Abstract
The γ-aminobutyric acid type A (GABAA) receptor assembles from individual subunits to form ligand-gated ion channels. Human (h) β3 subunits assemble to form homomeric surface receptors in somatic cells, but hβ1 subunits do not. We have identified three distinct sets of amino acid residues in the N-terminal extracellular domain of the hβ1 subunit, which when mutated to the homologous residue in hβ3 allow expression as a functional homomeric receptor. The three sets likely result in three modes of assembly. Mode 1 expression results from a single amino acid change at residue hβ1 Asp-37. Mode 2 expression results from mutations of residues between positions 44 and 73 together with residues between positions 169 and 173. Finally, mode 3 results from the mutations A45V and K196R. Examination of homology-based structural models indicates that many of the residues are unlikely to be involved in physical inter-subunit interactions, suggesting that a major alteration is stabilization of an assembly competent form of the subunit. These mutations do not, however, have a major effect on the surface expression of heteromeric receptors which include the α1 subunit.
The GABAA3 receptor is a member of the superfamily of fast acting ligand-gated ion channels, which includes the nicotinic acetylcholine, glycine, and serotonin receptors. They most likely originated from a single receptor subunit active as a homo-oligomer, which then evolved into a variety of subtypes and subunits (1). Individual subunits of these receptors have similar sequences and structural features such as four similarly distributed membrane-spanning regions and a characteristic cysteine loop (2). GABAA receptors are the major fast inhibitory neurotransmitter gated ion channels in the brain (3) and contain diversity of subunit isoforms as follows: α(1–6), β(1–3), γ(1–3), δ, ε, θ, and π (4–6). These and the related receptors formed from ρ subunits are most likely formed as pentamers of subunits (7, 8).
Studies have been made of the assembly of glycine and GABAA receptors, elucidating regions directing the protein interactions that underlie receptor assembly and revealing specific sequences involved in those interactions. These studies have shown that the N-terminal extracellular domain contains residues critical for the assembly of heteromeric receptors (9–15) and homomeric receptors (13, 16–18). Receptors are assembled in the endoplasmic reticulum and retained there by protein chaperones if subunits are incorrectly assembled or folded (19).
The GABAA receptor expresses in neurons as a heteromeric pentamer containing two or more different subunits (3). However, studies of homomeric receptors can reveal important requirements for assembly. Previous work has found that the β3 subunit can be expressed on the cell surface as a homomeric receptor after transfection into somatic cells (13), whereas the β2 and β1 subunits are expressed much more poorly (19, 20). An analysis of the mouse β2 subunit identified four residues in the N-terminal domain, which were required for surface assembly (13). In contrast, structural requirements for assembly of β1 homomeric receptors have not been examined. A small body of literature exists, describing expression and function of β1 homomeric receptors in Xenopus oocytes and somatic cell lines (21, 22). This literature, however, shows unclear and often contradictory results depending on expression system and species of subunit used. This study uses a somatic cell expression system (QT6 quail fibroblast cells) and compares hβ1 and hβ3 subunits to find assembly determinants in hβ3 that are not present in hβ1. The results indicate additional regions of the extracellular domain, which are important in conferring competence for assembly, and suggest that intrasubunit interactions can be of critical importance.
EXPERIMENTAL PROCEDURES
cDNA Constructs—A cDNA construct for the hβ1 subunit, GenBank™ accession number X14767, was obtained from David Weiss (University of Texas, San Antonio), originally cloned by Paul Whiting (23); and hβ3, accession number M82919, was obtained from Geoffrey White, (Neurogen, Bradford, CT). Construction of the Myc-tagged rat (r) α1 subunit was described previously (24). Restriction sites were introduced into each cDNA for making chimeras between hβ1 and hβ3. Mutagenesis was performed using QuikChange mutagenesis (Stratagene, La Jolla, CA) or pAltered Sites II (Promega, Madison, WI). The FLAG (F) epitope was inserted between amino acids 4 and 5 of the mature peptide for both hβ1 and hβ3 using the pAltered Sites II mutagenesis kit. FLAG-tagged constructs were transferred to pcDNA3 (Invitrogen). Site-specific mutagenesis was done using QuikChange.
Cell Culture—Quail fibroblasts (QT6 cells; initially provided by Dr. J. Merlie, Washington University) were maintained in Medium 199 (Earle's salts) containing 5% fetal bovine serum (Hyclone, Logan, UT), 10% tryptose phosphate broth (Invitrogen), 1% dimethyl sulfoxide (DMSO), penicillin (100 units/ml), and streptomycin (100 μg/ml) in a humidified atmosphere containing 5% CO2. Cells were passaged twice each week, maintaining subconfluent cultures.
ELISA and Transfection—Whole cell surface enzyme-linked immunosorbent assay (ELISA) was done as described by Bonnert et al. (5) with the following modifications. The primary antibodies, anti-FLAG M2 (Sigma) and anti-Myc (9E10, Invitrogen), were used at a concentration of 2 μg/ml. The secondary antibody, horseradish peroxidase-conjugated sheep anti-mouse (GE Healthcare) was used at a dilution of 1:100. Each subunit combination was transfected into 5 wells of a 24-well plate, of which three were for ELISA and two for assaying total protein. QT6 cells were transfected according to published methods (26). Cells were plated at a density of 100,000 cells per well. The next day, they were washed once with Dulbecco's modified Eagle's medium/F-12 + 5% fetal bovine serum, followed by the addition of 1.5 ml of fresh medium to each well. The precipitation reaction for each well contained 3 μg of cDNA, 7.5 μl of CaCl2 (2.5 m stock), distilled H2Ouptoa 75-μl volume, and 75 μl of 2× BES-buffered solution (50 mm BES, 280 mm NaCl, 1.5 mm Na2HPO4, pH 6.95). The mixture was precipitated for 5 min at room temperature, and 150 μl of the precipitation solution was added to each well. Cells were incubated overnight in 5% CO2. The next day, transfected cells were washed three times with complete growth medium. The 2nd day after transfection, each well was aspirated and blocked with 0.5 ml of 4% milk phosphate-buffered saline (MPBS: 4% powdered milk, 137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.3) for 30 min at room temperature. The blocking solution was then aspirated, and 0.5 ml of primary antibody in MPBS was added to three wells for ELISA. Following incubation for 1 h at room temperature, wells were washed once with MPBS. After that, 0.5 ml of MPBS containing secondary antibody was added to the wells for ELISA and incubated for 1 h at room temperature. Cells for protein analysis were maintained in MPBS until the final MPBS washes. Wells were aspirated and washed three times with MPBS and three times with phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.3). Next, 0.5 ml of ELISA reagent, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Vector Laboratories, Inc., Burlingame, CA) was added to the wells designated for ELISA and incubated for 30 min at room temperature protected from light. After incubation, 250 μl of reaction mix was transferred from each well to a 96-well plate; absorbance was read at 405 nm using a microplate reader (model 550, Bio-Rad).
ELISA results were calculated as absorbance/mg of protein. Values presented in this paper for homomeric expression are given as absorbance/mg normalized to hβ3F expression performed in parallel. Heteromeric assembly was determined by measuring the Myc epitope on the α1myc subunit and normalized to hβ3myc expression. The empty vector (pcDNA3) was transfected as a negative control in each experiment, and the signal was subtracted from the other data. Statistical analysis was performed using Systat 7 (Systat Software Inc., Point Richmond, CA).
Protein Assay—The remaining two transfected wells were assayed for protein using the bicinchoninic acid method (Pierce). Bicinchoninic acid reagent (0.5 ml) was added to the wells and incubated for 30 min at 37 °C. After incubation, the DNA was sheared using a 1-ml pipetter, and 250 μl from each well was transferred to a 96-well plate. The absorbance was read at 550 nm in a microplate reader. It was observed that when the protein content was less than 10 μg per well, the results were inconsistent. Therefore, if the hβ3F control protein content was below 10 μg, all data collected from that set of transfections were discarded. If a particular construct gave a value below 10 μg, data from that construct were discarded. There was no systematic difference between the protein values of cells transfected with the various constructs. Linearity curves for each assay were determined, and absorbance readings were accepted in that range. If a sample was predicted to fall outside the range of linearity, it was diluted with deionized water by half. Such dilution was observed not to affect the molar absorption coefficients of the chromogens.
Electrophysiology—Whole cell voltage clamp recordings were made from transiently transfected QT6 cells using standard methods (27), to determine whether the various constructs could assemble into functional receptors on the cell surface. Pentobarbital was dissolved in external solution and applied by a multichannel perfuser (SF-77B; Warner Instruments, Hamden, CT). The goal of these experiments was to examine the function of receptors on the cell surface, rather than to assay levels of expression. Accordingly, cells expressing high levels of transfected receptors were identified using beads with coupled anti-FLAG antibody (20). All constructs except hβ1F produced extensive bead binding. Cultures transfected with hβ1F only had a few cells per dish that bound beads. Previous studies have shown that untransfected cells show no responses to GABAergic drugs (20). Pentobarbital was used to activate homomeric receptors, as previous studies have shown that it activates homomeric hβ1 and hβ3 receptors expressed in Xenopus oocytes, whereas GABA does not (28).
Molecular Modeling—Modeling of the β subunits was done using the Deep View/Swiss-PDB viewer freeware version 3.7. Two copies of the hβ3 protein sequence from residues Ser-10 to Arg-216 were aligned against the acetylcholine-binding protein (AChBP) using the alignment proposed for hβ2 by Cromer et al. (29). The α-carbon atoms of the aligned residues were then threaded upon the crystal structure of the AChBP to obtain two, three-dimensional adjacent copies of the subunit. A few gaps with missing β3 residues occurred upon threading. The missing residues were built back into the subunits. The hβ1 subunit was then similarly aligned and threaded upon the rebuilt hβ3 model. Models of individual mutant subunits of hβ1 were also made. Viewing and graphic display of these models were done using the three-dimensional molecule viewer of the Vector NTI Advance version 10.0.1 software (Invitrogen).
RESULTS
Cell Surface Expression of hβ1 and hβ3—We used the binding of antibody to an extracellular epitope (FLAG) engineered into the N-terminal region of the hβ3 and hβ1 subunits to assay the surface expression of assembled receptors (inclusion of FLAG is indicated by, for example, hβ1F). The data are presented as the surface expression relative to that for hβ3F, measured in the same experiment. ELISA data indicate that hβ1F does not significantly express on the cell surface, expressing at only 3.9 ± 3.8% of the level of hβ3F (mean ± S.D.; n = 109 separate experiments) (Table 1). Co-transfection of hβ3 and hβ1F resulted in the rescue and surface expression of hβ1F (42 ± 20%, n = 5). Furthermore, co-expression of hβ3F and hβ1F resulted in identical amounts of FLAG epitope recognized on the surface as when hβ3F alone was expressed (99%; 111%, two experiments), suggesting that the heteropentamers have essentially equal total expression as the hβ3 homopentamers. Although it is known that the hβ1 subunit co-assembles with α subunits, co-assembly with hβ3 is suggestive that specific assembly determinants may be resolved because of the high homology between these subunits.
TABLE 1.
Surface expression of wild type hβ1 and hβ3 and major chimeric subunit constructs
| Construct | Expressiona | Nb | Pc |
|---|---|---|---|
| hβ1F | 3.9 ± 3.8 | (109) | X |
| hβ3F | 100 | (122) | d |
| X1F hβ1/hβ3 | 8.8 ± 4.0 | (5) | NSe |
| X1F hβ3/hβ1 | 74.8 ± 38.6 | (11) | d |
Mean ± S.D. was for expression as percentage of homomeric hβ3 expression measured in the same experiment
N indicates number of separate experiments
Results of analysis of variance with a Bonferroni post hoc correction. All pairwise comparisons for constructs included in the table were made, and the results presented are for difference to the expression of the construct indicated by X
p < 0.001
NS indicates p > 0.05
Sequence Homology between hβ1 and hβ3—N-terminal sequence alignment between hβ1 and hβ3 (Fig. 1) reveals two primary regions of divergence, one between residues 1 and 73 and the second between residues 169 and 201. There are 13 amino acid differences between residues in each region. Our hβ3 differs from the published sequence by containing Val-45 rather than Ala-45 as reported previously (30). This difference actually has some consequences for assembly, as shown below.
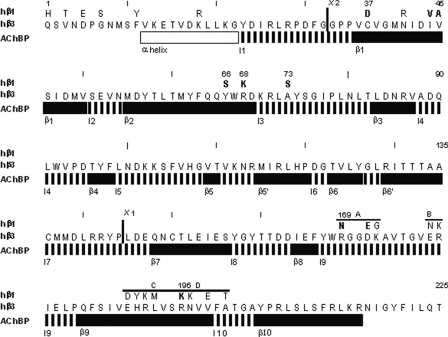
FIGURE 1.
The extracellular N-terminal regions of the hβ1 and hβ3 subunits. The amino acid sequence for the mature protein from residues 1 to 225 is shown for hβ3, whereas residues for hβ1 which differ are shown above the hβ3 sequence. Residues that were identified as important for assembly are indicated in boldface. Chimera sites are shown by heavy vertical lines. The location of the ABCD regions is also shown. The lines labeled AChBP show the protein secondary structure by homology to the AChBP (vertical hatching indicates loop, and black indicates β-sheet structure), with the name for the region shown below (e.g. β1 indicates β-sheet 1).
Fig. 1 also shows secondary structural features identified in the acetylcholine-binding protein (31). We used AChBP as a template for the homology model of the GABAA subunits we studied (29). The structural features seen in AChBP will be referred to as, for example, loop 10 and β-sheet 9.
Mutations of hβ1F Residues between 169 and 201—Previous studies have shown that the N terminus of the GABAA receptor contains major determinants for regulating assembly and surface expression. Taylor et al. (13) demonstrated that four amino acid substitutions (D171G, N173K, T179E, and K180R) in mouse (m) β2, which alter the residues to those found in mβ3, confer the ability to self-assemble. Accordingly, we initially examined the influence of mutations in hβ1 between residues 169 and 201, changing the residues in hβ1 to the homologous residues in hβ3. The mutated amino acids were grouped into four regions designated as A through D (Fig. 1). (The residues identified by Taylor et al. (13) fall in the A and B regions.) The possible roles of these residues in hβ1 were examined individually and in groups, with no significant increase in expression (supplemental Table). Finally, all the divergent amino acids between residues 169 and 201 were substituted, but no increase in expression was observed (3.3 ± 10%, n = 11). We confirmed that the four mutations of the mβ2 subunit identified by Taylor et al. (13) significantly increase surface expression of mβ2 (from 6 ± 6%, n = 8, to 20 ± 6%, n = 4). These results indicate that hβ1 assembly must be constrained differently than mβ2, and that other regions of the subunit are necessary to confer surface expression of hβ1 homomers.
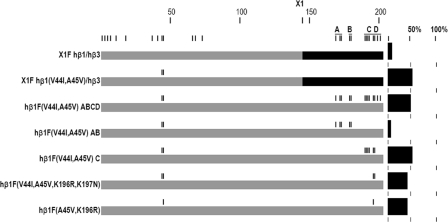
Chimeras of hβ1 and hβ3—Chimeras between hβ1 and hβ3 were then made to locate the sequences determining assembly. Key chimeras were made at amino acid 145 (X1) between hβ1 and hβ3. The location of the junction is shown in Fig. 1, and the chimeric subunit will be designated by, for example, X1 hβ1/hβ3 for the construct with hβ1 sequence from the N terminus to the join and hβ3 sequence to the C terminus. This position separates the two regions of sequence divergence (Fig. 1). Expression of X1F hβ1/hβ3 was low, similar to that of hβ1F (Table 1). The inverse chimera, X1F hβ3/hβ1, however, expressed as well as hβ3F. These results clearly show that the amino acid residues between positions 169 and 201 do not have to be changed for robust expression, if the appropriate sequence exists in the first 73 residues of the N terminus. The low expression of X1F hβ1/hβ3 confirms that the ABCD region is not sufficient to induce assembly. These results prompted two questions. First, which amino acids in X1F hβ3/hβ1 permit expression? Second, which amino acids in the X1F hβ1/hβ3 chimera, if changed, will allow for expression? One might expect that these two questions would lead to the same result. However, this experimental approach led to the discovery of distinct modes of expression for mutant hβ1 receptors, unmasking three groups of assembly determinants.
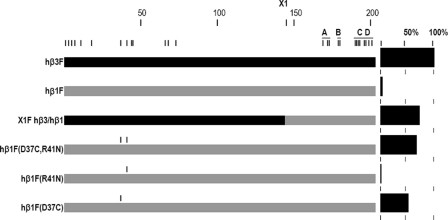
Mode 1 Expression, Mutations of hβ1(Asp-37)—In contrast to X1F hβ1/hβ3, which did not yield surface homomers, the inverse chimeric subunit X1F hβ3/hβ1 expressed as well as hβ3F (Table 1). Through the use of chimeric subunits and site-directed mutagenesis, the single residue at position 37 was found to play a major role in determining whether hβ1 subunits assemble on the cell surface as homomultimers (Fig. 2; Table 2). Hβ1F (D37C) expressed at 56%, a 14-fold increase over hβ1F. X1F hβ3(C37D)/hβ1 resulted in a significant loss of expression (to 18 ± 11%, n = 18) compared with X1F hβ3/hβ1, confirming the influence of residue 37.
FIGURE 2.
Graphical presentation of the results used to define mode 1 assembly. The figure shows the locations of critical residues in the primary sequence, in conjunction with the surface expression measured by ELISA. Construct names are indicated in the left column. A graphical representation of each construct appears in the center column, with the β3 portion of the protein shown in black and β1 in gray. Amino acid residues different between β3 and β1 are indicated by vertical marks at the top. Vertical marks above the constructs indicate the locations of substitutions. A scale at the top of the column indicates the approximate position in the primary sequence. The A–D regions are indicated by horizontal lines at the top. The right-hand column shows the results of ELISAs as percent of hβ3F expression.
TABLE 2.
Surface expression of constructs designed to examine mode 1 expression
| Constructa | Expressiona | Nb | Pc |
|---|---|---|---|
| hβ1F | 3.9 ± 3.8 | (109) | X |
| X1F hβ3/hβ1 | 74.8 ± 38.6 | (11) | d |
| hβ1F (D37C and R41N) | 69.1 ± 25.6 | (4) | d |
| hβ1F (R41N) | 2.9 ± 1.1 | (3) | NSe |
| hβ1F (D37C) | 56.4 ± 14.4 | (4) | d |
| hβ1F (D37A) | 78.8 ± 28.8 | (10) | d |
| hβ1F (D37K) | 60.2 ± 32.5 | (7) | d |
| hβ1F (D37E) | 45.6 ± 7.7 | (4) | d |
| hβ1F (D37N) | 78.7 ± 19.4 | (3) | d |
| hβ1F (D37G) | 10.1 ± 7.0 | (5) | NS |
Mean ± S.D. was for expression as percentage of homomeric hβ3 expression measured in the same experiment
N indicates number of separate experiments
Results of analysis of variance with a Bonferroni post hoc correction. All pairwise comparisons for constructs included in the table were made, and the results presented are for difference to the expression of the construct indicated by X
p < 0.001
NS indicates p > 0.05
We made several amino acid substitutions at residue 37, to explore the structural requirements of this residue that permit assembly. Other amino acids substituted at position 37 were alanine, glycine, asparagine, lysine, and glutamate. All substitutions except hβ1F (D37G) showed significantly more expression than hβ1F (Table 2). The expression of these substitutions did not differ significantly from that of hβ1 (D37C), except for the glycine substitution, which expressed as poorly as wild type hβ1. The physical characteristics of these substituted residues vary. Both alanine and glycine conserve the hydrophobic character of the cysteine residue. Asparagine is about the same volume as aspartate (96 and 91 Å3, respectively) and retains the polar nature but is uncharged. Finally, glutamate conserves the charge of aspartate in hβ1. From these observations we conclude that the presence of aspartate at position 37 has a specific, dominant-negative, effect on homomeric assembly of the hβ1 subunit.
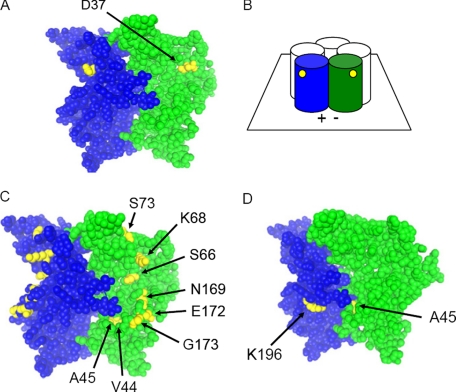
To visualize the location of Asp-37, we performed homology-based structural modeling of the extracellular domain of the hβ1 subunit. Crystallography of the AChBP (31) provides a structural model that can assist in understanding GABAA receptor assembly. The alignment used was that proposed for hβ2 by Cromer et al. (29). Two adjacent subunits were modeled to view both inter- as well as intrasubunit amino acid interactions.
The modeling predicts that Asp-37 is part of the β1-sheet on the exterior surface of the subunit, facing into the aqueous surroundings and not directly interacting with residues of adjacent subunits. Fig. 3A shows a molecular model of the Asp-37 residue situated in the middle of the exterior of the subunit, distant from the interface. It lies between residues Lys-68 and Glu-165 on the two flanking β-sheets, β2 and β8, respectively (supplemental Fig. 1). Accordingly, substitutions of Lys-68 and Glu-165 were made to test the hypothesis that Asp-37 interacts with these residues to suppress assembly. Neither K68L nor K68Q altered hβ1F expression (supplemental Table). Hβ1F (D37A and K68Q) was also tested to determine whether a mutated Lys-68 residue was generally detrimental to expression, but this construct expressed as well as hβ1 (D37A) (supplemental Table). However, the substitution E165N rescued assembly of hβ1F (73 ± 25%, n = 9). This residue is conserved in all human β subunits. The combination of D37A and E165N did not increase expression further (57 ± 12%, n = 4).
FIGURE 3.
Modeling the positions of identified residues. A, residues involved in mode 1 assembly. Two adjacent subunits illustrate the predicted position of Asp-37 in the homology model of the N-terminal extracellular portion of a subunit. The view is from the external solution, as schematized in B. Subunits are shown with the bottom near the membrane and the exterior surfaces showing. The plus and minus interfaces are indicated by (+) and (-). Individual subunits are colored blue and green as a space-filled model. Asp-37 is colored yellow and is shown to extend into the aqueous environment. It is situated distantly from the interface. C, residues involved in mode 2 assembly. Two adjacent subunits illustrate the relationships of Val-44, Ala-45, Ser-66, Lys-68, Ser-73, Asn-169, Glu-172, and Gly-173 to the interface and to each other. Ala-45 extends between subunits at the interface; Val-44 protrudes into the hydrophobic core of the subunit, and the remaining residues extend into the aqueous environment distant from the interface. D, residues involved in mode 3 assembly. Both residues are at the intersubunit interface and are likely to interact with residues on the adjacent subunit.
We then examined residues predicted to be neighbors of hβ1 Glu-165. Hβ1 Thr-161 and Lys-197 are predicted to be the nearest residues to Glu-165. However, mutations to Thr-161 and Lys-197 failed to have a significant influence on assembly (supplemental Table). These data suggest that an interaction between Asp-37 and Glu-165 reduces assembly, rather than an effect of Asp-37 on the interactions of Glu-165 with other neighboring residues. The nature of the interaction between these two residues is likely to be electrostatic.
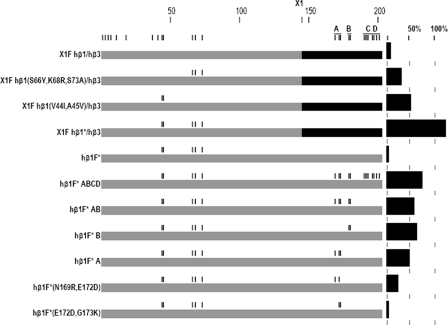
Mutations to X1F hβ1/hβ3 Indicate Additional Determinants for Assembly—We then examined the assembly-incompetent chimera, X1F hβ1/hβ3, to determine whether mutations in other regions could enhance surface expression. X1F hβ1/hβ3 was systematically mutated from amino acid 73 toward the N terminus to make it progressively more like hβ3. On the backbone of the chimera, two subsets of residues were shown to result in expression (S66Y, K68R, and S73A) and (V44I and A45V) (36 and 52%, respectively; see Fig. 4 and Table 3). The combination of all five substitutions (V44I, A45V, S66Y, K68R, and S73A) resulted in 110% expression of the X1 hβ1/hβ3 chimera, suggesting these two modes are additive for assembly. This set of mutations (V44I, A45V, S66Y, K68R, and S73A) on the β1 subunit, will be referred to as hβ1*.
FIGURE 4.
Graphical presentation of the results used to define mode 2 assembly. Residues from three distinct regions of the subunit were found to confer assembly competence: V44I and A45V; S66Y, K68R, and S73A; and N169R, E172D, and G173K. Hβ1* = hβ1(V44I, A45V, S66Y, K68R, and S73A). The results are presented as in Fig. 2.
TABLE 3.
Surface expression of constructs designed to examine mode 2 expression
| Construct | Expressiona | Nb | Pcb |
|---|---|---|---|
| hβ1F | 3.9 ± 3.8 | (109) | X |
| X1F hβ1/hβ3 | 8.8 ± 4.0 | (5) | NSd |
| X1F hβ1(S66Y, K68R, S73A)/hβ3 | 35.8 ± 16.1 | (6) | e |
| X1F hβ1(V44I, A45V)/hβ3 | 51.6 ± 27.1 | (10) | e |
| X1F hβ1*/hβ3f | 109.8 ± 23.3 | (8) | e |
| hβ1F* | 4.3 ± 4.9 | (5) | NS |
| hβ1F* A–Dg | 75.6 ± 28.0 | (10) | e |
| hβ1F* AB | 60.9 ± 33.0 | (14) | e |
| hβ1F* B | 65.3 ± 10.1 | (3) | e |
| hβ1F* A | 49.7 ± 25.9 | (21) | e |
| hβ1F* (N169R, E172D) | 25.9 ± 20.5 | (28) | e |
| hβ1F* (E172D, G173K) | 5.4 ± 2.5 | (5) | NS |
Mean ± S.D. was for expression as percentage of homomeric hβ3 expression measured in the same experiment
N indicates number of separate experiments
Results of analysis of variance with a Bonferroni post hoc correction. All pairwise comparisons for constructs included in the table were made, and the results presented are for difference to the expression of the construct indicated by X
p < 0.001
NS indicates p > 0.05
hβ1* is hβ1 (V44I, A45V, S66Y, K68R, and S73A)
hβ1 A is hβ1 (N169R, E172D, and G173K); hβ1 B is hβ1 (N179E and K180R); hβ1 C is hβ1 (D190E, Y191H, K192R, M193L, K196R, and K197N); hβ1 D is hβ1 (K196R, K197N, E199V, and T201A)
Mode 2 Assembly—Once X1F hβ1*/hβ3 was shown to express as well as hβ3F, mutagenesis was done to find the minimum complement of residues affecting assembly. Initially, we tested the role of amino acid differences in the extreme N-terminal sequence, by generating a chimera (X2) at residue 33 (Fig. 1). Neither X2F hβ3/hβ1* nor X2F hβ3/hβ1 ABCD expressed significantly better than hβ1F, suggesting that differences in the first 18 residues of the N terminus do not play a significant role in determining efficacy of assembly (supplemental Table). Once these residues were excluded, the remaining A–D regions were again investigated.
The combination of mutations to the A and B regions supported robust expression of hβ1F* (Table 3 and Fig. 4). Further subdivision of these regions indicated that mutations to either the A (residues hβ1 Asn-169 through Gly-173) or B (Asn-179 and Lys-180) region can confer partial assembly competence upon hβ1F*. We examined the A region in more detail, using the mutations N169R, E172D, and G173K. The data suggest that all three residues make some contribution, although the mutation N169R appears to be essential as hβ1F* (E172D and G173K) expresses at a level comparable with hβ1 (Fig. 4 and Table 3). However, the construct hβ1F* (N169R) also had very low expression (0.0%; 0.4%).
We then examined the effects of reducing the complement of mutations in hβ1* on the background of the N169R and E172D mutations. We found that all residues in hβ1* were necessary, and none of the reduced complements supported expression above the level of hβ1 (supplemental Table). These observations support the idea that the mutations V44I, A45V, S66Y, K68R, and S73A act in concert with mutations in the A region to enhance expression of the hβ1 subunit. This mode of expression was termed mode 2. There did not appear to be a significant interaction between residues involved in mode 1 and mode 2 assembly because the construct hβ1F* (D37C and R41N) expressed to essentially the same extent as hβ1F (D37C and R41N) (78 ± 18%, n = 6, compared with 69 ± 26%, n = 4).
Modeling of the residues conferring mode 2 assembly (Fig. 3C) reveals that each of them is distant from the subunit interface except for residue 45. Residues at positions 66, 68, 73, 169, 172, and 173 are all located on the outside surface of the subunit. Residue 44 protrudes into the β-barrel of the subunit, away from the interface. However, residue 45 extends between the subunits as part of the interface. The results of these modeling data suggest that both intrasubunit interactions (residues 44, 66, 68, 73, 169, 172, and 173) and intersubunit interactions (residue 45) stabilize the subunit structure.
Mode 3 Assembly—It was surprising that the studies of mode 2 assembly showed a lack of expression of the construct hβ1 (V44I, A45V, N169R, and E172D), because X1F hβ1(V44I and A45V)/hβ3 expressed well (Table 3). Accordingly, we examined the requirements for expression induced by the mutations V44I and A45V. In this case, expression was only found when the C and/or D regions were included (Table 4). Finer mapping with point mutations indicated that the combination of V45A and K196R contained the major portion of assembly determinants (Table 4 and Fig. 5). These observations support the idea that there is an alternative set of assembly determinants, underlying mode 3 expression.
TABLE 4.
Surface expression of constructs designed to examine mode 3 expression
| Construct | Expressiona | Nb | Pc |
|---|---|---|---|
| hβ1F | 3.9 ± 3.8 | (109) | X |
| X1F hβ1/hβ3 | 8.8 ± 4.0 | (5) | NSd |
| X1F hβ1(V44I, A45V)/hβ3 | 51.6 ± 27.1 | (10) | e |
| hβ1F (V44I, A45V) A–Df | 49.3 ± 24.0 | (13) | e |
| hβ1F (V44I, A45V) AB | 8.1 ± 9.0 | (4) | NS |
| hβ1F (V44I, A45V) C | 52.4 ± 15.8 | (16) | e |
| hβ1F (V44I, A45V) D | 34.3 ± 18.4 | (14) | e |
| Hβ1F (V44I, A45V, K196R, K197N) | 42.3 ± 16.2 | (9) | e |
| hβ1F (V44I, K196R, K197N) | 11.1 ± 5.5 | (7) | NS |
| hβ1F (A45V, K196R, K197N) | 22.0 ± 4.3 | (6) | g |
| hβ1F (A45V, K197N) | 8.6 ± 3.0 | (3) | NS |
| hβ1F (A45V, K196R) | 42.5 ± 6.8 | (4) | e |
Mean ± S.D. was for expression as percentage of homomeric hβ3 expression measured in the same experiment
N indicates number of separate experiments
Results of analysis of variance with a Bonferroni post hoc correction. All pairwise comparisons for constructs included in the table were made, and the results presented are for difference to the expression of the construct indicated by X
p < 0.001
NS indicates p > 0.05
hβ1 A is hβ1 (N169R, E172D, G173K); hβ1 B is hβ1 (N179E, K180R); hβ1 C ishβ1 (D190E, Y191H, K192R, M193L, K196R, K197N); hβ1 D is hβ1 (K196R, K197N, E199V, T201A)
p < 0.05
FIGURE 5.
Graphical presentation of the results used to define mode 3 assembly. Residues defining mode 3 are A45V and K196R. The results are presented as in Fig. 2.
The homology model suggests that the two residues underlying mode 3 assembly are located on opposite sides of the intersubunit interface (Fig. 3D). They are likely to be involved in intersubunit interactions (supplemental Fig. 2), although they do not interact with each other.
Reduction of hβ3 Expression—We attempted to reduce expression of the hβ3F subunit. Initially, we generated the mutated subunit hβ3 (G171D, K173N, E179T, and R180K) (13). Expression was reduced by about half (see Table 5). However, when coupled with the mutation V45A, expression of hβ3F was reduced to the level of hβ1F. The mutation hβ3 V45A, alone, produced a small reduction.
TABLE 5.
Surface expression of constructs designed to reduce expression of the hβ3F construct
| Construct | Expressiona | Nb | Pc |
|---|---|---|---|
| hβ3F | 100 | (122) | X |
| hβ3F (V45A) | 85.9 ± 24.6 | (8) | d |
| hβ3F DNTKe | 59.3 ± 17.8 | (13) | d |
| hβ3F (V45A, DNTK) | 12.6 ± 7.3 | (7) | d |
| hβ3F (C37D) | 92.5 ± 20.3 | (8) | NSf |
| Hβ3F (C37D, I44V, V45A, Y66S, R68K, A73S) | 67.6 ± 17.2 | (7) | d |
| hβ3F (C37D, R196K, N197K) AB | 57.9 ± 26.9 | (4) | d |
| hβ3F (C37D) AB | 45.0 ± 18.7 | (4) | d |
Mean ± S.D. was for expression as percentage of homomeric hβ3 expression measured in the same experiment
N indicates number of separate experiments
Results of analysis of variance with a Bonferroni post hoc correction. All pairwise comparisons for constructs included in the table were made, and the results presented are for difference to the expression of the construct indicated by X
p < 0.001
Hβ3 DNTK: hβ3 (G171D, K173N, E179T, R180K)
NS indicates p > 0.05
We then tested the effects of mutations to a number of residues involved in the modes for expression of hβ1F. The mutation C37D produced a small reduction (Table 5), in clear contrast to the greatly reduced expression of the X1F hβ3(C37D)/hβ1 chimera seen earlier. The combination of C37D with I44V, V45A, Y66S, R68K, A73S (i.e. the inverse of hβ1*) reduced expression by about one-third, whereas combinations of C37D and mutations to the A and B regions reduced expression by about one-half (Table 5).
Overall, these data suggest that assembly of hβ3 homopentamers is a robust phenomenon. It can most readily be disrupted by mutations in the A and B regions contained in loop 9. The disruptive effect of hβ1 Asp-37 seems to be largely overcome in hβ3 by other stabilizing interactions, in both the A and B regions of loop 9 and loop 10 and the residues between Ile-44 through Ala-73.
Functional Competence of Surface Receptors—We tested the ability of five subunits to assemble into homomeric surface receptors that could be activated by pentobarbital: hβ3F, hβ1F, hβ1F (D37A) (mode 1), hβ1F* A (mode 2), and hβ1F (A45V and K196R) (mode 3). The goal of these experiments was simply to determine whether the homomeric receptors could be activated, as a full physiological and pharmacological analysis is beyond the scope of this study. Three cells were tested for each construct. Cells were selected for study by the binding of beads coated with antibody to the FLAG epitope, to select for cells expressing a high density of the transfected subunits on the surface. When cells were selected for surface expression in this fashion, each construct resulted in cells that showed a response to both 100 and 1000 μm pentobarbital with an increase in current. Most constructs also showed a rebound current after removal of pentobarbital, which has been ascribed to the ability of pentobarbital to both activate and block the GABA receptor (32). Accordingly, each of these subunits can assemble to produce functional surface receptors. We believe that the finding that a few cells could express adequate levels of hβ1 to be detected by bead binding, and to produce a measurable response to pentobarbital, reflects cell-to-cell variability in levels of protein synthesis (and possibly chaperone proteins). The level of expression in the entire culture was very low for hβ1, as found for the ELISA results.
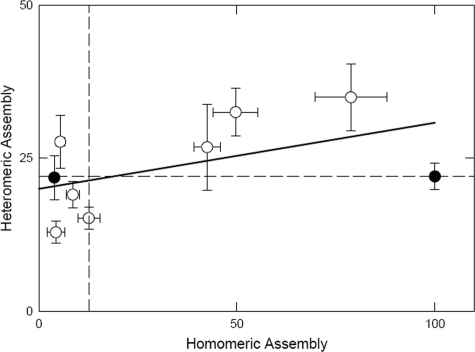
Heteromeric Assembly with the α1 Subunit—Both hβ1 and hβ3 subunits assemble with rat α1 subunits tagged with the Myc epitope (rα1myc) to the same extent (Table 6 and Fig. 6). We also examined the ability of selected mutated subunits to assemble with α1 to determine whether residues affecting homomeric assembly significantly affected heteromeric assembly. All tested subunits could assemble with rα1myc, and there was no correlation between the abilities of a subunit to assemble as a homomultimer or a heteromultimer (Fig. 6).
TABLE 6.
Heteromeric assembly of hβ1 mutant subunits with rα1 myc
| Constructa | Heteromeric expressionb | Nc | Homomeric expressiond | N |
|---|---|---|---|---|
| hβ3F | 22 ± 2.2 | (9) | 100 ± 0.9 | (122) |
| hβ1F | 22 ± 3.6 | (7) | 4 ± 0.4 | (109) |
| hβ1F (D37A) | 35 ± 5.5 | (6) | 79 ± 9 | (10) |
| hβ1F* | 13 ± 1.8 | (5) | 4 ± 2 | (5) |
| hβ1F*A | 32 ± 3.9 | (8) | 50 ± 6 | (21) |
| hβ1F*(E172D, G173K) | 28 ± 4.4 | (8) | 5 ± 1 | (5) |
| hβ1F A45V, K197N | 19 ± 2.2 | (5) | 9 ± 1.7 | (3) |
| hβ1F A45V, K196R | 27 ± 7.0 | (6) | 43 ± 3.4 | (4) |
| hβ3F V45A, DNTK | 15 ± 1.8 | (6) | 13 ± 2.8 | (7) |
Selected β subunit constructs were co-expressed with rat α1 subunits tagged with the Myc epitope
ELISA results for the Myc epitope mean ± S.D., expressed as percent of homomeric hβ3myc assembly
Number of separate experiments
Data for homomeric expression, extracted from earlier tables
FIGURE 6.
Heteromeric expression of mutated hβ1 subunits with rα1 myc. The figure shows the ability to express with rα1myc (given as the percentage of surface homomeric expression obtained with hβ3myc) plotted against the ability to express as a homomer (given as the percentage of surface homomeric expression with hβ3F). Each data point represents the mean, and bars show the mean ± S.E. Heteromeric expression levels of hβ1F and hβ3F with rα1myc are the same and are represented by the solid circles. The median expression levels are shown by the dashed lines. The solid line through the data represents the linear regression with a significance that is not different from zero (analysis of variance correlation coefficient of 0.151).
Assembly in TSA201 Cells—To determine that the results did not depend on cell type, we examined expression for selected constructs in the human embryonic kidney 293-derived cell line TSA201. We obtained similar results for wild type hβ1F (0.7 ± 1.0%, n = 5) in relation to hβ3F. Hβ1F (D37A) (mode 1) expressed at 32 ± 3% (n = 5) of hβ3F, hβ 1F*A (mode 2) at 15 ± 5% (n = 5), and hβ1F (A45V and K196R) (mode 3) at 12 ± 4% (n = 5). Although assembly in TSA201 cells was not as robust as in QT6 cells, expression of mutated subunits followed a similar pattern as in QT6. Analysis of variance statistical comparisons show all three constructs expressing better than hβ1F with a Bonferroni corrected P for differences of p <. 001 or better.
DISCUSSION
This study has shown that the hβ3 subunit expresses on the cell surface as a homopentamer in somatic cells, whereas hβ1 does not, and that mutagenesis of specific residues in hβ1 can result in homomeric expression.
There have been some differences in previous reports regarding the expression of homo-oligomeric receptors of β1 subunits. The β1 subunits of various species have been shown to express in Xenopus oocytes as homo-oligomers (21, 33–35). However, β1 expression in somatic cell lines has been more variable, and any reported expression has been low (15, 22, 33, 36).
We found that there are at least three sets of residues in the hβ1 subunit that when mutated can confer significantly increased surface expression on the hβ1 subunit. Functional assay of mutated subunits, from each mode of assembly, shows that surface expression results in receptors that can be activated by pentobarbital in a dose-dependent fashion. Therefore, ELISA data reflect functional assembled subunits.
We have used a homology-based structural model of the extracellular domain to infer the location of the identified residues, which produces some insights into possible mechanisms by which enhanced assembly is produced. Others have previously used the AChBP model in studies of assembly of nicotinic acetylcholine receptors (37) and heteromeric GABAA receptors (38).
Modes for Surface Expression of β1 Subunits—Mutations to hβ1 Asp-37 define mode 1. This residue is predicted to be located far from the intersubunit interface, and the results indicate that proper intrasubunit interactions are critical for subunit assembly. The effects of mutations to Asp-37 and Glu-165 indicate that side chain interactions may have profound effects on assembly, preventing the subunit from being able to adopt an appropriate conformation to allow efficient assembly. The nature of the interaction between Asp-37 and Glu-165 is not known, but it might involve electrostatic repulsion that impairs folding. Overall, mode 1 reflects an intrasubunit effect, which does not involve a change in any residues predicted to be involved in intersubunit interactions but permits appropriate folding. It might seem surprising that the residue found in the β2 subunit (alanine) conferred expression to hβ1 (D37A), whereas mouse β2 expresses poorly. It is likely that other residues in the A and B regions of the β2 subunit contribute to the poor expression (see below). Previous studies of the glycine receptor α1 subunit (18) have shown that Asn-38, homologous to Asp-37 in hβ1, is critical for assembly of the receptor. However, the underlying mechanism is likely to be different, as Asn-38 is required for a glycosylation step that is necessary for exit from the endoplasmic reticulum.
Mode 2 is characterized by seven mutations: V44I and A45V of β-sheet 1, S66Y and K68R of β-sheet 2, S73A of loop 3, and N169R and E172D of loop 9. Each of these secondary structures, except for loop 3, contains residues included in the negative interface of the subunit. However, only Ala-45 is likely to interact directly with residues of the adjacent subunit. Loop 9 contains both the A and B regions and clearly plays a major role in multimeric assembly, based on the present results and previous studies by Taylor et al. (13). Mutations of the B region (N179E and K180R) contribute to the subunit interface of loop 9 and can increase surface expression of the hβ1 subunit, as found for the mouse β2 subunit (13). We emphasized studies of residues in the A region, as they had not been previously examined, whereas the A and B regions appeared to contribute similar and nonadditive increases in expression. Various subsets of these seven residues were tested, but only the full combination was found to express at a level significantly greater than hβ1. Studies of the assembly of the α1 subunit of the glycine receptor have also implicated loop 3 as important for assembly (18). Overall, we interpret these results as indicating that mode 2 reflects a relatively distributed set of interactions that stabilize the subunit in a conformation that assembles efficiently into a homopentamer, with less contribution from changes in the residues directly involved in intersubunit interactions.
In contrast, mode 3 assembly seems to principally reflect roles for two residues directly involved in interactions at the subunit interface. The mutated residues (A45V and K196R) each are likely to interact with residues of the adjacent subunit. These interactions indicate assembly determinants of hβ3 that are lacking in hβ1, specifically hydrophobic and ionic bond anchors between subunits. No other studies of GABAA homomeric or heteromeric receptors have implicated these regions as important.
Comparison with Studies of mβ2 Receptor Expression—Taylor et al. (13) identified four residues in the β2 subunit, mutation of which could enhance surface expression. These residues are in the A and B regions, between amino acid residues 170 and 180. Our observations confirm the importance of residues in these regions for efficient assembly, and also identify additional portions of the β1 subunit that can significantly compromise assembly.
Reduction of hβ3 Receptor Expression—Reduction of hβ3 receptor expression was accomplished by utilizing the data acquired from our experiments on hβ1 assembly. Combining sequence elements from the several modes of assembly resulted in significant reductions in expression. These results confirm that assembly determinants have been identified and can be manipulated. Whereas residues involved in each mode of assembly could be combined to reduce assembly, the most dramatic reduction was observed with changes in β-sheet 1 and loop 9, specifically V45A and the mutations β3 (D171G, N173K, T179E, and K180R) of loop 9. In addition, combinations of C37D (mode 1) with changes in β-sheets 1 and 2 and the AB regions of loop 9 also gave significant reduction to hβ3 surface expression. A surprising observation is that the sequence variant of the hβ3 construct we studied, which has a valine at position 45, appears to make a significant contribution to the overall assembly competence of the hβ3 subunit. Although the single mutation hβ3 (V45A) produces a small reduction in expression, V45A results in a major reduction when assembly is compromised by other mutations. Overall, these observations, with the hβ3 subunit, reinforce the interpretation of the studies of the hβ1 subunit that multiple interactions within the extracellular domain of the subunit can establish or compromise the ability of the subunit to fold into a conformation supporting homomeric assembly.
Previous studies of heteromeric GABAA receptor assembly have also identified the β-sheet 2 (15, 25) and loop 9 (13) regions as critical for assembly. Although these studies focused on changes that affected assembly between α, β, and γ subunits, we have identified the same general regions for homomeric assembly. Specific residues may differ between interacting sites, but at least some of the structural regions have been conserved.
Expression of β1 Subunits in Heteromeric Receptors—We found that the β3 subunit could “rescue” expression of the β1 subunit. This observation suggests that the intersubunit interactions of the wild type β1 subunit are capable of supporting assembly. In addition, the GABAA α1 subunit, although it is not able to express as a homopentamer, can express as a heteropentamer with the β1 subunit. These observations suggest that other subunits may act as a crystallization surface for the normally incompetent hβ1 subunit, to stabilize assembly-competent conformations. Most GABAA receptors are composed of several different subunits (e.g. α1, β2, and γ2), and our data support the idea that the specific determinants for heteromeric assembly differ from those required for homomeric assembly.
Conclusions—In conclusion, these data show that GABAA β1 and β3 subunits can be a very useful model for probing determinants of assembly in a systematic fashion, in association with a structural model based on homology with the AChBP. Overall, our observations indicate that the tertiary structure of the β1 subunit is somewhat plastic, able to adopt assembly-competent or assembly-incompetent conformations. The observation that assembly can be driven by the presence of the β3 or α1 subunit suggests that the presence of a suitable template may stabilize the assembly-competent conformation. This idea is particularly suggested by the observation that the presence of the β3 subunit can overcome the effects of the presence of hβ1 Asp-37, as this residue is far from any predicted interfacial region. In general, the residues involved in the three modes of assembly indicate that the process of multimeric assembly depends on the overall result of multiple energetic contributions, rather than being the result of a collision between matching rigid surfaces. Each mode can independently confer a significant increase in surface expression, even though it seems likely that molecular interactions involved in each mode vary from removal of a dominant disruptive effect (mode 1), to a distributed intrasubunit interaction (mode 2), to a strengthening of intersubunit interactions (mode 3).
Supplementary Material
Acknowledgments
We thank Gustav Akk for critical review of the manuscript and Ping Li for electrophysiology.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 GM47969 and NS22356. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table and Figs. 1–3.
Footnotes
The abbreviations used are: GABAA, γ-aminobutyric acid type A; m, mouse; h, human; r, rat; F, FLAG; QT6, quail fibroblasts; ELISA, enzyme-linked immunosorbent assay; BES, N,N-bis(2-hydroxyethyl)-2-aminoethansulfonic acid; MPBS, milk phosphate-buffered saline; AChBP, acetylcholine-binding protein; X, chimera; hβ1*, hβ1 (V44I, A45V, S66Y, K68R, and S73A).
References
- 1.Ortells, M. O., and Lunt, G. G. (1995) Trends Neurosci. 18 121-127 [DOI] [PubMed] [Google Scholar]
- 2.Karlin, A., and Akabas, M. H. (1995) Neuron 15 1231-1244 [DOI] [PubMed] [Google Scholar]
- 3.Macdonald, R. L., and Olsen, R. W. (1993) Annu. Rev. Neurosci. 17 569-602 [DOI] [PubMed] [Google Scholar]
- 4.Davies, P. A., Hanna, M. C., Hales, T. G., and Kirkness, E. F. (1997) Nature 385 820-823 [DOI] [PubMed] [Google Scholar]
- 5.Bonnert, T. P., McKernan, R. M., Farrar, S., le Bourdelles, B., Heavens, R. P., Smith, D. W., Hewson, L., Rigby, M. R., Sirinathsinghji, D. J., Brown, N., Wafford, K. A., and Whiting, P. J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9891-9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sieghart, W., Fuchs, K., Tretter, V., Ebert, V., Jechlinger, M., Hoger, H., and Adamiker, D. (1999) Neurochem. Int. 34 379-385 [DOI] [PubMed] [Google Scholar]
- 7.Enz, R., and Cutting, G. R. (1998) Vision Res. 38 1431-1441 [DOI] [PubMed] [Google Scholar]
- 8.Amin, J., and Weiss, D. S. (1996) Proc. R. Soc. Lond. B Biol. Sci. 263 273-282 [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan, S., Nichols, C. J., Lawless, G. M., Olsen, R. W., and Tobin, A. J. (1999) J. Biol. Chem. 274 26633-26638 [DOI] [PubMed] [Google Scholar]
- 10.Klausberger, T., Fuchs, K., Mayer, B., Ehya, N., and Sieghart, W. (2000) J. Biol. Chem. 275 8921-8928 [DOI] [PubMed] [Google Scholar]
- 11.Sarto, I., Klausberger, T., Ehya, N., Mayer, B., Fuchs, K., and Sieghart, W. (2002) J. Biol. Chem. 277 30656-30664 [DOI] [PubMed] [Google Scholar]
- 12.Ehya, N., Sarto, I., Wabnegger, L., and Sieghart, W. (2003) J. Neurochem. 84 127-135 [DOI] [PubMed] [Google Scholar]
- 13.Taylor, P. M., Thomas, P., Gorrie, G. H., Connolly, C. N., Smart, T. G., and Moss, S. J. (1999) J. Neurosci. 19 6360-6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor, P. M., Connolly, C. N., Kittler, J. T., Gorrie, G. H., Hosie, A., Smart, T. G., and Moss, S. J. (2000) J. Neurosci. 20 1297-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollan, K., King, D., Robertson, L. A., Brown, K., Taylor, P. M., Moss, S. J., and Connolly, C. N. (2003) J. Biol. Chem. 278 4747-4755 [DOI] [PubMed] [Google Scholar]
- 16.Enz, R., and Cutting, G. R. (1999) Brain Res. 846 177-185 [DOI] [PubMed] [Google Scholar]
- 17.Xue, H., Zheng, H., Li, H. M., Kitmitto, A., Zhu, H., Lee, P., and Holzenburg, A. (2000) J. Mol. Biol. 296 739-742 [DOI] [PubMed] [Google Scholar]
- 18.Griffon, N., Buttner, C., Nicke, A., Kuhse, J., Schmalzing, G., and Betz, H. (1999) EMBO J. 18 4711-4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly, C. N., Krishek, B. J., McDonald, B. J., Smart, T. G., and Moss, S. J. (1996) J. Biol. Chem. 271 89-96 [DOI] [PubMed] [Google Scholar]
- 20.Ueno, S., Zorumski, C., Bracamontes, J., and Steinbach, J. H. (1996) Mol. Pharmacol. 50 931-938 [PubMed] [Google Scholar]
- 21.Cestari, I. N., Min, K. T., Kulli, J. C., and Yang, J. (2000) J. Neurochem. 74 827-838 [DOI] [PubMed] [Google Scholar]
- 22.Connolly, C. N., Wooltorton, J. R. A., Smart, T. G., and Moss, S. J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 9899-9904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadingham, K. L., Wingrove, P., Le Bourdelles, B., Palmer, K. J., Ragan, C. I., and Whiting, P. J. (1993) Mol. Pharmacol. 43 970-975 [PubMed] [Google Scholar]
- 24.Darbandi-Tonkabon, R., Hastings, W. R., Zeng, C. M., Akk, G., Manion, B. D., Bracamontes, J. R., Steinbach, J. H., Mennerick, S. J., Covey, D. F., and Evers, A. S. (2003) J. Biol. Chem. 278 13196-13206 [DOI] [PubMed] [Google Scholar]
- 25.Sarto, I., Wabnegger, L., Dogl, E., and Sieghart, W. (2002) Neuropharmacology 43 482-491 [DOI] [PubMed] [Google Scholar]
- 26.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1992) Short Protocols in Molecular Biology, 2nd Ed., pp. 9-5, John Wiley & Sons, Inc., New York
- 27.Li, P., Covey, D. F., Steinbach, J. H., and Akk, G. (2006) Mol. Pharmacol. 69 2015-2026 [DOI] [PubMed] [Google Scholar]
- 28.McCann, C. M., Bracamontes, J., Steinbach, J. H., and Sanes, J. R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5149-5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cromer, B. A., Morton, C. J., and Parker, M. W. (2002) Trends Biochem. Sci. 27 280-287 [DOI] [PubMed] [Google Scholar]
- 30.Wagstaff, J., Chaillet, J. R., and Lalande, M. (1991) Genomics 11 1071-1078 [DOI] [PubMed] [Google Scholar]
- 31.Brejc, K., van Dijk, W. J., Klaassen, R. V., Schuurmans, M., van der Oost, J., Smit, A. B., and Sixma, T. K. (2001) Nature 411 269-276 [DOI] [PubMed] [Google Scholar]
- 32.Serafini, R., Bracamontes, J., and Steinbach, J. H. (2000) J. Physiol. (Lond.) 524 649-676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishek, B. J., Moss, S. J., and Smart, T. G. (1996) Mol. Pharmacol. 49 494-504 [PubMed] [Google Scholar]
- 34.Sanna, E., Garau, F., and Harris, R. A. (1995) Mol. Pharmacol. 47 213-217 [PubMed] [Google Scholar]
- 35.Blair, L. A., Levitan, E. S., Marshall, J., Dionne, V. E., and Barnard, E. A. (1988) Science 242 577-579 [DOI] [PubMed] [Google Scholar]
- 36.Angelotti, T. P., and Macdonald, R. L. (1993) J. Neurosci. 13 1429-1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sine, S. M., Wang, H. L., and Bren, N. (2002) J. Biol. Chem. 277 29210-29223 [DOI] [PubMed] [Google Scholar]
- 38.Sarto-Jackson, I., Ramerstorfer, J., Ernst, M., and Sieghart, W. (2006) J. Neurochem. 96 983-995 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.