Abstract
The crenarchaeon Sulfolobus solfataricus uses arginine to produce putrescine for polyamine biosynthesis. However, genome sequences from S. solfataricus and most crenarchaea have no known homologs of the previously characterized pyridoxal 5′-phosphate or pyruvoyl-dependent arginine decarboxylases that catalyze the first step in this pathway. Instead they have two paralogs of the S-adenosylmethionine decarboxylase (AdoMetDC). The gene at locus SSO0585 produces an AdoMetDC enzyme, whereas the gene at locus SSO0536 produces a novel arginine decarboxylase (ArgDC). Both thermostable enzymes self-cleave at conserved serine residues to form amino-terminal β-domains and carboxyl-terminal α-domains with reactive pyruvoyl cofactors. The ArgDC enzyme specifically catalyzed arginine decarboxylation more efficiently than previously studied pyruvoyl enzymes. α-Difluoromethylarginine significantly reduced the ArgDC activity of purified enzyme, and treating growing S. solfataricus cells with this inhibitor reduced the cells' ratio of spermidine to norspermine by decreasing the putrescine pool. The crenarchaeal ArgDC had no AdoMetDC activity, whereas its AdoMetDC paralog had no ArgDC activity. A chimeric protein containing the β-subunit of SSO0536 and the α-subunit of SSO0585 had ArgDC activity, implicating residues responsible for substrate specificity in the amino-terminal domain. This crenarchaeal ArgDC is the first example of alternative substrate specificity in the AdoMetDC family. ArgDC activity has evolved through convergent evolution at least five times, demonstrating the utility of this enzyme and the plasticity of amino acid decarboxylases.
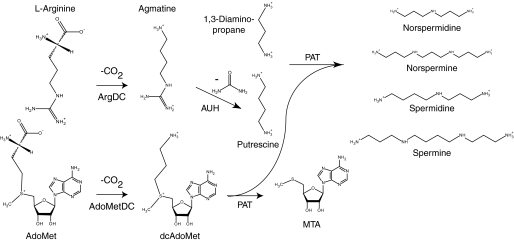
The Crenarchaeota include the most hyperthermophilic cultivated microorganisms. To help stabilize macromolecules in these extreme conditions, some hyperthermophiles produce unusually long chain or branched polyamines (1). Chromatographic analyses of extracts from the crenarchaeon Sulfolobus solfataricus identified significant amounts of the linear polyamines 1,3-diaminopropane, putrescine, sym-norspermidine (caldine), spermidine, and sym-norspermine (thermine), with traces of spermine (2, 3). The biosynthesis of the spermidine and spermine polyamines was predicted to follow a canonical eukaryotic pathway, where the decarboxylation of l-ornithine produces putrescine, and S-adenosyl-l-methionine (AdoMet)2 is decarboxylated to form the propylamine donor S-(5′-adenosyl)-3-methylthiopropylamine (dcAdoMet) (4). Subsequently, the AdoMet decarboxylase (AdoMetDC) (5) and propylamine transferase (6) enzymes were purified from S. solfataricus. The latter enzyme produces spermidine and spermine from putrescine and dcAdoMet, and it produces norspermidine and norspermine from 1,3-diaminopropane and dcAdoMet (Fig. 1).
FIGURE 1.
Proposed pathway for polyamine biosynthesis in S. solfataricus. AdoMetDC (EC 4.1.1.50) catalyzes dcAdoMet formation. ArgDC (EC 4.1.1.19) produces agmatine and CO2 from l-arginine. Agmatinase (AUH; EC 3.5.3.11) catalyzes the hydrolysis of agmatine to produce urea and putrescine. Finally, a propylamine transferase enzyme (PAT) transfers one or two aminopropyl groups from dcAdoMet to putrescine, producing spermidine or spermine. The same enzyme transfers aminopropyl groups to 1,3-diaminopropane, producing sym-norspermidine and sym-norspermine. Alternatively, it is possible S. solfataricus uses the N1-aminopropylagmatine pathway that was described for Thermus thermophilus (41). In that bacterium, PAT transfers a propylamine group to agmatine producing N1-aminopropylagmatine, which can be hydrolyzed to form spermidine by agmatinase. However, both the S. solfataricus and Pyrococcus furiosus PAT proteins efficiently use putrescine or diaminopropane as substrates (6, 7). The S. solfataricus agmatinase protein is more similar to the Pyrococcus horikoshii agmatinase, which hydrolyzes agmatine, than to the T. thermophilus homolog, which does not hydrolyze agmatine (9). Significant amounts of putrescine were detected in S. solfataricus extracts, supporting the canonical model shown here.
The complete genome sequences of 13 thermophilic crenarchaea encode orthologs of propylamine transferase (7) and two homologs of the archaeal type AdoMetDC (supplemental Table 1) (8). However, none of the genomes encodes a recognizable ornithine decarboxylase enzyme to produce putrescine. Instead they have homologs of the agmatine ureohydrolase enzyme (9), suggesting that putrescine is derived from l-arginine as in the Euryarchaeota. Some bacteria produce agmatine from arginine using a pyridoxal 5′-phosphate (PLP)-dependent arginine decarboxylase (ArgDC) (10). Alternatively, euryarchaea use a pyruvoyl-dependent ArgDC that is related to Lactobacillus sp. histidine decarboxylase (11). No crenarchaea have homologs of the PLP-dependent enzyme, and only three have homologs of the euryarchaeal enzyme. Therefore, we predicted that members of the Crenarchaeota have a new type of arginine decarboxylase that is evolutionarily unrelated to the previously characterized enzymes.
The two AdoMetDC proteins from S. solfataricus, SSO0536 and SSO0585, share 47% amino acid sequence identity (Fig. 2). mRNAs from the respective genes were previously shown to have different half-lives (7 min for SSO0536 and 17–20 min for SSO0585) (12). The same discrepancy was observed in Sulfolobus acidocaldarius, and the two genes could have different functions. Both AdoMetDC homologs from S. solfataricus were cloned in Escherichia coli, and the polyhistidine-tagged proenzymes were heterologously expressed and purified. These proteins self-cleaved at a conserved serine residue, forming aminoterminal β-subunits and active pyruvoyl groups on their carboxyl-terminal α-subunits. The His10-SSO0585 protein catalyzed the decarboxylation of AdoMet in a coupled assay with AdoMet synthetase enzyme, and it is probably the same protein that was previously purified (but not sequenced) by Cacciapuoti et al. (5). The His10-SSO0536 protein also self-cleaved, but it did not catalyze AdoMet decarboxylation. Instead, it efficiently catalyzed the decarboxylation of l-arginine. Therefore, this crenarchaeal protein represents a new class of arginine decarboxylases that developed through convergent evolution. A chimeric protein containing the β-subunit of SSO0536 and the α-subunit of SSO0585 functioned specifically as an ArgDC, correlating conserved changes in the amino-terminal part of this protein with the change in substrate specificity. The SSO0536 protein is the first member of the AdoMet decarboxylase family shown to catalyze another reaction. Phylogenetic analysis suggests that an ancestral AdoMetDC gene duplicated once in the crenarchaeal lineage. Subtle changes to the active site structure of one paralog enabled the evolution of this new arginine decarboxylase activity and displaced an ancestral euryarchaeal ArgDC.
FIGURE 2.
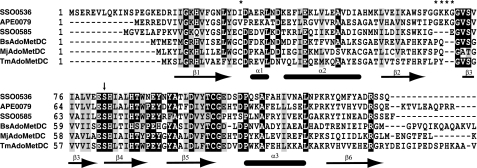
Protein sequence alignment of the S. solfataricus ArgDC protein (SSO0536) with a putative ArgDC from Aeropyrum pernix (APE0079; NP_146956.1), AdoMetDC from S. solfataricus (SSO0585), AdoMetDC from Bacillus subtilis (BsAdoMetDC; NP_390779.1), AdoMetDC from M. jannaschii (MjAdoMetDC; NP_247288.1), and AdoMetDC from T. maritima (TmAdoMetDC; NP_228464.1). The secondary structure of TmAdoMetDC is indicated as β-strands and α-helices below the alignment (from PDB 1VR7) (29). An arrow indicates the site of protein self-cleavage and pyruvoyl group formation at the conserved serine residue of the nascent α-subunit. Asterisks above the alignment indicate positions with amino acid replacements that correlate with substrate specificity. Sequences were aligned using the T-COFFEE program (version 4.96) (23).
EXPERIMENTAL PROCEDURES
Synthesis of dl-Difluoromethylarginine—2-Amino-5-carbamimidamido-2-(difluoromethyl)pentanoic acid (α-difluoromethylarginine (DFMA)) was synthesized by the guanylation of 2-(difluoromethyl)-dl-ornithine (DFMO) using a modification of the procedure described by Bey et al. (13). DFMO hydrochloride (1.0 g, 4.2 mmol) was dissolved in 2.5 ml of 2 m sodium hydroxide. S-Methylisothiourea hemisulfate (1.18 g, 8.5 mmol) was added with stirring at room temperature. The pH was adjusted to 10.5 with sodium hydroxide, and the solution was stirred for 5 days. The solution was adjusted to pH 7 with HCl and evaporated to dryness under vacuum at 60 °C. The residue was dissolved in 7 ml of water and applied to a Dowex 50 (H+) column (1 × 31 cm). The column was subsequently washed with 4 bed volumes of water and 2 volumes of 1 m ammonium hydroxide. DFMA was eluted with 2 m ammonium hydroxide. Ninhydrin-positive fractions were combined and evaporated to dryness under vacuum to produce 0.63 g of DFMA (2.8 mmol, 67%). Crystallization of the hydrochloride salt afforded hygroscopic white crystals. The 1H NMR and 39F NMR spectra were consistent with previous data (13). High resolution electrospray ionization mass spectrometry (ESI-MS) confirmed the chemical composition of the MH+ ion: expected 225.11576 m/z, observed 225.1160 m/z. DFMO was a gift from Dr. Patrick Woster (Wayne State University) and was originally produced by Dow Chemicals. All other chemicals were purchased from various distributors in reagent grade and used without further purification.
Strains and Growth Media—S. solfataricus P2 (DSM 1617) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen. Cultures were grown in complex DSMZ medium 182 containing yeast extract (2 g), KH2PO4 (3.1 g), (NH4)2SO4 (2.5 g), MgSO4·7H2O (0.2 g) and CaCl2·2H2O (0.25 g) in 1 liter adjusted to pH 3.7 with sulfuric acid. Defined growth medium contained (NH4)2SO4 (1.3 g), KH2PO4 (0.28 g), MgSO4·7H2O (0.28 g), CaCl2·2H2O (0.25 g), FeCl3·6H2O (0.02 g), and 100× mineral salt solution (10 ml) (14) in 1 liter adjusted to pH 3.5 with sulfuric acid (15). Defined medium was supplemented with either d-glucose (0.4%, w/v) or casein hydrolysate (0.4%, w/v). Batch cultures were grown in 75 ml of medium in 150-ml medium bottles, incubated in a convection oven at 85 °C attached to a linear motion shaker (80 rpm).
Polyamine Analysis—For analysis by gas chromatographymass spectrometry, cellular polyamines were converted to trifluoroacetamide derivatives. Cells (0.15 g wet mass) were suspended in 0.75 ml of water and lysed by sonication on ice. To the lysate was added 0.75 ml of trichloroacetic acid (10% w/v), and the mixture was centrifuged for 10 min at 18,000 × g. The pellet was reextracted with 1 ml of trichloroacetic acid, and the combined supernatant was extracted twice with 10 ml of diethyl ether. The aqueous solution was evaporated to dryness under vacuum, and the solids were suspended in 0.3 ml of trifluoroacetic anhydride for 30 min at 55 °C. The trifluoroacetyl polyamine derivatives were analyzed by gas chromatographymass spectrometry with chemical ionization in the positive ion mode. Mass spectra were consistent with those reported previously (2, 16).
For quantitative analysis by HPLC, cellular polyamines were converted to dansyl sulfonamide derivatives (17). Extracts of S. solfataricus cells were deproteinized with perchloric acid, and then the neutralized extracts were mixed with dansyl chloride. The derivatives were extracted into cyclohexane, evaporated to dryness by heating under N2, and dissolved in 50% acetonitrile. These samples were applied to a reversed phase HPLC column (Luna C18(2), 150 × 4.6 mm; Phenomenex) and eluted using a gradient from 50 to 100% acetonitrile in 0.02% trifluoroacetic acid. Dansyl sulfonamides were detected by their fluorescence with an excitation wavelength of 340 nm and an emission wavelength of 515 nm. These derivatives were identified by their co-elution with polyamine standards, and they were quantified by integrating the peak areas of the fluorescence chromatograms. 1,7-Diaminoheptane was used as an internal standard to measure polyamine extraction and derivatization efficiency (73–84% recovery).
Arginine Decarboxylase Assay—The rate of arginine decarboxylation was determined using a CO2 capture assay to detect the release of 14CO2 from 14C-labeled arginine (18). Standard reactions (100 μl) contained 12 mm citric acid, 26 mm sodium phosphate (pH 6.0), 10 mm l-arginine-HCl, 6.5–300 nCi of l-[1-14C]arginine (55 mCi mmol–1; American Radiolabeled Chemicals), and enzyme. After a 5-min incubation at 80 °C, the reactions were terminated by the addition of 100 μl of 4 m HCl and heated at 70 °C for 15 min. One unit of activity catalyzes the decarboxylation of 1 μmol of l-arginine/min. A nonlinear regression program (KaleidaGraph version 3.6) estimated steady-state kinetic parameters from the initial rate data fit to the hyperbolic Michealis-Menten-Henri equation.
Protein thermostability was tested by preincubating 2 μgof protein at temperatures from 4 to 90 °C for 10 min in citrate-phosphate buffer (pH 6.0). Arginine decarboxylase activity was determined at room temperature as described above. The temperature dependence of enzyme activity was determined by preincubating enzyme in reaction buffer at temperatures from 4 to 70 °C for 10 min. Reactions were initiated by the addition of arginine substrate and incubated at the same temperature.
The pH dependence of enzyme activity was analyzed at 70 °C in reactions containing 2 μg of purified enzyme, 10 mm l-arginine, 200 nCi of l-[U-14C]arginine (305 mCi mmol–1;GE Healthcare), and buffer (citrate-phosphate buffer (pH 2.8–6.0), 25 mm sodium potassium phosphate (pH 7.0–8.0), or 6 mm sodium borate (pH 10)).
Cloning and Molecular Biology—The MJ1208 gene was amplified by PCR using oligonucleotide primers 5MJ1208BN and 3MJ1208B (supplemental Table 2) and Methanocaldococcus jannaschii JAL-1 chromosomal DNA (19). The product was ligated in the NdeI and BamHI sites of vector pET-19b to produce vector pDG395 (supplemental Table 3). Primers 5SSO0585BN and 3SSO0585B were similarly used to clone the SSO0585 gene from S. solfataricus P2 chromosomal DNA producing vector pDG401. Primers 5SSO0536BN and 3SSO0536B were used to clone the S. solfataricus SSO0536 gene producing vector pDG398. Splicing overlap extension PCR was used to construct two chimeric genes (20). For the β536α585 chimera, the T7 promoter and SSO0536ChR primers were used to amplify the 5′-fragment from pDG401, and the SSO0585ChF and T7 terminator primers were used to amplify the 3′-fragment from pDG398. Both gel-purified PCR products were mixed together, the annealed product was extended with Vent DNA polymerase (New England Biolabs), and the chimeric template was amplified with the T7 promoter and T7 terminator primers. This chimeric product was ligated into the NdeI and BamHI sites of pET-19b to create vector pDG453. To produce the β585α536 chimera, the T7 promoter and SSO0585ChR primers were used to amplify the 5′-fragment from pDG398, and the SSO0536ChF and T7 terminator primers were used to amplify the 3′-fragment from pDG401. The products were spliced as described above and cloned into the NdeI and BamHI sites of pET-19b to create vector pDG483. Plasmids were propagated in E. coli DH5α cells (Invitrogen). Recombinant DNA was sequenced at the Institute for Cellular and Molecular Biology Core Laboratories DNA Sequencing facility (University of Texas, Austin, TX).
Protein Expression and Purification—The polyhistidine-tagged proteins His10-MJ1208, His10-SSO0536, His10-SSO0585, His10-β536α585, and His10-β585α536 were heterologously expressed in E. coli BL21 (DE3) strains transformed with the respective expression vectors (supplemental Table 3). The cells were grown with continuous shaking (250 rpm) at 37 °C in Luria-Bertani broth with ampicillin (100 μgml–1). Protein expression and cell lysis were performed by standard methods (18). Extracts were heated at 75 °C for 20 min and then centrifuged at 18,000 × g for 10 min. Ni2+ affinity chromatography separated the polyhistidine-tagged proteins. The concentrated, desalted proteins were further purified by strong anion exchange chromatography using a Mono Q column (5 × 50 mm, 10 μm; GE Healthcare). This column was equilibrated with buffer A containing 25 mm Tris-HCl (pH 7.6), and protein was eluted using a gradient to 100% buffer B (buffer A with 1 m NaCl). Purified protein was desalted using a HiTrap Sephadex G-25 column (5 ml; GE Healthcare) in 50 mm ammonium bicarbonate (pH 7.5) and was concentrated by ultrafiltration (3000 molecular weight cut-off).
Protein Size Measurements—Apparent masses of purified, denatured proteins were measured by SDS-PAGE using the Schägger and von Jagow Tris-Tricine system with 12% total and 3.3% cross-linked acrylamide (21). Protein bands were identified in the gels by silver staining (Pierce) or staining with Coomassie Brilliant Blue R dye. Measurements of native protein mass and Stokes radius were made by analytical size exclusion chromatography (18). Precipitation with trichloroacetic acid was used to concentrate proteins from collected fractions; these were analyzed by SDS-PAGE to determine the subunit composition of oligomers. For mass spectrometry, affinity-purified proteins were desalted into 50 mm ammonium bicarbonate (pH 7.5) using a Sephadex G-25 desalting column (5 ml; GE Healthcare). Protein ESI-MS was performed by the CRED Analytical Instrumentation Facility at the University of Texas at Austin.
S-Adenosylmethionine Decarboxylase Assay—AdoMet decarboxylase activity was measured in a coupled reaction using AdoMet synthetase to produce the S-adenosyl-l-[carboxyl-14C]methionine substrate. Reactions contained His10-MJ1208 protein (6 μg), 25 mm Hepes [4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid]KOH (pH 8), 50 mm KCl, 10 mm MgCl2, 2 mm l-methionine, 100 nCi of l-[1-14C]methionine (55 mCi mmol–1; American Radiolabeled Chemicals), and 9.5 mm ATP in a volume of 100 μl (19). After a 30-min incubation at 70 °C, purified decarboxylase enzyme (1 μg) was added, and reactions were incubated for 5 min. The reactions were terminated by the addition of 100 ml of 4 m HCl, and released 14CO2 was determined using a CO2 capture assay (8).
Identification of Alternative Substrates and Inhibitors—Substrate analogs and carbonyl-reactive compounds were preincubated with 1 μgofHis10-SSO0536 protein at 80 °C for 10 min before 0.2 mm arginine was added. After 5 min, the reactions were stopped, and the released 14CO2 was measured. The potential inhibitors tested included the arginine analogs l-arginine O-methyl ester, Nα-acetyl l-arginine, l-argininamide, l-canavanine, l-citrulline, d-arginine, l-homoarginine, NG-nitro-l-arginine methyl ester, NG-methyl l-arginine, l-ornithine, l-histidine, l-lysine, and l-methionine. Histidine, homoarginine, and canavanine were also tested as potential substrates in reactions without arginine. Primary amines in the reaction product were derivatized with naphthalene 2,3-dicarboxaldehyde and cyanide to produce fluorescent cyanobenz[f]isoindole derivatives, which were analyzed by HPLC as described previously (22). The nucleophiles O-methyl hydroxylamine hydrochloride and O-nitrobenzylhydroxylamine hydrochloride were also tested as inhibitors.
Phylogenetic Analysis of AdoMetDC Homologs—Amino acid sequences from 13 ArgDC and 19 AdoMetDC homologs were separately aligned using the T-Coffee program (version 4.96) (23). The alignments were manually combined using the CIN-EMA5 alignment editor program (24). From the full alignment, 118 positions that were confidently aligned were chosen for phylogenetic analysis. The proml program from the Phylip package (version 3.66; J. Felsenstein, University of Washington) was used to infer a maximum likelihood phylogeny from this alignment, with the Jones-Taylor-Thornton model of amino acid changes and a γ-distribution of rates (α = 2.4) approximated by three states. Bootstrap analysis was performed with 100 replicates. A similar phylogeny was inferred using the protdist and neighbor programs from the same software package. Full organism names and sequence accession numbers are listed in the supplemental materials.
RESULTS
Polyamines of S. solfataricus P2—Previous studies identified spermidine and norspermine as the predominant polyamines in Caldariella acidophila (now S. solfataricus DSM 5833) (2) and S. solfataricus P1 (3), with traces of norspermidine. However, cells in those experiment were grown on a complex medium containing yeast extract and casamino acids, which could have introduced exogenous polyamines. To identify the polyamines specifically produced by S. solfataricus P2, we grew the cells on a defined glucose minimal medium and extracted polyamines for gas chromatography-mass spectrometry analysis as the trifluoroacetyl derivatives (2). The polyamine pool comprised spermidine (76%), norspermine (10%), putrescine (8%), and norspermidine (5%). For comparison, cells grown with casamino acids instead of glucose contained higher total levels of polyamines, including spermidine (72%), norspermine (12%), norspermidine (11%), and putrescine (5%). Both cultures also contained high levels of the osmolyte trehalose (25). Therefore amino acids in the growth medium slightly changed the distribution and abundance of polyamines in S. solfataricus, although the types of polyamines detected were consistent with those reported previously.
Arginine Decarboxylase Activity in P2 Cells—A metabolic reconstruction of polyamine biosynthesis in S. solfataricus P2 from its genome sequence led us to predict that these cells decarboxylate arginine to produce agmatine, and hydrolyze the guanidinium group to produce putrescine (Fig. 1). To establish that arginine decarboxylase is expressed in this strain, we measured ArgDC activity at 80 °C in cell-free extracts of S. solfataricus grown to stationary phase in defined medium with glucose or casamino acids. Extract from glucose-grown cells catalyzed arginine decarboxylation with a specific activity of 160 pmol min–1 mg–1, compared with a specific activity of 400 pmol min–1 mg–1 for extract from cells grown on casamino acids. These cells catalyzed l-arginine decarboxylation at pH 6, consistent with the pathway proposed for polyamine biosynthesis in Fig. 1.
The physiological significance of arginine decarboxylase was tested by adding DFMA to S. solfataricus cultures growing in glucose minimal medium. DFMA irreversibly inhibits arginine decarboxylases, and DFMO irreversibly inhibits ornithine decarboxylases (26, 27). Cells inoculated into medium containing 2 mm DFMA had a prolonged lag phase of growth compared with cells inoculated into medium with 2 mm DFMO or no inhibitor. An actively growing culture that was split into three samples (DFMA, DFMO, or control) showed only a small decrease in the growth rate with DFMA. Cells growing in DFMA medium had a specific growth rate of 0.040 ± 0.0013 h–1, compared with cells in DFMO medium (0.045 ± 0.0034 h–1) or control medium (0.044 ± 0.0018 h–1). Cells grown with DFMA contained lower concentrations of putrescine and spermidine than cells grown in control or DFMO media (Table 1). However, cells grown in all three conditions had similar total polyamine contents. S. solfataricus cells adapted to DFMA by substantially increasing their relative levels of norspermine, which does not contain a putrescine core.
TABLE 1.
Polyamine contents of S. solfataricus cells
|
Samplea |
Polyaminesb |
||||
|---|---|---|---|---|---|
| Putrescine | Norspermidine | Spermidine | Norspermine | Total | |
| nmol mg-1 dry mass | |||||
| Untreated | 1.6 | 1.2 | 14.5 | 2.0 | 19.3 |
| DFMO | 1.4 | 1.0 | 13.2 | 1.9 | 17.4 |
| DFMA | 0.51 | 1.3 | 9.3 | 9.7 | 20.9 |
S. solfataricus P2 cells were grown in defined glucose medium at 85 °C to an optical density of 0.16 at 600 nm. The culture was divided into an untreated sample, a sample with 3 mm DFMO, and a sample with 3 mm DFMA. The cells were grown for an additional 50 h until the cultures reached optical densities of 1.1 (untreated and DFMO) or 0.8 (DFMA). Cells collected by centrifugation were stored at -20 °C.
Deproteinized cell extracts were mixed with dansyl chloride, and the sulfonamide derivatives were analyzed by HPLC as described under “Experimental Procedures” (17). The polyamine concentrations were determined using external standards and were corrected for losses during derivatization and extraction using 1,7-diaminoheptane as an internal standard.
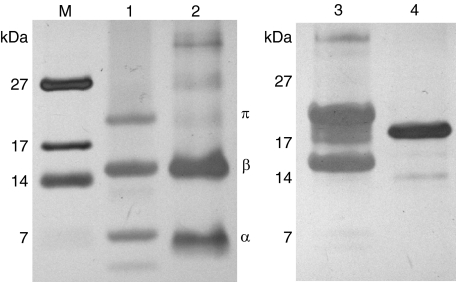
Expression, Purification, and Cleavage of AdoMetDC Homologs—Both AdoMetDC homologs from S. solfataricus were heterologously expressed in E. coli as soluble, heat-stable proteins. SDS-PAGE analysis of the affinity-purified His10-SSO0585 protein preparation showed three prominent bands with apparent molecular masses of 21, 15, and 5 kDa (Fig. 3). These bands corresponded to the proenzyme, β-subunit, and α-subunit, respectively, and densitometry indicated that 77% of the proenzyme had cleaved in the preparation shown. ESI-MS identified an ion series corresponding to the proenzyme (16,602 Da observed, 16,680 Da expected), the methionine aminopeptidase-cleaved proenzyme (16,546 Da observed, 16,549 Da expected) and the α-subunit (6,258 Da observed, 6,260 Da expected). Peaks corresponding to unidentified proteins with apparent masses of 11,758, 13,649, and 13,780 Da were also observed, at lower ion abundances. Size exclusion chromatography indicated that the His10-SSO0585 protein formed a dimeric complex with an apparent mass of 33 kDa and a Stokes radius of 26 Å.
FIGURE 3.
Proenzymes (π) of the heterologously expressed His10-SSO0356 and His10-SSO0585 proteins self-cleave to form amino-terminal subunits (β) and carboxyl-terminal subunits (α) with reactive pyruvoyl groups. Affinity-purified proteins were separated by SDS-PAGE and stained with silver. Lane M, polypeptide marker corresponding to the indicated molecular masses; lane 1, 0.9 μgofHis10-SSO0356; lane 2, 1.1 μgof His10-SSO0585; lane 3, 18 μg of His10-β536α585 chimera; lane 4, 9 μg of His10-β585α536 chimera.
The purified His10-SSO0536 protein preparation also formed three predominant bands on an SDS-polyacrylamide gel with apparent molecular masses of 21, 15, and 6 kDa. Up to 54% of the proenzyme was cleaved in preparations purified from heat-treated cell lysate; however, only 21% of the protein was cleaved in preparations from unheated lysates. ESI-MS analysis identified ion series corresponding to the proenzyme (17,937 Da observed, 18,014 Da expected) and the methionine aminopeptidase-cleaved proenzyme (17,880 Da observed, 17,883 Da expected), as well as the α-subunit (6,123 Da observed, 6,125 Da expected) and the β-subunit (11,759 Da observed, 11,760 Da expected for aminopeptidase-cleaved subunit). The His10-SSO0536 protein eluted from a size exclusion column with an apparent mass of 80 kDa and a Stokes radius of 35 Å, suggesting that it forms a tetramer. SDS-PAGE analysis of fractions eluting from the column showed that the oligomeric protein contained ∼50% uncleaved proenzyme. Although heating the unpurified proenzyme in E. coli lysate promoted cleavage, incubating the purified protein at 80 °C did not increase the portion of protein cleaved; nor did incubation with methoxyamine or O-nitrobenzylhydroxylamine at 80 °C for 20 min.
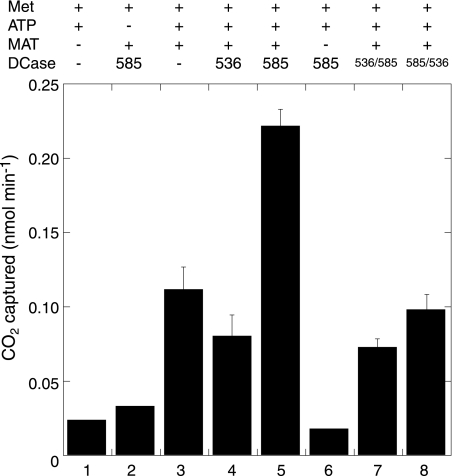
S-Adenosylmethionine Decarboxylase Activity of SSO0585—In a coupled reaction with AdoMet synthetase, the His10-SSO0585 protein catalyzed AdoMet decarboxylation with a net specific activity of 0.11 μmol min–1 mg–1 at 70 °C, corresponding to a rate of 2.3 min–1 (Fig. 4). This activity compares favorably with the AdoMet decarboxylase activity of the native enzyme purified previously from S. solfataricus (0.012 μmol min–1 mg–1) (5). In contrast, AdoMet decarboxylase activities were no higher than the background level in reactions containing His10-SSO0536. AdoMet decomposes through a number of spontaneous reactions (28); we have not characterized the volatile products of background activity detected in these assays.
FIGURE 4.
The SSO0585 protein has AdoMet decarboxylase activity. Reactions containing M. jannaschii AdoMet synthetase (MAT), ATP, l-[1-14C]methionine (Met), and buffer salts were preincubated for 30 min at 70 °C before the addition of decarboxylase (DCase), as indicated above the chart. Reactions 1–3 were control reactions omitting AdoMet synthetase and decarboxylase enzymes, ATP, or decarboxylase, respectively. Reaction 4 contained 1 μg of His10-SSO0536 protein, and reaction 5 contained 1 μg of His10-SSO0585 protein. Reaction 6 contained 1 μg of His10-SSO0536 protein but not AdoMet synthetase as a control reaction. Reaction 7 contained 5 μg of His10-β536α585 chimeric protein, and reaction 8 contained 5 μg of His10-β585α536 chimeric protein. Decarboxylase activities are shown with their S.D. values from triplicate experiments.
Arginine Decarboxylase Activity of SSO0536—The purified His10-SSO0536 protein catalyzed l-arginine decarboxylation with a specific activity of 0.5 units/mg at 70 °C. Agmatine was the only amine produced in reaction mixtures containing purified protein and l-arginine (data not shown). In standard assays, using either l-[U-14C]arginine or l-[1-14C]arginine as substrates for the enzyme, 14CO2 was trapped, confirming the decarboxylation reaction.
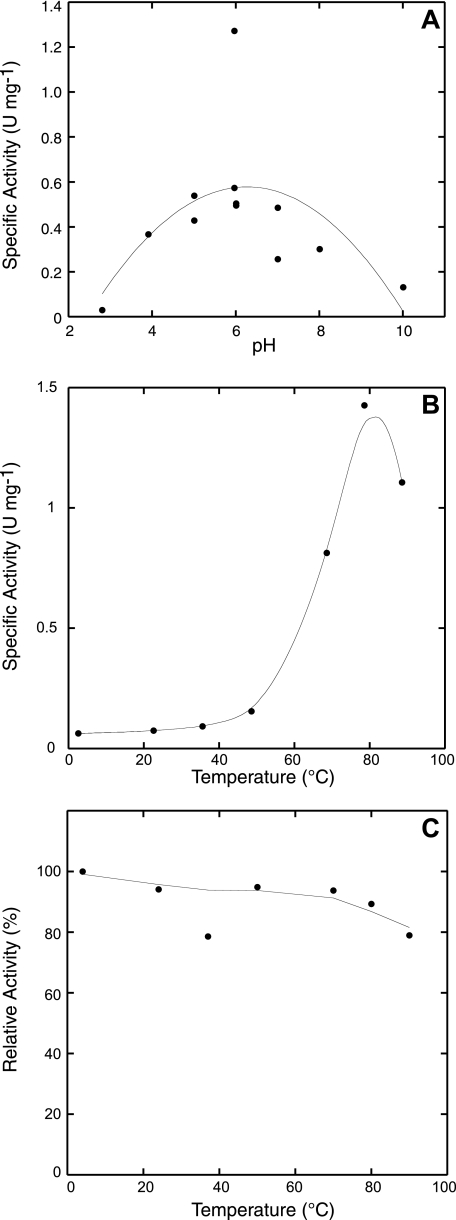
This enzyme had maximal activity at pH 6 (Fig. 5), and it was not stimulated by potassium chloride as observed for other decarboxylases. The SSO0536 decarboxylase was highly thermostable: after a 10-min incubation at 90 °C, the SSO0536 protein retained 80% of its original ArgDC activity. It had maximal decarboxylase activity at 80 °C, but remained active above the practical limits of the assay (Fig. 5). This reaction is first order with respect to arginine, so the Michaelis-Menten-Henri equation fit a plot of initial rates of decarboxylase activity with parameters Km of 150 ± 28 μm and Vmax of 1.1 ± 0.042 units min–1 mg–1 (Table 2). SDS-PAGE densitometry indicated that 46% of this protein was cleaved to form an active pyruvoyl group. Therefore, the enzyme had a turnover number (kcat) of 2.6 s–1, similar to the euryarchaeal enzyme. No arginine decarboxylase activity was detected in purified preparations of His10-SSO0585 (<0.001 units/mg activity).
FIGURE 5.
Arginine decarboxylase activity of the His10-SSO0536 protein. A, the purified enzyme has optimal activity at pH 6, consistent with a biosynthetic function. B, the protein has maximal arginine decarboxylase activity at 80 °C. C, the protein retained 80% arginine decarboxylase activity after preincubation at 90 °C for 10 min.
TABLE 2.
Decarboxylase activities in five classes of ArgDC
| Class | Enzyme | Cofactor | pH optimum | Km | kcat | kcat/Km |
|---|---|---|---|---|---|---|
| mm | s-1 | m-1 s-1 | ||||
| 1 | E. coli SpeA (10) | PLP | 8.4 | 0.03 | 19 | 6.2 × 105 |
| 2 | PBCV-1 DC (26) | PLP | 8.2 | 0.45 | 15 | 3.3 × 104 |
| 3 | E. coli AdiA (35) | PLP | 5.2 | 0.65 | 700 | 1.1 × 106 |
| 4 | M. jannaschii PvlArgDC (11) | Pyruvoyl | 6.0 | 7.1 | 2.7 | 3.8 × 102 |
| 4 | C. pneumoniae PvlArgDC (18) | Pyruvoyl | 3.4 | 5.0 | 6.9 | 1.4 × 103 |
| 5 | S. solfataricus SSO0536a | Pyruvoyl | 6.0 | 0.2 | 2.6 | 1.3 × 104 |
| 5 | S. solfataricus β536α585a | Pyruvoyl | NDb | 0.2 | 0.019 | 9.5 × 101 |
| 5 | Nonenzymatic Glyc (32) | None | 2 × 10-17 | |||
| 5 | Nonenzymatic Alad (33) | PLP | 7 × 10-5 |
Described in this work.
Not determined.
Nonenzymatic rate of glycine decarboxylation at 25 °C, pH 7.
Nonenzymatic rate of alanine decarboxylation catalyzed by 0.1 mm PLP at 25 °C, pH 5.
Preincubation of the SSO0536 protein with 1 mm O-methyl hydroxylamine or O-(4-nitrobenzyl) hydroxylamine reduced activity by 50%, consistent with the pyruvoyl group modifications observed previously. In contrast, 1 mm phenylhydrazine did not inactivate the enzyme. Incubation with 1 mm DFMO reduced activity by 20%, but incubation with 1 mm DFMA at 80 °C reduced activity by 64% over 15 min.
The most potent competitive inhibitor tested was l-argininamide, which almost completely abolished arginine decarboxylase activity at a 1 mm concentration. Incubation with 1 mm l-arginine methyl ester reduced activity by more than 70%, whereas 1 mm l-canavanine reduced activity by 46%. The same concentration of l-histidine, l-homoarginine, and Nα-acetyl-l-arginine reduced activity by 20–30%. No inhibition was observed with d-arginine, l-citrulline, l-lysine, NG-methyl-l-arginine, l-methionine, NG-nitro-l-arginine methyl ester, or l-ornithine. The SSO0536 protein also catalyzed the decarboxylation of l-canavanine with 40% relative activity compared with l-arginine. No other substrates were identified for this enzyme.
Modeling and Activity of a Chimeric Protein—The crystal structure of the (αβ)2 dimer of Thermotoga maritima AdoMetDC (TmAdoMetDC) showed that this protein is homologous to human AdoMetDC, despite their low sequence similarity (29). The TmAdoMetDC Ser63 site of protein cleavage and pyruvoyl group formation is highly conserved in this protein family, as are nearby active site residues Ser55, His68, and Cys83 (Fig. 2). From a model of human AdoMetDC bound to the methyl ester of AdoMet, Phe49, Cys83, and Glu72 (TmAdoMetDC positions) were predicted to be important for substrate binding and decarboxylation (30). These residues are conserved in both crenarchaeal AdoMetDC and ArgDC paralogs; therefore, other factors must determine the proteins' substrate specificities. From an alignment of crenarchaeal paralogs, we identified two regions where amino acid substitutions correlated with phylogenetically predicted protein function. Cys15 in most crenarchaeal and bacterial AdoMetDC proteins is replaced by a hydrophobic residue in ArgDC paralogs, and a series of charged residues is inserted in the loop between β-strands 2 and 3 of the ArgDC proteins.
In the TmAdoMetDC model, Cys15 is at the opposite end of the β-sheet from the pyruvoyl group active site of the monomer (22 Å apart), but in the dimeric model, this position is 16 Å from the pyruvoyl group of the adjacent protomer, which lies in a pocket at the dimer interface. This Cys15 residue could affect the position of β-strands 4 and 5, including Glu72, which forms hydrogen bonds to the AdoMet ribose hydroxyl groups (30). Similarly, the loop region between β-strands 2 and 3 is ∼13 Å from the pyruvoyl group of the adjacent TmAdoMetDC protomer, so residues inserted into this loop could directly contact the substrate.
Because both characteristic regions lie in the proteins' β-subunits, we constructed chimeric proteins by swapping the subunits. The His10-β536α585 protein self-cleaved to form subunits with apparent molecular masses of 21, 18, 15, and 7 kDa (Fig. 3), although it had a reduced cleavage efficiency (32%) compared with His10-SSO0536 protein. Purified His10-β536α585 catalyzed arginine decarboxylation with a Km identical to the wild-type protein (Table 2). The chimera's turnover was more than 100-fold lower, even after accounting for the reduced cleavage efficiency. The opposite chimera, His10-β585α536, self-cleaved with only 5% efficiency to produce subunits with apparent molecular masses of 18, 16, and 14 kDa. The purified protein had no detectable arginine decarboxylase activity. No AdoMetDC activity was detected in either chimeric protein preparation (Fig. 4), although that assay has a higher limit of detection than the ArgDC assay. We would not detect activity if the turnover was 100-fold lower, as observed for the His10-β536α585 chimera. Therefore, residues in the amino-terminal β-subunit of SSO0536 are sufficient to confer arginine decarboxylase activity on this AdoMetDC scaffold, although additional factors in the α-subunit are required for efficient cleavage and turnover.
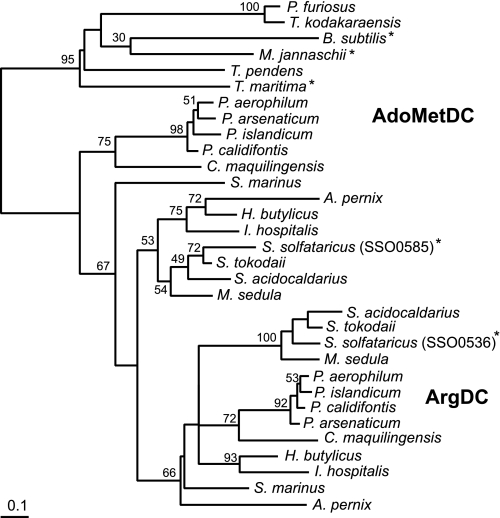
Evolution of Arginine Decarboxylase Activity in Crenarchaea—The phylogeny of the crenarchaeal homologs suggests that the ArgDC gene evolved from a single duplication of an ancestral AdoMetDC gene early in the Crenarchaeota (Fig. 6). Bootstrap analysis offers low support for most branches, due to the limited information content of the short protein sequence alignment. Three crenarchaeal genomes lack orthologs of the SSO0536 gene. Instead, the marine mesophiles Cenarchaeum symbiosum and Candidatus Nitrosopumilus maritimus as well as the thermophile Thermophilum pendens have homologs of the euryarchaeal pyruvoyl-dependent arginine decarboxylase (supplemental Table 1). All three organisms branch early in the Crenarchaeota, so the common archaeal ancestor could have had a homolog of the euryarchaeal ArgDC that was lost in most crenarchaea. Nevertheless, a phylogeny of euryarchaeal type arginine decarboxylase proteins groups the crenarchaeal homologs with two different branches of euryarchaea (data not shown). Therefore, we cannot rule out the acquisition of a euryarchaeal ArgDC gene by horizontal gene transfer to these microbes.
FIGURE 6.
Phylogeny of the paralogous crenarchaeal AdoMet and arginine decarboxylases, rooted using bacterial and euryarchaeal AdoMetDC sequences. This tree is consistent with an early gene duplication event in the crenarchaeal lineage, leading to the evolution of arginine decarboxylase activity. The phylogeny was inferred from an alignment of protein sequences using the protein maximum likelihood method. The scale bar indicates one amino acid substitution per 10 positions. Bootstrap values are shown near branches supported by a plurality of 100 trees. Another tree inferred by the protein distance and neighbor joining methods produced a similar topology. Enzymes whose functions have been experimentally confirmed are indicated by an asterisk. Boldface labels indicate the predicted functions of the two protein subfamilies. Full names of the organisms and their sequence accession numbers are listed in the supplemental materials.
DISCUSSION
Most organisms have an AdoMetDC that is required to produce the propylamine donor for long-chain polyamines. The sequences of AdoMetDC homologs have diverged significantly in the Bacteria, Archaea, and Eukarya. Yet the homologs adopt similar structures (29), and they share a common origin. Previously, all members of this pyruvoyl-dependent protein family were presumed to catalyze the decarboxylation of AdoMet. We have identified the first alternative reaction catalyzed by this protein scaffold.
An AdoMetDC was previously purified from S. solfataricus MT-4, and it was shown to be a thermostable, pyruvoyl-dependent enzyme with a relatively low specific activity (0.012 units/mg) (5). Although no sequence data were reported for this enzyme, its native molecular mass (32 kDa) and activity are similar to those of His10-SSO0585. The MT-4 protein was not resolved into its component subunits, and the heterologously expressed protein migrated anomalously by SDS-PAGE. Therefore, we cannot determine whether the native protein was fully cleaved. The SSO0585 sequence identifies it as member of the bacterial and archaeal type AdoMetDC family (Fig. 6) (8).
Human AdoMetDC co-crystal structures with AdoMet analogs show a limited number of interactions between the substrate's adenosine moiety and the protein active site residues (30). The Glu247 γ-carboxylate forms hydrogen bonds with the ribosyl 2′- and 3′-hydroxyl groups of AdoMet. Human Glu67, at the COOH-terminal end of the β-subunit, forms hydrogen bonds with the adenine base. Both residues are conserved in the two S. solfataricus homologs. Therefore, the specificity determinants in the S. solfataricus enzymes are ambiguous. ArgDC activity in the β536α585 chimera suggests that residues in the β-subunit are sufficient for arginine recognition, although more specific mutagenesis studies and structural models will be required to identify the new mode of substrate binding in the SSO0536 protein.
DFMA was shown to modify a cysteine thiol in the Chlorella virus PLP-dependent arginine decarboxylase (26). Although the mode of DFMA inhibition of the SSO0536 protein cannot be determined without additional experiments, the only cysteine in that protein, Cys102, is a likely target for adduct formation near the active site. Similar to E. coli polyamine-deficient mutants (31), DFMA-treated S. solfataricus had a reduced growth rate in minimal medium and a reduced putrescine content. 1,3-Diaminopropane can replace putrescine as the core polyamine for S. solfataricus, producing norspermidine and norspermine, yet the source of diaminopropane has not been identified. No diaminopropane was identified in polyamines extracted from these cells.
Polyamine-producing organisms have convergently evolved different ways to produce putrescine. Most eukaryotes and bacteria decarboxylate ornithine to directly form putrescine, whereas archaea and some bacteria decarboxylate arginine and hydrolyze agmatine to form putrescine. Although all known arginine decarboxylases use either a PLP or a pyruvoyl cofactor, ArgDC activity has evolved at least five times (Table 2). Three classes of PLP-dependent ArgDC enzymes share little sequence similarity, and the two classes of pyruvoyl-dependent ArgDC enzymes are also nonhomologous. The PLP-dependent ArgDCs have higher specificity constants than their pyruvoyl-dependent analogs. However, the differences in rate constants among the ArgDCs are small compared with the difference between catalyzed and uncatalyzed reaction rates. Amino acid decarboxylases have some of the highest catalytic rate enhancements observed in single substrate enzymes (Table 2) (32, 33).
Class 1 and 2 ArgDC enzymes belong to the ornithine/arginine/diaminopimelate decarboxylase family of PLP-dependent enzymes, which were called group IV amino acid decarboxylases by Sandmeier et al. (34). Class 1 ArgDC enzyme are exemplified by the E. coli biosynthetic PLP-dependent arginine decarboxylase (SpeA) (10). The Class 2 enzyme from Paramecium bursaria chlorella virus evolved from a eukaryotic ornithine decarboxylase (26). Class 3 ArgDC enzymes belong to the ornithine/lysine/arginine decarboxylase family that includes the E. coli inducible ArgDC (AdiA), which functions in an arginine-dependent acid resistance system (35). This class is part of the group III PLP-dependent amino acid decarboxylase family (34).
Both Class 4 and 5 ArgDCs self-cleave to form pyruvoyl cofactors. Euryarchaea use Class 4 enzymes for polyamine biosynthesis (11), but homologs have been recruited by Chlamydia spp. and other bacteria to function in a system analogous to the bacterial arginine-dependent acid resistance system (18). These proteins are homologous to the histidine decarboxylase from Lactobacillus sp. that also functions in acid resistance (36). This class of proteins forms trimers of cleaved protomers, each with an αββα sandwich fold. Two adjacent protomers contribute amino acid residues to each enzyme active site (37). Finally, the Class 5 enzymes described here evolved in the crenarchaea from an AdoMetDC scaffold. The bacterial and archaeal AdoMetDC proteins are dimers of two cleaved protomers that form a four-layer αββα sandwich fold (29, 30). Although gel filtration experiments indicated that the SSO0585 AdoMetDC is a dimer, the SSO0536 ArgDC appeared to be a tetramer. It is not clear how the quaternary structures of the proteins affect substrate specificity.
Hyperthermophiles contain relatively few PLP-dependent decarboxylases compared with mesophiles, although they have a full complement of PLP-dependent aminotransferases. M. jannaschii (optimum growth at 83 °C) has a PLP-dependent tyrosine decarboxylase for methanofuran coenzyme biosynthesis (38) and a PLP-dependent diaminopimelate decarboxylase for lysine biosynthesis (39). Crenarchaea that grow at even higher temperatures, such as Pyrobaculum aerophilum (100 °C growth optimum), appear to have no PLP-dependent decarboxylases (they use an α-aminoadipate pathway for lysine biosynthesis instead of a diaminopimelate pathway). At high temperatures, the selectivity of these enzymes for decarboxylation may decrease, leading to inactivating side reactions (40). Further studies on the temperature dependence of these side reactions are needed to compare the pyruvoyl and PLP-dependent decarboxylases.
Supplementary Material
Acknowledgments
We thank Drs. Marvin Hackert, Jon Robertus, Ian Molineux, and Gisela Kramer for helpful discussions. We are grateful to Dr. Patrick Woster for the generous gift of DFMO. The acquisition of mass spectra by LC-ESI-MS and the resulting determinations of the molecular weights of proteins were done by Dr. Herng-Hsiang Lo in the CRED Analytical Instrumentation Facility Core at the University of Texas (Austin, TX), supported by NIEHS, National Institutes of Health, center Grant ES07784.
This work was supported, in whole or in part, by National Institutes of Health Grant, NIAID, Public Health Service Grant AI064444. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3.
The amino acid sequence of these proteins can be accessed through NCBI Protein Database under NCBI accession number NP_248203.1 (MJ1208), NP_342065.1 (SSO0536), and NP_342108.1 (SSO0585).
Footnotes
The abbreviations used are: AdoMet, S-adenosyl-l-methionine; dcAdoMet, decarboxylated AdoMet; AdoMetDC, AdoMet decarboxylase; ArgDC, arginine decarboxylase; PLP, pyridoxal 5′-phosphate; DFMA, difluoromethylarginine; DFMO, difluoromethylornithine; ESI-MS, electrospray ionizationmass spectrometry; HPLC, high pressure liquid chromatography; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1.Terui, Y., Ohnuma, M., Hiraga, K., Kawashima, E., and Oshima, T. (2005) Biochem. J. 388 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Rosa, M., De Rosa, S., Gambacorta, A., Cartenì-Farina, M., and Zappia, V. (1976) Biochem. Biophys. Res. Commun. 69 253–261 [DOI] [PubMed] [Google Scholar]
- 3.Hamana, K., Hamana, H., Niitsu, M., Samejima, K., Sakane, T., and Yokota, A. (1994) Microbios 79 109–119 [PubMed] [Google Scholar]
- 4.De Rosa, M., De Rosa, S., Gambacorta, A., Cartenì-Farina, M., and Zappia, V. (1978) Biochem. J. 176 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cacciapuoti, G., Porcelli, M., De Rosa, M., Gambacorta, A., Bertoldo, C., and Zappia, V. (1991) Eur. J. Biochem. 199 395–400 [DOI] [PubMed] [Google Scholar]
- 6.Cacciapuoti, G., Porcelli, M., Cartenì-Farina, M., Gambacorta, A., and Zappia, V. (1986) Eur. J. Biochem. 161 263–271 [DOI] [PubMed] [Google Scholar]
- 7.Cacciapuoti, G., Porcelli, M., Moretti, M. A., Sorrentino, F., Concilio, L., Zappia, V., Liu, Z.-J., Tempel, W., Schubot, F., Rose, J. P., Wang, B.-C., Brereton, P. S., Jenney, F. E., and Adams, M. W. W. (2007) J. Bacteriol. 189 6057–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, A. D., Graham, D. E., Seeholzer, S. H., and Markham, G. D. (2000) J. Bacteriol. 182 6667–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goda, S., Sakuraba, H., Kawarabayasi, Y., and Ohshima, T. (2005) Biochim. Biophys. Acta 1748 110–115 [DOI] [PubMed] [Google Scholar]
- 10.Wu, W. H., and Morris, D. R. (1973) J. Biol. Chem. 248 1687–1695 [PubMed] [Google Scholar]
- 11.Graham, D. E., Xu, H., and White, R. H. (2002) J. Biol. Chem. 277 23500–23507 [DOI] [PubMed] [Google Scholar]
- 12.Andersson, A., Lundgren, M., Eriksson, S., Rosenlund, M., Bernander, R., and Nilsson, P. (2006) Genome Biol. 7 R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bey, P., Vevert, J.-P., Van Dorsselaer, V., and Kolb, M. (1979) J. Org. Chem. 44 2732–2742 [Google Scholar]
- 14.Mukhopadhyay, B., Johnson, E. F., and Wolfe, R. S. (1999) Appl. Environ. Microbiol. 65 5059–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972) Arch. Microbiol. 84 54–68 [DOI] [PubMed] [Google Scholar]
- 16.Shipe, J. R., Jr., Hunt, D. F., and Savory, J. (1979) Clin. Chem. 25 1564–1571 [PubMed] [Google Scholar]
- 17.Gaboriau, F., Havouis, R., Moulinoux, J.-P., and Delcros, J.-G. (2003) Anal. Biochem. 318 212–220 [DOI] [PubMed] [Google Scholar]
- 18.Giles, T. N., and Graham, D. E. (2007) J. Bacteriol. 189 7376–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, D. E., Bock, C. L., Schalk-Hihi, C., Lu, Z., and Markham, G. D. (2000) J. Biol. Chem. 275 4055–4059 [DOI] [PubMed] [Google Scholar]
- 20.Horton, R. M., Cai, Z., Ho, S. N., and Pease, L. R. (1990) BioTechniques 8 528–535 [PubMed] [Google Scholar]
- 21.Schägger, H., and von Jagow, G. (1987) Anal. Biochem. 166 368–379 [DOI] [PubMed] [Google Scholar]
- 22.Helgadóttir, S., Rosas-Sandoval, G., Söll, D., and Graham, D. E. (2007) J. Bacteriol. 189 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notredame, C., Higgins, D. G., and Heringa, J. (2000) J. Mol. Biol. 302 205–217 [DOI] [PubMed] [Google Scholar]
- 24.Lord, P. W., Selley, J. N., and Attwood, T. K. (2002) Bioinformatics 18 1402–1403 [DOI] [PubMed] [Google Scholar]
- 25.Nicolaus, B., Gambacorta, A., Basso, A. L., Riccio, R., De Rosa, M., and Grant, W. D. (1988) System. Appl. Microbiol. 10 215–217 [Google Scholar]
- 26.Shah, R., Coleman, C. S., Mir, K., Baldwin, J., Van Etten, J. L., Grishin, N. V., Pegg, A. E., Stanley, B. A., and Phillips, M. A. (2004) J. Biol. Chem. 279 35760–35767 [DOI] [PubMed] [Google Scholar]
- 27.Poulin, R., Lu, L., Ackermann, B., Bey, P., and Pegg, A. E. (1992) J. Biol. Chem. 267 150–158 [PubMed] [Google Scholar]
- 28.Hoffman, J. L. (1986) Biochemistry 25 4444–4449 [DOI] [PubMed] [Google Scholar]
- 29.Toms, A. V., Kinsland, C., McCloskey, D. E., Pegg, A. E., and Ealick, S. E. (2004) J. Biol. Chem. 279 33837–33846 [DOI] [PubMed] [Google Scholar]
- 30.Tolbert, W. D., Ekstrom, J. L., Mathews, I. I., Secrist, J. A., III, Kapoor, P., Pegg, A. E., and Ealick, S. E. (2001) Biochemistry 40 9484–9494 [DOI] [PubMed] [Google Scholar]
- 31.Tabor, H., Hafner, E. W., and Tabor, C. W. (1980) J. Bacteriol. 144 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snider, M. J., and Wolfenden, R. (2000) J. Am. Chem. Soc. 122 11507–11508 [Google Scholar]
- 33.Zabinski, R. F., and Toney, M. D. (2001) J. Am. Chem. Soc. 123 193–198 [DOI] [PubMed] [Google Scholar]
- 34.Sandmeier, E., Hale, T. I., and Christen, P. (1994) Eur. J. Biochem. 221 997–1002 [DOI] [PubMed] [Google Scholar]
- 35.Blethen, S. L., Boeker, E. A., and Snell, E. E. (1968) J. Biol. Chem. 243 1671–1677 [PubMed] [Google Scholar]
- 36.Riley, W. D., and Snell, E. E. (1968) Biochemistry 7 3520–3528 [DOI] [PubMed] [Google Scholar]
- 37.Tolbert, W. D., Graham, D. E., White, R. H., and Ealick, S. E. (2003) Struct. Fold. Des. 11 285–294 [DOI] [PubMed] [Google Scholar]
- 38.Kezmarsky, N. D., Xu, H., Graham, D. E., and White, R. H. (2005) Biochim. Biophys. Acta 1722 175–182 [DOI] [PubMed] [Google Scholar]
- 39.Ray, S. S., Bonanno, J. B., Rajashankar, K. R., Pinho, M. G., He, G., De Lencastre, H., Tomasz, A., and Burley, S. K. (2002) Structure 10 1499–1508 [DOI] [PubMed] [Google Scholar]
- 40.Toney, M. D. (2005) Arch. Biochem. Biophys. 433 279–287 [DOI] [PubMed] [Google Scholar]
- 41.Ohnuma, M., Terui, Y., Tamakoshi, M., Mitome, H., Niitsu, M., Samejima, K., Kawashima, E., and Oshima, T. (2005) J. Biol. Chem. 280 30073–30082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.