Abstract
The F0F1-ATP synthase couples the functions of H+ transport and ATP synthesis/hydrolysis through the efficient transmission of energy mediated by rotation of the centrally located γ, ε, and c subunits. To understand the γ subunit role in the catalytic mechanism, we previously determined the partial rate constants and devised a minimal kinetic model for the rotational hydrolytic mode of the F1-ATPase enzyme that uniquely fits the pre-steady state and steady state data (Baylis Scanlon, J. A., Al-Shawi, M. K., Le, N. P., and Nakamoto, R. K. (2007) Biochemistry 46 ,8785 -8797). Here we directly test the model using two single cysteine mutants, βD380C and βE381C, which can be used to reversibly inhibit rotation upon formation of a cross-link with the conserved γCys-87. In the pre-steady state, the γ-β cross-linked enzyme at high Mg·ATP conditions retained the burst of hydrolysis but was not able to release Pi. These data show that the rate-limiting rotation step, kγ, occurs after hydrolysis and before Pi release. This analysis provides additional insights into how the enzyme achieves efficient coupling and implicates the βGlu-381 residue for proper formation of the rate-limiting transition state involving γ subunit rotation.
The F0F1-ATP synthase, located in energy-transducing membranes, utilizes the electrochemical gradient of H+ or Na+ to synthesize ATP from ADP and Pi. Under anaerobic conditions, the bacterial enzyme can function as an ATPase, coupled to pump protons across the membrane to generate a ΔμH+. Proton transport is mediated by the membrane-embedded F0 sector (ab2c10) and is separated by a distance greater than 120 Å from the three catalytic sites where ATP hydrolysis/synthesis occurs on the membrane extrinsic F1 segment (α3β3γδε) (for reviews see Refs. 1-5). This distance indicates a complicated mechanism for effectual, efficient transmission between the two disparate functions. It is now well established that the enzyme functions as a molecular motor (6-8), transmitting energy between F1 and F0 through rotation of the central stalk subunits, γ, ε, and the ring of c subunits (for reviews see Refs. 9, 10).
The F1 complex can be reversibly stripped from the membrane and functions as a soluble ATPase. In describing the first high resolution x-ray structure, Abrahams et al. (11) named each catalytic β subunit for the nucleotide bound at that site as follows: βTP for the ATP analog AMPPNP,2 βDP for ADP-bound, and the structurally open βE empty site (Fig. 1). The three sites have vastly differing affinities for Mg·ATP (12), Kd1 = 10-10, Kd2 = 10-6, and Kd3 = 10-4 m-1 exhibiting negative cooperativity in nucleotide binding. At low substoichiometric Mg·ATP concentrations, characteristics of the basic catalytic reaction have been elucidated in unisite catalytic conditions (13, 14). At higher substrate concentrations, the enzyme enters the multisite hydrolysis mode, which is 105-106 times faster than unisite catalysis because of the high positive cooperativity among the sites with respect to promotion of catalysis (15-17). The Kd value for the binding of Mg·ATP to the low affinity catalytic site (12) matches the Km value for Mg·ATP hydrolysis (18, 19) and the Km value for rotation in the direct observation of single F1 molecules (20). Rotational catalysis requires the concerted, sequential participation of all three catalytic sites as they pass through the three different conformations (18, 21, 22). For these reasons, we have argued (23) that reversible hydrolysis/synthesis of ATP occurs in the βTP site, and product Pi and ADP are released after the site converts from βDP to βE (18). Further evidence using fluorescence resonance energy transfer confirmed that the βTP site is the high affinity site (24).
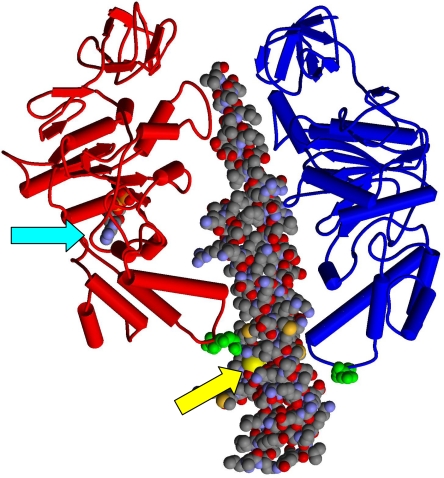
FIGURE 1.
The 380βDELSEED386 motifs relative to the catalytic sites. Two of the β subunit conformers, βDP (red) and βE (blue), and the coiled-coil region of the γ subunit (space-filling representation) from the x-ray structure of Abrahams et al. (69) are shown. ADP bound to βDP is shown in the space-filling model (cyan arrow). βAsp-380 and βGlu-381 (E. coli numbering) are shown in green space-filling representation near the “bottom” of the β subunits in the βDELSEED loop. γCys-87 is indicated by the yellow arrow.
Single molecule experiments have observed that each major 120° step in γ subunit rotation occurs in two substeps as follows: the 80° substep that is dependent on Mg·ATP concentration, followed by a 40° substep that is not affected by Mg·ATP concentration (25). Attempts to elucidate the role of rotation in the enzymatic mechanism using the single molecule approach include analysis of γ subunit substeps (20, 25) and concurrent visualization of the binding and release of fluorescent nucleotide (26). However, the structural and mechanistic coordination of γ subunit rotation and the kinetics of the elemental chemical hydrolysis reaction steps cannot be delineated by these studies, and the chemomechanical coupling of the enzyme remains not fully understood.
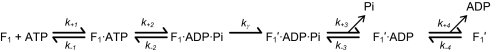
Using pre-steady state analysis, we recently determined the partial reactions of the multisite ATPase hydrolytic pathway and that the rate-limiting step occurs after the reversible hydrolysis/synthesis step and just prior to release of Pi (18). Scheme 1 adequately describes the multisite kinetic pathway (18):
SCHEME 1.
We conjectured that the rate-limiting step involves the 40° rotation, and therefore we called it kγ. The pre-steady state kinetics of this step were critical in allowing us to determine its order in the reaction scheme, which provided tremendous insight into the rotational catalytic mechanism. Even though we could directly determine several of the partial reaction rate constants, assignment of kγ to the rotation step in the presteady state was not confirmed.
In this study we test our model that the rate-limiting step kγ involves rotation of the γ subunit. To assay the rotation steps, we decided to use a rotor-stator, γ subunit to β subunit, disulfide bond, with the premise that a cross-link would block rotation and therefore stop the partial reactions involving rotation. Such disulfides have been previously observed between the native Cys at position 87 in the Escherichia coli γ subunit and Cys substitutions in the conserved 380βDELSEED386 motif, either at βAsp-380 (27) or βGlu-381 (28) (Fig. 1). For example, Garcia and Capaldi (29) reported that the γ-β-cross-linked F1 had nearly normal unisite kinetic behavior demonstrating that unisite catalysis is independent of the γ subunit rotation. We reasoned that cross-linking the rotor to the stator would lock the position, or at least greatly restrict the rotation of the γ subunit, and therefore prevent the kinetic steps that require rotation. We report here that our data do indeed show this to be the case. Preventing the rotation results in a slower pre-steady state burst of ATP hydrolysis and blocks the release of Pi. These data are consistent with the functional assignments of the catalytic sites where βTP is the high affinity site carrying out reversible hydrolysis/synthesis, and βDP is the site from which product Pi and ADP are released after it converts to βE (18). We also found that mild perturbations of the βDELSEED function by βE381C causes uncoupling of catalysis and rotation by affecting the establishment of the optimal catalytic conformation resulting in a bypass in the reaction pathway. In the alternative pathway, γ subunit rotation becomes disengaged from hydrolysis thus perturbing its ability to pump protons.
Evidence presented in this study suggests the 40° rotation is associated with kγ, and the enzyme needs to acquire the conformation after the 80° rotation substep to achieve positive cooperativity for the promotion of catalysis.
EXPERIMENTAL PROCEDURES
F0F1 Strains and Plasmids—The single cysteine mutations (βD380C and βE381C) were introduced into plasmid pUβSE derived from a modified pUC18 vector containing a portion of uncD (γ-subunit gene) between SacI and Eco47III restriction sites (30). Site-directed mutagenesis was performed with the QuickChange kit from Stratagene (La Jolla, CA), using the following primers: βE381C, 5′-CTGAAAGACATCATCGCCACCCTGGGTATGGATTGCCTTTCTGAAGAAGACAAACTGGTGG-3′, and βD380C, 5′-CGCCATCCTGGGTATGTGTGAACTTTCTGAAGAAGACAAACTGG-3′ (where the underlined letters show the converted β subunit codons GAA→TGC and GAT→TGT, respectively). Both mutations were isolated on the SacI to Eco47III fragment and ligated individually into the high copy number plasmid pBWU13 (31). Colonies were screened for the insert with a PCR approach using Taq Master Mix (Qiagen, Valencia, CA). Introduction of each mutation in the expression plasmid was verified by DNA sequencing. Mutant F0F1 complexes were expressed in the atp operon-deleted strain, DK8 (32).
The amino-terminal polyhistidine-tagged ε subunit (His-ε was expressed in E. coli strain BL21(DE3)pLysS (33) and purified as described previously (18). All molecular biology manipulations were done according to manufacturers' instructions or according to procedures described in Sambrook et al. (34).
Preparation and Characterization of Mutant F0F1 Enzymes—Oxidative phosphorylation-dependent growth of mutant strains was determined on minimal defined media containing 0.2% sodium succinate as a sole carbon source (31, 35).
Membrane vesicles containing F0F1 were prepared as described previously (36) from cells grown to mid-log phase in a minimal salt medium supplemented with 1.1% glucose, amino acids, thiamine, and 50 μg/ml ampicillin at 37 °C (31). F1 expression levels in the membrane preparations were analyzed using a quantitative immunoblot assay using polyclonal antibodies against E. coli F1 α subunits (kindly provided by Dr. Alan Senior, University of Rochester). Purified F1 was used as a standard as described previously (37). The formation of ATP- or NADH-dependent electrochemical gradients of protons across inside-out inner membrane vesicles was assessed by monitoring acridine orange fluorescence quenching at 530 nm as described previously (31, 38).
Preparation of F1 Enzymes—Cells for F1 purification were grown at 37 °C in minimal media supplemented with 1.1% glucose, amino acids, thiamine, and 50 μg/ml ampicillin. F1 was isolated and purified as described (14). The purity and subunit composition of F1 preparations were checked by SDS-PAGE analysis (39) and by comparison of steady state ATP hydrolysis rates with those reported previously (28, 33, 40).
Cross-linking of F1 Mutants—The βD380C mutant F1 was cross-linked by reacting with 5,5′-dithiobis-(2-nitrobenzoate) (DTNB) based on conditions described previously (40). The enzyme was thawed and stored at room temperature, diluted to ∼2.5 mg/ml, treated with 1 mm tris(2-carboxyethyl) phosphine hydrochloride (TCEP) for 20 min, and then passed over a Sephadex G-50 centrifuge column (41) to remove TCEP and nucleotides and to exchange the buffer into Buffer 1 (25 mm TES-KOH, pH 8.0, 1 mm MgSO4, 10% glycerol). Mutant F1 was incubated with 2.5 μm DTNB for 1 h to induce cross-linking. After incubation, the enzyme was again passed over a centrifuge column to remove oxidizing reagent and exchange the buffer into Buffer 2 (25 mm TES-KOH, 0.244 mm MgCl2, 0.2 mm EDTA, pH 7.5, at 25 °C) in preparation for pre-steady state measurements. The βE381C mutant was treated in the same manner as the βD380C mutant, but to induce the γ-β cross-link 50 μm CuCl2 was used for 1 h (29). Incubation with DTNB or CuCl2 overnight did not significantly increase cross-linking efficiency nor affect ATPase activity of wild-type F1. Interestingly, neither mutant F1 achieved a maximal cross-linking yield when treated with the oxidizing reagent optimized for the other.
Pre-steady State Multisite Hydrolysis of ATP—[γ-32P]ATP hydrolysis was measured in the millisecond time range using a Kintek RQF-3 rapid quench-flow apparatus (Austin, TX) with circulating water temperature control. The uncross-linked F1 was prepared for pre-steady state experiments by removing bound nucleotides and exchanging the enzyme into Buffer 2 as described previously (18).
In the rapid mixing device (1:1 mixing volume ratio), syringe A contained purified F1 (with an additional equimolar concentration of ε subunit) (18) in Buffer 2, and syringe B contained 25 mm TES-KOH, 0.2 mm EDTA, 0.5 mm [γ-32P]ATP, and 0.46 mm MgSO4, pH 7.5, at 25 °C, resulting in final concentrations of 104 and 50 μm for Mg·ATP and free Mg2+, respectively. The samples were processed according to the two-step procedure described previously (18) to minimize the background due to the high concentration of [γ-32P]ATP in the experiments. The amount of 32Pi generated was determined by Cerenkov counting in 0.2 m Tris base.
Unisite Binding and Hydrolysis of ATP—F1 was prepared as described above for the pre-steady state multisite experiments except that the final centrifuge column was equilibrated with unisite buffer (50 mm Tris base, 50 mm MOPS, 4.5 mm K2SO4 and 0.5 mm MgSO4 adjusted to pH 7.5 with H2SO4). Reactions were performed at 23 °C and were initiated by adding 15 μl of [γ-32P]ATP to F1 while rapidly mixing. The final molar mix ratio of ATP to F1 was ∼0.1. The reactions were quenched at varying times with 550 μl of 50 mm Tris-SO4, pH 8.0, 1 mm KH2PO4, 4.5 mm ATP, 2.8 mm MgSO4, 8.2% v/v HClO4. The total 32Pi generated was determined by the acid molybdate precipitation method of Sugino and Miyoshi (42) and Cerenkov counting in 15 ml of 0.2 m Tris base. The rate of [γ-32P]ATP binding was measured by the hexokinase trap method modified slightly from that described previously (29, 43). Briefly, the unisite reactions were started as described above, but at times shown 200 μl of hexokinase solution (unisite buffer with 2 mg/ml hexokinase (purified from Baker's yeast at ∼170 units/mg), 4.4 mg/ml glucose, and an extra 2.5 mm MgSO4) were added, and the reaction was allowed to proceed for 10 s before quenching with 2 n HCl containing 1 mm Pi. Samples were boiled for 7 min to hydrolyze the unreacted [γ-32P]ATP, and the 32Pi was removed by acid molybdate precipitation. The amount of glucose 6-[32P]phosphate formed was determined by measuring the radioactivity of the supernatant by Cerenkov counting in Tris base.
Pre-steady State Pi Release—Pi release was followed by the fluorescence intensity change of (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin)-labeled phosphate-binding protein (MDCC-PBP (44, 45)). Phosphate-binding protein was expressed from E. coli strain ANCC75 (leu purE trp his argG strA phoS64 met thi) harboring plasmid pSN5182/7 (kindly provided by Dr. Martin Webb, MRC, Mill Hill, London, UK) and prepared as described previously (45). Unreacted label was separated from MDCC-PBP by passage over a PD-10 desalting column (all chromatography materials were from Amersham Biosciences). Unlabeled phosphate-binding protein and Pi-insensitive MDCC-PBP were removed by passage over a Q-Sepharose column and a Mono-Q column, respectively (45). The final preparation had a 12-15-fold increase in fluorescence at 465 nm on binding Pi, indicating that the protein was largely Pi-free.
Removal of contaminating inorganic phosphate from buffers, the final enzyme preparation, and the stopped-flow spectrometer required a “Pi mop,” assembled as described by Nixon et al. (46). The Pi mop sequesters Pi into the stable ribose 5-phosphate molecule through a coupled enzyme system. Bacterial purine nucleoside phosphorylase and 7-methylguanosine converts phosphate and ribose to ribose 1-phosphate, which is then modified by phosphodeoxyribomutase (PDRM) in the presence of MnCl2 and α-d-glucose 1,6-bisphosphate to form the stable ribose 5-phosphate.
Purine nucleoside phosphorylase, 7-methylguanosine, and α-d-glucose 1,6-bisphosphate were purchased from Sigma. Purified PDRM was made by amplifying the E. coli PDRM gene from strain XL1 blue genomic DNA by PCR using primers 5′-ACTCCATGGAACGTGCATTTATTATGGTTCTGGACTCATTCGG-3′ and 5′-ATGCTCGAGTCAGAACATTTGCTTTGCCATATTCCATATCAG-3′. The primers included an NcoI site over the ATG start codon and an XhoI site down-stream of the native stop codon (the restriction sites are depicted by the underscored sequences). The open reading frame was verified by sequencing the entire insert, which was then ligated into the pHIS-Parallel1 vector (47) and used for expression in BL21(DE3) cells (Invitrogen). The PDRM protein was purified from cell lysates via an amino-terminal His6 affinity tag.
The kinetics of Pi release were followed in an Applied Photophysics SX.18MV-R stopped-flow spectrometer (Surrey, UK). The fluorescence change of MDCC-PBP was monitored at the excitation wavelength of 425 nm with a 455-nm emission cutoff filter. The syringe contents, listed in the figure legends, were essentially the same as for the rapid quench flow experiments except for the lack of radioactivity and the addition of 20 μm MDCC-PBP. The Pi binding response of MDCC-PBP fluorescence was calibrated using known Pi concentrations in the stopped flow at fixed photomultiplier voltage.
Kinetic Analysis—The experimental data fitting program, Scientist version 2.0 (Micromath Research, Inc., St. Louis), was used to fit the experimental data by numerical integration of differential equations described in detail previously (18). The calculated rate constants were evaluated based on the least squares regression value, R2, and model selection criteria) (see Ref. 48 and Baylis Scanlon et al. (18)) values from statistical analyses performed to determine the goodness-of-fit.
General Methods and Materials—Except where [γ-32P]ATP was used, steady state ATPase activities were determined as described previously (37) using the Taussky and Shorr colorimetric method (49). The nucleotide content of cross-linked and uncross-linked F1 was determined by an ion exchange high pressure liquid chromatography assay using a Titansphere TiO2 column (Alltech Assoc., Deerfield, IL) as described previously (18). Protein concentrations were determined using the method of Lowry et al. (50) and in most instances cross-checked by the Amido Black protein assay (51). The algorithm of Fabiato and Fabiato (52) was used to determine the concentrations of Mg·ATP and free Mg2+. Gel electrophoresis was performed according to Laemmli (39) using the NuPAGE gel system (Invitrogen) and Tris/glycine 4-12% gradient gels. [γ-32P]ATP was purchased from GE Biosciences. All other chemicals were the highest grade commercially available.
RESULTS
Characterization of Cys Mutants and Cross-linked Enzymes—It was previously shown that a disulfide bond can be induced between the native γ subunit Cys-87 and a Cys substitution in place of either β subunit Asp-380 (27, 40) or Glu-381 (28) (Fig. 1). The β subunit residues are part of the conserved 380βDELSEED386 motif, the loop near the carboxyl terminus that interacts directly with the γ subunit which, in part, establishes the conformational asymmetry of the catalytic sites during the course of rotational catalysis (53). Perturbations of the γ-β interaction affect steady state catalysis as well as coupling efficiency between ATP hydrolysis/synthesis and rotation (37, 54).
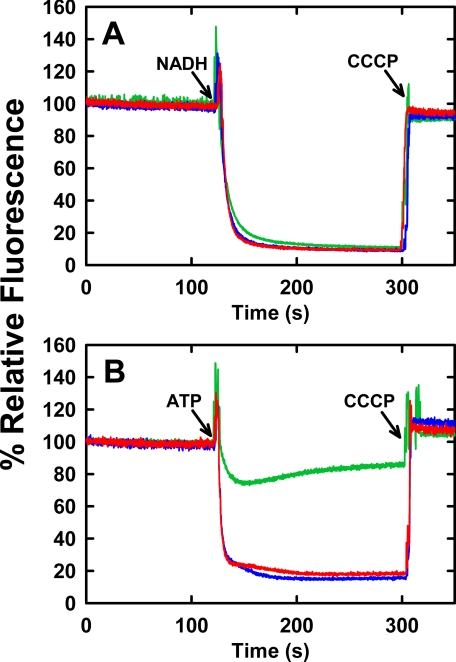
Growth yields of βE381C and βD380C mutant strains on minimally defined solid medium, with sodium succinate as the sole carbon source, were similar to wild type, indicating functional F0F1 enzymes competent enough to achieve net synthesis of ATP by oxidative phosphorylation. The expression levels of βE381C and βD380C F0F1 complexes in the inner membranes were measured quantitatively by immunoblot and were similar to wild-type F0F1 expression (Table 1). The steady state ATPase activities of F0F1-containing membranes of both mutants were also comparable with wild type (Table 1). Proton pumping of mutant F0F1 into inside-out inner membranes was measured by acridine orange fluorescence quenching and is shown in Fig. 2. The NADH-driven ΔpH for βE381C and βD380C membranes were similar to wild type (Fig. 2A) indicating the membranes from the mutant strains were not intrinsically more permeable to protons than wild-type membranes. This assay verifies that mutant F1 binds to F0 and is stable but is also used to determine the quality of the membrane preparations as a control for ATP-driven ΔpH. ATP-driven fluorescence quenching was only 25% for the βE381C membranes compared with 85 and 82% for wild type and βD380C, respectively. The βE381C mutant is able to sustain growth on succinate because its proton pumping ability, although impaired, is not completely abolished. The reduced ability of the βE381C mutant to form an ATP-driven pH gradient, despite similar expression level and ATPase activity to wild type, indicates that catalysis and transport are not efficiently coupled in this mutant. In contrast, the βD380C mutant enzyme is functionally similar to wild type in H+ pumping ability, and appears to have well coupled catalysis and transport.
TABLE 1.
ATP hydrolysis activities of membranes containing F0F1 and purified F1 before and after induction of the γ-β cross-link by oxidizing reagent
Conditions of assays are as described under “Experimental Procedures.”
|
Mutation |
F0F1
expressiona |
ATP
hydrolysisa |
|||||
|---|---|---|---|---|---|---|---|
|
Isolated F1 |
|||||||
| Membranesb | Untreatedc | After induction of γ-β cross-linkc,d | After treatment with 20 mm DTTc | Untreated Kme | Untreated kcat/Kme | ||
| % | μmol Pi/min/mg | s−1 | s−1 | s−1 | μm | s−1m1 | |
| Wild type | 12 ± 3 | 8.0 ± 0.9 | 95 ± 3 | NDf | NDf | 45 | 5.9 × 105 |
| βE381C | 20 ± 4 | 8.0 ± 1.9 | 120 ± 4 | 4.0 ± 0.9g | 130 ± 2 | 42 | 1.0 × 106 |
| βD380C | 15 ± 2 | 5.0 ± 0.9 | 84 ± 2 | 1.2 ± 0.02g | 77 ± 3 | 37 | 1.1 × 106 |
Expression of F0F1 on membranes was determined by quantitative immunoblot as outlined under “Experimental Procedures,” and values were expressed as % of total membrane protein, n = 4.
Membrane assays contained 1 μm carbonyl cyanide 3-chlorophenylhydrazone.
ATPase activities, under saturating ATP conditions, were measured in 50 mm HEPES-KOH, 10 mm ATP, 5 mm MgSO4, pH 7.5, at 30 °C, with an ATP-regenerating system of 5 mm phospho(enol)pyruvate and 50 μg/ml pyruvate kinase.
The γ-β cross-link was induced by treatment with 50 μm CuCl2 for βE381C-F1 and by treatment with 25 μm DTNB for βD380C-F1.
Values for Km and kcat were determined by measuring ATPase activity as a function of Mg·ATP concentration under conditions maintaining a low free Mg2+ concentration of 50 μm. The buffer consisted of 25 mm TES-KOH, 0.2 mm EDTA, and varying amounts of Na2-ATP (25 μm to 10 mm) and MgCl2 (0.269 mm to 4.57 mm), pH 7.5, at 25 °C, with an ATP-regenerating system of 2 mm phospho(enol)pyruvate, 50 μg/ml pyruvate kinase.
ND means experiment was not repeated here. Duncan et al. (27) have already shown there is no effect of either DTNB or DTT on wild-type activity.
Activity was fully inhibitable by 1 mm sodium azide, indicating the extent of uncross-linked enzyme.
FIGURE 2.
NADH- or ATP-dependent proton electrochemical gradient formation by mutant F0F1 in membranes. 200 μg of membrane vesicle protein from strain DK8, harboring plasmid pBWU13 wild type or carrying the mutations for βE381C or βD380C, were suspended in 2 ml of buffer containing 10 mm HEPES-KOH, 300 mm KCl, 5 mm MgCl2, 1 μm valinomycin, and 1 μm acridine orange at pH 7.5, with vigorous mixing. Fluorescence intensity at 530 nm (excitation at 460 nm) was monitored at 25 °C. A, NADH-driven quenching. Proton pumping was initiated at the indicated time (arrow) by addition of 1 mm NADH. B, ATP-driven quenching. Proton pumping was initiated at the indicated time (arrow) by addition of 1 mm ATP. 1 μm carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added, as indicated (arrows) to abolish the proton gradient and establish the maximum fluorescence value. The fluorescence traces are depicted as follows: wild type (blue), βD380C (red), βE381C (green). The traces are representative of those from several trials.
Steady state ATP hydrolysis rates of βE381C-F1 and βD380C-F1 were measured as a function of Mg·ATP concentration (10 μm to 4.3 mm) under conditions of a low free Mg2+ concentration of 50 μm, in the presence of an ATP-regenerating system, at 25 °C and pH 7.5. βE381C-F1 had a kcat of 41 s-1 and Km of 42 μm. βD380C-F1 had a kcat of 41 s-1 and Km of 37 μm (Table 1). These were similar to wild-type values measured under the same conditions (kcat of 26.6 s-1 and Km of 45 μm). The specificity constants (kcat/Km: βE381C-F1, 1.0 × 106 s-1 m-1; βD380C-F1, 1.1 × 106 s-1 m-1) were also similar to wild type (5.9 × 105 s-1 m-1), hence verifying that the mechanism of ATP hydrolysis in F1 is not drastically altered by the cysteine substitutions.
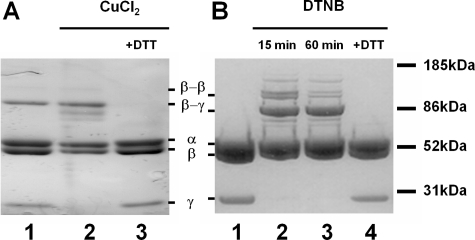
Formation of the Cross-link between γCys-87 and βE381C or βD380C—The disulfide bond forms to a slight degree without the presence of oxidizing agents in the βE381C mutant but not in the βD380C mutant (Fig. 3, A and B, lane 1). To maximize cross-linking, 50 μm copper chloride or 25 μm DTNB was used to induce cross-linking of γCys-87 to βE381C-F1 or βD380C-F1, respectively. A small percentage (∼0.5%) of the β subunits cross-link to form a homodimer (Fig. 3); however, this level is insignificant compared with the γ-β dimer, and it is not enough to affect the kinetic analyses described below. Consistent with previous studies (29, 55), the formation of the γ-β disulfide bond is effective in inhibiting the steady state turnover of both mutant enzymes (Table 1). There still remained ∼1% of the maximal activity for the cross-linked βD380C-F1 and ∼5% of activity for βE381C-F1. We questioned whether this slow rate of hydrolysis was because of the slow unisite mode of catalysis (29) or because of the small amount of uncross-linked enzyme. The residual activity for each enzyme was examined by treatment with 1 mm sodium azide (NaN3), as this compound apparently affects catalytic site cooperativity, and thus only inhibits F1-ATPase multisite activity with no effect on unisite activity (56). The NaN3 treatment verified that the activity measured after cross-linking was due almost entirely to uncross-linked enzyme. This corroborates observations that the level of inhibition of activity for γCys-87-βE381C-F1 correlated with the yield of cross-linking (29). Addition of 10 mm DTT removed the disulfide bond and restored steady state activity for both mutants (Table 1). In the case of βE381C, the activity level for the DTT-treated enzyme was higher than the initial rate because of complete reduction of the small fraction of cross-linked enzyme present prior to addition of CuCl2. There was no effect on wild-type steady state membrane ATPase activity by CuCl2, DTNB, or DTT, as shown previously (27).
FIGURE 3.
Reversible γ-β disulfide bond formation between native γCys-87 and βE381C or βD380C. A, βE381C-F1 was treated with 50 μm CuCl2, and aliquots were taken for SDS-PAGE and ATPase assays (activities listed in Table 1). Lane 1, initial sample, no treatment. Lane 2, βE381C-F1 after 1 h of incubation with CuCl2. Lane 3, βE381C-F1 incubated for 30 min with 10 mm DTT following the reaction with CuCl2. B, βD380C-F1 was treated with 25 μm DTNB and aliquots were taken for SDS-PAGE and ATPase assays (activities listed in Table 1). Lane 1, initial sample, pretreatment. Lane 2, βD380C-F1 after 15 min of incubation with DTNB. Lane 3, βD380C-F1 after 60 min of incubation with DTNB. Lane 4, βD380C-F1 incubated for 30 min with 10 mm DTT after the reaction with DTNB. Note that the gels are slightly overloaded to emphasize the complete disappearance of the γ subunit into the γ-β cross-linked product.
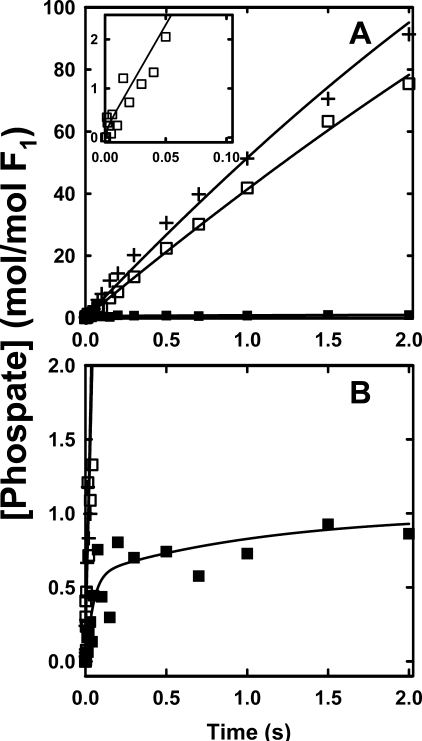
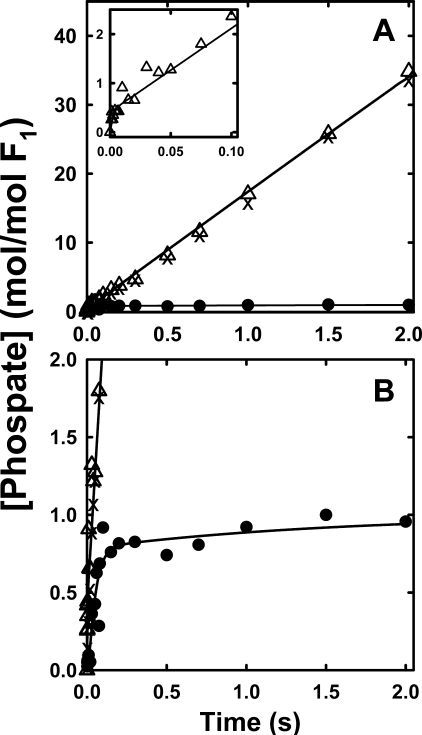
Unisite Binding and Hydrolysis of [γ-32P]ATP—The unisite kinetic activity was examined to determine the elementary rate constants, and thus whether either βDELSEED mutation or γ-β cross-link perturbed the mechanism of the enzymatic reaction at the high affinity catalytic site, which is believed to be the βTP conformer (18, 24). Unisite parameters for βE381C-F1, both cross-linked and uncross-linked, were determined by Garcia and Capaldi (29), and their rate constants are listed in Table 2. Their omission of values for k+2 and k-2 necessitated a refitting of their primary data, which was achieved without much change in the other rate constants. They noted an increased rate of ATP binding (k+1) compared with their measurements for wild-type F1, although the value was comparable with other previously published results for the wild-type enzyme (14), also listed in Table 2 for reference. The βE381C-F1 enzyme had an ATP off rate, k-1, 24-fold faster than wild type, and a lower affinity for ATP. It also showed slightly increased rates of unisite activity compared with wild type, due mostly to higher rates of ATP hydrolysis (k+2) and Pi release (k+3), although importantly, the hydrolysis/synthesis ratio, K2, was essentially unchanged either by the mutation or by cross-link formation.
TABLE 2.
Rate constants and kinetically calculated binding constants of cross-linked and noncross-linked enzymes
| k+1 | k−1 | K1 | k+2 | k−2 | K2 | kγ | k+3 | k−3 | K−3 | k+4 | k−4 | K−4 | Turnover (Vmax) | R2 | Model selection criteria | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m−1s−1 | s−1 | m−1s−1 | s−1 | s−1 | s−1 | s−1 | m−1s−1 | m−1 | s−1 | m−1s−1 | m1 | s−1 | ||||
| Unisite | ||||||||||||||||
| Wild typea | 1.1 × 105 | 2.5 × 10−5 | 4.4 × 109 | 0.12 | 0.043 | 2.8 | 1.2 × 10−3 | 4.8 × 10−4 | 0.4 | 1.6 × 10−3 | 1.8 × 102 | 1.1 × 105 | ||||
| βE381Cb | 5.3 × 105 | 6.0 × 10−4 | 8.8 × 108 | 1.2c | 1.2c | 1.0 | 2.2 × 10−2 | NDd | ND | 2.2 × 10−3 | ND | ND | 0.998c | 3.23c | ||
| βE381C(γ-β)b | 6.1 × 105 | 3.1 × 10−2 | 2.0 × 107 | 1.3c | 0.5c | 2.6c | 2.8 × 10−2 | ND | ND | 7.7 × 10−3 | ND | ND | 0.998c | 3.96c | ||
| βD380C | 6.5 × 105 | 7.3 × 10−2 | 8.9 × 106 | 1.8 | 1.2 | 1.5 | 2.5 × 10−3 | ND | ND | ND | ND | ND | 0.997 | 3.31 | ||
|
βD380C(γ-β) |
1.8 × 105 |
2.4 × 10−2 |
7.5 × 106 |
0.54 |
0.15 |
3.6 |
6.4 × 10−4 |
ND |
ND |
ND |
ND |
ND |
0.998 |
4.27 |
||
| Multisite | ||||||||||||||||
| Wild-typee | 4.0 × 107 | 1.9 × 103 | 2.1 × 104 | 140 | 120 | 1.2 | 29 | 210 | 13 | 0.062 | 360 | 4.2 × 106 | 1.2 × 104 | 27 | 0.997 | 5.10 |
| βE381C | 5.5 × 107 | 3.7 × 103 | 1.5 × 104 | 150 | 110 | 1.4 | 426 | 220 | 17 | 0.077 | 540 | 8.2 × 106 | 1.5 × 104 | 41 | 0.998 | 5.47 |
| βE381C(γ-β) | 2.2 × 105 | 6.1 × 10−3 | 3.6 × 107 | 120 | 80 | 1.5 | 0 | 0 (1.5)f | 0 | 0 | 0 | 0 | 0 | (0.44)f | 0.958 | 1.77 |
| βD380C | 4.4 × 107 | 2.1 × 103 | 2.1 × 104 | 270 | 150 | 1.8 | 38 | 267 | 26 | 0.1 | 826 | 4.0 × 105 | 4.8 × 102 | 41 | 0.999 | 6.30 |
| βD380C(γ-β) | 2.3 × 105 | 5.2 × 10−3 | 4.4 × 107 | 63 | 17 | 3.8 | 0 | 0 (0.8)f | 0 | 0 | 0 | 0 | 0 | (0.16)f | 0.972 | 2.60 |
Fig. 4 shows the hydrolysis of substoichiometric amounts of [γ-32P]ATP by βD380C-F1 and γ-β cross-linked βD380C-F1. The total 32Pi (enzyme-bound 32Pi plus 32Pi released to the medium) was determined by directly quenching samples with acid. The glucose/hexokinase trap additionally measured [γ-32P]ATP bound to F1. These two experiments allow derivation of ATP binding (k+1), ATP release (k-1), the ATP hydrolysis/synthesis rates (k+2, k-2), and Pi release (k+3) from fits of the data using a minimal unisite reaction scheme (ATP binding through Pi release) (14). The elemental rate constants are listed in Table 2. The parameters for both the βD380C-F1 and γ-β cross-linked βD380C-F1 enzymes were very similar to those measured for the wild-type enzyme, with the exception of faster rates for ATP release (k-1), resulting in a lower affinity for ATP, even greater than the effect described above for the βE381C mutant. Results from cold chase experiments corroborated this finding where addition of excess unlabeled ATP failed to promote ATP hydrolysis in βD380C-F1 previously incubated with [γ-32P]ATP, and actually decreased bound [γ-32P]ATP (data not shown). This is consistent with a fast ATP off rate, k-1, which allows the excess cold chase of ATP to displace the enzyme-bound [γ-32P]ATP. Importantly, the ATP hydrolysis/synthesis equilibrium constant, K2, was not changed by the βD380C mutation nor by the γ-β cross-linked enzyme, with values close to unity in accordance with Boyer's binding change mechanism (57, 58). In summary, unisite analysis shows that neither Cys mutation disrupts the mechanism of ATP hydrolysis at the high affinity catalytic site, although there is a decreased affinity for ATP. In addition, the βD380C unisite rate constants are essentially unaltered upon formation of the γ-β cross-link, in agreement with measurements of the βE381C enzyme (29), and verifying that the mechanism of unisite hydrolysis of ATP at the high affinity catalytic site does not involve γ subunit rotation.
FIGURE 4.
Unisite hydrolysis of βD380C-F1 and γ-β cross-linked βD380C-F1. F1 was prepared either by exchanging into unisite buffer or by treatment with DTNB and then exchanging into unisite buffer, as described under “Experimental Procedures.” Hydrolysis was initiated by adding 0.1 μm [γ-32P]ATP to 1 μm F1 (with equimolar ε added) in unisite buffer (50 mm Tris-OH, 50 mm MOPS, 4.5 mm K2SO4, and 0.5 mm MgSO4 adjusted to pH 7.5 with H2SO4 at 25 °C). A, the early time points. B, complete time course for the βD380C-F1 acid-quenched hydrolysis (•) and bound [γ-32P]ATP (○) measured in parallel by the hexokinase trap method. C, early time points. D, complete time course for the γ-β cross-linked βD380C-F1 reactions. At the times indicated samples were quenched directly with acid (▪), and bound [γ-32P]ATP (□) was measured in parallel by the hexokinase trap method. The lines indicated are the best fits to the data, and the parameters are listed in Table 2.
Pre-steady State Multisite Hydrolysis of [γ32P]ATP—We previously determined the partial reaction rate constants of the catalytic pathway involving γ subunit rotation by analyzing the pre-steady state kinetics of multisite ATP hydrolysis (18). Our data showed that rapid mixing with high Mg·ATP concentrations put the enzyme immediately into a rotational catalytic mode. Similar conditions were used in this study, for the same rationale. Briefly, nucleotide occupancy of all three catalytic sites is needed to ensure the enzyme enters the rotational catalytic mode (12, 20), and low free Mg2+ conditions are used to avoid the inhibitory effects of high concentrations of this ion (59, 60). Reaction conditions used for the time course of ATP hydrolysis are a compromise to satisfy these requirements, but also necessitate use of radioactive [γ-32P]ATP because of the small amounts of substrate hydrolyzed in the first few turn-overs. We also wished to limit the total radioactivity to keep the background at a reasonable level. The final concentrations after mixing are 0.5 μm F1, 0.352 mm MgCl2, and 0.25 mm [γ-32P]ATP to give [Mg2+free] = 49 μm and [Mg·ATP] = 105 μm at pH 7.5 and 25 °C. This Mg·ATP concentration is ∼2.5-fold higher than the Km value for steady state ATP hydrolysis (42 and 37 μm for βE381C and βD380C, respectively, and 45 μm for wild-type F1, see Table 1). Based on the affinity constants from Senior and co-workers (61, 62), and our previous work monitoring pre-steady state nucleotide binding (18), this is an adequate concentration of Mg·ATP to fill two sites completely and the third site partially.
We previously showed that the rate-limiting step, kγ, can be distinguished by pre-steady state analysis of Mg·ATP hydrolysis in the millisecond time regime (18). The pre-steady state hydrolysis of βE381C-F1 and βD380C-F1 before and after treatment with the oxidizing reagent and the curve fits to the data are shown in Fig. 5 and 6, respectively. After preparation of the F1 for the reaction, the nucleotide contents of each sample were determined to be close to 4.0 mol of nucleotides bound per mol of F1, and are listed in the appropriate figure legends. We assume that 3 mol of nucleotide are bound to the noncatalytic α subunits, and 1 mol of ADP is bound to the βDP site (18).
FIGURE 5.
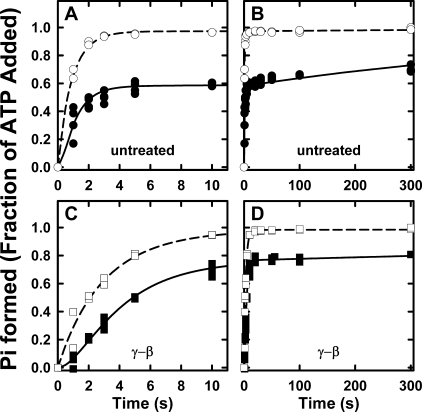
Pre-steady state hydrolysis of βE381C-F1 before and after induction of γ-β cross-link. A, complete time course up to 2.0 s. Inset, early time points up to 0.1 s of uncross-linked sample. B, complete time course with expanded y axis to show the γ-β cross-linked sample. Purified F1 was prepared as described under “Experimental Procedures” giving 4.0 ± 0.1 mol of nucleotide bound per mol of F1; equimolar ε was added, and the sample was diluted to 1 μm in 25 mm TES-KOH, 0.244 mm MgCl2, 0.2 mm EDTA, pH 7.5, at 25 °C and loaded into syringe A of the chemical quench instrument. Syringe B contained 25 mm TES-KOH, 0.46 mm MgCl2, 0.20 mm EDTA, 0.50 mm [γ-32P]ATP, pH 7.5, at 25 °C (final concentrations: 0.5 μm F1, 0.352 mm MgCl2, 0.25 mm [γ-32P]ATP; [Mg2+]free = 49 μm, [Mg·ATP] = 105 μm). 32Pi produced is reported in moles of Pi per mol of F1 for untreated F1 (□), F1 incubated with 50 μm CuCl2 to form the γ-β cross-linked sample (▪), and F1 incubated with 10 mm DTT after CuCl2 treatment (+). The solid lines are the best fits of each data set to our model, using the parameters listed in Table 2.
FIGURE 6.
Pre-steady state hydrolysis of βD380C-F1 before and after induction of γ-β cross-link. A, complete time course up to 2.0 s. Inset, early time points up to 0.1 s of uncross-linked sample. Note the y axis scales compared with Fig. 5. B, complete time course with expanded y axis to show the γ-β cross-linked sample. Preparation of F1 and reaction conditions are essentially the same as the experiments in Fig. 5, except that βD380CF1 is treated with 25 μm DTNB to induce the γ-β cross-link formation. Untreated F1 (○) initially contained 4.0 ± 0.1 nucleotides; F1 after DTNB incubation induced the γ-β cross-link (•), containing 3.9 ± 0.1 nucleotides; and DTT-treated F1 reduced the γ-β cross-link (×). The solid lines are the best fits of each data set to our model, using the parameters listed in Table 2.
As described under “Experimental Procedures” and explained in our previous analysis (18), the pre-steady state kinetics of ATP hydrolysis provide adequate constraints to define a unique model for the partial reaction pathway. The same model (see Scheme 1 and Table 2) was used to fit the kinetics of the cross-linked enzymes. Rate constants for the fits are listed in Table 2. Our rate constants for ATP binding for wild-type and both untreated mutants are consistent with the ATP kon value of 2.6 × 107 m-1s-1 determined from single molecule experiments (20). The total time courses of the reactions (Fig. 5A and Fig. 6A) show the cross-link was effective in inhibiting the steady state turnover in both mutants (closed symbols). The very slow hydrolysis and Pi production by the cross-linked enzymes, observed in the longer time course, are completely azide-inhibitable and because of the small amount of uncross-linked complex. The steady state hydrolysis was restored upon addition of 10 mm DTT for both enzymes (“+” in Fig. 5 and and “×” in Fig. 6). The βE381C enzyme hydrolytic activity, after addition of DTT, was increased to a rate ∼10% faster than the untreated sample (Fig. 5A), because of the presence of ∼10% of cross-linked enzyme in the untreated sample (Fig. 3A, lane 1). The steady state hydrolysis rates under the quench flow conditions are consistent with those under maximal ATP hydrolysis conditions, listed in Table 1, such that the activity of the DTT-treated sample is ∼10% faster than untreated sample.
Further examination of the untreated βE381C enzyme (before addition of CuCl2) yields important insights into the coupling function of the enzyme. As seen in the inset of Fig. 5A, depicting the early time course of the reaction, the untreated βE381C has no initial burst of hydrolysis and therefore no substantially slower step after hydrolysis, which would cause product accumulation. The rate constants for the data fits are listed in Table 2, and comparison of the βE381C enzyme to wild type shows similar rates for ATP binding and unbinding (k+1 and k-1), which reflects the affinity for ATP of the low affinity catalytic site, βE (see Introduction). The hydrolysis and synthesis rates (k+2 and k-2) are also similar, yielding a K2 value close to unity and showing the hydrolysis reaction at the active site is essentially unperturbed. The major functional difference of the βE381C mutant becomes evident when comparing rates of kγ, previously defined as the rate-limiting step and attributed, at least in part, to γ subunit rotation (18). The βE381C enzyme has an extremely fast kγ, which is not rate-limiting to the reaction. This is very different from the wild-type enzyme, which displays an initial burst of hydrolysis (18), and can only be fit by a reaction scheme that includes a relatively slow step after hydrolysis and before product release (18). The βE381C enzyme proceeds through a different catalytic pathway where kγ is not the rate-limiting step, which we hypothesize is not associated with γ subunit rotation (see “Discussion”). Our results confirm the importance of the γ-βDELSEED interaction in coupling efficiency.
In contrast to the βE381C mutant, the pre-steady state activity of the uncross-linked βD380C-F1 is very similar to wild type with an initial burst of hydrolysis in the first few milliseconds, after which the enzyme enters the steady state Fig. 6A (inset). The parameters for the fits are also very similar to the wild-type rate constants as follows: 1) the affinity for ATP, giving a Kd(ATP) for the third site similar to the Km value for ATP hydrolysis; 2) the hydrolysis/synthesis ratio, K2, close to 1, and 3) the value of kγ, the rate-limiting step of the reaction.
Fig. 5B and Fig. 6B each have an expanded y axis to allow clear illustration of the time course of Mg·ATP hydrolysis by the cross-linked enzymes. Both enzymes exhibit a burst of hydrolysis despite the γ-β rotation-inhibiting cross-link. Thereafter, the hydrolysis is severely abrogated. The initial burst is slower but of a similar amplitude as wild type, showing that inhibition of rotation does not prevent the first turnover of Mg·ATP hydrolysis. Moreover, the burst amplitude shows that simultaneous hydrolysis at multiple sites does not occur, and thus hydrolysis follows a sequential mechanism involving only a single site at any instant during catalysis.
It is also important to note that the rate constants for the fits of both cross-linked enzymes are similar suggesting that the functional uncoupling between catalysis and proton pumping observed in the steady state for the βE381C enzyme (Fig. 2B) is because of perturbation of γ subunit rotation, which presumably does not occur in the cross-linked enzyme (see “Discussion”). This is consistent with our previous thermodynamic analyses that suggested that the transition state for the steady state reaction occurs during γ subunit rotation between hydrolysis and release of Pi (23, 30).
The cross-linked enzymes had an ATP on rate (k+1), which was 2 orders of magnitude slower, and an ATP off rate (k-1), which was 5 orders of magnitude slower compared with the untreated enzyme. The hydrolysis/synthesis ratio, K2, remained close to that for wild type, still showing no great perturbation of the catalytic mechanism. Importantly the rates of ATP hydrolysis (k+2) and resynthesis (k-2) are similar to the untreated mutant enzymes and wild type under multisite conditions, indicating that the positive cooperativity for ATP hydrolysis (16) remains essentially intact despite inhibition of γ rotation.
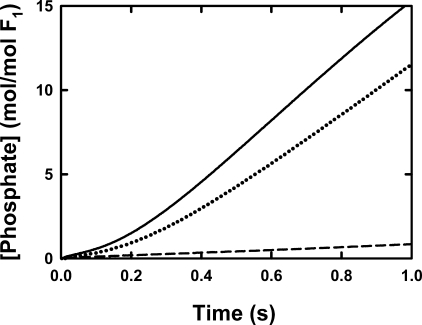
Pi Release for βD380C-F1—We will now focus on the βD380C enzyme because it is better coupled and therefore is more representative of the coupled pathway. We showed previously that the order of kγ in the reaction was critical and occurs after hydrolysis but before Pi release (18). The prediction from our model was that all steps after rotation should be inhibited in the γ-β cross-linked enzyme. To test this directly, we followed the release of Pi from the βD380C enzyme by measuring the fluorescence intensity change of the MDCC-labeled phosphate-binding protein (MDCC-PBP, see “Experimental Procedures” and Ref. 45). This protein binds Pi very fast (kon = 1.36 × 108 m-1s-1) with high affinity (Kd is ∼0.1 μm) and is an accurate probe for Pi in solution at low concentrations (63). The release of Pi from βD380C-F1 was monitored by 20 μm MDCC-PBP in the fluorescence stopped-flow spectrometer (see “Experimental Procedures”) using essentially the same reaction conditions as the quench flow experiment. Calibration experiments demonstrate that the fluorescence response of MDCC-PBP to rapid addition of 1-20 μm Pi is within the 1-2-ms dead time of the stopped-flow instrument (data not shown). Ideally, the rate of Pi release from F1 would be directly determined from this experiment; however, upon further examination it was evident that the PBP interacts both directly and indirectly with F1, resulting in a slight inhibition of ATP hydrolysis (data not shown). Although this negates quantitative analysis of the data, qualitative analysis can still yield valuable information. The Pi release data are valid up to ∼16 μm of released Pi because of the limiting amount of MDCC-PBP. Around this point, the fluorescence signal begins to saturate and no longer follows the hydrolysis kinetics. It is clear that upon formation of the rotation-inhibiting γ-β cross-link, the enzyme is prevented from releasing product Pi (Fig. 7). There is a very slow residual rate of Pi release, but this is because of a small amount of uncross-linked enzyme in the sample. Upon addition of DTT, the rate of Pi release increases and approaches the initial rate before treatment with DTNB. In this particular case the DTT-treated enzyme had a slightly slower steady state rate because of incomplete reduction of the γ-β cross-link. This demonstrates the reversible nature of the blockage of Pi release and that it is dependent on disulfide bond formation.
FIGURE 7.
Pre-steady release of Pi by βD380C-F1. Details are given under “Experimental Procedures” for the stopped-flow set up. F1 was prepared as described under “Experimental Procedures,” giving 4.0 ± 0.1 mol of nucleotide bound per mol of F1, equilibrated with 25 mm TES-KOH, 0.244 mm MgCl2, 0.2 mm EDTA, pH 7.5, at 25 °C, diluted to 1 μm with the same buffer, and loaded in syringe A. The ATP and MgCl2 concentrations in syringe B resulted in a final free Mg2+ of ∼50 and 105 μm Mg·ATP. The stopped-flow syringe also contained 20 μm MDCC-PBP. The traces of MDCC-PBP fluorescence show Pi released from the enzyme and were calibrated by measuring the fluorescence response to known concentrations of Pi. The response loses linearity above ∼16 μm because of saturation of Pi binding. Each trace is the average of at least three stopped-flow mixing experiments. The noise in the averaged data is within the thickness of the lines shown. The solid line shows the Pi release from the untreated F1; the dashed line depicts that from the γ-β cross-linked F1; and the dotted line is the Pi release from the partially reduced cross-linked sample after incubation with 5 mm DTT.
In the cases of the noncross-linked enzymes, we note that there is no burst of Pi release as shown for βD380C-F1 (Fig. 7), but rather a lag before Pi is released, which indicates a delay between hydrolysis of Mg·ATP (Fig. 6A) and the release of Pi, and it emphasizes the hydrolytic step is kinetically distinct from kγ and Pi release. Above all, these results show that γ subunit rotation, although not essential for the first cycle of ATP hydrolysis, is absolutely integral for decreasing the enzyme affinity for Pi, thus enabling F1 to release Pi and enter into the steady state mode of hydrolysis.
DISCUSSION
γ-β Cross-link Blocks the Rotation Step, kγ—The γ87Cys-β cross-link is effective in inhibiting steady state activity for both the βD380C and βE381C enzymes (Table 1, Figs. 5 and 6) under high Mg·ATP conditions, consistent with the notion that inhibition of rotation prevents the enzyme from entering the steady state mode. It is obvious from the multisite fit parameters in Table 2 that the steps most affected by cross-linking are kγ and steps subsequent to kγ in the reaction scheme. kγ is arrested, and aside from a low rate of Pi release, which was shown to be due to a very small fraction of uncross-linked enzyme, the data could be well fit to a minimal model with no steps subsequent to the hydrolytic step (k+2). Pi release is the major energy-producing process in the hydrolysis mode based on evidence from the synthesis direction that productive Pi binding requires an input of energy (23), and it does not occur without the F0 transport moiety and a protonmotive force (12-14, 64, 65). Concurring with this is the unidirectional rotation of F1, which does not rotate in reverse under high Mg·ATP conditions (6, 66). Product Pi is not released from the γ-β cross-linked enzyme, as demonstrated by use of the PBP-MDCC Pi detection system (Fig. 7), indicating that the γ subunit cannot rotate to carry out the power stroke that is required to release Pi. Clearly the enzyme is stuck at the hydrolysis step. Additional evidence that kγ involves γ subunit rotation step is provided by unisite analysis, which does not require a rate-limiting step to fit the data, and shows no change in the rate constants upon γ-β cross-link formation (Table 2) (29).
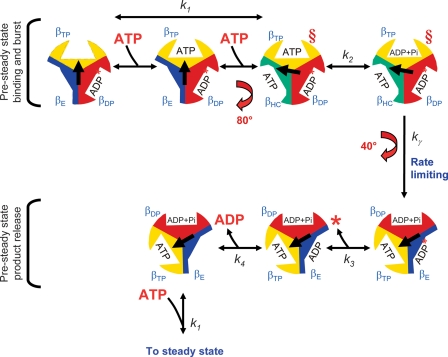
γ-β Cross-linked Enzymes Assume the 80° Conformation and Allow Assignment of Catalytic Sites—The γ-β cross-linked enzymes retain the burst of ATP hydrolysis (Fig. 5B and Fig. 6B), showing that the enzyme can hydrolyze substrate during the first cycle despite the rotation-inhibiting cross-link. However, direct effects on the rotational behavior of single molecules of F1 incubated with the slowly hydrolyzable substrate, ATPγS (25, 26), and of F1 complexes containing the hydrolysis-impeding βE190D mutation (21), show that hydrolysis occurs during the dwell time after the 80° γ subunit rotation, and before the 40° γ subunit rotation. We can reconcile these results by considering the conformational state of the cross-linked enzyme. The data are consistent with the γ-β cross-link rigidly locking the enzyme in a conformation similar to that achieved after the 80° rotation, depicted by the red § in Fig. 8. This is in agreement with the conclusion from the fluorescence study by Yasuda et al. (67) that the crystal structures solved by the Walker group (68-71) are more similar to the 80° conformation of the enzyme.
FIGURE 8.
Graphical representation of the pre-steady state to steady state pathway. See text for discussion of the model. The relative arrangement of the γ subunit conformations are as viewed from the bottom of the complex from the membrane. The conformations of the catalytic sites are labeled as βDP, βTP, βE, and βHC (the half-closed conformation described in Menz et al. (77)). The α subunits were omitted for clarity. The central arrow represents the relative position of the γ subunit during the course of multisite catalysis. The intermediate rotation of 80° and the completion of rotation to the next 120° position corresponds to the dwell positions observed by Yasuda et al. (20). The red § depict the steps in the pathway we propose are closest to the conformation of the cross-linked enzyme. We propose that unisite hydrolysis of ATP by the wild-type enzyme occurs in a conformation similar to that depicted prior to the 80° rotation step where ATP is bound only to the βTP site, and no nucleotide is bound to the βE site. In the steady state, the position of the γ subunit is offset 120° for each cycle in the steady state. The asterisk indicates that there is no Pi release in the first pre-steady state cycle (see Ref. 18 for discussion).
In fact, the conformation of the cross-linked enzyme allows us to distinguish between unisite and multisite rotational catalysis. Under multisite conditions, when rotation is inhibited by the γ-β cross-link, the rate of the hydrolytic burst is slower (Fig. 5B and Fig. 6B), due to 2 orders of magnitude slower ATP binding (k+1) and 5 orders of magnitude slower ATP unbinding (k-1), compared with the uncross-linked and wild-type enzymes (Table 2). We note that these rates for k+1 and k-1 are similar to all enzymes in unisite conditions; therefore, unisite experiments and cross-linked multisite behavior reflect binding of Mg·ATP to the same site, βTP. This is in contrast to the rates of ATP binding and unbinding in the multisite uncross-linked and wild-type enzymes that we have shown previously are because of binding to the low affinity βE site (18).
Effect of βDELSEED Mutations on the High Affinity Catalytic Site, βTP—The parameter fits to the unisite data of the βE381C and βD380C enzymes elucidate slight but significant differences between the two mutants. Based on the unisite experiments (Table 2), the βE381C mutant has an affinity for ATP that is similar to wild type, whereas the effect of the βD380C mutation is to decrease the affinity of the βTP catalytic site for ATP by about 500-fold. Upon formation of the γ-β cross-link, differences between the mutants are no longer apparent. Both cross-linked enzymes exhibit a decreased affinity for Mg·ATP at the βTP site, compared with wild type, because both have 2 orders of magnitude lower affinity (k+1/k-1) in unisite conditions (Table 1).
The two βDELSEED Cys mutations examined here have increased unisite rates of ATP hydrolysis, k+2, and resynthesis, k-2 (Table 2); however, they do not have a drastic effect on altering the catalytic mechanism, because the catalytic equilibrium constant, K2, and the specificity constant, kcat/Km, remain unaltered from wild-type values. The βE381C and βD380C mutations appear to affect the conformation of the high affinity catalytic site, βTP, as indicated by the lower affinity in unisite conditions and in the cross-linked enzyme under multisite conditions. In contrast analysis of multisite catalysis shows these mutations do not seem to have much influence on βE (Table 2) because k+1 and k-1 of uncross-linked and wild type enzymes are not affected. These results demonstrate the role of specific βDEL-SEED residues, and their interaction with the γ subunit, in establishing the proper conformation of the catalytic site at the proper time during rotational catalysis. This conclusion was previously made by analysis of γ subunit mutations (37) and is particularly interesting considering that the βDELSEED motif is physically distant from the nucleotide-binding sites (Fig. 1).
Role of the βGlu-381 of the DELSEED Motif in Chemomechanical Coupling—The marked functional differences between the two βDELSEED single Cys mutants become more apparent in the multisite mode of rotational catalysis and are particularly interesting because of their adjacent positions in the sequence. The βD380C mutant has very similar characteristics to wild type, and the transmission of coupling energy between the two distinct functions of proton translocation and ATP hydrolysis/synthesis is intact, i.e. it is a well coupled enzyme. This mutant can be used as a close representation of the activity and behavior of the wild-type enzyme. In contrast to the βD380C mutant, the βE381C mutant behaves in a manner dissimilar to the wild-type enzyme with characteristics indicating that catalysis and rotation are not efficiently coupled in this enzyme. Interestingly, Garcia and Capaldi (29) used the βE381C enzyme and must have been measuring an uncoupled enzyme. This would not affect their conclusions as most of their measurements were of unisite kinetics and transmission of coupling information requires γ subunit rotation and consequently participation of all three catalytic sites (12). Indeed, functional differences in coupling between the βE381C and βD380C mutants are alleviated when the γ-β cross-link is induced, validating the conclusion that the high energy transition state involves the power stroke rotation of the γ subunit, and specific interactions at the γ-β interface are critical for proper transmission of coupling energy (37).
The multisite kinetic rate constants of the uncross-linked βE381C-F1 were very similar to wild type, with the exception of kγ (Table 2). This enzyme hydrolyzes ATP faster than wild type because of a much faster kγ (Table 2), which is no longer the rate-limiting step of the reaction. The power stroke is not fully engaged in this mutant as it likely slips frequently and goes through a different pathway, allowing the enzyme to release Pi without complete rotation of γ. An analogy for this fast yet unproductive hydrolysis is the revving of a car engine with a partially disengaged or slipping clutch. The βDELSEED loop in a sense is the clutch motif that engages the γ subunit to cause rotation.
The uncoupling between the catalytic reaction and the rate-limiting rotation step, kγ, is likely the cause of inefficient transmission of information to the F0 transport sector (Fig. 2B). We propose that the alternative uncoupled pathway does not involve full rotation of the γ subunit. The γ and ε subunits are not mutated and neither is the F0 complex; therefore, for each ATP hydrolyzed, a complete 120° rotation of γ should drive the H+ transport. The 80° ATP-dependent rotation step is unaffected in the βE381C mutant, based on the similar rates of ATP binding and unbinding (k+1 and k-1) and ATP hydrolysis and resynthesis (k+2, k-2). In contrast, kγ, the rate-limiting step correlated with the 40° substep, is severely perturbed such that it is 2 orders of magnitude faster and appears to be the step that slips. Complete inhibition of rotation by the γCys-87-βE381C cross-link prevents Pi release and abrogates enzyme activity after the first cycle. The alternative pathway is also inhibited by the cross-link showing that partial rotation is necessary for activity. In the alternative pathway, it is likely that the slip of the rotation allows the γ subunit to fall back to the position prior to the 80° substep for a new cycle of hydrolysis (23). We speculate the mechanism for this involves Pi release from the βTP site instead of the βDP site (Fig. 8), without the 40° rotation of γ, thereby altering the timing and coupling between rotation and Pi release.
The much higher rate constant for kγ implies a lower transition state for the alternative uncoupled pathway so the βE381C mutation has made this bypass pathway in the reaction more likely. This is reminiscent of the γM23K mutation, which established that an alternative pathway of hydrolysis can exist (72). Thermodynamic analysis of the γM23K mutation revealed that inefficient communication of coupling energy, from the catalytic sites to the F0 transport sector, was because of increased energy of interaction between γ and β possibly caused by an extra ionized hydrogen bond between the mutant and βGlu-381 (37). The γM23K mutant impeded kγ, the rate-limiting transition state, and resulted in a slip or branch in the pathway allowing unproductive dissipation of energy (30, 37, 73). Importantly, this resulted in a 50-fold increase in the rate of Pi release (73), emphasizing that the very tight association between kγ and Pi release is required for efficient coupling.
βE381C-F1 may rotate under a load as seen by Hara et al. (74) who observed similar torque generation by the Bacillus PS3 thermophilic enzyme with an Ala mutation at the β381 position. Hara et al. (74) suggested that the side chain charges of the βDELSEED motif do not play a role in torque generation and proposed a stearic role for the loop in driving the γ subunit rotation. However, it is expected that a slipping mutation would either generate full torque or not at all during a slip. In the single molecule studies, slip events cannot be distinguished. In other words, the limitation of these measurements is that the catalytic reaction, especially release of products, cannot be observed concurrent with observations of rotation.
Implications for Our Model of Rotational Catalysis—Our model remains essentially the same as that published previously (18). We have modified the graphical representation to show only the pre-steady state steps of the reaction, and to illustrate enzyme rotation as viewed from the “bottom” or direction of the membrane for consistency with model illustrations presented by investigators measuring single particle rotation behavior (Fig. 8). Our analysis in this study clarifies the roles of each site where binding of ATP to βE promotes the hydrolysis/synthesis rates occurring in βTP, which occurs even in the cross-linked enzymes. βDP retains products of hydrolysis from the previous cycle and releases them after it changes conformation to βE. Our results indicate that perturbation of the 40° rotation step leading to product release is responsible for the alternative uncoupled pathway of the βE381C enzyme. This is consistent with observations of others (21, 26, 75) that ATP bound to the βE site in the first cycle, when γ is at 0°, is hydrolyzed during the second 120° cycle, specifically after γ has rotated through 200° (120° + 80°) (Fig. 8). However, our model differs from theirs in that we propose that ADP is released after βDP converts to βE (after completion of the third cycle when γ has rotated through 360°). This is in keeping with the results of Weber et al. (12) that the three catalytic sites are almost always occupied with nucleotide during steady state. A temperature-sensitive, ADP-dependent reaction intermediate for the Bacillus PS3 thermophilic enzyme was recently detected at low temperatures. This intermediate occurred before the 80° substep during the ATP binding dwell time and was assigned to ADP release (76). These results are in agreement with our model where ADP is released after the γ subunit has completed a 360° rotation cycle, before binding of another ATP molecule to that site.
A recent study of the timing of Pi release and γ subunit rotation (75), in contrast to our data, suggests that Pi is released before the 40° rotational substep, based on the effects on the substeps upon incubation with Pi. Although we agree with their conclusion that the Pi release reaction is directly coupled to the 40° rotation, our data suggest that Pi is not released before the rate-limiting rotation step kγ. We could only fit the pre-steady state data with kγ preceding k+3 (18). In addition, if Pi were released before the kγ step, we would expect that K2 would be dramatically altered. Blocking kγ by the cross-link did not perturb the K2 ratio; therefore, there must be an intervening energy-producing step between hydrolysis, k+2, and Pi release, k+3.
This analysis further elucidates the role of each partial rotation in the multisite reaction. The 40° rotational substep is associated with kγ, the rate-limiting step, and the 80° substep with the conformation required for the promotion of catalysis. Several lines of evidence lead to these conclusions. kγ was previously determined to occur after hydrolysis and before Pi release (18), and single molecule studies place the 40° substep after hydrolysis (25). Here we showed kγ, and steps subsequent to kγ, were severely perturbed when rotation was blocked by the γ-β cross-link under multisite conditions. In the unisite mode, which does not involve γ subunit rotation, there was no change in the rate constants upon formation of the γ-β cross-link. Analysis of the βE381C uncoupling mutant revealed that kγ was affected by the cysteine replacement, consistent with previous studies that implicated the specific interaction between this residue and the γ subunit at Met-23 and Arg-242 for proper formation of the rate-limiting transition state hypothesized to involve γ subunit rotation (30, 37). Essentially in each case where kγ is perturbed, K2 is unaffected showing kγ is independent of hydrolysis. The 80° rotational substep allows the enzyme to achieve the conformation required for the promotion of catalysis. This was shown by the cross-linked enzymes, which in multisite conditions show unperturbed positive catalytic cooperativity with promoted rates of hydrolysis and resynthesis of ATP (k+2 and k-2) similar to uncross-linked enzymes and wild type. Further evidence comes from the uncross-linked F1 with the βE381C mutation in which the ATP binding/unbinding (k+1, k-1) and ATP hydrolysis/resynthesis (k+2, k-2) parameters are unaltered, even though kγ was strongly perturbed. Together, these results link the 80° ATP-dependent substep to the promoted catalytic rates.
Acknowledgments
We thank Julie Teater and Dorothy Zhang for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM50957 from the USPHS. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AMPPNP, adenosine 5′-(β,γ-imido)triphosphate; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); DTT, dl-dithiothreitol; MDCC-PBP, 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin)-labeled phosphate-binding protein; PDRM, phosphodeoxyri-bomutase; TCEP, tris(2-carboxyethyl)phosphine hydrochloride; TES, 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Boyer, P. D. (1997) Annu. Rev. Biochem. 66717 -749 [DOI] [PubMed] [Google Scholar]
- 2.Nakamoto, R. K., Ketchum, C. J., Kuo, P. H., Peskova, Y. B., and Al-Shawi, M. K. (2000) Biochim. Biophys. Acta 1458289 -299 [DOI] [PubMed] [Google Scholar]
- 3.Boyer, P. D. (2002) FEBS Lett. 512 29-32 [DOI] [PubMed] [Google Scholar]
- 4.Weber, J., and Senior, A. E. (2003) FEBS Lett. 54561 -70 [DOI] [PubMed] [Google Scholar]
- 5.Junge, W., Panke, O., Cherepanov, D. A., Gumbiowski, K., Muller, M., and Engelbrecht, S. (2001) FEBS Lett. 504152 -160 [DOI] [PubMed] [Google Scholar]
- 6.Noji, H., Yasuda, R., Yoshida, M., and Kinosita, K. (1997) Nature 386299 -302 [DOI] [PubMed] [Google Scholar]
- 7.Tanabe, M., Nishio, K., Iko, Y., Sambongi, Y., Iwamoto-Kihara, A., Wada, Y., and Futai, M. (2001) J. Biol. Chem. 27615269 -15274 [DOI] [PubMed] [Google Scholar]
- 8.Nishio, K., Iwamoto-Kihara, A., Yamamoto, A., Wada, Y., and Futai, M. (2002) Proc. Natl. Acad. Sci. U. S. A. 9913448 -13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida, M., Muneyuki, E., and Hisabori, T. (2001) Nat. Rev. Mol. Cell Biol. 2 669-677 [DOI] [PubMed] [Google Scholar]
- 10.Nakamoto, R. K., Baylis Scanlon, J. A., and Al-Shawi, M. K. (2008) Arch. Biochem. Biophys. 476 43-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrahams, J. P., Buchanan, S. K., van Raaij, M. J., Fearnley, I. M., Leslie, A. G. W., and Walker, J. E. (1996) Proc. Natl. Acad. Sci. U. S. A. 939420 -9424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber, J., Wilke-Mounts, S., Lee, R. S.-F., Grell, E., and Senior, A. E. (1993) J. Biol. Chem. 26820126 -20133 [PubMed] [Google Scholar]
- 13.Grubmeyer, C., Cross, R. L., and Penefsky, H. S. (1982) J. Biol. Chem. 25712092 -12100 [PubMed] [Google Scholar]
- 14.Al-Shawi, M. K., and Senior, A. E. (1992) Biochemistry 31878 -885 [DOI] [PubMed] [Google Scholar]
- 15.Grubmeyer, C., and Penefsky, H. S. (1981) J. Biol. Chem. 2563728 -3734 [PubMed] [Google Scholar]
- 16.Cross, R. L., Grubmeyer, C., and Penefsky, H. S. (1982) J. Biol. Chem. 25712101 -12105 [PubMed] [Google Scholar]
- 17.Wise, J. G., Latchney, L. R., Ferguson, A. M., and Senior, A. E. (1984) Biochemistry 231426 -1432 [DOI] [PubMed] [Google Scholar]
- 18.Baylis Scanlon, J. A., Al-Shawi, M. K., Le, N. P., and Nakamoto, R. K. (2007) Biochemistry 468785 -8797 [DOI] [PubMed] [Google Scholar]
- 19.Weber, J., Bowman, C., and Senior, A. E. (1996) J. Biol. Chem. 27118711 -18718 [DOI] [PubMed] [Google Scholar]
- 20.Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., and Itoh, H. (2001) Nature 410898 -904 [DOI] [PubMed] [Google Scholar]
- 21.Ariga, T., Muneyuki, E., and Yoshida, M. (2007) Nat. Struct. Mol. Biol. 14 841-846 [DOI] [PubMed] [Google Scholar]
- 22.Ren, H., Bandyopadhyay, S., and Allison, W. S. (2006) Biochemistry 456222 -6230 [DOI] [PubMed] [Google Scholar]
- 23.Al-Shawi, M. K., Ketchum, C. J., and Nakamoto, R. K. (1997) Biochemistry 3612961 -12969 [DOI] [PubMed] [Google Scholar]
- 24.Mao, H. Z., and Weber, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 10418478 -18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimabukuro, K., Yasuda, R., Muneyuki, E., Hara, K. Y., Kinosita, K., and Yoshida, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 10014731 -14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishizaka, T., Oiwa, K., Noji, H., Kimura, S., Muneyuki, E., Yoshida, M., and Kinosita, K. (2004) Nat. Struct. Mol. Biol. 11142 -148 [DOI] [PubMed] [Google Scholar]
- 27.Duncan, T. M., Zhou, Y., Bulygin, V., Hutcheon, M. L., and Cross, R. L. (1995) Biochem. Soc. Trans. 23 736-741 [DOI] [PubMed] [Google Scholar]
- 28.Aggeler, R., Haughton, M. A., and Capaldi, R. A. (1995) J. Biol. Chem. 2709185 -9191 [DOI] [PubMed] [Google Scholar]
- 29.García, J. J., and Capaldi, R. A. (1998) J. Biol. Chem. 27315940 -15945 [DOI] [PubMed] [Google Scholar]
- 30.Ketchum, C. J., Al-Shawi, M. K., and Nakamoto, R. K. (1998) Biochem. J. 330707 -712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriyama, Y., Iwamoto, A., Hanada, H., Maeda, M., and Futai, M. (1991) J. Biol. Chem. 26622141 -22146 [PubMed] [Google Scholar]
- 32.Klionsky, D. J., Brusilow, W. S. A., and Simoni, R. D. (1984) J. Bacteriol. 1601055 -1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews, S. H., Peskova, Y. B., Polar, M. K., Herlihy, V. B., and Nakamoto, R. K. (2001) Biochemistry 4010664 -10670 [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 35.Senior, A. E., Latchney, L. R., Fersuson, A., and Wise, J. G. (1984) Arch. Biochem. Biophys. 228 49-53 [DOI] [PubMed] [Google Scholar]
- 36.Futai, M., Sternweis, P. C., and Heppel, L. A. (1974) Proc. Natl. Acad. Sci. U. S. A. 712725 -2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Shawi, M. K., Ketchum, C. J., and Nakamoto, R. K. (1997) J. Biol. Chem. 2722300 -2306 [DOI] [PubMed] [Google Scholar]
- 38.Shin, K., Nakamoto, R. K., Maeda, M., and Futai, M. (1992) J. Biol. Chem. 26720835 -20839 [PubMed] [Google Scholar]
- 39.Laemmli, U. K. (1970) Nature 227680 -685 [DOI] [PubMed] [Google Scholar]
- 40.Duncan, T. M., Bulygin, V. V., Zhou, Y., Hutcheon, M. L., and Cross, R. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 9210964 -10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penefsky, H. S. (1979) Methods Enzymol. 56527 -530 [DOI] [PubMed] [Google Scholar]
- 42.Sugino, Y., and Miyoshi, Y. (1964) J. Biol. Chem. 2392360 -2364 [PubMed] [Google Scholar]
- 43.Senior, A. E., Lee, R. S.-F., Al-Shawi, M. K., and Weber, J. (1992) Arch. Biochem. Biophys. 297340 -344 [DOI] [PubMed] [Google Scholar]
- 44.White, H. D., Belknap, B., and Webb, M. R. (1997) Biochemistry 3611828 -11836 [DOI] [PubMed] [Google Scholar]
- 45.Brune, M., Hunter, J. L., Howell, S. A., Martin, S. R., Hazlett, T. L., Corrie, J. E. T., and Webb, M. R. (1998) Biochemistry 3710370 -10380 [DOI] [PubMed] [Google Scholar]
- 46.Nixon, A. E., Hunter, J. L., Bonifacio, G., Eccleston, J. F., and Webb, M. R. (1998) Anal. Biochem. 265299 -307 [DOI] [PubMed] [Google Scholar]
- 47.Sheffield, P. J., Garrard, S. M., and Derewenda, Z. S. (1999) Protein Expression Purif. 15 34-39 [DOI] [PubMed] [Google Scholar]
- 48.MicroMath_Research (1995) MicroMath Scientist: For Experimental Data Fitting/Microsoft Windows, Version 2.0, MicroMath Research, St. Louis
- 49.Taussky, H. H., and Shorr, E. (1953) J. Biol. Chem. 202675 -685 [PubMed] [Google Scholar]
- 50.Lowry, O. H., Rosebrough, N. J., Farr, A. C., and Randall, R. J. (1951) J. Biol. Chem. 193265 -275 [PubMed] [Google Scholar]
- 51.Kaplan, R. S., and Pedersen, P. L. (1985) Anal. Biochem. 15097 -104 [DOI] [PubMed] [Google Scholar]
- 52.Fabiato, A., and Fabiato, F. (1979) J. Physiol. (Paris) 75463 -505 [PubMed] [Google Scholar]
- 53.Kaibara, C., Matsui, T., Hisabori, T., and Yoshida, M. (1996) J. Biol. Chem. 2712433 -2438 [DOI] [PubMed] [Google Scholar]
- 54.Nakamoto, R. K., Al-Shawi, M. K., and Futai, M. (1995) J. Biol. Chem. 27014042 -14046 [DOI] [PubMed] [Google Scholar]
- 55.Aggeler, R., and Capaldi, R. A. (1992) J. Biol. Chem. 26721355 -21359 [PubMed] [Google Scholar]
- 56.Noumi, T., Maeda, M., and Futai, M. (1987) FEBS Lett. 213381 -384 [DOI] [PubMed] [Google Scholar]
- 57.Boyer, P. D. (1975) FEBS Lett. 58 1-6 [DOI] [PubMed] [Google Scholar]
- 58.Boyer, P. D. (1993) Biochim. Biophys. Acta 1140215 -250 [DOI] [PubMed] [Google Scholar]
- 59.Kanazawa, H., Horiuchi, Y., Takagi, M., Ishino, Y., and Futai, M. (1980) J. Biochem. (Tokyo) 88 695-703 [DOI] [PubMed] [Google Scholar]
- 60.Al-Shawi, M. K., Parsonage, D., and Senior, A. E. (1988) J. Biol. Chem. 26319633 -19639 [PubMed] [Google Scholar]
- 61.Löbau, S., Weber, J., and Senior, A. E. (1997) J. Biol. Chem. 2723648 -3656 [DOI] [PubMed] [Google Scholar]
- 62.Weber, J., and Senior, A. E. (2001) J. Biol. Chem. 27635422 -35428 [DOI] [PubMed] [Google Scholar]
- 63.Brune, M., Hunter, J. L., Corrie, J. E., and Webb, M. R. (1994) Biochemistry 338262 -8271 [DOI] [PubMed] [Google Scholar]
- 64.Al-Shawi, M. K., and Senior, A. E. (1988) J. Biol. Chem. 26319640 -19648 [PubMed] [Google Scholar]
- 65.Al-Shawi, M. K., Parsonage, D., and Senior, A. E. (1990) J. Biol. Chem. 2654402 -4410 [PubMed] [Google Scholar]
- 66.Yasuda, R., Noji, H., Kinosita, K., and Yoshida, M. (1998) Cell 931117 -1124 [DOI] [PubMed] [Google Scholar]
- 67.Yasuda, R., Masaike, T., Adachi, K., Noji, H., Itoh, H., and Kinosita, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 1009314 -9318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stock, D., Leslie, A. G. W., and Walker, J. E. (1999) Science 2861700 -1705 [DOI] [PubMed] [Google Scholar]
- 69.Abrahams, J. P., Leslie, A. G. W., Lutter, R., and Walker, J. E. (1994) Nature 370621 -628 [DOI] [PubMed] [Google Scholar]
- 70.Gibbons, C., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2000) Nat. Struct. Biol. 71055 -1061 [DOI] [PubMed] [Google Scholar]
- 71.Braig, K., Menz, R. I., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2000) Structure (Lond.) 8 567-573 [DOI] [PubMed] [Google Scholar]
- 72.Nakamoto, R. K., Ketchum, C. J., and Al-Shawi, M. K. (1999) Annu. Rev. Biophys. Biomol. Struct. 28205 -234 [DOI] [PubMed] [Google Scholar]
- 73.Al-Shawi, M. K., and Nakamoto, R. K. (1997) Biochemistry 3612954 -12960 [DOI] [PubMed] [Google Scholar]
- 74.Hara, K. Y., Noji, H., Bald, D., Yasuda, R., Kinosita, K., and Yoshida, M. (2000) J. Biol. Chem. 27514260 -14263 [DOI] [PubMed] [Google Scholar]
- 75.Adachi, K., Oiwa, K., Nishizaka, T., Furuike, S., Noji, H., Itoh, H., and Yoshida, M. (2007) Cell 130309 -321 [DOI] [PubMed] [Google Scholar]
- 76.Watanabe, R., Iino, R., Shimabukuro, K., Yoshida, M., and Noji, H. (2008) EMBO Rep. 9 84-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menz, R. I., Walker, J. E., and Leslie, A. G. W. (2001) Cell 106331 -341 [DOI] [PubMed] [Google Scholar]