Abstract
Here we describe a novel role for the phosphatidylinositol 3-kinase/AKT pathway in mediating induction of interleukin-6 (IL-6) in response to IL-1. Pharmacological inhibition of phosphatidylinositol 3-kinase (PI3K) inhibited IL-6 mRNA and protein production. Overexpression of either dominant-negative AKT or IκB kinase α mutant, IKKαT23A, containing a mutation in a functional AKT phosphorylation site, shown previously to be important for NFκB activation, completely abrogated IL-6 promoter activation in response to IL-1. However, mutation of the consensus NFκB site on the IL-6 promoter did not abrogate promoter activation by IL-1 in contrast to the AP-1 site mutation. IL-1 induces phosphorylation of IKKα on the NFκB inducing kinase (NIK) phosphorylation sites Ser176/Ser180 and on the Thr23 site, and although phosphorylation of IKKαT23 is inhibited both by LY294002 and wortmannin, phosphorylation of Ser176/Ser180 is not. Neither inhibition of PI 3-kinase/AKT nor IKKαT23A overexpression affected IκBα degradation in response to IL-1. Only partial inhibition by dominant-negative AKT and no inhibitory effect of IKKαT23A was observed on an IL-6 promoter-specific NFκB site in contrast to significant inhibitory effects on the AP-1 site. Taken together, we have discovered a novel PI 3-kinase/AKT-dependent pathway in response to IL-1, encompassing PI 3-kinase/AKT/IKKαT23 upstream of AP-1. This novel pathway is a parallel pathway to the PI 3-kinase/AKT upstream of NFκB and both are involved in IL-6 gene transcription in response to IL-1.
Interleukin-6 (IL-6)2 is a pleiotropic cytokine that plays a crucial role in immune and inflammatory responses. Among its functions, IL-6 is involved in induction of the hepatic acute phase response, bone metabolism, reproduction, neoplasia and aging (1, 2). Sometimes viewed as an anti-inflammatory cytokine, prolonged IL-6 production may also cause disease and injury. Inhibition of IL-6 production in males reduces the risk of chemically induced hepatocellular carcinoma (3). IL-6 is a recognized modifier gene of intestinal tumorigenesis (4) and serves as a growth factor for pre-malignant enterocytes that give rise to colitis-induced cancer (5–7).
Increased mucosal IL-6 production was shown to cause local inflammation associated with Crohn disease and ulcerative colitis (inflammatory bowel disease) (8–11). Although the intestinal epithelium is not the main source of IL-6, intestinal macrophages and CD4+T-cells also increase their production of IL-6 and its soluble receptor leading to IL-6 trans-signaling via glycoprotein 130 during inflammatory bowel disease (10). In the intestinal mucosa during sepsis, endotoxemia, and severe injury, the enterocyte increases its production of IL-6 (12). Although in the short term the beneficial effects of IL-6 includes enterocyte acute phase protein induction, mucosal protein synthesis and IgA production in Peyers patch B cells elevated IL-6 may impair mucosal integrity (13, 14).
The primary inflammatory cytokine, interleukin-1 (IL-1), tumor necrosis factor (TNF), platelet-derived growth factor, bacterial lipopolysaccharide, acute viral infections, and transforming growth factor (TGFβ) each induce IL-6 expression. IL-1 is the major proinflammatory cytokine responsible for mediating several physiological responses such as fever, activation of lymphocytes, and induction of acute phase protein synthesis (15). Recent findings suggest that cellular responses to IL-1 are mediated by cascades of intracellular events including activation of mitogen-activated protein kinases (MAPKs) involved in the activation of AP-1 and IκB kinases (IKKs) involved in the activation of NF-κB (16, 17) (Fig. 9).
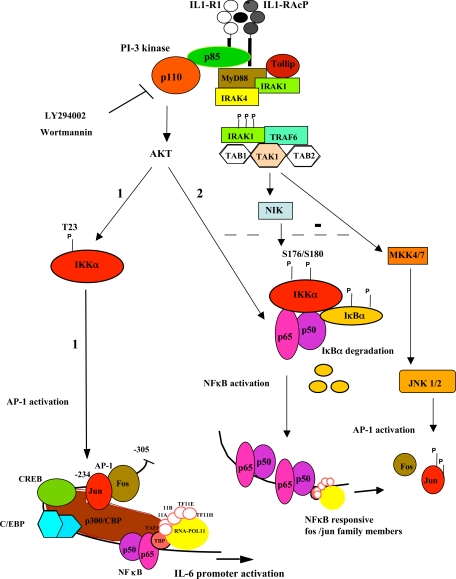
FIGURE 9.
PI 3-kinase/AKT-dependent pathways involved in IL-1 induction of IL-6. The current model for the binding of IL-1 to its receptor, IL-1R1, and co-receptor, IL-1 receptor accessory protein IL1-RAcP (16). Formation of the signaling module containing MyD88, phosphorylated interleukin-1 receptor-associated kinase (IRAK), and TRAF6 (TNF receptor associated factor), is essential for PI 3-kinase recruitment and AKT activation (21, 73, 74, 81). The kinase TAK1 (TGFβ-activated kinase) activates the NIK-IκB-NFκB pathway and the MAP kinase cascade (MKK4/7 to JNK/AP-1 activation as well as the MKK3/6 to p38 activation (not shown) (55, 75, 82–84). Possible negative regulation in the Caco-2 cell line between AKT and JNK pathways is shown as a broken line (85, 86). We have identified 2 separate PI-3 kinase-dependent pathways from the IL1-R1 complex to the activation of IL-6 gene transcription. 1, a novel pathway leading to AP-1-dependent induction of IL-6 via IKKαT23. 2, an NFκB-dependent pathway, which indirectly induces IL-6, likely by inducing factors such as AP-1 family members, which are necessary for IL-6 gene transcription in response to IL-1.
Induction of IL-6 by lipopolysaccharide, from Gram-negative bacteria, lipoteichoic, LTA, from Gram-positive bacteria, and lysophosphatidic acid, a naturally occurring phospholipid have been shown to be phosphatidylinositol 3-kinase (PI3K)-dependent (18, 19). Little attention, however, has focused on PI 3-kinase as a downstream effector of IL-1 (20, 21). The Ser/Thr kinase AKT/protein kinase B has been identified as an important target of PI 3-kinase. AKT provides a potent cell survival signal that is likely involved in its transformation and growth-promoting properties (22, 23). NFκB is an important downstream target of AKT (24). NFκB is typically composed of a dimer between p50 and the transactivating subunit p65 (RelA). IKKα and IKKβ are two important kinases required for NFκB activation. The canonical NFκB pathway, triggered in response to microbial and viral infections as well as proinflammatory cytokines, involves IKKα or IKKβ-mediated phosphorylation of the inhibitor, IκBα, followed by its subsequent ubiquitination, degradation, and entry of p50/p65 into the nucleus (25). This pathway is cell type-specific and depends on the levels of IKKα or -β within the cell (26).
IKKα is the predominant form of the IKK complex activated in response to IL-1 (27). PI 3-kinase/AKT is involved in the activation of IKKα. AKT binds to and increases the activity of IKKα (28). The PI 3-kinase/AKT-mediated phosphorylation of IKKα on Thr23 in response to TNFα results in the activation of canonical NFκB through liberation of IκBα (29). IKKα is also activated by autophosphorylation and phosphorylation on Ser176 and Ser180 by its upstream activator, NIK (30, 31). Both AKT and NIK acting in parallel contribute to NFκB activation in response to TNFα (29). IKKα also mediates activation of the non-canonical pathway, which is involved in B cell maturation and lymphoid organogenesis, through AKT-dependent phosphorylation of the p100 NFκB precursor (32).
Activation of AP-1 occurs primarily through signaling pathways terminating in a group of serine-threonine kinases, MAP kinases that act separately on its components. The predominant form of AP-1 in most cells are heterodimers of fos and jun, which have high affinity for binding to an AP-1 site (33). The MAP kinase, ERK, phosphorylates Elk-1, which is part of a complex that binds to the serum response element in the fos promoter. JNK phosphorylates c-jun, Elk-1, and ATF-2 and is regulated itself by MAP/ERK kinase-4 (MEK4/SEK1) and MEK7, which are dual specificity kinases mediating phosphorylation on the TPY motif (34). p38 kinase phosphorylates Elk-1 and ATF-2 (35). AP-1 activates a broad range of genes designed to protect cells from adverse environmental conditions. Activation of PI 3-kinase by IL-1 is sufficient for full activation of AP-1 but not NFκB (21). AP-1 activation by epidermal growth factor, 12-O-tetradecanoylphorbol-13-acetate (TPA), as well as by 5-MCDE another tumor promoter, and the tax oncoprotein is PI 3-kinase dependent (36–39). Furthermore, it has been demonstrated that the PTEN phosphatase down-regulates AP-1 via PI 3-kinase/AKT inhibition (40).
This investigation addresses the role of the PI 3-kinase signaling pathway in IL-1 induction of the IL-6 gene in the Caco-2 cell, a colon carcinoma with enterocyte-like characteristics. We provide evidence for two PI 3-kinase/AKT-dependent pathways to induction of the IL-6 gene in response to IL-1. The first is a newly discovered pathway encompassing PI3K/AKT/IKKα upstream of AP-1. This IL-1 responsive pathway targets the IL-6 promoter AP-1 site and mutation of this site and not the NFκB site significantly reduces activation of the gene by IL-1. The second PI 3-kinase/AKT-dependent pathway is upstream of NFκB and both pathways, in parallel, are necessary for induction of IL-6 gene transcription in response to IL-1. A novel PI 3-kinase-dependent pathway involving AKT/IKKα upstream of AP-1 suggests a more widespread effect of IKKα on gene expression than can be attributed to activation of NFκB and is further evidence of cross-talk between two transcription factors known to be involved in growth and tumor development. With increasing association of IL-6 and chronic disease, discovery of novel signaling pathways can lead to the design of therapeutic interventions.
EXPERIMENTAL PROCEDURES
Cell Culture—Caco-2 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (<0.05 units endotoxin), 10 μg/ml apotransferrin, and antibiotics (100 mg/liter) streptomycin and penicillin.
Antibodies, Immunoblotting, Kinase Assay—The anti-AKT antibody (Cell Signaling) detects total levels of endogenous AKT1, AKT2, and AKT3 proteins, rabbit polyclonal anti-AKTpS473 (BioSource) detects AKT1, AKT2, and AKT3 when phosphorylated at Ser473. Mouse monoclonal anti-AKTpS473 (587F11) (Cell Signaling) was also used for Western blotting. The GSK3β antibody (Cell Signaling) detects total levels of endogenous GSK3β. The phospho-GSK3 antibody (Ser21/9) (Cell Signaling) detects endogenous levels of GSK3 only when phosphorylated at Ser21 of GSK3α or Ser9 of GSK3β. IKKα was detected using rabbit polyclonal IKKα (Cell Signaling). IKKα was first immunoprecipitated and blotted for the presence of the phosphorylated forms, pIKKSer176/Ser180 (rabbit polyclonal antibody, BioSource) or pIKKα/βThr23 rabbit polyclonal antibody from Santa Cruz Biotechnology. Western blots were carried out as previously described (41).
Nonradioactive AKT kinase assay kit (Cell Signaling) components, immobilized AKT antibody (anti-phospho-AKT Ser473), phospho-GSK3 α/β antibody, GSK3 fusion protein (paramyosin fused to GSK3 α/β cross-tide corresponding to residues surrounding GSK3 α/β serine 21/9) were employed according to the manufacturer's instructions. Briefly, lysates were immunoprecipitated overnight with immobilized anti-AKT, followed by washes and resuspension of the beads in 50 μl of assay buffer and incubation with GSK3 α/β cross-tide (1 μg) and 10 mm ATP (1 μl) for 30 min at 30 °C. Reaction was terminated with SDS sample buffer, electrophoresed on a 10% SDS gel, and blotted for anti-phospho-GSK3 α/β 21/9, and later for endogenous AKT.
Co-immunoprecipitation of IKKα and AKT—Lysates from control and IL-1-treated cells were incubated overnight with control rabbit IgG, anti-IKKα antibody (Cell Signaling), or AKT antibody. After several washes, immunoprecipitated proteins were run on a 10% SDS gel followed by Western blotting (41). IKKα immunoprecipitates were blotted for the presence of both AKT and IKKα and AKT immunoprecipitates were blotted for the presence of both IKKα and AKT. mRNA Isolation and Reverse Transcriptase-PCR—Total RNA was isolated using Stat 60 (AMS biotechnology). Reverse transcriptase-PCR was carried out as previously described in detail (42) using the following primers and probes: hIL-6–87F, CCAGTACCCCCAGGAGAAGAT; hIL-6-157R, CGTTCTGAAGAGGTGAGTGGC; and TaqMan probe: hIL-6-110T, CAAAGATGTAGCCGCCCCACACAGAC. Amplification of 18 S RNA as an internal standard was performed in the same reaction with the alternatively labeled Vic probe. IL-6 mRNA was normalized to the 18 S mRNA levels. Samples were assayed in duplicate.
IL-6 Assay—Tissue culture supernatants were assayed using an ELISA from Endogen (sensitivity 1 pg/ml).
Plasmids and Transfections—All AKT expression plasmids were a gift from Dr. J. R. Woodgett, Department of Molecular and Medical Genetics, Ontario Cancer Institute, University of Toronto. The dominant-negative AKT (dnAKT) (AKT AAA) plasmid in pcDNA3 has Thr179, Thr308, and Ser473 residues all substituted by alanine rendering it unable to bind ATP or be activated. The constitutively active AKT (caAKT) in pcDNA3 is linked to the viral gag sequence, which targets it to the cell membrane. The pIL-6-luc651 plasmid, containing a 651-bp fragment of the human IL-6 gene promoter located directly upstream of the transcription start site and the 3 mutated plasmids, pIL-6-651mAP-1, pIL-6-651mC/EBP-β, and pIL-6-651mNFκB were a generous gift from Dr. Oliver Eickelberg, University Hospital, Basel, Switzerland. Transcription factor binding site mutations were as described below.
AP-1–283 to 276 5′-TGAGTCAC-3′ was changed to 5′-TGCAGCAC-3′; C/EBP-β at –154 to –146, 5′TTGCACAAT-3′ was changed to 5′-CCGTTCAAT-3′; and the NFκB consensus sequence from –72 to –63, 5′-GGGATTTTCC-3′, was changed to 5′-CTCATTTTCC-3′. These mutations have previously been shown to abrogate transcription factor binding (43). The IKKα and IKKαT23A were obtained from Dr. David Donner (Indiana University School of Medicine). The IgκB and IL-6-specific NFκB reporter plasmids were obtained from Dr. Guy Haegeman (LMMP, University of Ghent, Belgium). The constitutively active CAAX-p110 plasmid was obtained from Dr. Lou Cantley, BIDMC, HMS. The AP-1 reporter plasmid, a 12-O-tetradecanoylphorbol-13-acetate responsive AP-1 site was from Dr. Mike Greenberg, Children's Hospital Boston containing 5X (3′-TGAGTCAC-5′) identical to the IL-6 AP-1 sequence (33).
Transient Transfections—Caco-2 cells were plated to 50% confluence in transfection media (growth media minus antibiotics) in 24-well plates. After overnight attachment, cells were transfected with reporter plasmid (0.25 μg) together with β-galactosidase reporter plasmid (0.25 μg) for transfection efficiency, in the presence or absence of expression plasmids (0.125 μg/well). 18 h following transfection, plates were either left untreated (C) or treated overnight with IL-1β (0.5 μg/ml). All plates were harvested 18 h after treatments and assayed for luciferase activity (Invitrogen, Bright Glo) and β-galactosidase (Invitrogen). Results are expressed as normalized values, luciferase/β-galactosidase (41).
Trans-AM NFκB ELISA and Inhibitors—p50 and p65 binding to the WT IgκB binding site (GGGACTTTCC) was measured by ELISA using antibodies to the activated form of the proteins. This site differs from the WT IL-6-kB site by one base at position 5, which is replaced by a C in the IL-6-κB site (GGGACTTTCC). This nucleotide is crucial for the binding of a repressor protein RBP-Jκ (44).
The IKK-2 complex inhibitor was from Calbiochem with IC50 3–12 μm. It acts as a potent reversible ATP-competitive inhibitor for the IKKβ homodimer and IKK α/β heterodimer. LY294002 and wortmannin, selective and potent PI 3-kinase inhibitors, were from Calbiochem (45).
Statistical Analysis—Analysis of variance was used to compare control versus IL-1 treated with Tukey test comparisons of all groups. t test (paired) two-tailed analysis was also used in parallel to compare 2 treatment groups. Experiments were performed at least 3 times with high reproducibility.
RESULTS
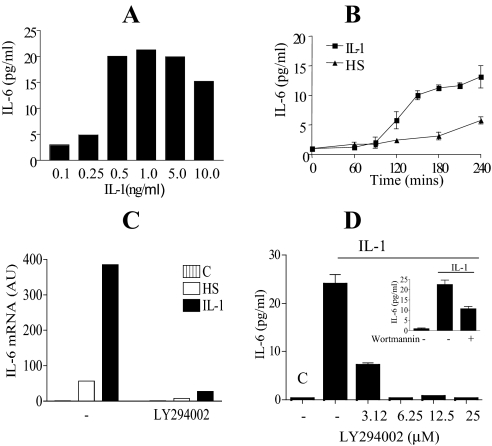
Interleukin-1 Induction of IL-6 Production in Caco-2 Cells Is PI 3-Kinase Dependent—To determine the optimal dose of IL-1 needed to induce maximal IL-6 secretion, Caco-2 cells were exposed to dose levels of IL-1 ranging from 0 to 10 ng/ml. Maximum IL-6 levels in the media was observed with dose levels between 0.5 and 5 ng/ml (Fig. 1A), demonstrating a 10-fold increase from 2 to 20 pg/ml. A dose level of 0.5 ng/ml of IL-1 was subsequently used. A time course is presented in Fig. 1B demonstrating increasing levels of IL-6 secreted into the tissue culture media from 2 to 4 h. Because both IL-1 and IL-6 are mediators of the acute phase response, including fever production, and elevated temperature has previously been demonstrated to induce IL-6 gene expression (46), we compared the effects of heat shock (HS) to IL-1 induction of IL-6. A 43 °C heat shock by itself increased IL-6 levels from 2 to 5.8 pg/ml with significant increases observed only at the end of the 4-h time course (Fig. 1B). IL-1 induces 6-fold more IL-6 mRNA than HS, reflecting the larger increase in IL-6 protein production by IL-1 (Fig. 1, B and C).
FIGURE 1.
Interleukin 1β induction of IL-6 mRNA and IL-6 release in Caco-2 cells is mediated by a PI 3-kinase-dependent pathway. A, dose response for IL-1 treatment of Caco-2 cells (0–10 ng/ml). Semi-confluent cells were treated with IL-1 and the culture supernatant harvested and assayed by ELISA for IL-6. Data are presented as duplicate experiments measured in triplicate. B, time course of IL-6 produced in response to IL-1 or HS. Cells were treated with 0.5 ng/ml of IL-1 or heat shocked at 43 °C for 1 h and returned to 37 °C. Culture supernatant was harvested at the indicated time points up to 4 h and assayed for IL-6 by ELISA. Mean ± S.E. (n = 3) from three separate experiments. C, cells were treated with IL-1 (0.5 ng/ml) or heat shocked (HS) at 43 °C for 1 h and returned to 37 °C, in the presence or absence of the PI 3-kinase inhibitor LY294002 (25 μm) and harvested at To (C) and at 2 h following initial treatment. Stat-60 was used for mRNA extraction. Quantitative reverse transcriptase-PCR for IL-6 mRNA analysis is presented, mean ± S.E. (n = 2). D, Caco-2 cells were left untreated (C; control), or IL-1 (0.5 ng/ml) alone treated. Treatments were given in the presence or absence of the PI 3-kinase inhibitor LY294002 (3.12 to 25 μm) or wortmannin (100 nm, inset) administered 10 min prior to IL-1 treatment. Tissue culture media were harvested 4 h after IL-1 treatment and assayed for IL-6 by ELISA. Mean ± S.E. (n = 3) from three separate experiments are presented.
One of the downstream kinases stimulated by IL-1 is PI 3-kinase. We next investigated if this pathway was important in the induction of IL-6 by IL-1. The PI 3-kinase inhibitor LY294002 inhibited IL-6 mRNA in response to IL-1 and HS and significantly inhibited IL-1 induction of IL-6 secretion into the media in a dose responsive manner (Fig. 1, C and D). Significant inhibition was observed even at the low dose of 3.12 μm. In addition, another highly specific PI 3-kinase inhibitor, wortmannin, significantly inhibited IL-1 induction of IL-6 secretion, further supporting a role for the PI 3-kinase pathway in IL-6 induction by IL-1.
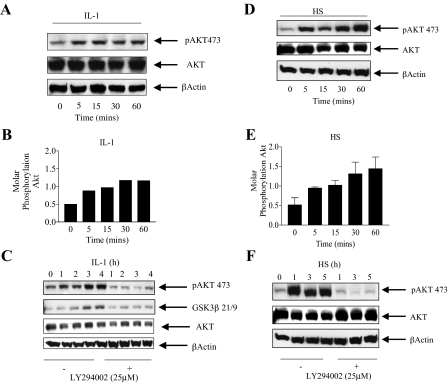
AKT Is Activated by Interleukin-1 and Heat Shock in a PI 3-Kinase-dependent Manner—One of the major downstream targets of PI 3-kinase is the AKT kinase. Stress stimuli, including heat shock, as well as growth factor stimulation have been shown to activate AKT in a PI 3-kinase dependent manner (47). AKT activation is followed by phosphorylation on its 3 major phosphorylation sites, Thr179, on its ATP site, Thr308 in the activation loop, and Ser473 in the kinase domain (23). We therefore investigated whether IL-1 and HS induced phosphorylation of the serine 473 AKT site, the site most commonly used to demonstrate activated AKT.
A short time course (0–60 min) was performed to examine the pattern of AKT Ser473 phosphorylation by IL-1 (Fig. 2A). Significant activation of AKT as detected by phosphorylation on Ser473 is detected by 5 min and is maximal by 30 min to 1 h representing a 2-fold increase in molar phosphorylation (ratio of phosphorylated to non-phosphorylated AKT) (Fig. 2B). In Fig. 2C an extended time course up to 4 h is shown in the absence and presence of the PI 3-kinase inhibitor LY294002. AKT Ser473 phosphorylation and kinase activation, as measured by GSK3β S21/9 phosphorylation, was sustained up to 4 h. At the end of the 4-h time course the molar phosphorylation ratio for AKT was 0.71 ± 0.16 (n = 4) (Fig. 2C). Near complete inhibition of AKT Ser473 and GSK3β phosphorylation, by LY294002, over the entire period of the time courses was observed demonstrating that activation of AKT in response to IL-1 is PI 3-kinase dependent.
FIGURE 2.
Interleukin-1 and heat shock activate AKT in a PI 3-kinase dependent manner. A, Caco-2 cells were treated with IL-1 (0.5 ng/ml) and harvested at the indicated time points. Lysates (70 μg) were run on 10% SDS-PAGE followed by immunoblotting with phospho-specific AKT Ser473. The lysate blots were stripped using 5 ml of Pierce stripping reagent and reprobed using an anti-AKT antibody and an antibody for β-actin. B, the intensity of phosphorylation of AKT over a 1-h time course was estimated using NIH Image software and is expressed as the ratio of phosphorylated protein over total AKT. C, Caco-2 cells were treated with IL-1 (0.5 ng/ml) for up to 4 h in the presence or absence of the PI 3-kinase inhibitor (LY294002, 25 μm) incubated 30 min prior to IL-1 treatment. Cells were harvested and lysed at the indicated hourly time points. AKT was immunoprecipitated and the kinase assay performed using GSK3β peptide. The product of the reaction and the initial lysate were blotted as in A. D, cells were heat shocked (43 °C for 1 h) and harvested at the indicated time points during HS. Cells were lysed, and blotted for AKT Ser473 as in A. E, the intensity of phosphorylation of AKT over the 1-h time course for HS treatment was estimated using NIH Image software as in B. Mean ± S.E. (n = 3) from four separate experiments are presented. F, cells were HS at 43 °C for 1 h in the presence or absence of the PI 3-kinase inhibitor (LY294002, 25 μm) incubated 30 min prior to HS and returned to 37 °C for an additional 4 h. Cells were harvested and lysed at the indicated hourly time points. AKT Ser473 phosphorylation was detected as in A.
Lysates from cells exposed to a heat shock of 43 °C were next blotted for activated AKT (Ser473 phosphorylation) and total AKT. A HS of 43 °C induced a rapid onset of AKT activation, within 5 min, representing a 3-fold increase in molar phosphorylation by 1 h (Fig. 2, D and E). This was sustained over a 5-h time course (Fig. 2F) with an ending molar phosphorylation ratio of 1.33 ± 0.28 (n = 4). AKT activation by HS was also blocked by LY294002 and therefore is PI 3-kinase dependent.
Taken together, the data shows that activation of AKT by IL-1 is maximal by 1 h and correlates with the initiation of IL-6 secretion into the media. Inhibition of PI 3-kinase by LY294002 inhibited both AKT activation and IL-6 secretion. Similar increases in AKT activation in response to HS were insufficient to induce comparable IL-6 secretion to that induced by IL-1.
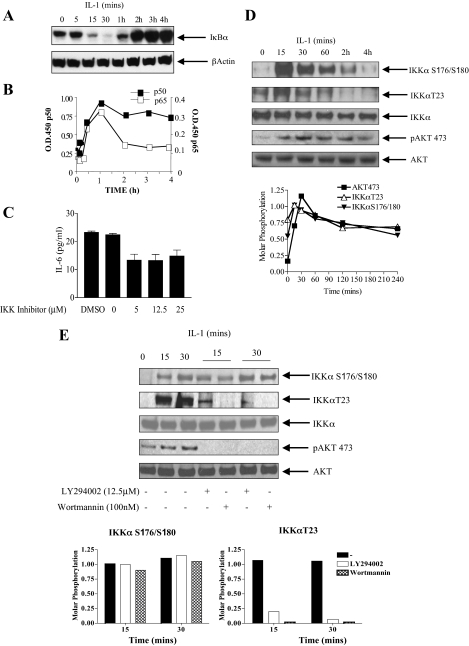
Canonical NFκB Activation Occurs following AKT Activation in Response to IL-1, the IKK Complex Is Required for IL-1 Induction of IL-6—Because NFκB is a well known downstream target of AKT in response to IL-1 we next examined the time course of activation of NFκB in terms of IκBα degradation and DNA binding of p50/p65. In Fig. 3A degradation of IκBα in response to IL-1β occurs between 15 and 30 min following stimulation and this was unaffected by PI 3-kinase inhibition by either LY294002 or wortmannin (see Fig. 7B). This was followed by a 3-fold increase in the binding of p50 by 1 h and a 4-fold induction in the binding of p65. Binding of p65 decreased thereafter, whereas p50 binding was sustained for up to 4 h (Fig. 3B). Taken together with Fig. 2A degradation of IκBα occurs immediately following initial AKT activation at 5 min. There is also a correlation between the initial and sustained p50 subunit of NFκB binding to DNA and the pattern of AKT activation.
FIGURE 3.
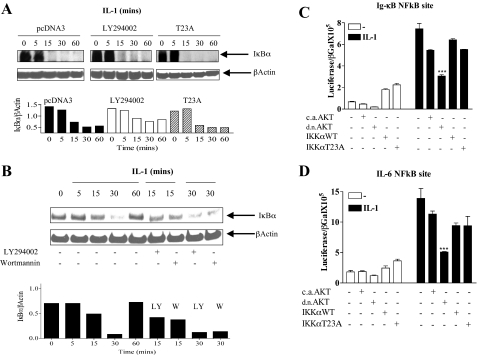
Canonical NFκB activation correlates with AKT phosphorylation on phospho-Ser473; requirement for the IKK complex in IL-6 production. A, cells were treated with IL-1 (0.5 ng/ml) and harvested at the indicated time points. Lysates were run on 10% SDS-PAGE and blotted for IκBα. B, nuclear extracts prepared from IL-1-treated cells at the indicated time points were assayed for the binding of p50 and p65 to an NFκB consensus binding site in a Trans-AM ELISA system according to the manufacturer's instructions. Means from duplicate measurements from two experiments are presented. C, cells were treated with IL-1 (0.5 ng/ml) for 4 h in the presence or absence of an IKK complex inhibitor (5–25 μm) (Calbiochem) incubated 30 min prior to IL-1 stimulation. Tissue culture supernatants were harvested and assayed for IL-6 by ELISA. Mean ± S.E. (n = 3) from three separate experiments are presented. D, cells were treated with IL-1 at the indicated time points and total IKKα was immunoprecipitated overnight. Immunoprecipitates were run on a 10% SDS gel and blotted separately for phospho-IKKα Ser176/Ser180, phospho-IKKαT23, and total IKKα. Lysates were run in parallel and blotted for phospho-AKTS473 and total AKT. Molar phosphorylation of IKKα on Ser176/Ser180 and Thr23, together with Akt Ser473 is presented (lower panel). E, effect of the PI 3-kinase inhibitors LY294002 (12.5 μm) and wortmannin (100 nm) on the phosphorylation of IKKα S176/180, IKKαT23, and AKTS473 following IL-1 stimulation at 15 and 30 min. Molar phosphorylation of IKKα on Ser176/Ser180 and Thr23 is presented (lower panel).
FIGURE 7.
AKT is required for NFκB activation in response to IL-1, independent of IKKαT23 and IκBα degradation. A, cells were transfected with empty vector, pcDNA3, or IKKαT23A. Eighteen hours after transfection cells were serum starved. Cycloheximide (50 μg/ml) was added to prevent protein resynthesis; 1 h thereafter cells were treated with IL-1β. Cells were either untreated or pretreated with the PI 3-kinase inhibitor LY294002 (25 μm) prior to IL-1 (0.5 ng/ml) treatment. Lysates harvested at the indicated time points were blotted for the presence of IκBα. β-Actin is included as a loading control and the ratio of IκBα to β-actin (NIH Image analysis) is plotted below for the three treatment groups. B, cells were treated with IL-1β in the presence or absence of LY294002 (12.5 μm) or wortmannin (100 nm) pretreated 10 min prior to IL-1 treatment. Cell lysates were harvested at the indicated time points and blotted for IκBα. Blots were stripped and reprobed for β-actin as a loading control. The ratio of IκBα to β-actin (NIH Image analysis) is plotted below for the treatment groups. C, Caco-2 cells were transfected with luciferase reporter plasmids containing a 3× NFκB consensus site from the IgκB gene (0.25 μg); or D, the IL-6 promoter NFκB site (0.25 μg), in the presence or absence of one of the following expression plasmids (0.125 μg), caAKT, dnAKT, IKKαWT, or IKKαT23A. β-Galactosidase (0.25 μg) was included for transfection efficiency. Eighteen hours after transfection, cells were starved for 3 h and treated overnight with IL-1β (0.5 ng/ml). Cells were harvested and lysates were assayed for luciferase and β-galactosidase. Reporter activation is expressed as the ratio of luciferase/β-galactosidase. Mean ± S.E. (n = 3). ***, p < 0.001 IL-1-treated NFκB reporter transfectants compared with the co-transfected dnAKT.
In Fig. 3C, a cell permeable aminoacetamide compound and potent inhibitor of IKK complex activity and NFκB activation was used. At dose levels between 5 and 25 μm this inhibitor lowered IL-6 secretion into the media by 50% in response to IL-1, suggesting that the NFκB pathway contributes to IL-6 production in response to IL-1.
To further explore the possibility that AKT might be involved in the activation of the NFκB pathway in response to IL-1 we examined the time course of activation of IKKα in the presence and absence of the PI 3-kinase inhibitors. Phosphorylation of Ser176/Ser180 (NIK phosphorylation sites) and Thr23 (AKT phosphorylation site) have been shown previously to indicate IKKα activation that precedes IκBα degradation and NFκB activation (25, 29–31). Phosphorylation of IKKα on Ser176/Ser180 and Thr23 in response to IL-1 as detected by Western analysis using phospho-specific antibodies was maximal by 15–30 min and decreased thereafter to baseline levels (Fig. 3D). Phosphorylation of IKKα at 15–30 min correlates with the degradation of IκBα at the same time point in Fig. 3A. A comparison of molar phosphorylation ratios of AKT activation on Ser473 and IKKα on Ser176/ Ser180 and Thr23 (Fig. 3D, lower) demonstrates that increases in AKT phosphorylation at 15 min coincides with maximal IKKα phosphorylation and that although AKT molar phosphorylation is maximal at 30 min, thereafter there is a similar decline in the molar phosphorylation of the 2 kinases.
We next demonstrate in Fig. 3E that neither wortmannin nor LY294002, two potent PI 3-kinase inhibitors, at concentrations that inhibit PI 3-kinase in cell based assays (57), affect the phosphorylation of IKKαS176/180, while significantly inhibiting the phosphorylation of IKKαT23 and, as expected, the phosphorylation of AKT Ser473. Taken together, this suggests that PI 3-kinase/AKT is upstream of IKKαT23 (and not IKKαS176/180) and, as is the case with TNFα, is involved in a parallel pathway with NIK to activate IKKα in response to IL-1.
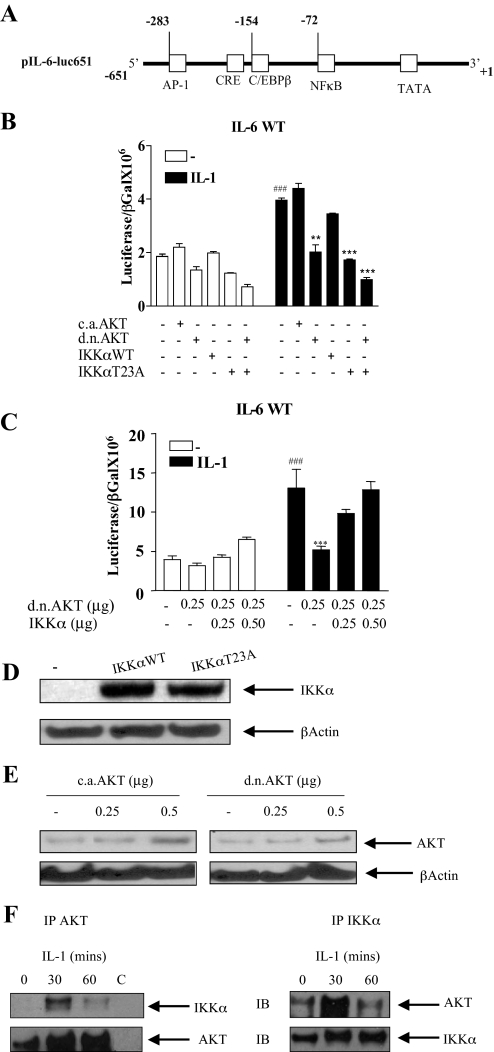
AKT and IKKα Are Necessary for IL-6 Promoter Induction in Response to IL-1—Expression of IL-6 is tightly regulated at the level of transcription. To further explore the role of AKT in the induction of IL-6 by IL-1, the wild type IL-6 promoter reporter construct (pIL-6-luc651), containing a 651-base pair fragment of the IL-6 promoter, in front of the luciferase gene was used (Fig. 4A). This contains all the elements necessary for its induction by a variety of stimuli. There is a multiple response element consisting of a CRE followed by a consensus binding site for C/EBPβ/NF-IL-6 at –173 to –145, which is involved in tissue-specific transcription of the IL-6 gene (87). There is an NFκB binding site from –73 to –63, and an AP-1 site from –283 to –277 (48).
FIGURE 4.
Interleukin 1 induction of IL-6 promoter activation is dependent on AKT and on the Thr23 AKT phosphorylation site on IKKα. A, parental IL-6 promoter construct, pIL-6-luc651 containing 651 bases from the transcription start site. Relevant transcription factor binding sites are shown, including NFκB, AP-1, and C/EBPβ. B, cells were transfected with the IL-6 promoter luciferase construct pIL-6-luc651 (0.25 μg) in the presence or absence of a constitutively active AKT (caAKT) (0.125 μg), dominant-negative AKT (dnAKT) (0.125 μg), IKKαWT (0.125 μg) or IKKαT23A (0.125 μg). β-Galactosidase (0.25 μg) was co-transfected for transfection efficiency. Eighteen hours after transfection cells were starved for 3 h and then either untreated (–) or treated with IL-1 (0.5 ng/ml) overnight. Cells were harvested and lysates were assayed for luciferase and β-galactosidase. Results are expressed as the ratio of luciferase over β-galactosidase. Mean ± S.E. (n = 3). ###, p < 0.001 untreated pIL-6-luc651 alone transfected compared with IL-1 treated. Comparison of IL-1-treated transfectants; **, p < 0.01, pIL-6-luc651 alone transfected compared with co-transfection with dnAKT; ***, p < 0.001 pIL-6-luc651 alone transfected compared with co-transfection with IKKαT23A or IKKαT23A + dnAKT. C, cells were transfected with the IL-6 promoter luciferase construct pIL-6-luc651 (0.50 μg) in the presence or absence of dnAKT (0.125 μg) or IKKα WT (0–0.50 μg) together with β-galactosidase (0.25 μg) for transfection efficiency. Results are expressed as the ratio of luciferase over β-galactosidase. Mean ± S.E. (n = 3). ###, p < 0.001 pIL-6-luc651 alone transfected, untreated compared with IL-1 treated. IL-1-treated transfectants, ***, p < 0.001, pIL-6-luc651 alone transfected compared with co-transfection with dnAKT. D, Western blot of lysates from Caco-2 cells transfected with empty vector (–) or plasmids expressing IKKα WT or IKKαT23A (0.25 μg), using polyclonal IKKα antibody and a β-actin antibody for loading control. E, Western blot of lysates from Caco-2 cells transfected with empty vector (–) or plasmids expressing caAKT or dnAKT (0.25 and 0.5 μg) using an antibody against total AKT and β-actin for loading control. F, AKT association with IKKα in response to IL-1. Caco-2 cells were either untreated or treated with IL-1 (0.5 ng/ml) at the indicated time points. Lysates were immunoprecipitated (IP) overnight with anti-IKKα antibody, AKT antibody, or rabbit IgG (C, control). Washed immunoprecipitates were run on 10% SDS-PAGE and the AKT immunoprecipitates (left panel) and IKKα immunoprecipitates (right panel) were blotted for the presence of both IKKα and AKT.
NFκB is a well known downstream target of AKT, and one of its activating kinases, IKKα, is the predominant form of the IKK complex activated in response to IL-1 (27). IKKα contains a functional AKT phosphorylation site on Thr23 (29) and this is a necessary step for the degradation of IκBα and NFκB activation in response to TNFα. We therefore investigated if co-transfection of either dnAKT or IKKαT23A would affect IL-6 promoter induction by IL-1. Caco-2 cells were transfected with the IL-6 promoter together with, caAKT, dnAKT (mutated in the 3 phosphorylation sites, Thr179, Thr308, and Ser473), IKKαWT, or IKKαT23A (containing the AKT site mutation at Thr23).
The pIL-6-luc651 construct was activated 2-fold by IL-1 (Fig. 4B). Co-transfection of caAKT only slightly enhanced baseline and IL-1 induction of the IL-6 promoter, whereas dnAKT completely abrogated its induction by IL-1 (compare the seventh to ninth bars). Although IKKαWT did not, by itself, activate the IL-6 promoter (fourth bar) nor did it enhance its induction by IL-1 (compare the seventh to 10th bars), IKKαT23A, inhibited the induction by IL-1 similar to that observed by dnAKT (compare the seventh bar to the ninth and 11th bars). In Fig. 4C, overexpression of IKKαWT was sufficient to reverse the inhibitory effects of dnAKT on IL-6 promoter activation (compare the sixth bar to the seventh and eighth bars). Taken together, this demonstrates that AKT and IKKα are part of a common pathway to IL-6 gene transcription in response to IL-1.
To control for transfection efficiency and to ensure equal expression of plasmids in transfected cells, Western blots were performed to compare levels of expression. Western blotting confirmed similar levels of expression of IKKαWT and IKKαT23A in the transfected cells (Fig. 4D). Elevated and comparable levels of expression in transfected cells of both caAKT and dnAKT with an increasing dose of plasmid were also observed (Fig. 4E).
To determine whether AKT and IKKα associate in vivo in response to IL-1 we performed cross-coimmunoprecipitation experiments in the presence or absence of IL-1. These demonstrated that AKT and IKKα, as is the case in response to TNFα (29), associated in vivo in the cell in response to IL-1 with maximal association at 30 min (Fig. 4F). Significant decreases in association of AKT and IKKα were observed by 1 h in both pull-downs correlating with the decrease in phosphorylation of IKKα observed in Fig. 3D. No detectable AKT or IKKα was detectable in immunoprecipitates using control rabbit IgG (see Fig. 4F).
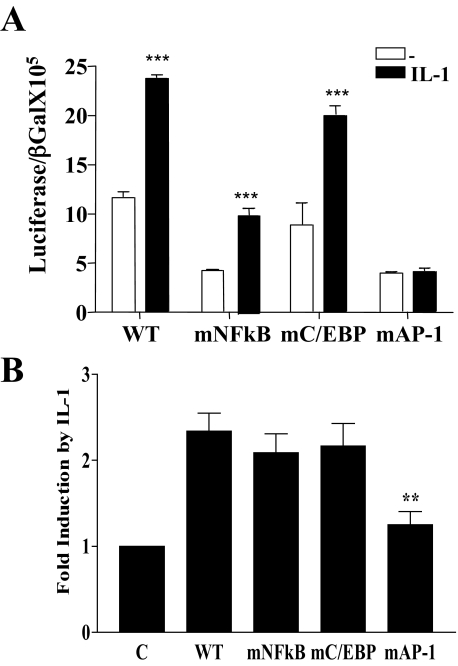
Mutation of the NFκB Site Affects Baseline IL-6 Promoter Activation While Its Induction by IL-1 Is Dependent on an Intact AP-1 Site—To distinguish which of the promoter elements contained within the pIL-6-luc651 was most important in baseline or IL-1 activation of the gene, we compared baseline and IL-1 activation of three mutant IL-6 promoter constructs to the WT pIL-6-luc651. The mutated promoters, pIL-6-luc651mNFκB, pIL-6-luc651mC/EBP, and pIL-6-luc651mAP-1, contained mutations in their respective transcription factor binding sites that have been shown previously to abrogate their binding (43) (Fig. 4A). In one representative of at least three experiments (Fig. 5A), mutation of the NFκB site significantly reduced baseline IL-6 promoter activation with no significant baseline effect when the C/EBPβ site was mutated (Fig. 5A). Together with WT pIL-6-luc651, both the mNFκB and mC/EBP IL-6 promoter constructs were significantly activated by IL-1 in marked contrast to the near absence of activation of the pIL-6-luc651mAP-1 (AP-1 mutant) IL-6 promoter reporter.
FIGURE 5.
Mutation of the consensus NFκB site affects baseline IL-6 promoter activity, whereas mutation of the AP-1 site significantly impairs its induction by IL-1. Caco-2 cells were transfected with the IL-6 promoter luciferase constructs (0.25 μg), pIL-6-luc651WT (WT), pIL-6-luc651 mutated in the NFκB binding site, pIL-6-luc651 (mNFκB), pIL-6-luc651 mutated in the consensus binding site for C/EBPβ, pIL-6-luc651 (mC/EBPβ), or pIL6–651 mutated in the AP-1 site, pIL-6-luc651 (mAP-1). A β-galactosidase reporter (0.25 μg) was included for transfection efficiency. Eighteen hours after transfection, cells were starved for 3 h and were either untreated or IL-1 treated (0.5 ng/ml) overnight. Lysates were assayed for luciferase and β-galactosidase. A, baseline and IL-1 stimulation of each construct with promoter activation expressed as the ratio of luciferase/β-galactosidase. Mean ± S.E. (n = 3). ***, p < 0.001, pIL-6-luc651WT and mutants, mNFκB, mC/EBP, untreated compared with IL-1 treated. B, -fold induction by IL-1 with untreated, control values set at 1, mean ± S.E. (n = 3). ***, p < 0.001, IL-6-651mAP-1 transfectants compared with pIL-6–651WT transfectants treated with IL-1.
In an average of three separate experiments shown in Fig. 5B and expressed as -fold activation by IL-1, the AP-1 site mutant was consistently less responsive to IL-1 compared with the WT, pIL-6-luc651mNFκB (NFκB mutant), or pIL-6-luc651mC/EBP (C/EBP mutant) plasmids (Fig. 5B). These experiments demonstrate that AP-1 is the primary target site for activation of the IL-6 gene in response to IL-1. Mutation of the consensus C/EBP site had no effect on either baseline or IL-1 induction of the pIL-6-luc651 construct. As the IL-6 promoter mutated in the NFκB site does not bind NFκB, the 2-fold IL-1 induction is due to other factors that bind to the remaining intact sites on this mutated promoter of which AP-1 is the most likely, as the AP-1 site is the only transcription factor binding site (of the three transcription factor sites investigated (NFκB, C/EBP, and AP-1 sites) (Fig. 5B) that is necessary for full IL-1 induction. Taken together, these experiments demonstrated that baseline activation depends on intact NFκB and AP-1 sites and that maximal IL-1 induction depends only on an intact AP-1 site.
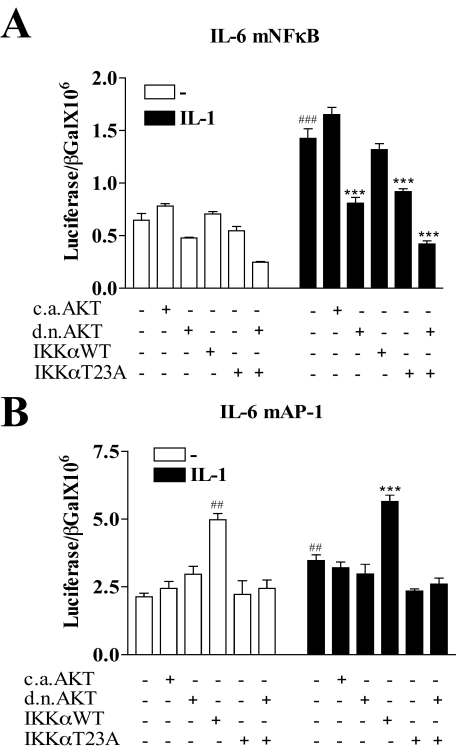
An Intact AP-1 Site and Not an Intact NFκB Site Is Necessary for Inhibition by dnAKT and IKKαT23A—To further explore the downstream target of the AKT/IKKα pathway on IL-6 gene transcription we next investigated the effect of overexpression of dnAKT and IKKαT23A on two of the IL-6 promoter constructs employed above, the pIL-6-luc651mNFκB (NFκB mutant) and pIL-6-luc651mAP-1 (AP-1 mutant). Similar to the WT promoter (Fig. 4B) the pIL-6-luc651mNFκB, containing the NFκB site mutation (Fig. 6A), demonstrated a 2-fold induction by IL-1 as well as almost complete inhibition of the IL-1 response by either dnAKT or IKKαT23A (compare the seventh bar to the ninth and 11th bars). This suggests that an intact NFκB site on the IL-6 promoter is not necessary for inhibition by either dnAKT or IKKαT23A.
FIGURE 6.
Mutation of the IL-6 promoter NFκB site neither abrogates IL-1 induction nor the inhibitory effect of transfected dominant-negative AKT or IKKαT23A. Caco-2 cells were transfected with the IL-6 promoter luciferase reporter plasmids (0.25 μg) containing either a mutated NFκB site, pIL-6–651mNFκB(A), or a mutated AP-1 site, pIL-6–651mAP-1 (B), in the presence or absence of one of the following expression plasmids (0.125 μg), caAKT, dnAKT, IKKαWT, or IKKαT23A. β-Galactosidase (0.25 μg) was co-transfected for transfection efficiency. Eighteen hours after transfection, cells were starved for 3 h and treated overnight with IL-1β (0.5 ng/ml). Cells were harvested and lysates were assayed for luciferase and β-galactosidase. Promoter activation is expressed as the ratio of luciferase/β-galactosidase. Mean ± S.E. (n = 3). ###, p < 0.001; ##, p < 0.01, pIL-6-luc651 control, untreated transfectants compared with IL-1 treated; ***, p < 0.001, pIL-6-luc651, IL-1 treated compared with co-transfected dnAKT or IKKαT23A (n = 3).
The IL-6 promoter containing the AP-1 site mutation, pIL-6-luc651mAP-1 (Fig. 6B), demonstrated near complete loss of promoter induction by IL-1. There was no significant inhibition of baseline activity (Fig. 6B) either by dnAKT or IKKαT23A (compare the first bar to the third and fifth bars). Furthermore, IL-1 induction of pIL-6-luc651mAP-1 activation was not inhibited by dnAKT but was inhibited by IKKαT23A suggesting that an intact AP-1 site was necessary for inhibition by dnAKT but not IKKαT23A. Taken together, this suggests that an intact AP-1 site is necessary for inhibition by at least dnAKT and IKKαT23A may, in addition to AP-1, be targeting another transcription factor necessary for IL-1 induction of the IL-6 promoter.
Interestingly, we observe a significant 2-fold baseline activation of the pIL-6luc651mAP-1 promoter by IKKαWT but not by IKKαT23A, demonstrating that IKKα overexpression can compensate for the loss of an IL-1 effect from AP-1 site mutation. This suggests involvement of the AKT/IKKα pathway in the activation of another transcription factor that may be negatively regulated by AP-1 binding. These results suggest that AP-1 is a likely downstream target of AKT/IKKα in response to IL-1 on the intact IL-6 promoter and that NFκB may yet be the target of AKT/IKKα on promoters not carrying an AP-1 site.
Requirement for AKT but Not the IKKαT23 AKT Phosphorylation Site for NFκB Activation in Response to IL-1—In Fig. 7A we investigated the effect of either pharmacological inhibition of PI 3-kinase/AKT, by LY294002 or IKKαT23A overexpression on canonical NFκB activation by Western blotting for the presence of IκBα. Neither IKKαT23A overexpression nor exposure to LY294002 prevented significant loss of IκBα, suggesting that AKT/IKKα is not upstream of IκBα degradation in the NFκB activation pathway. This is further confirmed in Fig. 7B (in the absence of cycloheximide) where again no significant inhibition of the degradation of IκBα was observed in the presence of either LY294002 or wortmannin.
To further define the role of NFκB and determine whether AKT and/or IKKαT23 are necessary for NFκB activation downstream of IκBα degradation we performed luciferase reporter assays employing two separate reporter plasmids, each containing three NFκB binding sites. The first reporter plasmid contains a 3× NFκB binding site from the IgκB gene (Fig. 7C), the second reporter plasmid contained a 3× NFκB site from the IL-6 gene itself (Fig. 7D). A representative experiment from three separate experiments is presented, each with consistent results. As expected, IL-1 strongly activated the NFκB reporters. A 15-fold induction of the IgκBNFκB reporter, and a 7-fold induction of the IL-6-specific NFκB reporter was observed in response to IL-1 (Fig. 7, C and D). Although caAKT did not, by itself, activate the NFκB reporter, dnAKT significantly reduced baseline (compare the first bar to the third bar) and its induction by IL-1 (compare the sixth bar to the eighth bar). This suggests that AKT is necessary for NFκB activation in response to IL-1. This is also reflected in Fig. 7D employing the IL-6 promoter-specific NFκB site (compare the sixth bar to the eighth bar).
This experiment also demonstrated that IKKαWT overexpression alone is capable of activating the NFκB luciferase reporters up to 3-fold (compare the first bar to the fourth bar, Fig. 7, C and D) and this is independent of the Thr23 phosphorylation site as IKKαT23A is also capable of activation. Similarly, and contrary to its inhibitory effect on IL-1 induction of pIL-6-luc651WT, IKKαT23A overexpression had no significant effect on IL-1 induction of the NFκB reporter plasmids (compare the sixth bars to the ninth and 10th bars, Fig. 7, C and D). Taken together, this suggests that although IKKα is capable of activating NFκB by itself, AKT is necessary for NFκB activation in response to IL-1 independent of IKKα.
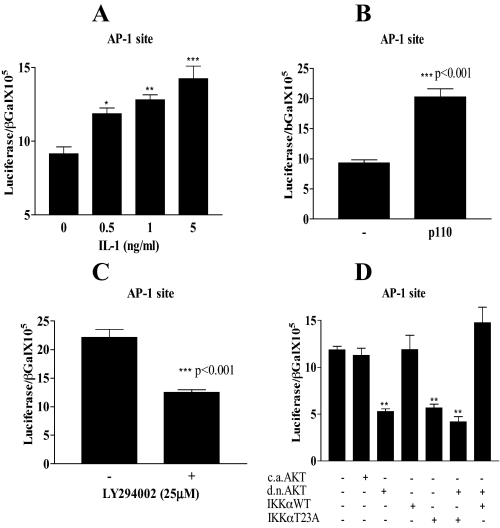
PI 3-Kinase AKT/IKKα Thr23 Is Upstream of AP-1—To determine whether AP-1 is the downstream target of AKT/IKKαT23 on the IL-6 promoter, we employed an AP-1 luciferase reporter plasmid containing 5 copies of the IL-6 AP-1 site (Fig. 8). We observed a significant dose-dependent activation of this AP-1 reporter in response to IL-1 with increasing activation up to 5 ng/ml (Fig. 8A). In the HT29 colon carcinoma cell line the AP-1 reporter could be activated by PI 3-kinase catalytic domain p110 overexpression (Fig. 8B). The AP-1 reporter displayed elevated constitutive activation in Caco-2 cells, which was inhibited by the PI 3-kinase inhibitor LY294002 (Fig. 8C).
FIGURE 8.
AP-1 is the target transcription factor of PI 3-kinase/AKT/IKKα. In A, Caco-2 cells were transfected with the IL-6 promoter-specific AP-1 site luciferase reporter (0.25 μg) together with β-galactosidase (0.25 μg). Eighteen hours after transfection, cells were treated with IL-1 (0, 0.5, 1, and 5 ng/ml) overnight. B, IL-6 promoter-specific AP-1 site reporter (0.25 μg) was transfected alone or co-transfected with p110 subunit of PI 3-kinase (0.125 μg) together with β-galactosidase (0.25 μg) in HT29 cells. C, Caco-2 cells were transfected with the IL-6 promoter-specific AP-1 site reporter (0.25 μg). Eighteen hours after transfection cells were either untreated or treated overnight with the PI 3-kinase inhibitor LY294002 (25 μm). D, Caco-2 cells were transfected with the IL-6 promoter-specific AP-1 reporter (0.25 μg), alone or co-transfected with caAKT, dnAKT, IKKαWT, or IKK T23A (0.125 μg) and combinations of either, dnAKT + IKKαT23A or dnAKT + IKKαWT together with β-galactosidase. Eighteen hours after transfection, cells were starved for 3 h and treated overnight with IL-1 (0.5 ng/ml). Cells were harvested 18 h following treatments and lysates were assayed for luciferase and β-galactosidase. Reporter activation for A–D is expressed as the ratio of luciferase/β-galactosidase. Mean ± S.E., n = 3. **, p < 0.01 AP-1 luciferase alone transfected compared with co-transfection with dnAKT, IKKαT23A, or both.
In Fig. 8D in the presence of IL-1 (0.5 ng/ml), the dose used to induce maximal IL-6 secretion in Caco-2 cells, dnAKT overexpression abrogated the induction by IL-1, and lowered constitutive activation by over 50% (compare the first bar to the third bar). Similar effects were observed with IKKαT23A overexpression (compare the first bar to the fifth bar). Overexpression of either caAKT or IKKα WT alone had no significant effect on AP-1 luciferase activation, whereas both together significantly enhanced constitutive activation (data not shown) suggesting a common pathway. This is further supported by the demonstration that overexpression of IKKα WT reversed the inhibitory effect of dnAKT as also observed on the intact IL-6 promoter (Figs. 4C and 8D). This supports a functional role for AKT/IKKα in the regulation of constitutive and IL-1-stimulated AP-1. The pattern of inhibition of the AP-1 reporter gene both by dnAKT and IKKαT23A reflects the pattern observed on the WT IL-6 promoter as well as that observed on the IL-6 promoter containing a mutated NFκB site. Taken together, this suggests that AKT/IKKα targets AP-1 on the IL-6 gene and unravels a novel signaling pathway encompassing PI 3-kinase/AKT/IKKα and AP-1.
Fig. 9 is a model of the PI 3-kinase/AKT-dependent pathways involved in induction of the IL-6 gene in response to IL-1. The first (1) involves AKT directly targeting IKKα on its Thr23 phosphorylation site, a necessary step toward AP-1-mediated transcription. In the second pathway (2) AKT directly targets NFκB, which is indirectly involved in the induction of the IL-6 gene, likely by binding to other NFκB responsive promoters and inducing the transcription of other factors with a transcriptional role such as fos/jun, which are involved in IL-6 gene expression in response to IL-1.
DISCUSSION
We demonstrate for the first time the requirement for PI 3-kinase/AKT in the induction of the IL-6 gene in response to IL-1. This pathway has 2 components. The first pathway targets IKKα, one of the kinases involved in the activation of NFκB and is upstream, not of NFκB, but of AP-1. The second pathway directly targets NFκB, which is likely binding to and activating not the consensus NFκB site on the IL-6 promoter but the promoters of other genes involved directly or indirectly in the induction of IL-6 by IL-1.
This is the first demonstration that AP-1 is a downstream target of IKKα mediated by AKT and defines a new IL-1 responsive pathway. This novel pathway is important not only in terms of the regulation of the IL-6 gene by IL-1, but also in terms of further evidence of cross-talk between two transcription factors known to be involved in growth and tumor development. It also lends support to recent findings showing evidence for a wider role for IKKα in gene regulation other than in the activation of non-canonical NFκB (49–52).
Future investigations will determine which components of the AP-1 complex are activated by IL-1 to bind to the AP-1 site in the IL-6 promoter in the Caco-2 cell line, and whether IKKα targets one of these directly. The role of the MAP kinase pathway in IL-6 induction has been extensively investigated in the Caco-2 cell line (53). With the use of specific pharmacological inhibitors it has been shown that all three MAP kinases (ERK, JNK, and p38) are activated in response to IL-1 and are involved in the induction of IL-6 (54). Interestingly, however, the JNK inhibitor SP 600125 was reported to be significantly less potent than the other MAP kinase inhibitors in the inhibition of IL-6 secretion in the Caco-2 cell (53).
As JNK is one of the major kinases that phosphorylates c-Jun leading to the activation of AP-1, there are likely physiological circumstances where its activation is compromised. For example, binding of vaccinia-related kinase 2 to the JIP scaffolding protein prevented the recruitment of JNK and caused down-regulated IL-1 responsive AP-1 transcription (55). Additionally it has been reported that AKT phosphorylates kinases upstream of JNK activation, i.e. SEK1 (MKK4) and ASK1 leading to inactivation of JNK (85, 86). Therefore if PI 3-kinase/AKT can lead to the loss of JNK activation an alternative pathway to the activation of AP-1 might be via IKKα. Other studies have demonstrated that although JNK is the most important MAPK involved in IL-6 production by renal epithelial cells, the regulation of IL-6 gene transcription by JNK is independent of the AP-1 binding site but rather involves interference with other signaling pathways such as NFκB and ERK (56).
Inhibition of IL-6 mRNA and protein production by LY294002 suggested the involvement of PI 3-kinase in the induction of IL-6 in response to IL-1. To confirm the involvement of PI 3-kinase the more specific inhibitor wortmannin was also shown to inhibit IL-6 production at concentrations previously shown to inhibit PI 3-kinase in cell based assays (57). Maximal activation of AKT by 1 h was followed by elevations in IL-6 mRNA and increasing IL-6 secretion into the media from 2 to 4 h (Fig. 1B). The IL-6 promoter contains, among others, binding sites for several AKT responsive transcription factors including NFκB, CREB, and AP-1 (43, 48, 58). Consistent with previous investigators, a highly consistent and statistically significant 2–3-fold activation of the WT IL-6 promoter by IL-1 was observed in Caco-2 cells (44, 59). Although caAKT overexpression, by itself, was not sufficient to activate the IL-6 promoter, dnAKT completely abrogated the promoter response to IL-1 demonstrating for the first time that AKT is necessary for IL-1 activation of the gene.
A well characterized and commercially available IKK complex inhibitor that inhibits the classical NFκB pathway significantly reduced IL-6 secretion in response to IL-1 implying a role for NFκB. We explored the possibility that NFκB might be the downstream target of PI 3-kinase/AKT. In this regard, the PI 3-kinase/AKT-mediated transactivation of the p65 and p50 subunits of NFκB in response to IL-1 has previously been demonstrated (24, 36, 60, 61). IKKα, previously shown to be the predominant form of the IKK complex activated in response to IL-1, contains 2 serine residues previously shown to be phosphorylated by NIK, Ser176/Ser180 (25), as well as a functional AKT phosphorylation site at Thr23 shown to be necessary for NFκB activation in response to TNFα (29). This phosphorylation site had not previously been tested on any signaling pathway downstream of IL-1. Maximal phosphorylation of these sites in response to IL-1 followed the initial activation of AKT in response to IL-1 but only the Thr23 site and not the Ser176/Ser180 site was inhibited by LY294002 and wortmannin suggesting that AKT was likely upstream of IKKαT23 activation with parallel activation by NIK.
Unexpectedly, mutation of the consensus NFκB site did not abrogate induction of the IL-6 promoter in response to IL-1 nor did it abrogate the inhibitory effects of either dnAKT or IKKαT23A (Fig. 6A), however, baseline promoter activation was significantly impaired (Fig. 5A). Inhibition of the NFκB reporter plasmids (Fig. 7, C and D) by dnAKT, but not IKKαT23A, suggested that AKT was necessary for NFκB activation but that it was not upstream of IKKαT23. The lack of an effect of LY294002 or wortmannin on the degradation of IκBα or on the phosphorylation of IKKα Ser176/Ser180 together with the sustained pattern of AKT phosphorylation correlating with the pattern of p50 binding to the NFκB response element suggested that AKT might have an effect on p50 binding, this awaits further investigation.
IKKα overexpression by itself, either WT or IKKαT23A, was sufficient to activate the NFκB reporters 2–3-fold suggesting that activation of the kinase by autophosphorylation was sufficient to activate NFκB without any requirement for AKT. IKKα has been detected both in the nucleus and cytoplasm of cells consistent with roles for IKKα in gene expression other than in the liberation of IκBα and activation of NFκB (51).
Taken together, this suggested that there was likely another transcription factor target of the AKT/IKKα pathway on the IL-6 promoter. Our data supports the model that the role of NFκB in the induction of the IL-6 gene in response to IL-1 is likely indirect, perhaps involving activation of NFκB responsive AP-1 family members. AKT is also likely involved in this pathway. Thus a positive feedback loop of canonical NFκB activation on AP-1 family members to increase IL-1 responsive IL-6 gene transcription might occur. At least one AP-1 family member, JunB has an NFκB responsive promoter. Furthermore Elk-1, a TCF family member and an important regulator of c-fos transcription has an NFκB site on its promoter. In fact through NFκB-dependent AP-1 activation, NFκB could indirectly control the expression of an AP-1 target gene such as IL-6, as it does vascular endothelial growth factor, by increasing the levels of family members, AP-1 making them more available for up-regulation by MAP kinase (see model in Fig. 9).
Mutation of the IL-6 promoter AP-1 site significantly reduced induction of the IL-6 promoter by IL-1. Consistent with this, other investigators have found that the IL-6 promoter AP-1 site is necessary for promoter activation in response to several stimuli including TGFβ and the Kaposi sarcoma herpes virus (43, 62). In our study, mutation of the IL-6 promoter AP-1 site unmasks additional regulation of NFκB not observed in the WT IL-6 promoter (Fig. 6B). In this regard a 2-fold activation of this mutant IL-6 promoter by IKKαWT, not seen in the intact promoter, is also seen on the NFκB reporters (Fig. 7, B and C) and suggests negative cross-talk between IKKα/AP-1 and IKKα/NFκB pathways. In addition, the IL-6 promoter AP-1 site demonstrated high constitutive activation in Caco-2 cells, suggesting that the AP-1 site placed in the context of the IL-6 promoter may be subject to negative regulation (63). Negative cross-talk has also been described in liver tumor cells in response to TGFβ where transient NFκB activation inhibits AP-1 DNA binding and signaling with important consequences for tumor progression (64). There is evidence in the literature of both positive and negative cross-talk between the NFκB and AP-1 pathways and many genes require the simultaneous activation of both transcription factors working cooperatively (64–66).
Although p110 PI 3-kinase overexpression as well as IL-1 treatment activated the IL-6-specific AP-1 site reporter plasmid (Fig. 8A), the elevated constitutive activation likely masked a more robust IL-1 response. Nevertheless, inhibition of PI 3-kinase by LY294002 as well as overexpression of either dnAKT or IKKαT23A significantly lowered constitutive AP-1 activation. Furthermore, overexpression of IKKαWT reversed the inhibitory effect of dnAKT. Taken together, this is direct evidence that AP-1 is a major downstream target of AKT/IKKα and is likely part of a novel IL-1 responsive signaling pathway to the induction of IL-6 gene transcription. Although other investigators have found evidence for the requirement of the IKK complex, as a whole, including IκBα in AP-1 function, in response to lipopolysaccharide (66), this is the first report of a direct role for AKT/IKKα in AP-1 function. Interestingly in IKKα knock-out mouse embryonic fibroblasts there was evidence of decreased induction by serum stimulation of both JunB and JunD (59). A link between IKKα and c-fos activation in response to epidermal growth factor has been demonstrated to involve the phosphorylation of histone H3 on the c-fos promoter (67). Future work will address if IKKα directly phosphorylates AP-1 family members or is upstream of another kinase in AP-1 activation.
In our study there was 20-fold more IL-6 mRNA produced than was translated into protein, suggesting a translational block on IL-6 mRNA in Caco-2 cells. However, previous studies have shown that the 3′-untranslated region of the IL-6 mRNA is rich in AU sequences involved in mRNA stability by IL-1, lipopolysaccharide, and TNFα in osteoblasts (68). These sequences may not be functional in the Caco-2 cell line where at 4 h post-IL-1 treatment most of the IL-6 mRNA was down-regulated suggesting rapid degradation. A role for the AKT/IKKα pathway in regulation of Tor kinase has recently been established with resulting increases in protein synthesis rates (52). It will be of interest to determine whether AKT/IKKα might also be involved in IL-6 translational regulation in response to IL-1.
IKKα has been shown recently to phosphorylate the CREB co-activator CBP and mediates cytokine-induced phosphorylation and subsequent acetylation of specific residues in histone H3 on NFκB responsive promoters (69). IKKα phosphorylation of CBP, which increases its histone acetyltransferase activity can switch its binding preference from one transcription factor to another with consequences for cell growth (51). Enhanced IL-6 promoter activation is associated with CBP/p300 binding to p65/NFκB as well as to CREB with associated increases in histone acetylation (70). One might speculate that IL-1 signaling via AKT/IKKα could switch the binding preference of p300 from NFκB to AP-1. Future investigations will address this as well as the role of AKT/IKKα and CREB in IL-1 induction of IL-6.
There have been several studies describing a causative role for IL-6 in colon tumor development and progression (71–74). One of these involves an IL-6 gene variant with high sensitivity to IL-1β (75, 76). In colon cancer cells harboring this variant IL-6 may be particularly effective in advancing the adenoma/carcinoma sequence. Interestingly, a dominant-negative mutant of c-Jun exhibited a significant antitumor effect in colon cancer demonstrating the possibility of AP-1-based gene therapy as a novel treatment of colorectal cancer (77–80). Understanding the regulation of NFκB and AP-1 and their cross-talk in the regulation of their target genes such as IL-6 may lead to the development of novel therapeutics for the control of inflammatory diseases of the mucosa.
Acknowledgments
We greatly appreciate the support of Dr. Ross Stein, Dr. Rudolph Tanzi, Dr. Jerrold Rosenbaum, and Dr. Marcie Glicksman for infrastructural support and Hyun-Hee Cho for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AG20181. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IL, interleukin; NIK, NFκB inducing kinase; CBP, CREB-binding protein; TNF, tumor necrosis factor; MAPK, mitogen-activated protein kinase; IKK, IκB kinases; PI 3-kinase, phosphatidylinositol 3-kinase; ERK, extracellular signal-regulated kinase; GSK, glycogen synthase kinase; ELISA, enzyme-linked immunosorbent assay; dn, dominant negative; ca, constitutively active; HS, heat shock; TGF, transforming growth factor; WT, wild type; CREB, cAMP-response element-binding protein; 5-MCDE, 5-methylchrysene-1,2-diol-3, 4-epoxide.
References
- 1.Hirano, T. (1998) Int. Rev. Immunol. 16 249–284 [DOI] [PubMed] [Google Scholar]
- 2.Nishimoto, N., and Kishimoto, T. (2006) Nat. Clin. Pract. Rheumatol. 2 619–626 [DOI] [PubMed] [Google Scholar]
- 3.Naugler, W. E., Sakurai, T., Kim, S., Maeda, S., Kim, K., Elsharkawy, A. M., and Karin, M. (2007) Science 317 121–124 [DOI] [PubMed] [Google Scholar]
- 4.Rakoff-Nahoum, S., and Medzhitov, R. (2007) Science 317 124–127 [DOI] [PubMed] [Google Scholar]
- 5.Greten, F. R., Eckmann, L., Greten, T. F., Park, J. M., Li, Z. W., Egan, L. J., Kagnoff, M. F., and Karin, M. (2004) Cell 118 285–296 [DOI] [PubMed] [Google Scholar]
- 6.Lin, W. W., and Karin, M. (2007) J. Clin. Investig. 117 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karin, M., Lawrence, T., and Nizet, V. (2006) Cell 124 823–835 [DOI] [PubMed] [Google Scholar]
- 8.Ruffolo, C., Scarpa, M., Faggian, D., Romanato, G., De Pellegrin, A., Filosa, T., Prando, D., Polese, L., Scopelliti, M., Pilon, F., Ossi, E., Frego, M., D'Amico, D. F., and Angriman, I. (2007) J. Gastrointest. Surg. 11 16–21 [DOI] [PubMed] [Google Scholar]
- 9.Sher, M. E., D'Angelo, A. J., Stein, T. A., Bailey, B., Burns, G., and Wise, L. (1995) Am. J. Surg. 169 133–136 [DOI] [PubMed] [Google Scholar]
- 10.Mitsuyama, K., Sata, M., and Rose-John, S. (2006) Cytokine Growth Factor Rev. 17 451–461 [DOI] [PubMed] [Google Scholar]
- 11.Atreya, R., and Neurath, M. F. (2005) Clin. Rev. Allergy Immunol. 28 187–196 [DOI] [PubMed] [Google Scholar]
- 12.Swank, G. M., and Deitch, E. A. (1996) World J. Surg. 20 411–417 [DOI] [PubMed] [Google Scholar]
- 13.Beagley, K. W., Eldridge, J. H., Lee, F., Kiyono, H., Everson, M. P., Koopman, W. J., Hirano, T., Kishimoto, T., and McGhee, J. R. (1989) J. Exp. Med. 169 2133–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, W., Smail, N., Wang, P., and Chaudry, I. H. (1998) J. Surg. Res. 79 39–46 [DOI] [PubMed] [Google Scholar]
- 15.Stylianou, E., and Saklatvala, J. (1998) Int. J. Biochem. Cell Biol. 30 1075–1079 [DOI] [PubMed] [Google Scholar]
- 16.Wietek, C., and O'Neill, L. A. (2007) Trends Biochem. Sci. 32 311–319 [DOI] [PubMed] [Google Scholar]
- 17.Martin, M. U., and Wesche, H. (2002) Biochim. Biophys. Acta 1592 265–280 [DOI] [PubMed] [Google Scholar]
- 18.Dahle, M. K., Overland, G., Myhre, A. E., Stuestol, J. F., Hartung, T., Krohn, C. D., Mathiesen, O., Wang, J. E., and Aasen, A. O. (2004) Infect. Immun. 72 5704–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou, C. H., Wei, L. H., Kuo, M. L., Huang, Y. J., Lai, K. P., Chen, C. A., and Hsieh, C. Y. (2005) Carcinogenesis 26 45–52 [DOI] [PubMed] [Google Scholar]
- 20.Marmiroli, S., Bavelloni, A., Faenza, I., Sirri, A., Ognibene, A., Cenni, V., Tsukada, J., Koyama, Y., Ruzzene, M., Ferri, A., Auron, P. E., Toker, A., and Maraldi, N. M. (1998) FEBS Lett. 438 49–54 [DOI] [PubMed] [Google Scholar]
- 21.Reddy, S. A., Huang, J. H., and Liao, W. S. (1997) J. Biol. Chem. 272 29167–29173 [DOI] [PubMed] [Google Scholar]
- 22.Alessi, D. R., and Cohen, P. (1998) Curr. Opin. Genet. Dev. 8 55–62 [DOI] [PubMed] [Google Scholar]
- 23.Coffer, P. J., Jin, J., and Woodgett, J. R. (1998) Biochem. J. 335 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane, L. P., Shapiro, V. S., Stokoe, D., and Weiss, A. (1999) Curr. Biol. 9 601–604 [DOI] [PubMed] [Google Scholar]
- 25.Hacker, H., and Karin, M. (2006) Sci. STKE 2006 re13. [DOI] [PubMed] [Google Scholar]
- 26.Gustin, J. A., Ozes, O. N., Akca, H., Pincheira, R., Mayo, L. D., Li, Q., Guzman, J. R., Korgaonkar, C. K., and Donner, D. B. (2004) J. Biol. Chem. 279 1615–1620 [DOI] [PubMed] [Google Scholar]
- 27.Solt, L. A., Madge, L. A., Orange, J. S., and May, M. J. (2007) J. Biol. Chem. 282 8724–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustin, J. A., Korgaonkar, C. K., Pincheira, R., Li, Q., and Donner, D. B. (2006) J. Biol. Chem. 281 16473–16481 [DOI] [PubMed] [Google Scholar]
- 29.Ozes, O. N., Mayo, L. D., Gustin, J. A., Pfeffer, S. R., Pfeffer, L. M., and Donner, D. B. (1999) Nature 401 82–85 [DOI] [PubMed] [Google Scholar]
- 30.Ling, L., Cao, Z., and Goeddel, D. V. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3792–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senftleben, U., Cao, Y., Xiao, G., Greten, F. R., Krahn, G., Bonizzi, G., Chen, Y., Hu, Y., Fong, A., Sun, S. C., and Karin, M. (2001) Science 293 1495–1499 [DOI] [PubMed] [Google Scholar]
- 32.Xiao, G., Fong, A., and Sun, S. C. (2004) J. Biol. Chem. 279 30099–30105 [DOI] [PubMed] [Google Scholar]
- 33.Shaulian, E., and Karin, M. (2002) Nat. Cell Biol. 4 E131–E136 [DOI] [PubMed] [Google Scholar]
- 34.Manning, A. M., and Davis, R. J. (2003) Nat. Rev. Drug Discov. 2 554–565 [DOI] [PubMed] [Google Scholar]
- 35.Robinson, M. J., and Cobb, M. H. (1997) Curr. Opin. Cell Biol. 9 180–186 [DOI] [PubMed] [Google Scholar]
- 36.Sizemore, N., Leung, S., and Stark, G. R. (1999) Mol. Cell. Biol. 19 4798–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, C., Ma, W. Y., and Dong, Z. (1996) Mol. Cell. Biol. 16 6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, J., Chen, H., Tang, M. S., Shi, X., Amin, S., Desai, D., Costa, M., and Huang, C. (2004) J. Cell Biol. 165 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peloponese, J. M., Jr., and Jeang, K. T. (2006) J. Biol. Chem. 281 8927–8938 [DOI] [PubMed] [Google Scholar]
- 40.Koul, D., Shen, R., Shishodia, S., Takada, Y., Bhat, K. P., Reddy, S. A., Aggarwal, B. B., and Yung, W. K. (2007) Mol. Cell Biochem. 300 77–87 [DOI] [PubMed] [Google Scholar]
- 41.Cahill, C. M., Tzivion, G., Nasrin, N., Ogg, S., Dore, J., Ruvkun, G., and Alexander-Bridges, M. (2001) J. Biol. Chem. 276 13402–13410 [DOI] [PubMed] [Google Scholar]
- 42.Okuno, Y., Huettner, C. S., Radomska, H. S., Petkova, V., Iwasaki, H., Akashi, K., and Tenen, D. G. (2002) Blood 100 4420–4426 [DOI] [PubMed] [Google Scholar]
- 43.Eickelberg, O., Pansky, A., Mussmann, R., Bihl, M., Tamm, M., Hildebrand, P., Perruchoud, A. P., and Roth, M. (1999) J. Biol. Chem. 274 12933–12938 [DOI] [PubMed] [Google Scholar]
- 44.Plaisance, S., Vanden Berghe, W., Boone, E., Fiers, W., and Haegeman, G. (1997) Mol. Cell. Biol. 17 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlahos, C. J., Matter, W. F., Hui, K. Y., and Brown, R. F. (1994) J. Biol. Chem. 269 5241–5248 [PubMed] [Google Scholar]
- 46.Jiang, Q., Detolla, L., Singh, I. S., Gatdula, L., Fitzgerald, B., van Rooijen, N., Cross, A. S., and Hasday, J. D. (1999) Am. J. Physiol. 276 R1653–R1660 [DOI] [PubMed] [Google Scholar]
- 47.Bang, O. S., Ha, B. G., Park, E. K., and Kang, S. S. (2000) Biochem. Biophys. Res. Commun. 278 306–311 [DOI] [PubMed] [Google Scholar]
- 48.Vanden Berghe, W., Vermeulen, L., De Wilde, G., De Bosscher, K., Boone, E., and Haegeman, G. (2000) Biochem. Pharmacol. 60 1185–1195 [DOI] [PubMed] [Google Scholar]
- 49.Albanese, C., Wu, K., D'Amico, M., Jarrett, C., Joyce, D., Hughes, J., Hulit, J., Sakamaki, T., Fu, M., Ben-Ze'ev, A., Bromberg, J. F., Lamberti, C., Verma, U., Gaynor, R. B., Byers, S. W., and Pestell, R. G. (2003) Mol. Biol. Cell 14 585–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo, J. L., Tan, W., Ricono, J. M., Korchynskyi, O., Zhang, M., Gonias, S. L., Cheresh, D. A., and Karin, M. (2007) Nature 446 690–694 [DOI] [PubMed] [Google Scholar]
- 51.Huang, W. C., Ju, T. K., Hung, M. C., and Chen, C. C. (2007) Mol. Cell 26 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dan, H. C., Adli, M., and Baldwin, A. S. (2007) Cancer Res. 67 6263–6269 [DOI] [PubMed] [Google Scholar]
- 53.Garat, C., and Arend, W. P. (2003) Cytokine 23 31–40 [DOI] [PubMed] [Google Scholar]
- 54.Yang, H. T., Cohen, P., and Reist, C. (2008) Cell. Signal. 20 375–380 [DOI] [PubMed] [Google Scholar]
- 55.Blanco, S., Sanz-Garcia, M., Santos, C., and Lazo, P. (2008) PLoS ONE 3 1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Haij, S., Bakker, A. C., van der Geest, R. N., Haegeman, G., Vanden Berghe, W., Aarbiou, J., Daha, M. R., and van Kooten, C. (2005) J. Am. Soc. Nephrol. 16 1603–1611 [DOI] [PubMed] [Google Scholar]
- 57.Davies, S., Reddy, H., Caivano, M., and Cohen, P. (2000) Biochem. J. 351 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanden Berghe, W., Plaisance, S., Boone, E., De Bosscher, K., Schmitz, M. L., Fiers, W., and Haegeman, G. (1998) J. Biol. Chem. 273 3285–3290 [DOI] [PubMed] [Google Scholar]
- 59.Stein, B., and Yang, M. X. (1995) Mol. Cell. Biol. 15 4971–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo, F., and Wu, S. (2000) Inflammation 24 305–316 [DOI] [PubMed] [Google Scholar]
- 61.Koul, D., Yao, Y., Abbruzzese, J. L., Yung, W. K., and Reddy, S. A. (2001) J. Biol. Chem. 276 11402–11408 [DOI] [PubMed] [Google Scholar]
- 62.An, J., Lichtenstein, A. K., Brent, G., and Rettig, M. B. (2002) Blood 99 649–654 [DOI] [PubMed] [Google Scholar]
- 63.Samuel, S., Twizere, J. C., and Bernstein, L. R. (2005) Biochem. J. 388 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arsura, M., Panta, G. R., Bilyeu, J. D., Cavin, L. G., Sovak, M. A., Oliver, A. A., Factor, V., Heuchel, R., Mercurio, F., Thorgeirsson, S. S., and Sonenshein, G. E. (2003) Oncogene 22 412–425 [DOI] [PubMed] [Google Scholar]
- 65.Fujioka, S., Niu, J., Schmidt, C., Sclabas, G. M., Peng, B., Uwagawa, T., Li, Z., Evans, D. B., Abbruzzese, J. L., and Chiao, P. J. (2004) Mol. Cell. Biol. 24 7806–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krappmann, D., Wegener, E., Sunami, Y., Esen, M., Thiel, A., Mordmuller, B., and Scheidereit, C. (2004) Mol. Cell. Biol. 24 6488–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anest, V., Cogswell, P. C., and Baldwin, A. S., Jr. (2004) J. Biol. Chem. 279 31183–31189 [DOI] [PubMed] [Google Scholar]
- 68.Patil, C., Zhu, X., Rossa, C., Jr., Kim, Y. J., and Kirkwood, K. L. (2004) Immunol. Investig. 33 213–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto, Y., Verma, U. N., Prajapati, S., Kwak, Y. T., and Gaynor, R. B. (2003) Nature 423 655–659 [DOI] [PubMed] [Google Scholar]
- 70.Vanden Berghe, W., De Bosscher, K., Boone, E., Plaisance, S., and Haegeman, G. (1999) J. Biol. Chem. 274 32091–32098 [DOI] [PubMed] [Google Scholar]
- 71.Brozek, W., Bises, G., Girsch, T., Cross, H. S., Kaiser, H. E., and Peterlik, M. (2005) Eur. J. Cancer 41 2347–2354 [DOI] [PubMed] [Google Scholar]
- 72.Kinoshita, T., Ito, H., and Miki, C. (1999) Cancer 85 2526–2531 [DOI] [PubMed] [Google Scholar]
- 73.Schneider, M. R., Hoeflich, A., Fischer, J. R., Wolf, E., Sordat, B., and Lahm, H. (2000) Cancer Lett. 151 31–38 [DOI] [PubMed] [Google Scholar]
- 74.Matsuo, K., Oka, M., Murase, K., Soda, H., Isomoto, H., Takeshima, F., Mizuta, Y., Murata, I., and Kohno, S. (2003) J. Int. Med. Res. 31 69–75 [DOI] [PubMed] [Google Scholar]
- 75.Hefler, L. A., Grimm, C., Lantzsch, T., Lampe, D., Leodolter, S., Koelbl, H., Heinze, G., Reinthaller, A., Tong-Cacsire, D., Tempfer, C., and Zeillinger, R. (2005) Clin. Cancer Res. 11 5718–5721 [DOI] [PubMed] [Google Scholar]
- 76.Belluco, C., Olivieri, F., Bonafe, M., Giovagnetti, S., Mammano, E., Scalerta, R., Ambrosi, A., Franceschi, C., Nitti, D., and Lise, M. (2003) Clin. Cancer Res. 9 2173–2176 [PubMed] [Google Scholar]
- 77.Suto, R., Tominaga, K., Mizuguchi, H., Sasaki, E., Higuchi, K., Kim, S., Iwao, H., and Arakawa, T. (2004) Gene Ther. 11 187–193 [DOI] [PubMed] [Google Scholar]
- 78.Neumann, D., Lienenklaus, S., Rosati, O., and Martin, M. U. (2002) Eur. J. Immunol. 32 3689–3698 [DOI] [PubMed] [Google Scholar]
- 79.Wang, K. Z., Wara-Aswapati, N., Boch, J. A., Yoshida, Y., Hu, C. D., Galson, D. L., and Auron, P. E. (2006) J. Cell Sci. 119 1579–1591 [DOI] [PubMed] [Google Scholar]
- 80.Brikos, C., Wait, R., Begum, S., O'Neill, L. A., and Saklatvala, J. (2007) Mol. Cell. Proteomics 6 1551–1559 [DOI] [PubMed] [Google Scholar]
- 81.Watters, T. M., Kenny, E. F., and O'Neill, L. A. (2007) Immunol. Cell Biol. 85 411–419 [DOI] [PubMed] [Google Scholar]
- 82.O'Neill, L. A., and Dinarello, C. A. (2000) Immunol. Today 21 206–209 [DOI] [PubMed] [Google Scholar]
- 83.O'Neill, L. (2000) Biochem. Soc. Trans. 28 557–563 [DOI] [PubMed] [Google Scholar]
- 84.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. (1999) Nature 398 252–256 [DOI] [PubMed] [Google Scholar]
- 85.Park, H. S., Kim, M. S., Huh, S. H., Park, J., Chung, J., Kang, S. S., and Choi, E. J. (2002) J. Biol. Chem. 277 2573–2578 [DOI] [PubMed] [Google Scholar]
- 86.Song, J. J., and Lee, Y. J. (2005) J. Cell Biol. 170 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ray, A., Sassone-Corsi, P., and Sehgal, P. (1989) Mol. Cell Biol. 9 5537–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]