Abstract
The heavy chain of cytoplasmic dynein contains four nucleotide-binding domains referred to as AAA1–AAA4, with the first domain (AAA1) being the main ATP hydrolytic site. Although previous studies have proposed regulatory roles for AAA3 and AAA4, the role of ATP hydrolysis at these sites remains elusive. Here, we have analyzed the single molecule motility properties of yeast cytoplasmic dynein mutants bearing mutations that prevent ATP hydrolysis at AAA3 or AAA4. Both mutants remain processive, but the AAA4 mutant exhibits a surprising increase in processivity due to its tighter affinity for microtubules. In addition to changes in motility characteristics, AAA3 and AAA4 mutants produce less maximal force than wild-type dynein. These results indicate that the nucleotide binding state at AAA3 and AAA4 can allosterically modulate microtubule binding affinity and affect dynein processivity and force production.
Cytoplasmic dynein is a molecular motor that moves toward the minus-end of microtubules. Underscoring its biological significance, dynein has been implicated in numerous microtubule-related functions, including cargo transport, mitotic spindle positioning, and nuclear segregation (see Ref. 1 for a review). Like many other biological motors, cytoplasmic dynein uses chemical energy derived from ATP hydrolysis to perform mechanical work. However, in contrast to other cytoskeletal motors of the kinesin and myosin superfamilies, dynein has multiple ATP binding sites. This poses the question of how dynein makes use of these multiple ATP sites and whether they might be involved in the regulation of the motor.
Dyneins are AAA+ ATPases, a superfamily of enzymes that have a diverse array of functions ranging from protein unfolding to membrane trafficking (see Refs. 2 and 3) for reviews). Despite their varied functions, AAA+ ATPases all share a similar core architecture with conserved Walker-A (P loop) and Walker-B (phosphate sensor) motifs in their nucleotide binding domains (4, 5). Most AAA+ proteins oligomerize into hexameric, ringlike structures that act upon their substrates. In some cases, the identical AAA+ subunits may fire stochastically (e.g. ClpX (6)), whereas in other cases, sequential hydrolysis around the ring may occur (e.g. helicases (7)). Dynein is unusual in having multiple AAA+ domains concatenated in a single polypeptide chain that folds into a ringlike structure (8–10). The first four AAA+ domains (AAA1–AAA4) are capable of binding nucleotide (11, 12), whereas the last two AAA+ domains (AAA5–AAA6) are highly divergent, no longer bind nucleotide, and appear to serve a structural role in completing the ring. Between AAA4 and AAA5, an antiparallel coiled-coil stalk emerges with a microtubule binding domain at the tip. NH2-terminal to the first AAA domain is a “linker” domain that is thought to swing with respect to the stalk, possibly constituting the dynein power stroke (9).
The roles of the four functional AAA domains have been investigated by biochemical and mutagenesis studies. AAA1, the site of ATP-vanadate photocleavage (13), is generally acknowledged to be the major site of ATP hydrolysis and primary driver of the power stroke. Mutagenesis of this site greatly decreases ATP turnover (14, 15), abolishes motility (14), and eliminates the conformational change of the linker domain (16). The roles of the other sites remain less well understood. Single molecule studies suggest that a single ATP binding event may suffice for dynein to take a step along the microtubule (17). However, mutagenesis of the Walker A domain of AAA3 (predicted to interfere with nucleotide binding) greatly decreases microtubule-stimulated ATPase and microtubule gliding activity and causes “rigor-like” binding with the microtubule (14, 18, 19). Mutagenesis studies of AAA2 and AAA4 suggest they may have more minor roles. Collectively, these results suggest that AAA2–AAA4 assume some sort of regulatory role, but the details of how they participate in the dynein mechanism remain unclear.
Although prior in vitro motility studies have been performed on dynein ATP site mutations (14, 18, 19), they have focused upon Walker A mutations that disrupt ATP binding and have not examined the processive movement of a two-headed dynein. Here, we used previously developed single molecule motility assays (17, 20) to investigate the role of ATP hydrolysis at AAA3 and AAA4 on processivity and force production. We find that dynein bearing a Walker B mutation (that specifically disrupts ATP hydrolysis) at AAA3 is still processive, despite a severe effect on ATPase activity and motor velocity. Surprisingly, the AAA4 Walker B mutant displays enhanced processivity that is most likely mediated by an increase in microtubule binding affinity. We also show that AAA3 and AAA4 mutants can only generate 2-fold lower forces than wild-type dynein. Thus, the nucleotide binding state at AAA3 and AAA4 can regulate dynein microtubule affinity, processivity, and force-generating ability.
EXPERIMENTAL PROCEDURES
Protein Expression and Preparation—A 330-kDa artificially dimerized expression construct of cytoplasmic dynein (glutathione S-transferase-Dyn1331kDa) was prepared and purified from Saccharomyces cerevisiae as previously described (17). Single point mutations (E2488Q (AAA3) and E2819Q (AAA4)) were introduced by the QuikChange mutagenesis kit (Stratagene). All constructs contained an NH2-terminal IgG binding domain and a Tev protease cleavage site for purification, a green fluorescent protein tag for specific attachment to surfaces, and a COOH-terminal HaloTag (DHA; Promega) for fluorescent labeling. Prior to single molecule analyses, dynein was further purified by binding 50 μl of ∼300 μg/ml affinity-purified dynein to 10 μl of 500 μg/ml taxol-stabilized microtubules in the absence of ATP, centrifuging the microtubules, and then releasing from microtubules with 10 mm MgATP.
Single Molecule Total Internal Reflection Fluorescence Microscopy—Dynein was labeled with halotetramethylrhodamine (Promega) in the HaloTag domain and assayed for motility on Cy5-labeled axonemes, as previously described (17). Single molecules of dynein were visualized by a custom-built total internal reflection microscope using objective style total internal reflection fluorescence and an argon laser with 514 nm illumination at 3 milliwatts. Images were collected with an intensified CCD camera every 2 s for 5–10 min. Velocities and run lengths were determined by kymograph analysis in ImageJ and corrected for photobleaching rates and axoneme length as previously described (17).
Measurement of ATPase Activity—Basal and microtubule-stimulated ATPase activities were measured by the EnzChek phosphate assay kit (Invitrogen). Assays were performed in motility buffer (30 mm Hepes, pH 7.4, 50 mm KAc, 2 mm Mg(Ac)2, and 1 mm EGTA) with 0–15 μm taxol-stabilized microtubules and 5–10 nm dynein. Reactions were initiated with the addition of MgATP to a final concentration of 0–5 mm, and the absorbance at 360 nm was monitored by a spectrophotometer for 5–10 min. Protein concentrations of dynein were determined on SDS-polyacrylamide gels stained with SYPRO-Red (Invitrogen), with a known concentration of β-actin used as a standard.
Optical Trapping—All experiments were performed with a custom-built force feedback-enhanced optical trapping microscope, as previously described (20). Carboxylated latex beads (0.92-μm diameter; Invitrogen) coated with anti-green fluorescent protein antibodies were mixed with dynein in an assay solution containing 30 mm HEPES (pH 7.4), 50 mm KAc, 2 mm Mg(Ac)2, 1 mm EGTA, 0.5 mg/ml casein, 4.5 mg/ml glucose, 10 mm dithiothreitol, and an oxygen scavenging system of glucose oxidase and catalase (21). Stall force measurements and nucleotide-dependent movement studies were performed with 1 mm MgATP, whereas nucleotide-independent movement studies were performed in the presence of 10 units/ml apyrase. Dynein-coated beads were flowed into a standard flow chamber with adhered tetramethylrhodamine-labeled sea urchin axonemes, and bead displacements were recorded with a quadrant photodiode at 2 kHz.
RESULTS
Construction and Purification of ATPase Mutant Dyneins—The native dynein heavy chain gene consists of a ∼470-kDa polypeptide with NH2-terminal cargo binding and dimerization domains and a COOH-terminal motor domain. We previously engineered a minimal dynein dimer that contains a 330-kDa minimal motor domain fused at its NH2 terminus to glutathione S-transferase, which self-associates to form a dimer (17). This construct (referred to as “wild-type dynein” in this study), which does not bind the dynein light or intermediate chains and has very low, substoichiometric amounts of the yeast Lis1 homologue, Pac1, shows robust processive movement in a single molecule fluorescence assay.
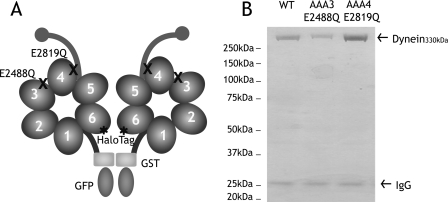
In order to specifically disrupt ATP hydrolysis at sites AAA3 and AAA4, point mutations changing an essential glutamate to a glutamine were introduced into the Walker B motifs (AAA3-E/Q and AAA4-E/Q; Fig. 1A). This glutamate residue is highly conserved, and glutamate-to-glutamine mutations disrupt nucleotide hydrolysis, but not nucleotide binding, in many other AAA proteins (6, 22). AAA2 does not have the conserved Walker B glutamate and thus was not investigated by mutagenesis in this study. Recombinant dyneins, without or with AAA3 or AAA4 point mutations, were purified from S. cerevisiae with an NH2-terminal affinity tag and then labeled with tetramethylrhodamine at the COOH-terminal HaloTag (Fig. 1B).
FIGURE 1.
Construction and purification of dynein ATP hydrolysis mutants. A, a minimal S. cerevisiae cytoplasmic dynein that demonstrates processive motility was engineered as described previously (17). A glutathione S-transferase tag (GST) was incorporated at the NH2 terminus for the dimerization of the two heads of cytoplasmic dynein, whereas a HaloTag was fused to the COOH terminus for fluorescent labeling of the heads. In this paper, this construct is referred to as “wild-type dynein.” Highly conserved glutamate residues in the Walker B motif of domains AAA3 (E2488) and AAA4 (E2819) were mutated to glutamine to block ATP hydrolysis. B, Coomassie Blue-stained polyacrylamide gel of recombinant cytoplasmic dynein purified from S. cerevisiae by affinity purification. 330-kDa recombinant dynein is purified with minor amounts of 26-kDa IgG from the affinity matrix. WT, wild type.
Single Molecule Motility of Dynein Mutants—To measure the motility of individual dynein molecules, we observed tetramethylrhodamine-labeled dynein moving along axonemes by total internal reflection fluorescence microscopy (17) (supplemental Movies 1–3). Contrary to bulk gliding assays, this method provides a direct determination of velocities and run lengths by observing single attachment, movement, and detachment events.
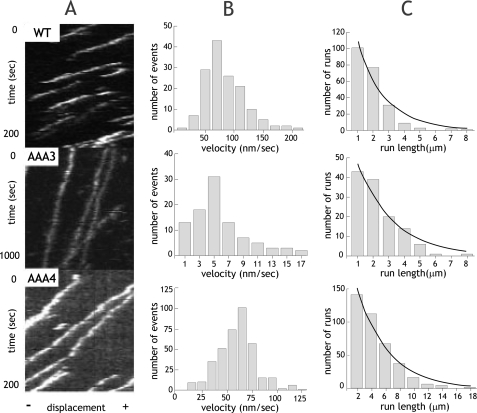
At 1 mm ATP, single wild-type, AAA3-E/Q and AAA4-E/Q dynein molecules demonstrated processive movement (continuous lines in kymographs in Fig. 2A). However, the velocity of the AAA3-E/Q mutant was substantially decreased (4.6 ± 3.7 nm/s) compared with wild-type motors (73.9 ± 34.2 nm/s; Fig. 2B). In contrast, AAA4-E/Q demonstrated only a modest decrease in velocity (60.6 ± 18.9 nm/s; Fig. 2B). The relative severity of these defects is reflected in the in vivo mutant phenotypes of these dyneins, where severe nuclear segregation defects are observed for the AAA3-E/Q but not the AAA4-E/Q mutant (Fig. S1) (18). These velocity decreases are similar to those reported for AAA3 and AAA4 Walker A mutant dynein monomers assayed for microtubule gliding in vitro (14), but the results here also demonstrate that these mutants still retain processive behavior.
FIGURE 2.
Single molecule processivity of dynein ATP hydrolysis mutants. A, kymographs of single molecules of wild-type (WT) or ATP hydrolysis mutants. The x axis represents the length of an axoneme, and the y axis shows time. B, velocity histograms of wild-type and ATP hydrolysis mutants. The mean velocities ± S.D. are 73.9 ± 34.2 nm/s, 4.6 ± 3.7 nm/s, and 60.6 ± 18.9 nm/s (n = 221, 117, and 384) for wild type, AAA3-E/Q, and AAA4-E/Q, respectively. C, run length histograms of wild-type and ATP hydrolysis mutants are distributed in a single exponential decay. Run lengths were corrected for photobleaching and average axoneme length, and calculations for correct binning were performed as previously described (17). Run lengths (±S.E. as estimated by bootstrapping (17)) are 2.25 ± 0.14, 1.79 ± 0.18, and 4.38 ± 0.45 μm for wild type, AAA3-E/Q, and AAA4-E/Q, respectively.
To further evaluate processivity, run lengths were measured by kymograph analysis (Fig. 2C). The lengths of dynein runs were exponentially distributed, with the exponential decay constant representing the mean run length (23). The AAA3-E/Q mutant displayed a slight decrease in run length (1.79 ± 0.18 μm) compared with wild-type dynein (2.25 ± 0.14 μm). In contrast, AAA4-E/Q mutants had a surprising 2-fold increase in run length (4.39 ± 0.45 μm). The frequency of extremely long runs (10–20 μm) further demonstrated the pronounced gain in processivity.
We wished to exclude the possibility that the longer run length of AAA4-E/Q was due to aggregation, since an aggregate might have more attachments to the microtubule and thereby detach less frequently. To test whether one or multiple dyneins are present in the moving spots, we examined their photobleaching behavior (Fig. S2). We found that all of the moving molecules (n = 47, 25, and 45 for wild-type, AAA3-E/Q, and AAA4-E/Q, respectively) showed only one- or two-step photobleaching, as expected for single dynein dimers. There was no significant difference in photobleaching between wild-type and mutant dyneins, ruling out the possibility that protein aggregation accounts for the increased run length of AAA4-E/Q or decreased velocity of AAA3-E/Q.
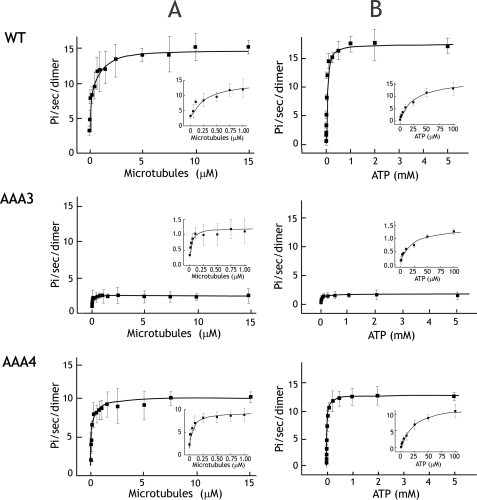
Microtubule-stimulated ATPase Activity of Dynein ATPase Mutants—To better understand the single molecule behaviors of the AAA3 and AAA4 ATP hydrolysis mutants, we measured their basal and microtubule-stimulated ATPase activities (Fig. 3A). In accordance with velocity reductions in the single molecule assay, the microtubule-stimulated ATPase rates, kcat, of AAA3 and AAA4 mutants, were reduced 10- and 1.5-fold, respectively, compared with wild-type dynein (Table 1). The basal ATPase rates were reduced by a similar margin. Thus, microtubule stimulation was comparable (3–4-fold) for the mutants and wild-type dynein, implying that the loss of motility in the mutants is not due to a lack of microtubule stimulation. In summary, these results suggest that trapping AAA4 and particularly AAA3 in an ATP state decreases ATP turnover at AAA1, the main hydrolytic site of dynein.
FIGURE 3.
Microtubule-stimulated ATPase activity of dynein ATP hydrolysis mutants. A, microtubule-stimulated ATPase activity of wild type and AAA mutants at 2 mm ATP. The insets show detailed views of microtubule-stimulated ATPase activity at low microtubule concentrations. Km,MT values for wild type (WT), AAA3-E/Q, and AAA4-E/Q are 0.31, 0.03, and 0.069 μm. respectively. B, the ATP dependence of microtubule-stimulated ATPase activity measured with 5 μm taxol-stabilized microtubules. Insets show detailed views of the curve at low ATP concentrations. Km,ATP values for wild type, AAA3-E/Q, and AAA4-E/Q are 25.2, 24.6, and 24.7 μm, respectively. Each dot represents the mean ± S.D. from three measurements of one preparation. Mean values from three preparations are presented in Table 1.
TABLE 1.
Motility and ATPase activity of AAA hydrolysis mutants
Data was collected from three independent preparations of dynein for each construct. Reported values are mean and S.E. for three independent preparations. For velocity and run length data, >100 molecules were measured for each preparation.
|
Velocity |
Run length |
MT-stimulated ATPase |
Basal ATPase kcat |
|||
|---|---|---|---|---|---|---|
| kcat | Km,MT | Km,ATP | ||||
| nm/s | μm | s-1 | μm | μm | s-1 | |
| Wild type | 72.5 ± 5.5 | 1.99 ± 0.16 | 14.1 ± 0.36 | 0.59 ± 0.28 | 26.2 ± 0.8 | 3.74 ± 0.35 |
| AAA3 (E2488Q) | 5.1 ± 0.9 | 1.78 ± 0.10 | 1.38 ± 0.14 | 0.03 ± 0.01 | 23.7 ± 0.7 | 0.30 ± 0.05 |
| AAA4 (E2819Q) | 62.5 ± 1.6 | 4.55 ± 0.39 | 10.6 ± 0.72 | 0.09 ± 0.03 | 25.3 ± 2.4 | 3.36 ± 0.59 |
The AAA3 and AAA4 mutants also exhibited a striking increase in their binding affinity for microtubules. A ∼20-fold increase in Km,MT3 for microtubule-stimulated ATPase activity was observed for AAA3-E/Q (Km,MT = 0.03 μm). AAA4-E/Q also exhibited a ∼5-fold (Km,MT = 0.09 μm) increase in microtubule-binding affinity, and this tighter interaction with the microtubule might explain the increased processivity of the AAA4-E/Q mutant. Interestingly, the relative increase in microtubule affinity for the AAA4-E/Q mutant appeared to be specific for the dimeric dynein construct. In a dynein monomer (lacking the NH2-terminal glutathione S-transferase), the Km,MT for AAA4-E/Q (1.7 μm) and wild type (2.2 μm) were comparable (data not shown), suggesting that the mutation may affect microtubule affinity by increasing coordination between the two heads of dynein.
We also determined the Km,ATP by measuring microtubule-stimulated ATPase activity at different ATP concentrations (Fig. 3B). If ATP hydrolysis occurs at multiple AAA domains, we would expect the data to show a biphasic fit, with at least two binding constants for ATP, as was suggested motility studies other AAA ATPases (24) and motility studies with mammalian dynein-dynactin complexes (25). However, our data were well fit by a Michaelis-Menten equation (Fig. 3B), implying that a single nucleotide binding site dominates the ATPase reaction. In addition, we find no significant difference between the ATP binding affinities of wild-type and AAA mutant dyneins, implying that blocking ATP hydrolysis at AAA3 and AAA4 does not significantly affect ATP binding at AAA1.
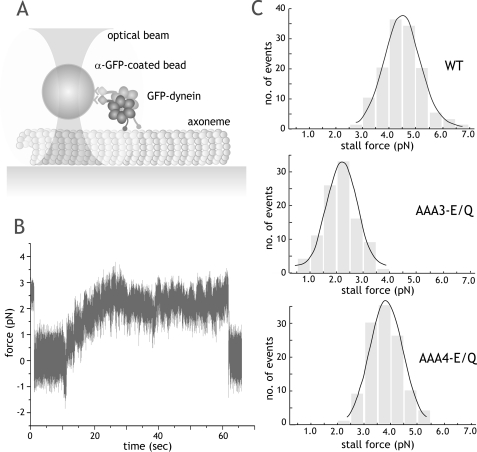
Stall Force Measurements of ATPase Mutant Dyneins—We next determined whether ATP hydrolysis at AAA3 and AAA4 contributed to the force generation of dynein. For these experiments, we bound wild-type and mutant green fluorescent protein-tagged dyneins to latex beads, which could be captured by a fixed optical trap (Fig. 4A). To ensure that bead movements were due to a single dynein molecule, the dynein-to-bead ratio was adjusted so that the fraction of moving beads was <0.3 (representing a >99% probability that movements were due to a single molecule (26)). Dynein mutants exhibited similar behavior to wild-type dynein, moving the bead away from the trap center and then stalling, often for several minutes, under a maximum opposing load (Fig. 4B).
FIGURE 4.
Stall force measurements of ATPase mutant dynein in the optical trap. A, schematic representation of the fixed optical trap setup used for stall force measurements in this paper. B, a representative trace of a single AAA4-E/Q dynein motor moving against force in a fixed optical trap at 1 mm ATP (trap stiffness (k) = 0.034 pN/nm). The trace shows a long stall event of ∼1 min, followed by release, which is typical of both wild-type and mutant dynein. C, stall force distributions of wild-type and ATPase mutant dyneins. Stall forces (mean ± S.D.) are 4.5 ± 1.3 pN (n = 132), 2.6 ± 1.2 pN (n = 100), and 3.7 ± 1.2 pN (n = 115) for wild type (WT), AAA3-E/Q, and AAA4-E/Q, respectively. Black lines, Gaussian fit of the data.
Both AAA3-E/Q and AAA4-E/Q (2.6 and 3.7 pN, respectively; Fig. 4C) exhibited lower stall forces compared with wild-type dynein (4.5 pN; p < 0.0001, Student's t test). These experiments show that the ATP hydrolysis mutants remain processive under load but fail to achieve the same stall forces as wild-type dynein.
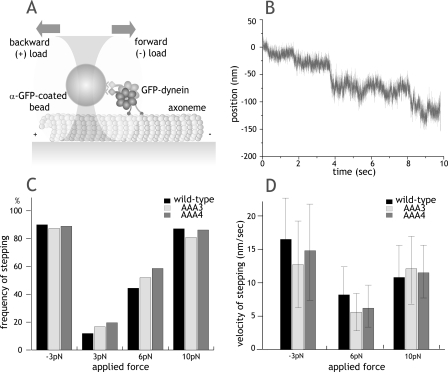
Nucleotide-independent Movement of Dynein Induced by Force—We have recently shown that a pull from an optical trap will cause dynein to step processively along a microtubule in the absence of nucleotide (20). In this experiment, tension applied from the optical trap causes the rear dynein head to detach from the microtubule and then rebind to a new site further along the microtubule. We applied this assay to gauge the microtubule-binding affinities of AAA mutants under an applied load (Fig. 5A).
FIGURE 5.
Nucleotide-independent movement of dynein induced by force. A, schematic of force-induced stepping experiments. Forward force is defined as the direction in which dynein normally moves (toward the microtubule minus-end). B, example trace of nucleotide-free, force-induced stepping for AAA4 mutants in a force feedback trap with 6 pN of backward load. k = 0.062 pN/nm. C, frequency of nucleotide-independent dynein movement after applying constant forward (–3 pN) or backward load (3, 6, and 10 pN). n > 25 molecules were tested at each force for each construct. Movement was scored within a 10-s time window of pulling on a dynein-coated bead attached to the microtubule. D, velocity of force-induced dynein movement with forward (–3 pN) or backward (6 and 10 pN) load (mean ± S.D.). Velocities at 3 pN backward load were not measured due to the small fraction of moving motors. GFP, green fluorescent protein.
Although ATP-driven velocity differed between wild-type, AAA3-E/Q, and AAA4-E/Q dyneins, all three motors behaved in an indistinguishable manner in this nucleotide-independent, force-driven stepping assay. The frequency (Fig. 5C) and velocity (Fig. 5D) were very similar for all three dyneins at different applied loads. Both AAA mutants also showed the same intrinsic asymmetry to force-induced movement as found for wild-type dynein, with only 3 pN of force required to induce robust movement in the forward direction and 10 pN of force to induce robust movement in the backward direction (Fig. 5C).
This assay primarily tests the microtubule binding affinity in the apo state, and the results reveal that the Walker B mutations in AAA3 and AAA4 do not affect the microtubule-binding domain under conditions where the motor is not undergoing cycles of ATP hydrolysis.
DISCUSSION
In this work, we have explored the roles of nucleotide hydrolysis at the dynein “regulatory” AAA domains, AAA3 and AAA4. This study differs from prior biochemical work on the AAA domains (14), which employed nonprocessive dynein monomers and mutated the Walker A motif, which is expected to interfere with nucleotide binding. Our results show that blocking nucleotide hydrolysis at AAA3 and AAA4 significantly affects motor velocity, processivity, and force production but not nucleotide affinity at AAA1 and microtubule binding affinity in the apo state. These studies provide new insight into how the nucleotide states of AAA3 and AAA4 can affect the main hydrolytic site (AAA1) and the mechanics of the motor.
The principal consequence of blocking ATP hydrolysis at AAA4 is on motor processivity, resulting in a 2-fold increase in the run length. The processivity of molecular motors is thought to be mediated by alternating catalysis of the two heads (27), resulting in hand-over-hand motion (28). A processive run is terminated when both motor domains simultaneously detach from the microtubule, which would be more likely if both heads are in a weak binding state. Here, we show that blocking ATP hydrolysis at the AAA4 domain increases the binding affinity for microtubules in the presence of ATP for the dimeric dynein construct. This higher microtubule binding affinity is probably responsible for the longer run length of AAA4-E/Q, since the tighter interaction would likely equate to a lower probability of dissociation from the track. AAA3-E/Q has an even higher affinity for microtubules, although its run length is similar to wild-type. However, if one calculates mean attachment times, AAA3-E/Q is attached much longer (360 s) than both wild-type (30 s) and AAA4-E/Q (72 s), showing that higher microtubule affinity is also reflected in the motility characteristics of AAA3-E/Q.
In previous studies, motors with increased processivity have been made by engineering the dimerization domain of the motor (29) or microtubule-interacting elements (30). Our finding highlights a novel example where engineering an ATPase domain, which has no known interactions with microtubules, causes an increase in processivity. ATP hydrolysis at AAA3 and AAA4 could affect microtubule binding affinity either by a direct allosteric effect communicated through the coiled-coil stalk to the microtubule binding domain or by affecting the kinetic cycle of AAA1, such that the motor spends a longer time in nucleotide states associated with tight microtubule binding. In addition, the pronounced difference of microtubule binding affinity between dimeric wild-type and AAA4-E/Q dynein, but not monomeric wild-type and AAA4-E/Q dynein, suggests the possibility that kinetic coupling is increased between the two heads of dynein in AAA4-E/Q mutants. Further studies will be required to distinguish between these mechanisms.
Our data also reveal that blocking ATP hydrolysis at AAA3 or AAA4 affects the catalytic and mechanical force production activities of dynein. This is particularly evident for AAA3-E/Q, which reduces the overall ATPase rate and movement velocity by an order of magnitude, effects that are most likely mediated by repressing ATP turnover at AAA1. Previous studies have shown that a Walker A mutation (which interferes with nucleotide binding) in AAA3 similarly reduces the ATPase and microtubule gliding velocity of Dictyostelium dynein by ∼20-fold (14). Taken together, these results suggest that both ATP binding and hydrolysis at AAA3 are important for fast nucleotide turnover at AAA1, implying allosteric communication between different AAA sites.
Although these and other mutagenesis studies show that the nucleotide state of AAA3 and AAA4 can affect overall dynein activity, the predominant nucleotide state of AAA3 and AAA4 has yet to be determined. The nucleotide turnover rate at AAA3 and AAA4 also remains unknown, although prior dwell time analysis (17) and the single site fit of the ATPase data in this study suggest that ATP hydrolysis at AAA3 (and AAA4) does not occur during every cycle of ATP turnover at AAA1. Thus, AAA3 and AAA4 may function as regulatory sites for AAA1, perhaps analogous to the main (D2) and regulatory (D2) catalytic sites of p97, another AAA+ protein (31–33). To answer such questions and better understand the interplay of the four AAA nucleotide sites, new tools will be needed to directly probe the nucleotide state of each AAA domain in the native dynein enzyme.
Supplementary Material
Acknowledgments
We thank Derek Applewhite and Erin Reicha (students of the 2006 MBL Physiology Course for initial experimental work, which was instrumental for launching this project). We also thank Roberto Albanese for preparation of media and Andrew Carter, Arne Gennerich, and Ahmet Yildiz for technical assistance and valuable discussions.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grant T32 GM07810 (to C. C.). This work was also supported by the Howard Hughes Medical Institute (to R. D. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies S1–S3.
Footnotes
The abbreviations used are: MT, microtubule; pN, piconewtons.
References
- 1.Vallee, R. B., Williams, J. C., Varma, D., and Barnhart, L. E. (2004) J. Neurobiol. 58 189–200 [DOI] [PubMed] [Google Scholar]
- 2.Vale, R. D. (2000) J. Cell Biol. 150 F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogura, T., and Wilkinson, A. J. (2001) Genes Cells 6 575–597 [DOI] [PubMed] [Google Scholar]
- 4.Neuwald, A. F., Aravind, L., Spouge, J. L., and Koonin, E. V. (1999) Genome Res. 9 27–43 [PubMed] [Google Scholar]
- 5.Erzberger, J. P., and Berger, J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35 93–114 [DOI] [PubMed] [Google Scholar]
- 6.Martin, A., Baker, T. A., and Sauer, R. T. (2005) Nature 437 1115–1120 [DOI] [PubMed] [Google Scholar]
- 7.Enemark, E. J., and Joshua-Tor, L. (2006) Nature 442 270–275 [DOI] [PubMed] [Google Scholar]
- 8.Samso, M., Radermacher, M., Frank, J., and Koonce, M. P. (1998) J. Mol. Biol. 276 927–937 [DOI] [PubMed] [Google Scholar]
- 9.Burgess, S. A., Walker, M. L., Sakakibara, H., Knight, P. J., and Oiwa, K. (2003) Nature 421 715–718 [DOI] [PubMed] [Google Scholar]
- 10.Mizuno, N., Narita, A., Kon, T., Sutoh, K., and Kikkawa, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons, I. R., Gibbons, B. H., Mocz, G., and Asai, D. J. (1991) Nature 352 640–643 [DOI] [PubMed] [Google Scholar]
- 12.Ogawa, K., Kamiya, R., Wilkerson, C. G., and Witman, G. B. (1995) Mol. Biol. Cell 6 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, B. H., and Gibbons, I. R. (1987) J. Biol. Chem. 262 8354–8359 [PubMed] [Google Scholar]
- 14.Kon, T., Nishiura, M., Ohkura, R., Toyoshima, Y. Y., and Sutoh, K. (2004) Biochemistry 43 11266–11274 [DOI] [PubMed] [Google Scholar]
- 15.Takahashi, Y., Edamatsu, M., and Toyoshima, Y. Y. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12865–12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kon, T., Mogami, T., Ohkura, R., Nishiura, M., and Sutoh, K. (2005) Nat. Struct. Mol. Biol. 12 513–519 [DOI] [PubMed] [Google Scholar]
- 17.Reck-Peterson, S. L., Yildiz, A., Carter, A. P., Gennerich, A., Zhang, N., and Vale, R. D. (2006) Cell 126 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reck-Peterson, S. L., and Vale, R. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Silvanovich, A., Li, M. G., Serr, M., Mische, S., and Hays, T. S. (2003) Mol. Biol. Cell 14 1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gennerich, A., Carter, A. P., Reck-Peterson, S. L., and Vale, R. D. (2007) Cell 131 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yildiz, A., Forkey, J. N., McKinney, S. A., Ha, T., Goldman, Y. E., and Selvin, P. R. (2003) Science 300 2061–2065 [DOI] [PubMed] [Google Scholar]
- 22.Schwacha, A., and Bell, S. P. (2001) Mol. Cell 8 1093–1104 [DOI] [PubMed] [Google Scholar]
- 23.Block, S. M., Goldstein, L. S., and Schnapp, B. J. (1990) Nature 348 348–352 [DOI] [PubMed] [Google Scholar]
- 24.Hattendorf, D. A., and Lindquist, S. L. (2002) EMBO J. 21 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross, J. L., Wallace, K., Shuman, H., Goldman, Y. E., and Holzbaur, E. L. (2006) Nat. Cell. Biol. 8 562–570 [DOI] [PubMed] [Google Scholar]
- 26.Svoboda, K., and Block, S. M. (1994) Cell 77 773–784 [DOI] [PubMed] [Google Scholar]
- 27.Farrell, C. M., Mackey, A. T., Klumpp, L. M., and Gilbert, S. P. (2002) J. Biol. Chem. 277 17079–17087 [DOI] [PubMed] [Google Scholar]
- 28.Yildiz, A., Tomishige, M., Vale, R. D., and Selvin, P. R. (2004) Science 303 676–678 [DOI] [PubMed] [Google Scholar]
- 29.Tomishige, M., Klopfenstein, D. R., and Vale, R. D. (2002) Science 297 2263–2267 [DOI] [PubMed] [Google Scholar]
- 30.Thorn, K. S., Ubersax, J. A., and Vale, R. D. (2000) J. Cell Biol. 151 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, X., Shaw, A., Bates, P. A., Newman, R. H., Gowen, B., Orlova, E., Gorman, M. A., Kondo, H., Dokurno, P., Lally, J., Leonard, G., Meyer, H., van Heel, M., and Freemont, P. S. (2000) Mol. Cell 6 1473–1484 [DOI] [PubMed] [Google Scholar]
- 32.Huyton, T., Pye, V. E., Briggs, L. C., Flynn, T. C., Beuron, F., Kondo, H., Ma, J., Zhang, X., and Freemont, P. S. (2003) J. Struct. Biol. 144 337–348 [DOI] [PubMed] [Google Scholar]
- 33.DeLaBarre, B., and Brunger, A. T. (2003) Nat. Struct. Biol. 10 856–863 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.