Abstract
The photoreceptor phosphodiesterase (PDE6) regulates the intracellular levels of the second messenger cGMP in the outer segments of cone and rod photoreceptor cells. PDE6 contains two regulatory GAF domains, of which one (GAF A) binds cGMP and regulates the activity of the PDE6 holoenzyme. To increase our understanding of this allosteric regulation mechanism, we present the 2.6Å crystal structure of the cGMP-bound GAF A domain of chicken cone PDE6. Nucleotide specificity appears to be provided in part by the orientation of Asn-116, which makes two hydrogen bonds to the guanine ring of cGMP but is not strictly conserved among PDE6 isoforms. The isolated PDE6C GAF A domain is monomeric and does not contain sufficient structural determinants to form a homodimer as found in full-length PDE6C. A highly conserved surface patch on GAF A indicates a potential binding site for the inhibitory subunit Pγ. NMR studies reveal that the apo-PDE6C GAF A domain is structured but adopts a significantly altered structural state indicating a large conformational change with rearrangement of secondary structure elements upon cGMP binding. The presented crystal structure will help to define the cGMP-dependent regulation mechanism of the PDE6 holoenzyme and its inhibition through Pγ binding.
Light detection by rod and cone photoreceptors of vertebrate retina is a complex signal transduction process that utilizes cGMP as an intracellular messenger in the conversion of photon stimulation into an electrical response. Upon light stimulation, the phototransduction cascade begins with activation of light-sensitive rhodopsin, which activates the G-protein transducin, whose Gα subunit binds and sequesters an inhibitory subunit Pγ of the photoreceptor phosphodiesterase (PDE6).4 This in turn leads to the activation of the catalytic subunit of PDE6 and hydrolysis of cGMP (1, 2). By lowering the intracellular cGMP levels, activated PDE6 causes hyperpolarization of the photoreceptor cell via cGMP-gated ion channels. cGMP levels are quickly restored by synthesis through calcium-sensitive photoreceptor guanylyl cyclases to re-establish the dark-adapted state. As a rapid turnover of cGMP is mandatory for effective visual function, PDE6 is well adapted for this purpose.
Rod photoreceptors are at least a hundred times more sensitive to light than cone photoreceptors. Nevertheless, cone photoreceptors are able to respond more quickly and terminate the light signal more rapidly (3). Structural differences between rod and cone isoforms of PDE6 are thought to be critical determinants of the photoreceptor responses as they display subunit organizations that differ between rod and cone isoforms. Rod PDE6 contains two distinct catalytic subunits (α and β) and two inhibitory Pγ subunits to form a heterotetramer (Pαβγ2), whereas cone PDE6 contains two identical cone-specific catalytic subunits α′ and two cone-specific Pγ subunits to form Pα′2γ2 (4). Most previous biochemical studies have utilized the heteromeric Pαβγ2 holoenzyme from rod photoreceptors. Less is known about the cone photoreceptor isoform of PDE6.
PDE6 is one of 11 vertebrate cyclic nucleotide PDE families. Of these, PDE2, -5, -6, -10, and -11 contain tandem GAF domains (GAF A and GAF B) that are located N-terminally to the more conserved catalytic domains. The acronym GAF is derived from the first identified GAF domain-containing protein families: mammalian cGMP-regulated PDEs, Anabaena adenylyl cyclases, and the Escherichia coli transcription factor FhlA (5). GAF domains are small molecule binding domains that are structurally related to Per-ARNT-Sim domains (6, 7). GAF domains are conserved over more than 3 billion years and function as signal transduction modules in light detection of plants (8), sodium sensing (9), and may in some cases even display enzymatic activity (10).
At this point, the details of the regulatory mechanism(s) and the exact roles of the GAF A and GAF B domains of PDE6 remain elusive. GAF A from PDE6C has been shown to bind cGMP (11), but whether cGMP binding activates the catalytic domain through an allosteric conformational change as reported for PDE2A and PDE5A (12, 13) remains to be determined. Binding of cGMP to rod PDE6 has been shown to increase the binding affinity of Pγ to the holoenzyme (14, 15). Cross-linking studies of rod Pγ to the catalytic PDE6 αβ-heterodimer revealed that Pγ binds in an extended manner, with the N-terminal, polycationic region binding in the vicinity of GAF A, and the C-terminal region in the vicinity of the catalytic domain (16) as confirmed by NMR (17). Pγ binding blocks catalytic activity and also increases the affinity of GAF A for cGMP. As no recombinant PDE6 holoenzyme has been successfully expressed, we have utilized the recombinantly expressed GAF A domain of chicken PDE6C in an attempt to gain insights into the structural nature and function of the allosteric cGMP-binding site and of the oligomerization of the PDE6 holoenzyme.
In this study, we report a 2.6 Å crystal structure of the GAF A domain from chicken cone PDE6. Although the overall fold is similar to other known GAF domains, a detailed comparison of the cGMP-binding pockets of PDE6C GAF A and PDE2A GAF B reveals significant differences, indicating that the PDE GAF domains have a variety of ways to bind the identical ligand, an observation that is consistent with their widespread occurrence as ligand-binding domains in signal transduction pathways (5, 18). The monomeric structure of GAF A stands in contrast to the dimeric crystal structure of PDE2A GAF AB, in which GAF A is part of the dimerization interface of a parallel dimer (19). Furthermore, combined NMR and circular dichroism studies reveal that the ligand-free form of PDE6C GAF A adopts a thermodynamically less stable structural state that is significantly altered compared with the cGMP-bound state.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification—cDNA for full-length chicken PDE6C was a generous gift from Dr. Susan Semple-Rowland (University of Florida). GAF A (residues 55–225 plus the C-terminal LE(H6) tag) was expressed in C41 E. coli cells (a derivative strain of BL21(DE3) (20)) and grown in LB media or M9 minimal media plus Se-Met for Se-Met-labeled protein (21) at 37 °C and 200 rpm to an A600 of ∼0.5–0.6, at which point isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.2 mm was added, and the temperature was lowered to 16 °C for 18–24 h.
Purification was performed at 4 °C. Four buffers were prepared: lysis (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 5 mm imidazole, 20% glycerol, 1 mm phenylmethylsulfonyl fluoride); wash-1 (50 mm Tris-HCl, pH 8.0, 15 mm imidazole, 20% glycerol, 400 mm NaCl, 2 mm MgCl2); wash-2 (same as wash-1, but 10 mm NaCl); and elution (50 mm Tris-HCl, pH 8.0, 250 mm imidazole, 20% glycerol, 10 mm NaCl, 2 mm MgCl2). After centrifugation at 3,000 × g, cells were resuspended in lysis buffer and lysed in a microfluidizer at 10,000 p.s.i. After centrifugation, the lysate was applied to a nickel-nitrilotriacetic acid gravity column (15 ml). The column was washed with wash-1 and wash-2 buffers. Protein was eluted with elution buffer and then applied to 25 ml of epoxy-Sepharose cGMP affinity resin (column size of 25 mm) and washed with high salt buffer (0.5 m NaCl, 20 mm Tris-HCl, pH 7.5, 0.1 mm EDTA, 1 mm β-mercaptoethanol) and low salt buffer (same as high salt buffer but with 0.2 m NaCl). The protein was eluted with low salt buffer (plus 1 mm cGMP (Sigma)). Usual yields of purified protein were ∼5 mg from 9 liters of LB culture. The protein was determined to be >95% pure by SDS-PAGE.
Crystallization, Structure Determination, and Refinement—The purified GAF A domain was concentrated in Centricon 30 units (Millipore) to 5 mg/ml and exchanged into 20 mm Tris-HCl, pH 7.5, 20 mm NaCl, 0.1 mm EDTA, 1 mm β-mercaptoethanol, 1 mm cGMP. Crystals were grown at room temperature in sitting drops with 0.5 ml of precipitant (1.5–1.8 m ammonium sulfate, 0.1 m sodium acetate, pH 4.5–5.0) in the reservoir and equal volumes (2–10 μl) of protein and precipitant in the drop. Crystals grew to maximum dimensions of ∼600 × 300 × 250 μm within several weeks and were then frozen in liquid nitrogen after soaking for 2–3 min in cryo-buffer (20% v/v glycerol, 1.5 or 1.6 m ammonium sulfate, 100 mm sodium acetate, pH 4.5 or 5.0, 1 mm cGMP, 1 mm β-mercaptoethanol, 0.1 mm EDTA). A 2.5 Å SAD peak Se-Met data set was collected from a crystal in cryo-buffer (pH 5.0, 1.6 m ammonium sulfate) at 12,662.5 eV (0.97912 Å) (1 eV = 1.602 × 10–19 J) at the National Synchrotron Light Source beamline X4A. Nine of the 10 selenium atoms (five seleniums for each of two monomers in asymmetric unit) were determined with the program SOLVE (22). Two sets of four selenium positions each were found related in the density by non-point group symmetry and used for averaging and solvent flattening in a 3.0 Å RESOLVE map (23). A model was built manually using the COOT program (24) and refined using a 2.6 Å native sulfomethionine data set collected from a crystal in cryo-buffer (pH 4.5, 1.5 m ammonium sulfate) with the program CNS, using restrained individual B factors (25). Further refinement was performed in REFMAC using TLS refinement and riding hydrogens (26). The final model consists of residues 55–225 with an Rfree factor of 25.7% and Rwork factor of 22.1%. The space group was P3221, and 87.6% of the residues in the final model were in the most favored area of the Ramachandran plot (additional structural statistics are provided in supplemental Table S1). The atomic coordinates are deposited in the Protein Data Bank (code 3dba).
Preparation of Isotopically Labeled PDE6 GAF A and NMR Experiments—Uniformly 15N-labeled PDE6C GAF A for NMR studies was grown in M9 minimal media with 15N-NH4Cl (Cambridge Isotope Laboratories). Cell lysis and nickel affinity column chromatography were carried out as described above. The nickel column eluate was concentrated and injected onto a Superdex 75 column. A single peak was eluted with 150 mm NaCl, 25 mm sodium phosphate, pH 7.0, 0.1 mm EDTA. Upon fractionation, dithiothreitol to a final concentration of 1 mm was added. NMR samples were concentrated to ∼150 μm, and 10% D2O was added. cGMP to a final concentration of 1 mm was added for the spectrum of the cGMP-bound form. 1H,15N HSQC spectra were collected at 20 °C on a Varian 800-MHz INOVA spectrometer equipped with 1H-13C,15N-cold probe. All spectra were processed and analyzed using NMRDraw (27) and NMRView (28).
Determination of the Rotational Correlation Time—To determine the rotational correlation time of 0.7 mm PDE6C GAF A in the presence of cGMP, we measured 15N spin relaxation parameters (R1, R2) and the steady-state heteronuclear 1H,15N NOEs using standard pulse sequences (29) at 37 °C on a Bruker 500 MHz DMX spectrometer. 15N-T1 relaxation delays were 10, 50, 100, 200, 400, 600, 800, and 1000 ms (underlined delays were duplicated). 15N-T2 relaxation delays were 17.2, 34.4, 51.7, 69.0, 86.2, 103.5, and 120.7 ms (underlined delays were duplicated). Only well resolved resonances were used for the dynamics analysis (85 resonances) by NMRView (28) and Modelfree 4.15 (30).
Circular Dichroism Studies—Circular dichroism studies were performed on a 62A DS spectrometer (Aviv Biomedical). Spectra were collected with 10–20 μm PDE6C GAF A in 100 mm NaCl, 25 mm sodium phosphate, pH 7.0, 0.1 mm dithiothreitol in the absence and presence of 0.5 mm cGMP (cuvette path length, 0.1 cm). To monitor the temperature-dependent denaturation through thermal scans, ellipticity at the α-helical signal of 222 nm as a function of temperature was measured in the range of 10–85 °C with steps every 3 °C and a 60-s equilibration time at each step.
RESULTS
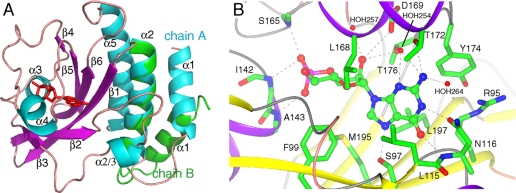
Overall GAF Domain Fold—The crystal structure of the chicken PDE6C GAF A domain, determined at a resolution of 2.6 Å, displays a typical GAF domain fold with a six-stranded antiparallel β-sheet arranged in the strand order 3-2-1-6-5-4 (Fig. 1A). The β-sheet provides the floor, and the two helices α3 and α4 comprise the roof of a deeply buried cGMP-binding pocket. The helical dipole of α3, which is between β3 and β4 in the primary structure, points toward the phosphate group of the bound cGMP molecule. α4, which is between β4 and β5 in sequence, sits atop the cGMP molecule. Helices α1 and α2 precede the first β-strand and, together with α5, form a three-helix bundle that packs against the β-sheet on the opposite side of the cGMP-binding pocket. Strands β2 and β3 are connected by an 18-residue loop that includes a short α-helix (α2/3). Excluding helix α1, both monomers in the asymmetric unit superimpose well with a root mean square deviation of 0.5 Å over 149 equivalent C-α atoms.
FIGURE 1.
A, overall structure of PDE6C GAF A. Ribbon diagram of chain A. Secondary structure elements are labeled. α-Helices are shown in cyan; β-strands in purple, and loops in tan. cGMP is shown as red sticks. Chain B was superimposed (residues 92–225) on chain A, and the first two helices (residues 55–91) from chain B are shown in green. This figure was generated with PyMOL (DeLano Scientific). B, cGMP-binding site of chicken PDE6C GAF A. The backbone is shown as a ribbon and side chains contacting the cGMP as sticks. Shown are five side chain contacts (Ser-97, Asn-116, Asp-169, Thr-172, and Thr-176), four backbone contacts (Asn-116, Ile-142, Ala-143, and Ser-165), five hydrophobic contacts (Phe-99, Leu-115, Tyr-174, Leu-168, and Leu-197), and three contacts with waters (HOH-254, -257, and -264). Arg-95 and Met-195 do not make contacts with cGMP but are discussed in the text. This figure was generated with CCP4MG (47, 48).
The crystal structure of chicken PDE6C GAF A indicates that GAF A does not form a homodimer. The asymmetric unit of the crystal contains two monomers that do not dimerize with each other or with monomers related by lattice symmetry. Notably, helix α1 is in a different orientation in the two monomers (Fig. 1A) because of a distinct packing environment of each monomer and lack of a dimerization interface to restrain the helix. To confirm the monomeric state in solution, we determined the rotational correlation time τc of cGMP-bound 15N-PDE6C GAF A through 15N relaxation measurements by NMR. The calculated value for τc of 12.4 ns at 37 °C is in a range typical for proteins of similar size (∼20 kDa), therefore confirming that GAF A is monomeric in solution at millimolar concentrations. Apo and cGMP-bound PDE6C GAF A elute at the same elution volume and have identical elution peak shape in analytical size exclusion chromatograms (supplemental Fig. S1). Together, the data indicate that GAF A is intrinsically a monomer and that cGMP binding does not alter the oligomeric state of GAF A.
cGMP-binding Site of PDE6C GAF A—Cyclic GMP is bound in the anti-conformation (Fig. 1B), similar to cGMP in PDE2A GAF B (19) and cAMP in the tandem GAF domains of the cyanobacterial adenylyl cyclase cyaB2 (31) and PDE10A GAF B (32). Similar to the previously solved cyclic nucleotide-binding GAF domains, nearly the entire 493 Å2 of solvent-accessible surface area of the nucleotide is buried in PDE6C, indicating that the GAF domain is in a closed state that forms upon cGMP binding.
Because of a lower overall B factor, we use chain A to describe the binding pocket. The electron density map shows that the 3′,5′-cyclic phosphate group is in the chair conformation (supplemental Fig. S2). The N-terminal end of helix α3 points toward the phosphate group, bringing the negatively charged phosphate group adjacent to the partial positive charge of the helix dipole. The guanine ring makes several contacts to the protein and is well defined through a series of protein-ligand interactions (Fig. 1B and supplemental Fig. S3). Asn-116 provides two important cGMP-specific protein-ligand hydrogen bonds. The carbonyl oxygen of its side chain makes a 2.7-Å hydrogen bond to the protonated N-1 of the guanine ring, and its backbone amide makes a 2.9 Å hydrogen bond to the O-6 carbonyl oxygen of cGMP. Furthermore, Asn-116 is locked into position by a 3.0-Å salt bridge between the partial negative charge of its carbonyl oxygen and the guanidine group of Arg-95. Finally, Asn-116 may provide yet another specificity determinant through a water-mediated hydrogen bond to the N-2 amino group (via HOH-264). Another hydrogen bond to the N-2 amino group of the guanine ring is provided by the side chain of Thr-172, which is 3.4 Å away. This side chain also provides two water-mediated hydrogen bonds (via HOH-254) to the N-2 amino group and the N-3 ring nitrogen. Ser-97 provides an additional 2.7 Å hydrogen bond to N-7 of the guanine ring of cGMP. In addition to the described protein-ligand hydrogen bond network, hydrophobic interactions play an important role in the definition of the cGMP-binding pocket and nucleotide orientation. The guanine ring is sandwiched between the side chains from Phe-99, Leu-115, Leu-168, Tyr-174, and Leu-197.
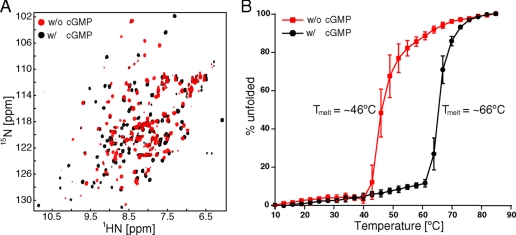
Conformational Change upon Binding of cGMP—We are able to purify and concentrate the recombinant PDE6C GAF A domain in the absence of cGMP to millimolar concentrations but have been unsuccessful in producing diffraction-quality crystals. Therefore, we utilized two-dimensional NMR spectroscopy to investigate the nature of the conformational change between the ligand-free and cGMP-bound forms. An overlay of 1H,15N HSQC spectra of 15N-labeled PDE6C GAF A in the absence and presence of cGMP (Fig. 2A) reveals that PDE6C GAF A is folded in both cases. However, the two states adopt significantly different conformations, as an overwhelming majority of the amide resonances is perturbed by more than a line width. The resonances in the spectrum of apo-PDE6C GAF A vary widely in their intensities, indicative of exchange among multiple states, with a subset of peaks broadened beyond detection.
FIGURE 2.
A, 1H,15N HSQC spectral overlay of PDE6C GAF A in the absence (red) and presence (black) of cGMP. B, percent unfolded plotted as a function of temperature in °C for PDE6C GAF A in the absence (red) and presence (black) of cGMP. Ellipticity at 222 nm was measured, and values at 10 and 85 °C were set arbitrarily as 0% unfolded and 100% unfolded, respectively. Values of the average from three independent experiments are shown. Error bars indicate standard deviation.
Circular dichroism spectra of both states of PDE6C GAF A are virtually indistinguishable and both have a strong signal for α-helical secondary structure, indicated by minima at 208 and 218 nm. The negligible difference in the spectra suggests that the two states do not differ significantly in their secondary structural element content. To assess protein stability, we measured the ellipticity at the α-helical signal of 222 nm as a function of temperature (Fig. 2B). The observed 20 °C increase in the melting temperature upon addition of cGMP indicates considerable thermodynamic stabilization of PDE6C GAF A through cGMP binding. This supports the hypothesis that both states differ in their global tertiary structure rather than through local unfolding of secondary structure elements. Nevertheless, the conformational change does not lead to an overall altered hydrodynamic behavior as analytical size exclusion chromatography reveals no observable difference in peak position or shape (supplemental Fig. S1).
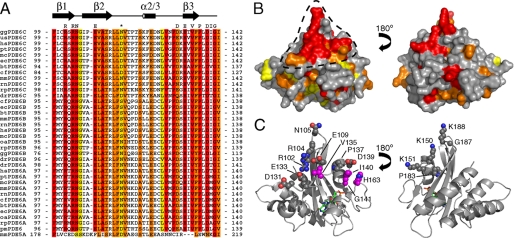
Conserved Interaction Surface—An alignment of PDE6 GAF A sequences from cone and rod photoreceptors reveals that stretches of the sequences between β1 and α3, which encompass β2, α2/3, and β3, are strongly conserved with a large number of strictly conserved residues (Fig. 3A). When plotted onto the crystal structure of PDE6C GAF A, several of these conserved residues form a contiguous solvent-accessible surface patch on the PDE6C GAF A structure (Fig. 3B). This surface patch contains several charged residues. Arg-102 and Arg-104 are surrounded by Glu-109, Asp-131, and Glu-133. Val-135 and Pro-137 extend the surface toward another negatively charged residue, Asp-139 (Fig. 3C, left panel). Ile-140, Gly-141, and His-163, which all have been previously proposed to be in the vicinity of the Pγ-binding site (33, 34), are also within the conserved surface or in close proximity to it (highlighted in magenta in Fig. 3C). An elongated surface with numerous negatively charged and hydrophobic residues can be drawn from Asp-131 to His-163 and may be responsible for the interaction with the polycationic region of Pγ. A second smaller conserved surface patch is located on the opposite side of GAF A. It contains several positively charged amino acids (Fig. 3C, right panel) indicating that it is less likely to be the interaction surface for Pγ.
FIGURE 3.
A, sequence alignment of PDE6A, -6B, and -6C from different species. The region between β1 and α3 is shown. Red, identical; orange, strongly similar; yellow, weakly similar. Species are as follows: gg, Gallus gallus (chicken); us, Uta stansburiana (common side-blotched lizard); hs, Homo sapiens (human); pt, Pan troglodytes (chimpanzee); mc, Macaca mulatto (rhesus monkey); ec, Equus caballus (Przewalski's horse); bt = Bos taurus (cattle); cf, Canis familiaris (dog); mm, Mus musculus (mouse); rn, Rattus norvegicus (rat); rp, Rana pipiens (leopard frog); dr, Danio rerio (zebrafish); oa, Ornithorhynchus anatinus (platypus); pm, Petromyzon marinus (sea lamprey). The alignment was generated with ClustalW (49). The asterisk indicates the discussed residue Asn-116. Residues highlighted above alignment form a conserved surface patch and are shown as spheres in C. Sequence of mouse PDE5A is shown for comparison. B, surface representation of PDE6C GAF A. Colors are based on sequence conservation as in A. Discussed surface patch is indicated by black dashed line. C, ribbon representation of PDE6C GAF A. Side chain atoms of residues, which are part of the conserved surface in B, are shown as spheres with nitrogen atoms in blue and oxygen atoms in red. Residues that were previously implicated in Pγ binding are highlighted with magenta-colored carbon atoms. cGMP is shown as sticks with green carbon atoms.
DISCUSSION
Comparison of Overall GAF Domain Fold—The overall fold of the PDE6C GAF A domain is very similar to the topology of previously characterized cyclic nucleotide-binding GAF domains. The largest difference between the published GAF domain structures is the length and sequence of the β2-β3 loop, as was previously discussed (31). This stretch contains a short helical turn in PDE6C, termed α2/3, which is positioned to stabilize Asn-116, a critical cGMP-contacting residue. In particular, the side chains of Ser-121 and Asn-126 make hydrogen bonds to the backbone carbonyl of Asn-116 and may help to lock its side and main chains into position to provide the nucleotide-selective hydrogen bonds. A solution structure of PDE5A GAF A revealed that the cGMP-selective GAF domain of PDE5A contains a similar helical turn (35).
Comparison of cGMP-binding Pockets of PDE6C and PDE2A—Systematic alanine mutations of binding residues of PDE2A GAF B revealed that three guanine-binding side chains (Asp-439, Phe-438, and Thr-488) were required for the ∼30-fold higher affinity for cGMP over cAMP (36). The side chain of Asp-439 is hydrogen-bound to the guanine N-1, and its backbone amide is a hydrogen bond donor to the O-6 carbonyl of the guanine. Phe-438 is base-stacked with the guanine ring, whereas Thr-488 contacts the guanine N-2 via a water-mediated hydrogen bond. In PDE6C GAF A, these residues correspond to Asn-116, Leu-115, and Thr-172.
Asn-116 of PDE6C, the equivalent residue to Asp-439 of PDE2A, provides two protein-ligand hydrogen bonds (Fig. 1B) and therefore serves as a negative determinant against cAMP binding as both hydrogen bonds cannot be satisfied by the adenine ring. Surprisingly, a sequence alignment of GAF A domain sequences from PDE6A, -B, and -C of different species reveals that Asn-116 is variable within the PDE6 gene family and can be an Asn, an Asp, or a Ser (Fig. 3A). As all three side chains have oxygen atoms capable of serving as hydrogen bond acceptors, it is likely that Ser, Asn, and Asp each make similar side chain contacts to N-1 of the guanine ring. All other characterized cGMP-binding GAF domains (PDE2A GAF B, PDE5A GAF A, and PDE11A GAF A) have an Asp at the analogous position.
Leu-115 of PDE6C is equivalent to Phe-438 in PDE2A. Whereas Phe-438 likely provides aromatic π -π -base stacking as it is aligned in parallel with the guanine ring, Leu-115 of PDE6C provides hydrophobic interactions instead. The equivalent amino acid in rod PDE6 (isoforms A and B) is a Phe, whereas it is a Leu in most PDE6C sequences (Fig. 3A).
Thr-172 of PDE6C makes a direct hydrogen bond to the guanine N-2, whereas Thr-488, the equivalent residue in PDE2A, is too far for a direct hydrogen bond (3.8 Å) and makes a water-mediated hydrogen bond instead. Other side chain contacts with the cGMP molecule are similar in PDE6C and PDE2A.
Structural Determinants of Affinity and Specificity—GAF domains from different PDEs bind cGMP and cAMP with a wide variety of affinities and specificities. The tandem GAF domains of PDE2A and PDE5A have low nanomolar affinities for cGMP binding but vary in their selectivity against cAMP from 30-fold (PDE2A (36)) to several thousandfold (PDE5A (35, 37)). The tandem GAF domains from chicken PDE6C have a similar high affinity for cGMP (KD of ∼10 nm) but discriminate over 3 million-fold against cAMP (KD of ∼35 mm) (38). These binding constants imply that the cGMP selectivity of PDE GAF domains is achieved largely by modulating the strength of cAMP binding. The more selective a given PDE GAF domain is, the weaker its affinity for cAMP.
As there are no structures available for the cAMP-bound form of any cGMP-selective GAF domain, we can only infer reasons for the lower cAMP affinities. The GAF domains of PDE2A, PDE6C, and PDE5A all use a residue at the position analogous to Asn-116 in PDE6C to make cGMP-specific contacts. We propose that the difference in cAMP affinity (and therefore, selectivity) between PDE6C and PDE2A stems from the ability or inability of Asp-439/Asn-116 to swing out and make room to accommodate cAMP. Consistent with this hypothesis, the equivalent residue in the cAMP-binding PDE10A GAF B is an Asp whose side chain points away from the binding pocket allowing an Arg to make nucleotide-specific contact with cAMP. As noted above, Asn-116 in PDE6C is locked into position by its surrounding residues and helix α2/3. In contrast, the shorter sequence between strands β2 and β3 in PDE2A GAF B does not allow for formation of the α2/3 helix, and there are no analogous contacts with Asp-439, thereby allowing the residue to swing out and cAMP to bind more easily. PDE5A with its thousandfold cGMP preference also contains a α2/3 helix in similar orientation (35).
cGMP-dependent Conformational Change—To date, no high resolution structure of a cyclic nucleotide-binding GAF domain in the absence of its ligand has been reported. The NMR spectra presented here provide evidence for a change in both the conformation and flexibility of PDE6C GAF A upon cGMP binding. Our data indicate that the apo-GAF A domain adopts neither a preformed pocket nor an unstructured molten globule formation but rather a dynamic domain with a structured core, suggesting the existence of two distinct states.
Recently, our laboratories investigated the cGMP-induced conformational change of GAF A from PDE5A (35). In contrast to apo-GAF A of PDE6C, apo-GAF A of PDE5A is not stable in solution. The presence of GAF B stabilizes GAF A of PDE5A and enabled us to monitor the cGMP-dependent conformational change by NMR. Whereas apo-GAF A of PDE5A appears to sample multiple dynamic states, which results in practically no observed NMR resonances from GAF A, the core of apo-GAF A from PDE6C is stably structured and therefore appears to exist in a single state (Fig. 2A). This indicates that subtle structural and dynamic differences in the magnitude of the cGMP-dependent conformational changes between GAF domains may exist. These may account for the large variety of cyclic nucleotide affinities and different time scales of allosteric regulation. However, in both cases cGMP causes a large scale induced fit of the GAF domain upon binding.
Dimerization—All PDEs are known to form dimers although the functional significance of PDE dimerization remains elusive. The monomeric crystal structures of the catalytic domains from PDE2A (39), PDE5A (40, 41), and PDE10A (42) imply that the regulatory N-terminal region with tandem GAF domains provide dimerization contacts. Our crystal structure reveals that GAF A of PDE6C is not intrinsically dimeric, in contrast to what EM studies suggest for the full-length PDE6AB enzyme (43, 44). Although helix α1 is in approximately the same orientation in chain A of our PDE6C GAF A crystal structure and PDE2A GAF A (19) (supplemental Fig. S4), it is not involved in a similar dimerization interface as indicated by the lack of crystal packing contacts to restrain the conformation of the helix (Fig. 1A) and the total correlation time of 12.4 ns in solution. A previous report identified α1 as essential for affinity and selectivity of heterodimerization in rod PDE6AB (45). The fact that the presence of α1 is not sufficient for homodimerization of our cone PDE6C GAF A construct indicates that dimerization of the full-length enzyme is mediated by multiple regions and that additional sequences directly N- and C-terminal to our recombinant GAF A construct (e.g. residues 1–54 or GAF B) are necessary for dimerization of PDE6C. It is also likely that dimerization contacts vary between the heterodimeric rod PDE6AB and the homodimeric cone PDE6C. The diversity of dimerization modes of the GAF-containing PDEs indicates that generalizations about the dimerization properties of individual GAF domains cannot be made without high resolution structural data from each PDE.
Pγ-binding Site—Activity of the PDE6 holoenzyme is regulated by the inhibitory Pγ subunit, a small protein found in both rod and cone photoreceptor cells. The recently determined solution structure of bovine rod Pγ revealed a loose fold with transient structural features, characteristic of an intrinsically disordered protein (17). Pγ binds in an extended manner to rod PDE6AB, and residues within the polycationic region (residues 21–45) of Pγ bind in the vicinity to GAF A as determined by cross-linking studies (16). In particular, Met-138 and Gly-139 of bovine rod PDE6A were found to cross-link to the polycationic region (34) (residues correspond to Ile-140 and Gly-141 of chicken PDE6C). Mutations of Val-130 and His-158 (residues correspond to Val-135 and His-163 of chicken PDE6C) considerably impaired the interaction with Pγ (33). In the crystal structure of PDE6C, these residues are located within or in proximity to the highly conserved surface patch (Fig. 3C, left panel). The abundance of negatively charged residues in this region is suitable for an ionic interaction with the highly conserved polycationic region of Pγ.Pγ reportedly prefers the cGMP-bound state of GAF A (14, 46), but none of the residues within the surface patch are in close proximity to the binding pocket suggesting that the mechanism of positive cooperativity of cGMP and Pγ binding likely involves conformational rearrangements of secondary structure elements. Alternatively, the conserved surface may be involved in other intra- and intermolecular interactions.
Concluding Remarks—The crystal structure of PDE6C GAF A confirms in atomic detail that GAF A is the cGMP-binding site of PDE6. The structural data in this study and the observed large conformational change provide important clues to the molecular basis of cGMP binding and allosteric regulation of the PDE6 enzyme. The atomic details of its crystal structure, the small size of the isolated GAF domain, and the large scale recombinant expression and purification (with and without cGMP) make PDE6C GAF A amenable to study the interaction of PDE6 with Pγ and other binding partners.
Supplementary Material
Acknowledgments
X-ray diffraction data were collected at the National Synchrotron Light Source beamline X4A in the Brookhaven National Laboratories. NMR experiments were performed, in part, at the Environmental Molecular Science Laboratories (a national scientific user facility sponsored by the Department of Energy Biological and Environmental Research) located at Pacific Northwest National Laboratories and operated for Department of Energy by Battelle. We thank Dr. Peter Brzovic for collecting NMR spectra and Simeon Andrews for assisting with the initial characterization of the cGMP-dependent conformational change by NMR.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK21723 (to J. A. B.) and P01 HL44948 (to J. A. B. and R. E. K.) and by a Boehringer Ingelheim Fonds Ph.D. scholarship (to C. C. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Discussion,” additional references, Table S1, and Figs. S1–S4.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 3dba) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: PDE6, photoreceptor phosphodiesterase; PDE, 3′,5′-cyclic nucleotide phosphodiesterase; HSQC, heteronuclear single quantum correlation; SeMet, selenomethionine.
References
- 1.Cote, R. H. (2007) in Cyclic Nucleotide Phosphodiesterases in Health and Disease (Beavo, J. A., Francis, S. H., and Houslay, M. D., eds) pp. 165–193, CRC Press, Inc., Boca Raton, FL
- 2.Arshavsky, V. Y., Lamb, T. D., and Pugh, E. N., Jr. (2002) Annu Rev. Physiol. 64 153–187 [DOI] [PubMed] [Google Scholar]
- 3.Pugh, E. N., Jr., and Cobbs, W. H. (1986) Vision Res. 26 1613–1643 [DOI] [PubMed] [Google Scholar]
- 4.Artemyev, N. O., Surendran, R., Lee, J. C., and Hamm, H. E. (1996) J. Biol. Chem. 271 25382–25388 [DOI] [PubMed] [Google Scholar]
- 5.Aravind, L., and Ponting, C. P. (1997) Trends Biochem. Sci. 22 458–459 [DOI] [PubMed] [Google Scholar]
- 6.Martinez, S. E., Beavo, J. A., and Hol, W. G. (2002) Mol. Interv. 2 317–323 [DOI] [PubMed] [Google Scholar]
- 7.Zoraghi, R., Corbin, J. D., and Francis, S. H. (2004) Mol. Pharmacol. 65 267–278 [DOI] [PubMed] [Google Scholar]
- 8.Wagner, J. R., Brunzelle, J. S., Forest, K. T., and Vierstra, R. D. (2005) Nature 438 325–331 [DOI] [PubMed] [Google Scholar]
- 9.Cann, M. (2007) Mol. Microbiol. 64 461–472 [DOI] [PubMed] [Google Scholar]
- 10.Lin, Z., Johnson, L. C., Weissbach, H., Brot, N., Lively, M. O., and Lowther, W. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9597–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie, P. G., and Beavo, J. A. (1988) J. Biol. Chem. 263 8133–8141 [PubMed] [Google Scholar]
- 12.Martins, T. J., Mumby, M. C., and Beavo, J. A. (1982) J. Biol. Chem. 257 1973–1979 [PubMed] [Google Scholar]
- 13.Corbin, J. D., Blount, M. A., Weeks, J. L., 2nd, Beasley, A., Kuhn, K. P., Ho, Y. S., Saidi, L. F., Hurley, J. H., Kotera, J., and Francis, S. H. (2003) Mol. Pharmacol. 63 1364–1372 [DOI] [PubMed] [Google Scholar]
- 14.Cote, R. H., Bownds, M. D., and Arshavsky, V. Y. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 4845–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki, M., Li, N., Bondarenko, V. A., Yamazaki, R. K., Baehr, W., and Yamazaki, A. (2002) J. Biol. Chem. 277 40675–40686 [DOI] [PubMed] [Google Scholar]
- 16.Guo, L. W., Muradov, H., Hajipour, A. R., Sievert, M. K., Artemyev, N. O., and Ruoho, A. E. (2006) J. Biol. Chem. 281 15412–15422 [DOI] [PubMed] [Google Scholar]
- 17.Song, J., Guo, L. W., Muradov, H., Artemyev, N. O., Ruoho, A. E., and Markley, J. L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anantharaman, V., Koonin, E. V., and Aravind, L. (2001) J. Mol. Biol. 307 1271–1292 [DOI] [PubMed] [Google Scholar]
- 19.Martinez, S. E., Wu, A. Y., Glavas, N. A., Tang, X. B., Turley, S., Hol, W. G., and Beavo, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miroux, B., and Walker, J. E. (1996) J. Mol. Biol. 260 289–298 [DOI] [PubMed] [Google Scholar]
- 21.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L., and Clardy, J. (1993) J. Mol. Biol. 229 105–124 [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger, T. C. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905–921 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 27.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277–293 [DOI] [PubMed] [Google Scholar]
- 28.Johnson, B. A., and Blevins, R. A. (1994) J. Biomol. NMR 4 603–614 [DOI] [PubMed] [Google Scholar]
- 29.Farrow, N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., Forman-Kay, J. D., and Kay, L. E. (1994) Biochemistry 33 5984–6003 [DOI] [PubMed] [Google Scholar]
- 30.Mandel, A. M., Akke, M., and Palmer, A. G., III (1995) J. Mol. Biol. 246 144–163 [DOI] [PubMed] [Google Scholar]
- 31.Martinez, S. E., Bruder, S., Schultz, A., Zheng, N., Schultz, J. E., Beavo, J. A., and Linder, J. U. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3082–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handa, N., Mizohata, E., Kishishita, S., Toyama, M., Morita, S., Uchikubo-Kamo, T., Akasaka, R., Omori, K., Kotera, J., Terada, T., Shirouzu, M., and Yokoyama, S. (2008) J. Biol. Chem. 283 19657–19664 [DOI] [PubMed] [Google Scholar]
- 33.Muradov, H., Boyd, K. K., and Artemyev, N. O. (2004) Vision Res. 44 2437–2444 [DOI] [PubMed] [Google Scholar]
- 34.Muradov, K. G., Granovsky, A. E., Schey, K. L., and Artemyev, N. O. (2002) Biochemistry 41 3884–3890 [DOI] [PubMed] [Google Scholar]
- 35.Heikaus, C. C., Stout, J. R., Sekharan, M. R., Eakin, C. M., Rajagopal, P., Brzovic, P. S., Beavo, J. A., and Klevit, R. E. (2008) J. Biol. Chem. 283 22749–22759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, A. Y., Tang, X. B., Martinez, S. E., Ikeda, K., and Beavo, J. A. (2004) J. Biol. Chem. 279 37928–37938 [DOI] [PubMed] [Google Scholar]
- 37.Zoraghi, R., Bessay, E. P., Corbin, J. D., and Francis, S. H. (2005) J. Biol. Chem. 280 12051–12063 [DOI] [PubMed] [Google Scholar]
- 38.Huang, D., Hinds, T. R., Martinez, S. E., Doneanu, C., and Beavo, J. A. (2004) J. Biol. Chem. 279 48143–48151 [DOI] [PubMed] [Google Scholar]
- 39.Iffland, A., Kohls, D., Low, S., Luan, J., Zhang, Y., Kothe, M., Cao, Q., Kamath, A. V., Ding, Y. H., and Ellenberger, T. (2005) Biochemistry 44 8312–8325 [DOI] [PubMed] [Google Scholar]
- 40.Wang, H., Liu, Y., Huai, Q., Cai, J., Zoraghi, R., Francis, S. H., Corbin, J. D., Robinson, H., Xin, Z., Lin, G., and Ke, H. (2006) J. Biol. Chem. 281 21469–21479 [DOI] [PubMed] [Google Scholar]
- 41.Sung, B. J., Hwang, K. Y., Jeon, Y. H., Lee, J. I., Heo, Y. S., Kim, J. H., Moon, J., Yoon, J. M., Hyun, Y. L., Kim, E., Eum, S. J., Park, S. Y., Lee, J. O., Lee, T. G., Ro, S., and Cho, J. M. (2003) Nature 425 98–102 [DOI] [PubMed] [Google Scholar]
- 42.Chappie, T. A., Humphrey, J. M., Allen, M. P., Estep, K. G., Fox, C. B., Lebel, L. A., Liras, S., Marr, E. S., Menniti, F. S., Pandit, J., Schmidt, C. J., Tu, M., Williams, R. D., and Yang, F. V. (2007) J. Med. Chem. 50 182–185 [DOI] [PubMed] [Google Scholar]
- 43.Kajimura, N., Yamazaki, M., Morikawa, K., Yamazaki, A., and Mayanagi, K. (2002) J. Struct. Biol. 139 27–38 [DOI] [PubMed] [Google Scholar]
- 44.Kameni Tcheudji, J. F., Lebeau, L., Virmaux, N., Maftei, C. G., Cote, R. H., Lugnier, C., and Schultz, P. (2001) J. Mol. Biol. 310 781–791 [DOI] [PubMed] [Google Scholar]
- 45.Muradov, K. G., Boyd, K. K., Martinez, S. E., Beavo, J. A., and Artemyev, N. O. (2003) J. Biol. Chem. 278 10594–10601 [DOI] [PubMed] [Google Scholar]
- 46.Mou, H., and Cote, R. H. (2001) J. Biol. Chem. 276 27527–27534 [DOI] [PubMed] [Google Scholar]
- 47.Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K., and Noble, M. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1955–1957 [DOI] [PubMed] [Google Scholar]
- 48.Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A., and Noble, M. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2288–2294 [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.