reactive metabolites of acetaminophen in vivo in mice using two-dimensional gel electrophoresis and mass spectrometry. J Biol Chem. 1998;273:17940–17953. doi: 10.1074/jbc.273.28.17940. [DOI] [PubMed] [Google Scholar]
- 43.Isbell MA, Morin D, Boland B, Buckpitt A, Salemi M, Presley J. Identification of proteins adducted by reactive naphthalene metabolites in vitro. Proteomics. 2005;5:4197–4204. doi: 10.1002/pmic.200401278. [DOI] [PubMed] [Google Scholar]
- 44.Lin CY, Isbell MA, Morin D, Boland BC, Salemi MR, Jewell WT, Weir AJ, Fanucchi MV, Baker GL, Plopper CG, Buckpitt AR. Characterization of a structurally intact in situ lung model and comparison of naphthalene protein adducts generated in this model vs lung microsomes. Chem Res Toxicol. 2005;18:802–813. doi: 10.1021/tx049746r. [DOI] [PubMed] [Google Scholar]
- 45.Lin CY, Boland BC, Lee YJ, Salemi MR, Morin D, Miller LA, Plopper CG, Buckpitt AR. Identification of proteins adducted by reactive metabolites of naphthalene and 1-nitronaphthalene in dissected airways of rhesus macaques. Proteomics. 2006;6:972–982. doi: 10.1002/pmic.200500170. [DOI] [PubMed] [Google Scholar]
- 46.Lame MW, Jones AD, Wilson DW, Segall HJ. Protein targets of 1,4-benzoquinone and 1,4-naphthoquinone in human bronchial epithelial cells. Proteomics. 2003;3:479–495. doi: 10.1002/pmic.200390062. [DOI] [PubMed] [Google Scholar]
- 47.Lame MW, Jones AD, Wilson DW, Dunston SK, Segall HJ. Protein targets of monocrotaline pyrrole in pulmonary artery endothelial cells. J Biol Chem. 2000;275:29091–29099. doi: 10.1074/jbc.M001372200. [DOI] [PubMed] [Google Scholar]
- 48.Lame MW, Jones AD, Wilson DW, Segall HJ. Monocrotaline pyrrole targets proteins with and without cysteine residues in the cytosol and membranes of human pulmonary artery endothelial cells. Proteomics. 2005;5:4398–4413. doi: 10.1002/pmic.200402022. [DOI] [PubMed] [Google Scholar]
- 49.Koen YM, Hanzlik RP. Identification of seven proteins in the endoplasmic reticulum as targets for reactive metabolites of bromobenzene. Chem Res Toxicol. 2002;15:699–706. doi: 10.1021/tx0101898. [DOI] [PubMed] [Google Scholar]
- 50.Koen YM, Gogichaeva NV, Alterman MA, Hanzlik RP. A proteomic analysis of bromobenzene reactive metabolite targets in rat liver cytosol in vivo. Chem Res Toxicol. 2007;20:511–519. doi: 10.1021/tx6003166. [DOI] [PubMed] [Google Scholar]
- 51.Williams KE, Carver TA, Miranda JJ, Kautiainen A, Vogel JS, Dingley K, Baldwin MA, Turteltaub KW, Burlingame AL. Attomole detection of in vivo protein targets of benzene in mice: evidence for a highly reactive metabolite. Mol Cell Proteomics. 2002;1:885–895. doi: 10.1074/mcp.m200067-mcp200. [DOI] [PubMed] [Google Scholar]
- 52.Carbone DL, Doorn JA, Kiebler Z, Sampey BP, Petersen DR. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem Res Toxicol. 2004;17:1459–1467. doi: 10.1021/tx049838g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J Pharmacol Exp Ther. 2005;315:8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 54.Kapphahn RJ, Giwa BM, Berg KM, Roehrich H, Feng X, Olsen TW, Ferrington DA. Retinal proteins modified by 4-hydroxynonenal: identification of molecular targets. Exp Eye Res. 2006;83:165–175. doi: 10.1016/j.exer.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 55.Meier BW, Gomez JD, Zhou A, Thompson JA. Immunochemical and proteomic analysis of covalent adducts formed by quinone methide tumor promoters in mouse lung epithelial cell lines. Chem Res Toxicol. 2005;18:1575–1585. doi: 10.1021/tx050108y. [DOI] [PubMed] [Google Scholar]
- 56.Meier BW, Gomez JD, Kirichenko OV, Thompson JA. Mechanistic basis for inflammation and tumor promotion in lungs of 2,6-di-tert-butyl-4-methylphenol-treated mice: electrophilic metabolites alkylate and inactivate antioxidant enzymes. Chem Res Toxicol. 2007;20:199–207. doi: 10.1021/tx060214f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shibata T, Kondo M, Osawa T, Shibata N, Kobayashi M, Uchida K. 15-deoxy-delta 12,14-prostaglandin J2. A prostaglandin D2 metabolite generated during inflammatory processes. J Biol Chem. 2002;277:10459–10466. doi: 10.1074/jbc.M110314200. [DOI] [PubMed] [Google Scholar]
- 58.Shibata T, Yamada T, Ishii T, Kumazawa S, Nakamura H, Masutani H, Yodoi J, Uchida K. Thioredoxin as a molecular target of cyclopentenone prostaglandins. J Biol Chem. 2003;278:26046–26054. doi: 10.1074/jbc.M303690200. [DOI] [PubMed] [Google Scholar]
- 59.Aldini G, Carini M, Vistoli G, Shibata T, Kusano Y, Gamberoni L, Dalle-Donne I, Milzani A, Uchida K. Identification of actin as a 15-deoxy-Delta12,14-prostaglandin J2 target in neuroblastoma cells: mass spectrometric, computational, and functional approaches to investigate the effect on cytoskeletal derangement. Biochemistry. 2007;46:2707–2718. doi: 10.1021/bi0618565. [DOI] [PubMed] [Google Scholar]
- 60.Itoh K, Mochizuki M, Ishii Y, Ishii T, Shibata T, Kawamoto Y, Kelly V, Sekizawa K, Uchida K, Yamamoto M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi M, Ono H, Mihara K, Tauchi H, Komatsu K, Shibata T, Shimizu H, Uchida K, Yamamoto K. ATM activation by a sulfhydryl-reactive inflammatory cyclopentenone prostaglandin. Genes Cells. 2006;11:779–789. doi: 10.1111/j.1365-2443.2006.00976.x. [DOI] [PubMed] [Google Scholar]
- 62.Stamatakis K, Sanchez-Gomez FJ, Perez-Sala D. Identification of novel protein targets for modification by 15-deoxy-Delta12,14-prostaglandin J2 in mesangial cells reveals multiple interactions with the cytoskeleton. J Am Soc Nephrol. 2006;17:89–98. doi: 10.1681/ASN.2005030329. [DOI] [PubMed] [Google Scholar]
- 63.Adam GC, Sorensen EJ, Cravatt BF. Chemical strategies for functional proteomics. Mol Cell Proteomics. 2002;1:781–790. doi: 10.1074/mcp.r200006-mcp200. [DOI] [PubMed] [Google Scholar]
- 64.Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol. 2002;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- 65.Yates JR., III Mass spectral analysis in proteomics. Annu Rev Biophys Biomol Struct. 2004;33:297–316. 297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]
- 66.Dennehy MK, Richards KAM, Wernke GW, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2006;19:20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- 67.Shin NY, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem Res Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J, Li Q, Yang X, van Breemen RB, Bolton JL, Thatcher GR. Analysis of protein covalent modification by xenobiotics using a covert oxidatively activated tag: raloxifene proof-of-principle study. Chemical research in toxicology. 2005;18:1485–1496. doi: 10.1021/tx0501738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 70.Saxon E, Armstrong JI, Bertozzi CR. A “traceless” Staudinger ligation for the chemoselective synthesis of amide bonds. Org Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 71.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 72.Chavez J, Wu J, Han B, Chung WG, Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N′-aminooxymethylcarbonylhydrazino d-biotin. Anal Chem. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 73.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal Chem. 2007;79:3342–3354. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 74.Person MD, Monks TJ, Lau SS. An integrated approach to identifying chemically induced posttranslational modifications using comparative MALDI-MS and targeted HPLC-ESI-MS/MS. Chem Res Toxicol. 2003;16:598–608. doi: 10.1021/tx020109f. [DOI] [PubMed] [Google Scholar]
- 75.Person MD, Mason DE, Liebler DC, Monks TJ, Lau SS. Alkylation of cytochrome c by (glutathion-S-yl)-1,4-benzoquinone and iodoacetamide demonstrates compound-dependent site specificity. Chem Res Toxicol. 2005;18:41–50. doi: 10.1021/tx049873n. [DOI] [PubMed] [Google Scholar]
- 76.Cai H, Guengerich FP. Reaction of trichloroethylene oxide with proteins and dna: instability of adducts and modulation of functions. Chem Res Toxicol. 2001;14:54–61. doi: 10.1021/tx000185n. [DOI] [PubMed] [Google Scholar]
- 77.Burcham PC, Fontaine FR, Petersen DR, Pyke SM. Reactivity with Tris(hydroxymethyl)aminomethane confounds immunodetection of acrolein-adducted proteins. Chemical research in toxicology. 2003;16:1196–1201. doi: 10.1021/tx0341106. [DOI] [PubMed] [Google Scholar]
- 78.Burcham PC, Fontaine FR, Kaminskas LM, Petersen DR, Pyke SM. Protein adduct-trapping by hydrazinophthalazine drugs: mechanisms of cytoprotection against acrolein-mediated toxicity. Mol Pharmacol. 2004;65:655–664. doi: 10.1124/mol.65.3.655. [DOI] [PubMed] [Google Scholar]
- 79.Dalle-Donne I, Carini M, Vistoli G, Gamberoni L, Giustarini D, Colombo R, Maffei Facino R, Rossi R, Milzani A, Aldini G. Actin Cys374 as a nucleophilic target of alpha, beta-unsaturated aldehydes. Free Radic Biol Med. 2007;42:583–598. doi: 10.1016/j.freeradbiomed.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 80.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem Res Toxicol. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 81.Zhang WH, Liu J, Xu G, Yuan Q, Sayre LM. Model Studies on Protein Side Chain Modification by 4-Oxo-2-nonenal. Chem Res Toxicol. 2003;16:512–523. doi: 10.1021/tx020105a. [DOI] [PubMed] [Google Scholar]
- 82.Codreanu SG, Adams DG, Dawson ES, Wadzinski BE, Liebler DC. Inhibition of protein phosphatase 2A activity by selective electrophile alkylation damage. Biochemistry. 2006;45:10020–10029. doi: 10.1021/bi060551n. [DOI] [PubMed] [Google Scholar]
- 83.Mason DE, Liebler DC. Quantitative analysis of modified proteins by LC-MS-MS of peptides labeled with phenyl isocyanate. J Proteome Res. 2003;2:265–272. doi: 10.1021/pr0255856. [DOI] [PubMed] [Google Scholar]
- 84.Szapacs ME, Riggins JN, Zimmerman LJ, Liebler DC. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry. 2006;45:10521–10528. doi: 10.1021/bi060535q. [DOI] [PubMed] [Google Scholar]
- 85.Orton CR, Liebler DC. Analysis of protein adduction kinetics by quantitative mass spectrometry Competing adduction reactions of glutathione-S-transferase P1-1 with electrophiles. Chem Biol Interact. 2007 doi: 10.1016/j.cbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proceedings of the National Academy of Sciences, USA. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Talalay P, Fahey JW, Holtzclaw WD, Prestera T, Zhang Y. Chemoprotection against cancer by phase 2 enzyme induction. Toxicol Lett. 1995;82–83:173–179. doi: 10.1016/0378-4274(95)03553-2. [DOI] [PubMed] [Google Scholar]
- 88.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 89.Liu H, Bowes RC, III, van de WB, Sillence C, Nagelkerke JF, Stevens JL. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J Biol Chem. 1997;272:21751–21759. doi: 10.1074/jbc.272.35.21751. [DOI] [PubMed] [Google Scholar]
- 90.van de Water B, Wang Y, Asmellash S, Liu H, Zhan Y, Miller E, Stevens JL. Distinct endoplasmic reticulum signaling pathways regulate apoptotic and necrotic cell death following iodoacetamide treatment. Chem Res Toxicol. 1999;12:943–951. doi: 10.1021/tx990054q. [DOI] [PubMed] [Google Scholar]
- 91.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway: Identification of novel gene clusters for cell survival. J Biol Chem. 2002;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 92.Dinkova-Kostova AT, Holtzclaw WD, Kensler TW. The role of Keap1 in cellular protective responses. Chem Res Toxicol. 2005;18:1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- 93.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 94.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 99.Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci USA. 2001;98:3404–3409. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 102.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li X, Zhang D, Hannink M, Beamer LJ. Crystallization and initial crystallographic analysis of the Kelch domain from human Keap1. Acta Crystallogr D Biol Crystallogr. 2004;60:2346–2348. doi: 10.1107/S0907444904024825. [DOI] [PubMed] [Google Scholar]
- 104.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005 doi: 10.1021/tx0502138. in press. [DOI] [PubMed] [Google Scholar]
- 105.Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 106.Nemani R, Lee EY. Reactivity of sulfhydryl groups of the catalytic subunits of rabbit skeletal muscle protein phosphatases 1 and 2A. Arch Biochem Biophys. 1993;300:24–29. doi: 10.1006/abbi.1993.1004. [DOI] [PubMed] [Google Scholar]
- 107.Zhang L, Zhang Z, Long F, Lee EY. Tyrosine-272 is involved in the inhibition of protein phosphatase-1 by multiple toxins. Biochemistry. 1996;35:1606–1611. doi: 10.1021/bi9521396. [DOI] [PubMed] [Google Scholar]
- 108.Cavigelli M, Li WW, Lin A, Su B, Yoshioka K, Karin M. The tumor promoter arsenite stimulates AP-1 activity by inhibiting a JNK phosphatase. EMBO J. 1997;15:6269–6279. [PMC free article] [PubMed] [Google Scholar]
- 109.Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:E1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 110.Sommer D, Coleman S, Swanson SA, Stemmer PM. Differential susceptibilities of serine/threonine phosphatases to oxidative and nitrosative stress. Arch Biochem Biophys. 2002;404:271–278. doi: 10.1016/s0003-9861(02)00242-4. [DOI] [PubMed] [Google Scholar]
- 111.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 112.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IkappaB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 113.Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 114.Ji C, Kozak KR, Marnett LJ. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J Biol Chem. 2001;276:18223–18228. doi: 10.1074/jbc.M101266200. [DOI] [PubMed] [Google Scholar]
- 115.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EF, Janssen-Heininger YM. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci USA. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reynaert NL, Ckless K, Korn SH, Vos N, Guala AS, Wouters EF, van der Vliet A, Janssen-Heininger YM. Nitric oxide represses inhibitory kappaB kinase through S-nitrosylation. Proc Natl Acad Sci USA. 2004;101:8945–8950. doi: 10.1073/pnas.0400588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee JH, Koo TH, Yoon H, Jung HS, Jin HZ, Lee K, Hong YS, Lee JJ. Inhibition of NF-kappa B activation through targeting I kappa B kinase by celastrol, a quinone methide triterpenoid. Biochem Pharmacol. 2006;72:1311–1321. doi: 10.1016/j.bcp.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 118.Erve JC, Barofsky E, Barofsky DF, Deinzer ML, Reed DJ. Alkylation of Escherichia coli thioredoxin by S-(2-chloroethyl)glutathione and identification of the adduct on the active site cysteine-32 by mass spectrometry. Chem Res Toxicol. 1995;8:934–941. doi: 10.1021/tx00049a006. [DOI] [PubMed] [Google Scholar]
- 119.Kaetzel RS, Stapels MD, Barofsky DF, Reed DJ. Alkylation of protein disulfide isomerase by the episulfonium ion derived from the glutathione conjugate of 1,2-dichloroethane and mass spectrometric characterization of the adducts. Arch Biochem Biophys. 2004;423:136–147. doi: 10.1016/j.abb.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 120.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 121.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 122.Asif AR, Armstrong VW, Voland A, Wieland E, Oellerich M, Shipkova M. Proteins identified as targets of the acyl glucuronide metabolite of mycophenolic acid in kidney tissue from mycophenolate mofetil treated rats. Biochimie. 2006 doi: 10.1016/j.biochi.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 123.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 126.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 127.Said MR, Begley TJ, Oppenheim AV, Lauffenburger DA, Samson LD. Global network analysis of phenotypic effects: Protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:18006–18011. doi: 10.1073/pnas.0405996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome biology. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liebler DC. The poisons within: application of toxicity mechanisms to fundamental disease processes. Chemical research in toxicology. 2006;19:610–613. doi: 10.1021/tx0600292. [DOI] [PubMed] [Google Scholar]
- 131.James LP, Alonso EM, Hynan LS, Hinson JA, Davern TJ, Lee WM, Squires RH. Detection of acetaminophen protein adducts in children with acute liver failure of indeterminate cause. Pediatrics. 2006;118:e676–681. doi: 10.1542/peds.2006-0069. [DOI] [PubMed] [Google Scholar]
- 132.Davern TJ, 2nd, James LP, Hinson JA, Polson J, Larson AM, Fontana RJ, Lalani E, Munoz S, Shakil AO, Lee WM. Measurement of serum acetaminophen-protein adducts in patients with acute liver failure. Gastroenterology. 2006;130:687–694. doi: 10.1053/j.gastro.2006.01.033. [DOI] [PubMed] [Google Scholar]