Abstract
The gelsolin family of proteins is a major class of actin regulatory proteins that sever, cap, and nucleate actin filaments in a calcium-dependent manner and are involved in various cellular processes. Typically, gelsolin-related proteins have three or six repeats of gelsolin-like (G) domain, and each domain plays a distinct role in severing, capping, and nucleation. The Caenorhabditis elegans gelsolin-like protein-1 (gsnl-1) gene encodes an unconventional gelsolin-related protein with four G domains. Sequence alignment suggests that GSNL-1 lacks two G domains that are equivalent to fourth and fifth G domains of gelsolin. In vitro, GSNL-1 severed actin filaments and capped the barbed end in a calcium-dependent manner. However, unlike gelsolin, GSNL-1 remained bound to the side of F-actin with a submicromolar affinity and did not nucleate actin polymerization, although it bound to G-actin with high affinity. These results indicate that GSNL-1 is a novel member of the gelsolin family of actin regulatory proteins and provide new insight into functional diversity and evolution of gelsolin-related proteins.
Actin cytoskeleton is essential for a wide variety of cellular functions, such as cell motility, phagocytosis, cell division, and muscle contraction. A tremendous number of molecules regulate the function of actin cytoskeleton. Regulation of polymerization and depolymerization of actin is crucial for the function of the actin cytoskeleton. Gelsolin-related proteins and actin depolymerizing factor (ADF)2/cofilin are the two major classes of actin filament-severing proteins that enhance actin filament turnover by severing and depolymerizing actin filaments and are involved in a number of cell biological processes (for reviews, see Refs. 1 and 2).
The actin cytoskeleton is highly differentiated into sarcomeric structures in striated muscle, and polymerization and depolymerization of actin must be precisely regulated in order to assemble and maintain striated myofibrils. Functional significance of ADF/cofilin in organized assembly of actin filaments in striated muscle has been demonstrated (1). A muscle-specific ADF/cofilin isoform, M-cofilin/cofilin-2, is expressed in mammalian striated muscle (3, 4). A mutation in the human cofilin-2 gene causes nemaline myopathy (5). In the nematode Caenorhabditis elegans, UNC-60B, a muscle-specific ADF/co-filin isoform, is required for organized assembly of actin filaments in body wall muscle (6, 7) and cooperates with UNC-78/actin-interacting protein 1 to promote actin filament disassembly (8, 9). Gelsolin-related proteins are also expressed in striated muscle, but their function in muscle is not clearly understood. Gelsolin localizes to the thin filaments in vertebrate striated muscle (10, 11) and ascidian muscle (12, 13), suggesting that actin severing activity of gelsolin is inhibited or that actin filaments are protected from severing. In Drosophila melanogaster and C. elegans, mutations of Flightless-1, a gelsolin-related protein with N-terminal leucine-rich repeats (14), cause disorganization of actin filaments in striated muscle (15, 16). However, significance of actin severing activity of Flightless-1 has not been demonstrated.
Gelsolin strongly severs actin filaments, caps the barbed ends and nucleates actin polymerization in a calcium-dependent manner (17–19). The gelsolin family proteins have repeats of homologous domains of 100–120 amino acids, which are designated as gelsolin-like (G) domains. Many gelsolin-related proteins, including gelsolin, villin, Flightless-1, and adseverin/scinderin, have six G domains, whereas Physarum fragmin, Dictyostelium severin, and vertebrate CapG have three G domains (see Refs. 1 and 17–19). Thus, it has been speculated that gelsolin-related proteins with six G domains have evolved from gene duplication of the three-G-domain proteins (20). However, recently, unconventional gelsolin-related proteins with two, four, or five G domains have been discovered (21–23). Actin binding protein29 (ABP29) from Lilium pollen has only two G domains; nevertheless it has severing, nucleating, and capping activities (23). However, biochemical properties of other unconventional gelsolin-related proteins are not clearly understood.
C. elegans has three genes that encode gelsolin-related proteins. fli-1 encodes a homolog of Flightless-1 (24). FLI-1 is widely expressed in many tissues, and fli-1 mutations cause a number of developmental defects (15). Viln-1 (C10H11.1) encodes a villin-like protein with six G domains and a C-terminal villin headpiece, but its function is currently under investigation. K06A4.3 encodes a gelsolin-related protein with four G domains. Because K06A4.3 was most closely related to conventional gelsolin among the three genes, this gene has been designated as gsnl-1 (gelsolin-like protein-1) in this study. We are particularly interested in the function of gsnl-1, because biochemical properties of a gelsolin-related protein with four G domains have not been characterized, and mRNA of gsnl-1 is enriched in body wall muscle (25), suggesting that the GSNL-1 protein is a strong candidate of a muscle-specific regulator of actin reorganization. Our biochemical analysis indicates that GSNL-1 is a novel gelsolin-like actin severing and capping protein but, unlike gelsolin, stays bound to the side of actin filaments, binds to G-actin in a 1:1 molar ratio, and does not nucleate actin polymerization. These results provide a new aspect of functional diversity of gelsolin-related proteins.
EXPERIMENTAL PROCEDURES
Proteins—Rabbit muscle actin was purified from acetone powder as described (26). Pyrene-labeled rabbit muscle G-actin was prepared as described (27). Alexa488-labeled rabbit muscle G-actin (1.5 labels/molecule on amines) was purchased from Invitrogen. Rhodamine-labeled rabbit muscle G-actin (0.5 labels/molecule on amines) was purchased from Cytoskeleton, Inc. Gelsolin was purified from newborn calf serum (N4637, Sigma) as reported by Kurokawa et al. (28) with slight modifications. After gelsolin was eluted from DEAE-cellulose, it was further purified by anion exchange chromatography using Mono Q (4.6/100PE column, Amersham Biosciences). Gelsolin was dialyzed against F-buffer (0.1 m KCl, 20 mm Hepes-NaOH, pH 7.5, 2 mm MgCl2) containing 50% glycerol and stored at -20 °C. Bacterially expressed C. elegans UNC-60B was prepared as described previously (29). Bacterially expressed chicken CapZ (a gift of Dr. Takashi Obinata, Chiba University, Chiba, Japan) was prepared as described previously (30).
Expression and Purification of GSNL-1—A full-length cDNA clone for GSNL-1 (yk1613a08) was obtained from Dr. Yuji Kohara (National Institute of Genetics, Mishima, Japan). A full-length coding region of the GSNL-1 cDNA was amplified with PCR and cloned into pGEX-2T using an infusion cloning kit (BD Biosciences). The sequence of the insert was verified by DNA sequencing. The Escherichia coli strain BL21(DE3) was transformed with pGEX-GSNL-1 and cultured in M9ZB (18.7 mm NH4Cl, 22 mm KH2PO4, 42.3 mm Na2HPO4, 1% Tryptone, 85.6 mm NaCl, 1 mm MgSO4, and 0.4% glucose) containing 50 μg/ml ampicillin at 37 °C until A600 reached 0.6 cm-1. Then the culture was cooled to room temperature, and expression was induced by adding 0.1 mm isopropyl-1-thio-β-d-galactopyranoside for 2 h at room temperature. The cells were harvested by centrifugation at 5000 × g for 10 min and homogenized by a French Pressure cell at 360–580 kg/cm2 in phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 1.4 mm KH2PO4, 8 mm Na2HPO4). The homogenates were centrifuged at 20,000 × g for 15 min, and the supernatants were applied to a glutathione Uniflow (Clontech) column (1.5-ml bed volume). After washing with phosphate-buffered saline, 15 units of thrombin (Roche Applied Science) was added to cleave the N-terminal glutathione S-transferase tag, and GSNL-1 eluted from the column. GSNL-1 was dialyzed against F-buffer containing 50% glycerol and stored at -20 °C. Protein concentration was determined with a BCA protein assay (23225, Pierce).
Actin Filament Severing and Capping Assays with Fluorescence Microscopy—Observation of actin filament severing activity by fluorescence microscopy was performed as described previously (31–33) with slight modifications. Previously, we used anti-biotin antibody (Invitrogen) to immobilize biotin-labeled actin on the glass surface. However, this antibody has been discontinued by the company, and we found that several other commercially available anti-biotin antibodies were not very efficient in tethering actin filaments. Instead, unlabeled actin (1.6 μm) and Alexa488-labeled actin (0.4 μm) were co-polymerized and attached to a glass coverslip using heavy meromyosin (MH01, Cytoskeleton Inc.). Other procedures were the same as our previous reports.
Capping activity of GSNL-1 was monitored as described previously (32). Briefly, Alexa488-labeled actin filaments were incubated with GSNL-1 in a perfusion chamber for 3 min, then 0.4 μm rhodamine-labeled G-actin (AR05, Cytoskeleton Inc.) was infused and allowed to elongate from free barbed ends for 3 min. Unincorporated actin was washed with anti-bleaching buffer containing 0.2 μm cytochalasin D, and micrographs of Alexa488- and rhodamine-actin from the same field were taken. Filaments were observed by epifluorescence using a Nikon TE2000 inverted microscope with a 60× Plan Apo objective (oil, NA = 1.4), and images were captured by a SPOT RT Monochrome CCD camera (Diagnostic Instruments) and processed by IPLab (BD Biosciences Bioimaging) and Adobe Photoshop CS3.
Light Scattering and Fluorescence Assays for Actin Depolymerization and Polymerization—Kinetics of actin depolymerization induced by gelsolin or GSNL-1 were monitored by light-scattering measurements. 10 μm actin was polymerized in the presence of 100 nm CapZ for 2 h at room temperature in F-buffer containing 0.1 mm CaCl2. CapZ-F-actin was diluted to 0.5 μm actin in F-buffer containing 0.1 mm CaCl2 or 5 mm EGTA in the presence of 1 μm latrunculin A (Biomol) and various concentrations of gelsolin or GSNL-1. Intensity of light scattering was monitored at a wavelength of 400 nm and at an angle of 90° with a PerkinElmer Life Sciences LS50B fluorescence spectrophotometer.
Kinetics of actin polymerization were monitored by measuring fluorescence of pyrene-labeled actin. 20 μm pyrene-labeled G-actin (20% labeled) was diluted to 2.5 μm with G buffer (0.2 mm ATP, 0.2 mm CaCl2, 0.2 mm dithiothreitol, 2 mm Tris-HCl, pH 8) in the presence of gelsolin or GSNL-1. After 5 min, salt and buffer were adjusted to final concentrations of 0.1 m KCl, 2 mm MgCl2, 0.1 mm CaCl2, 20 mm Hepes-NaOH, pH 7.5, and actin was diluted to 2 μm. Fluorescence of pyrene (excitation at 366 nm and emission at 384 nm) was monitored for 20 min with a PerkinElmer Life Sciences LS50B fluorescence spectrophotometer.
To determine the critical concentration for actin polymerization, varying concentrations (0.1–1 μm) of pyrene-labeled G-actin (20% labeled) was polymerized for 18 h at room temperature in the presence of a constant concentration of GSNL-1 or CapZ in F-buffer containing 0.1 mm CaCl2 or 5 mm EGTA. Final fluorescence intensity of pyrene (excitation at 366 nm and emission at 384 nm) was measured.
F-actin Sedimentation Assay—Varying concentrations of gelsolin or GSNL-1 were added to 5 μm F-actin in 100 mm KCl, 2 mm MgCl2, 0.2 mm dithiothreitol, 20 mm Hepes-NaOH, pH 7.5, containing 0.1 mm CaCl2 or 0.1 mm EGTA. 1 mm EGTA was used for gelsolin. After incubation for 1 h at room temperature, the mixtures were ultracentrifuged at 436,000 × g for 15 min at 20 °C. Supernatant and pellet fractions were adjusted to the same volumes and subjected to SDS-PAGE and staining with Coomassie Brilliant Blue R-250. Gels were scanned by a UMAX Powerlook III scanner at 300 dots per inch, and band intensity was quantified using Image J.
Actin Monomer Binding Assays—Nondenaturing polyacrylamide gel electrophoresis was performed as described (34). G-actin and GSNL-1 were incubated in G-buffer for 30 min at room temperature. The samples were supplemented with 0.25 volume of a loading buffer (50% glycerol, 0.05% bromphenol blue) and electrophoresed using a Bicine/triethanolamine buffer system. The proteins were visualized by staining with Coomassie Brilliant Blue R-250 (National Diagnostics). To analyze stoichiometry of actin and GSNL-1, a mixture of 10 μm actin and 10 μm GSNL-1 was applied to nondenaturing polyacrylamide gel electrophoresis, and protein composition of each band was examined by SDS-PAGE. Core regions of the observed six bands were excised and cut into small pieces. After washing with deionized water, 50 μl of SDS sample buffer (2% SDS, 80 mm Tris-HCl, 5% β-mercaptoethanol, 15% glycerol, 0.05% bromphenol blue) was added. The gel pieces were then extensively sonicated and heated at 98 °C for 5 min. The extracted proteins were resolved by SDS-PAGE, and relative band density of actin and GSNL-1 was compared with a standard (a 1:1 mixture of actin and GSNL-1).
The change in the fluorescence of pyrene-labeled G-actin was used to detect binding of GSNL-1 to G-actin. A dissociation constant (Kd) for binding of GSNL-1 with G-actin was determined by a modification of the method that was developed for 7-chloro-4-nitrobenz-2-oxa-1,3-diazole (NBD)-labeled actin by Carlier et al. (35). Varying concentrations of GSNL-1 (0.05–2 μm) were incubated with 1 μm G-actin (20% pyrene-labeled) in G-buffer at room temperature for 30 min. Then the pyrene fluorescence (F) (excitation at 366 nm and emission at 384 nm) was measured, and relative fluorescence (E) was calculated as
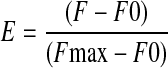 |
(Eq. 1) |
where F0 is fluorescence of 1 μm actin alone, and Fmax is the maximal fluorescence when GSNL-1 saturated binding to 1 μm actin.
Second, the data were fitted to Equation 2,
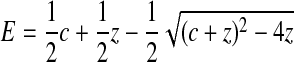 |
(Eq. 2) |
where
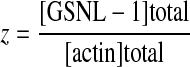 |
(Eq. 3) |
and
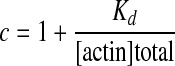 |
(Eq. 4) |
curve fitting was performed with SigmaPlot 10 (SYSTAT Software, Inc.).
RESULTS
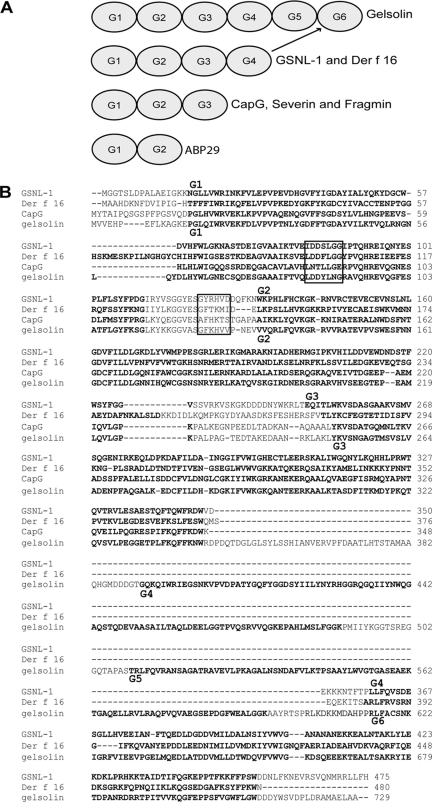
GSNL-1 Has Four Gelsolin-like Domains—The C. elegans gene K06A4.3 encodes a 55-kDa protein that has four gelsolin-like (G) domains (Fig. 1A). Therefore, we designated this gene as gsnl-1. Previously, Der f 16, an allergen from the house dust mite, has been reported to have four G domains (22). However, gelsolin-related proteins typically have three or six G domains, and a protein with four G domains has not been biochemically characterized. Sequence alignment by the ClustalW2 method (36) indicates that GSNL-1 is highly homologous to Der f 16 (31% identity) and that the first three G domains (G1-G3) are very similar to the corresponding domains of human gelsolin, a six-G-domain protein, and CapG, a three-G-domain protein with no severing activity (Fig. 1B). Two regions of gelsolin, LDDYLNG in G1 and GFKHVV in a linker between G1 and G2 (boxed in Fig. 1B), are important for its actin filament severing activity. CapG does not sever actin filaments because these sequences are not conserved (37). GSNL-1 has very similar sequence to gelsolin in these two regions (boxed in Fig. 1B), suggesting that GSNL-1 possesses severing activity. Interestingly, the ClustalW2 method aligned the fourth G domain (G4) of GSNL-1 with the sixth G domain (G6) of gelsolin (Fig. 1B). Indeed, individual comparison of GSNL-1 G4 with gelsolin G4, G5, or G6 indicated that GSNL-1 G4 is most closely related to gelsolin G6 (data not shown). One notable difference between gelsolin and GSNL-1 was that gelsolin has a flexible linker of ∼50 amino acids between G3 and G4 that acts like a hinge during calcium activation (38), whereas GSNL-1 has only a very short linker of ∼10 amino acids between G3 and G4 (Fig. 1B), suggesting that gelsolin and GSNL-1 undergo different conformational changes upon calcium activation. Thus, GSNL-1 is an unconventional gelsolin-related protein that apparently lacks two G domains that are equivalent to G4 and G5 of gelsolin.
FIGURE 1.
GSNL-1 is an unconventional member of the gelsolin family. A, schematic representation of domain structures of gelsolin-related proteins including gelsolin, C. elegans GSNL-1, mite Der f 16, CapG, Dictyostelium severin, Physarum fragmin, and Lilium ABP29. Gelsolin-like (G) domains are numbered from G1 to G6 in the order of appearance from the N termini but not necessarily representing sequence homology. Indeed, G4 of GSNL-1 is most closely related to G6 of gelsolin. B, sequence alignment of GSNL-1 (GenBank™ accession number NM_073047) mite Der f 16 (GenBank™ accession number AF465625), human CapG (GenBank™ accession number M94345), and human gelsolin (GenBank™ accession number NM_198252). G domains as predicted by SMART (60) are highlighted in bold. Sequences that are important for severing activity of gelsolin (37) are indicated by boxes. G domains of GSNL-1 and gelsolin are numbered on the top and the bottom of the alignment, respectively.
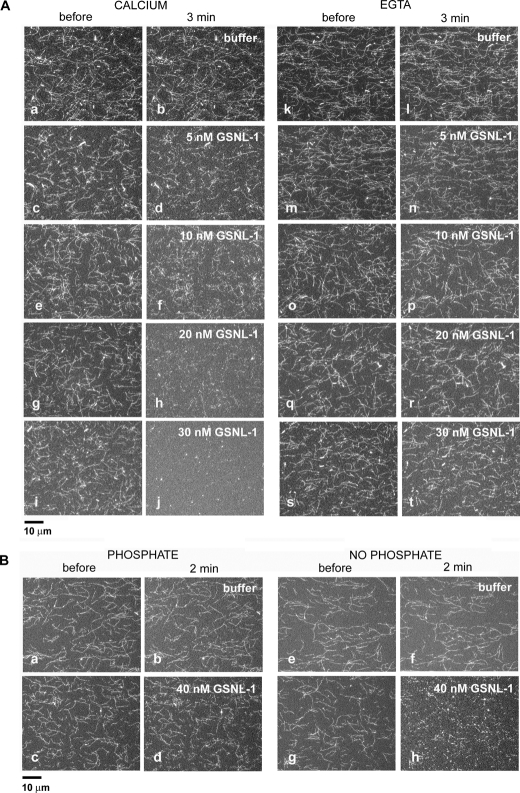
GSNL-1 Severs Actin Filaments in a Calcium-dependent Manner—To elucidate whether GSNL-1 possesses actin filament severing activity, a microscopic perfusion assay (31–33) was employed. Alexa488-labeled F-actin was attached to a heavy meromyosin-coated coverslip, and GSNL-1 was infused and incubated for 3 min. Incubation of actin filaments with buffer only in the presence (Fig. 2A, a and b) or absence (Fig. 2A, k and l) of Ca2+ caused no major alteration in the morphology of the actin filaments. In the presence of 100 μm Ca2+, GSNL-1 caused fragmentation of actin filaments (Fig. 2A, c and d, e and f, g and h, i and j). Fragmentation was detected at 5 nm GSNL-1 (Fig. 2A, c and d). As the concentrations of GSNL-1 were increased, actin filaments became shorter and nearly completely disassembled at 30 nm GSNL-1 (Fig. 2A, i and j). Similar actin filament severing activity of GSNL-1 was observed when calcium concentration was decreased to 1 μm (data not shown). However, when calcium was removed by EGTA, 5–30 nm GSNL-1 did not sever actin filaments (Fig. 2A, m and n, o and p, q and r, s and t). Thus, these microscopic assays demonstrate that GSNL-1 severs actin filaments in a calcium-dependent manner.
FIGURE 2.
GSNL-1 severs actin filaments. A, Alexa488-labeled actin filaments were tethered to a glass coverslip and treated with buffer alone (a, b, k, and l) or varying concentrations of GSNL-1 in the same buffer (c–j and m–t) for 3 min. The buffer in a–j contained 0.1 mm CaCl2, and the buffer in k–t contained 5 mm EGTA. The filaments were observed before (a, c, e, g, i, k, m, o, q, and s) and 3 min after the incubation (b, d, f, h, j, l, n, p, r, and t), and micrographs of the same fields were taken. Bar, 10 μm. B, Alexa488-labeled actin filaments were preincubated with buffer containing 10 mm potassium phosphate (a and c) or no phosphate (e and g). Then buffer alone containing 10 μm CaCl2 and no phosphate (b and f) or the same buffer containing 40 nm GSNL-1(d and h) was infused and incubated for 2 min. The filaments were observed before (a, c, e, and g) and after the incubation (b, d, f, and h), and micrographs of the same fields were taken. Bar, 10 μm.
Allen et al. (39) reported that gelsolin, the six-G-domain protein, severs ADP-actin more strongly than ADP-Pi actin filaments. Therefore, we tested whether GSNL-1 also preferentially severs ADP-actin. Inorganic phosphate (Pi) reversibly binds to ADP-actin filaments with a millimolar affinity (40). Actin filaments in a perfusion chamber were preincubated with a buffer with or without 10 mm potassium phosphate for 5 min, then they were incubated with 40 nm GSNL-1 in the absence of Pi for 2 min. In the absence of GSNL-1, Pi caused no major alteration in the actin filaments (compare Fig. 2B, a and b, and e and f). GSNL-1 only weakly severed the filaments that were preincubated with Pi (Fig. 2B, c and d), whereas it strongly severed filaments that were preincubated in a buffer without Pi (Fig. 2B, g and h). These results indicate that GSNL-1 preferentially severs ADP-bound actin filaments in a similar manner to gelsolin (39).
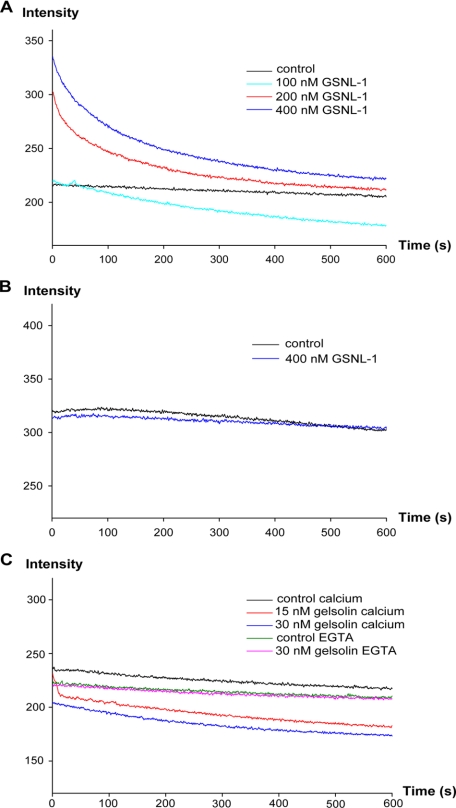
Next, we compared the severing activities of GSNL-1 and gelsolin by measuring the kinetics of actin depolymerization (Fig. 3). CapZ-capped actin filaments were diluted to 0.5 μm actin in the presence of GSNL-1 or gelsolin. Latrunculin A was included in the reactions to sequester actin monomers. Because 0.5 μm actin is below the critical concentration (Cc) at the pointed end, severing should increase the number of pointed ends and accelerate the rate of depolymerization. In the presence of calcium, GSNL-1 increased the light scattering signal at time 0 and gradually decreased the signal over time (Fig. 3A). Because we manually assembled the reactions in a cuvette and set in a fluorometer, there was a delay of 10–15 s before time 0. Therefore, we interpreted that the initial increase is due to rapid association of GSNL-1 to the side of actin filaments before time 0 (see below) and that the following decrease is caused by filament severing in a similar manner to ADF/cofilin (41). In the absence of calcium, 400 nm GSNL-1 did not cause significant alteration in the depolymerization kinetics (Fig. 3B). In contrast, 30 nm gelsolin decreased the initial level of light scattering in the presence of calcium (Fig. 3C, blue curve). By reducing gelsolin to 15 nm, we were able to detect a rapid decrease in light scattering within 10 s (Fig. 3C, red curve). In the presence of EGTA 30 nm gelsolin did not alter the depolymerization kinetics (Fig. 3C, green and purple curves). These results suggest that gelsolin instantaneously severs actin filaments. Our direct comparison of GSNL-1 and gelsolin revealed two major differences; 1) GSNL-1 severs actin filaments more slowly and requires higher concentrations than gelsolin, and 2) GSNL-1 initially associates with the side of filaments and gradually severs them, whereas gelsolin instantaneously severs filaments.
FIGURE 3.
Effect of GSNL-1 on actin depolymerization. CapZ-capped actin filaments (CapZ: actin = 1:100) were diluted to 0.5 μm actin in the presence of 1 μm latrunculin A, and varying concentrations of GSNL-1 or gelsolin and changes in the intensity of light scattering were monitored for 10 min. GSNL-1 in the presence of 0.1 mm CaCl2 (A), GSNL-1 in the absence of calcium (5 mm EGTA) (B), and gelsolin in the presence and absence of calcium (C) were examined.
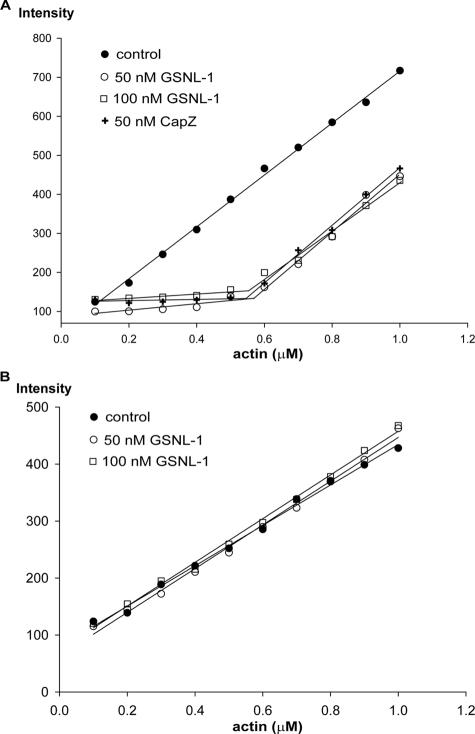
GSNL-1 Caps the Barbed Ends of Actin Filaments—Gelsolin caps the barbed ends of actin filaments, thus preventing polymerization and depolymerization from these ends (42, 43). To determine whether GSNL-1 caps the barbed ends, we examined the effect of GSNL-1 on the Cc of actin. Cc at the barbed end is ∼0.1 μm, whereas Cc at the pointed end is ∼0.6 μm (44). Therefore, if the barbed ends are capped, Cc of total actin will be close to the Cc value at the pointed end. When actin alone was polymerized, amounts of F-actin as measured by pyrene fluorescence, linearly increased at above 0.2 μm actin in the presence (Fig. 4A, black circles) or absence of calcium (Fig. 4B, black circles). In the presence of calcium and GSNL-1, the critical concentration was shifted, and the amounts of F-actin increased at above 0.6 μm actin, whereas at below 0.6 μm actin, fluorescence of pyrene remained constant (Fig. 4A, open circles and squares). CapZ, which is known to cap the barbed ends (45, 46), similarly shifted the critical concentration to ∼0.6 μm (Fig. 4A, crosses). In the presence of EGTA, GSNL-1 had no effect on the critical concentration (Fig. 4B, open circles and squares). These results strongly suggest that GSNL-1 shifts the critical concentration by capping the barbed ends in a calcium-dependent manner.
FIGURE 4.
Effect of GSNL-1 on the critical concentration of actin. Varying concentrations (0.1–1 μm) of G-actin (20% pyrene-labeled) were polymerized alone or in the presence of 50 or 100 nm GSNL-1 or 50 nm CapZ. After 18 h at room temperature, the final pyrene fluorescence was measured. A, actin alone or actin with GSNL-1 or CapZ in the presence of 0.1 mm CaCl2. B, actin alone or actin with GSNL-1 in the absence of calcium (5 mm EGTA).
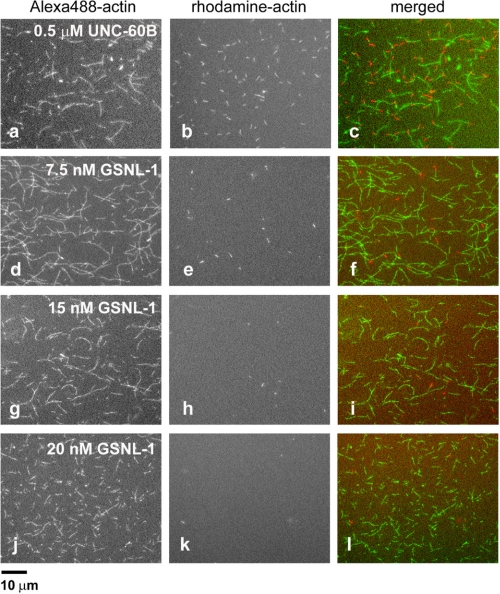
To obtain more direct evidence of the capping activity of GSNL-1, a microscopic assay was used. Alexa488-labeled actin filaments were incubated with GSNL-1 and subsequently with 0.4 μm rhodamine-labeled G-actin for 5 min. Under these conditions, rhodamine-actin should only polymerize from the barbed ends, because the concentration of actin monomers is below Cc at the pointed end (0.6 μm). UNC-60B, a C. elegans ADF/cofilin protein that severs filaments but does not cap filament ends (32), did not block elongation of rhodamine-actin at filament ends (Fig. 5, a–c). When actin filaments were incubated with 7.5 nm GSNL-1, the number of filaments that incorporated rhodamine-actin was significantly decreased (Fig. 5, d–f). Incubation of filaments with 15 or 20 nm GSNL-1 nearly completely abolished incorporation of rhodamine-actin (Fig. 5, g–i and j–l). Taken together, these data show that GSNL-1 caps the barbed ends.
FIGURE 5.
Direct observation of barbed-end capping by GSNL-1. Alexa488-labeled actin filaments were incubated with 0.5 μm UNC-60B or 7.5, 15, or 20 nm GSNL-1 for 3 min and then incubated with 0.4 μm rhodamine-labeled G-actin for 5 min. Alexa488-actin (a, d, g, and j), rhodamine-actin (b, e, h, and k), and merged images (Alexa488 in green and rhodamine in red) (c, f, i, and l) are shown. Bar, 10 μm.
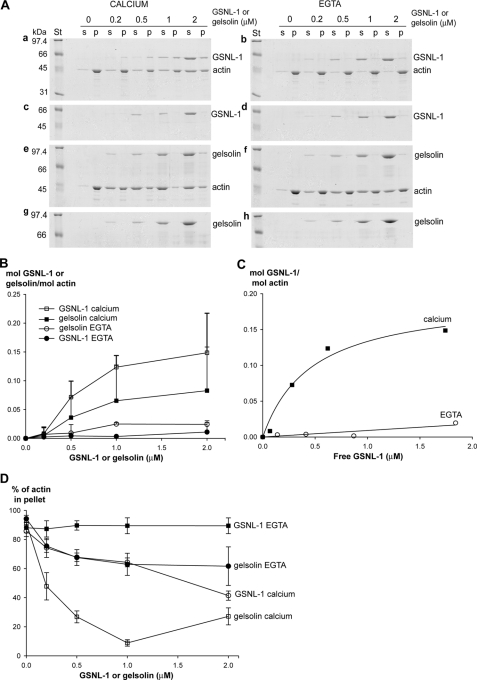
GSNL-1 Binds to Actin Filaments—Our light scattering measurements (Fig. 3A) suggested that GSNL-1 increases light scattering at time 0 by binding to the side of actin filaments. This activity was further examined by high speed sedimentation. Increasing concentrations from 0.2 to 2 μm GSNL-1 or gelsolin was mixed with a constant concentration of F-actin (5 μm). After 1 h of incubation at 25 °C, actin filaments were sedimented by ultracentrifugation (436,000 × g for 15 min), and co-sedimentation of GSNL-1 or gelsolin was examined by SDS-PAGE (Fig. 6A). In the presence of calcium, significant portions of GSNL-1 co-sedimented with actin (Fig. 6A, a), whereas negligible amounts of GSNL-1 sedimented in the absence of actin (Fig. 6A, c). Densitometric analysis of the gels suggest that GSNL-1 binds to F-actin in a saturable manner (Fig. 6B). Assuming that GSNL-1 in the supernatants was free and GSNL-1 in the pellets bound to F-actin, a dissociation constant (Kd) was estimated to be 0.47 ± 0.17 μm, and the binding saturated at ∼0.2 mol GSNL-1/mol actin (Fig. 6C). When calcium was removed by EGTA, GSNL-1 did not co-sediment with F-actin (Fig. 6A, b). In contrast, co-sedimentation of gelsolin with F-actin in the presence of calcium was much less than that of GSNL-1 (Fig. 6, Ae, and B). Instead, gelsolin greatly decreased the amounts of pellet-able actin (Fig. 6, Ae and D), most likely by strongly severing F-actin into short oligomers and capping barbed end to prevent re-annealing. GSNL-1 also decreased pellet-able actin in the presence of calcium, but this activity was much weaker than that of gelsolin (Fig. 6, Aa and D). In the presence of EGTA gelsolin did not significantly co-sediment with actin, although residual activity to decrease pellet-able actin was detected (Fig. 6, Af and D). Because of the large changes in the amounts of pellet-able actin, direct comparison of affinity of GSNL-1 and gelsolin with F-actin was difficult by this method. Nonetheless, these results are consistent with the kinetic measurement of actin depolymerization in Fig. 3 and suggest that GSNL-1 remains bound to the side of F-actin and only weakly severs F-actin, whereas gelsolin immediately severs actin filaments.
FIGURE 6.
GSNL-1 remains bound to F-actin. 5 μm F-actin was incubated with GSNL-1 or gelsolin (0–2 μm) for 1 h at room temperature and ultracentrifuged (436,000 × g, 15 min), and the supernatants (s) and the pellets (p) were analyzed by SDS-PAGE (A). The same experiments were performed for GSNL-1 and gelsolin in the absence of F-actin to estimate actin-independent precipitation. A, experiments were performed in the presence of 0.1 mm CaCl2 (a, c, e, and g) or EGTA (0. 1 mm for GSNL-1 and 1 mm for gelsolin) (b, d, f, and h). Molecular weight standards (St) are shown on the left of each gel. B–D, the band intensities were quantified, and the results are shown. B, molar ratios of GSNL-1 or gelsolin to actin in the pellets are plotted as a function of total concentrations of GSNL-1 or gelsolin in the presence and absence of calcium. Actin-independent sedimentation of GSNL-1 or gelsolin were quantified and subtracted from the experimental data in the presence of actin. Data are the average ± S.D. (for clarity, negative error bars are not shown) of three experiments. C, the results of GSNL-1 are re-plotted as a function of free GSNL-1 assuming that GSNL-1 in the supernatants was free. D, percentages of actin in the pellet fractions are plotted as a function of total concentration of GSNL-1 or gelsolin in the presence or absence of calcium. Data are average ± S.D. of three experiments.
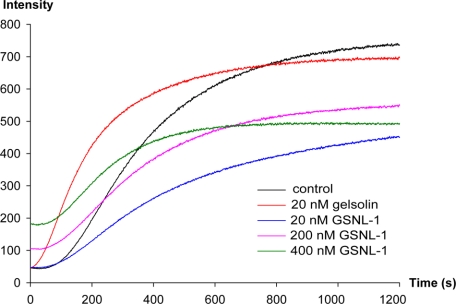
GSNL-1 Does Not Nucleate Actin Polymerization—Gelsolin nucleates actin polymerization (43). Therefore, we tested the effect of GSNL-1 on polymerization of actin monomers. Pyrene-labeled G-actin was polymerized in the presence or absence of GSNL-1 or gelsolin, and the kinetics of polymerization was monitored by changes in the fluorescence of pyrene. In the absence of GSNL-1 or gelsolin, polymerization proceeded with a sigmoidal curve as characterized by an initial lag phase due to slow nucleation (Fig. 7, black curve). When actin was polymerized in the presence of 20 nm gelsolin, initial polymerization was accelerated, and a lag phase was diminished (Fig. 7, orange curve), indicating that gelsolin nucleated polymerization as reported previously (42). Interestingly, the same concentration (20 nm) of GSNL-1 had no detectable effect on the initial lag phase and slowed the following elongation phase (Fig. 7, blue curve). Higher concentrations (200 and 400 nm)of GSNL-1 had similar effects except that the initial fluorescence was increased (Fig. 7, purple and green curves). This increase is unlikely due to polymerization before salt addition at time 0, because if filaments were formed, exposed pointed ends should nucleate polymerization, and the initial lag phase should be eliminated. Rather, this change could be the result of GSNL-1 binding to G-actin (see below). These results strongly suggest that GSNL-1 does not nucleate actin polymerization but slows down elongation by blocking the fast-growing barbed ends.
FIGURE 7.
GSNL-1 does not nucleate actin polymerization. 2.5 μm G-actin (20% pyrene-labeled) was incubated with GSNL-1 or gelsolin at room temperature for 5 min, and then salt was added to initiate polymerization at time 0 (final actin concentration was 2 μm).
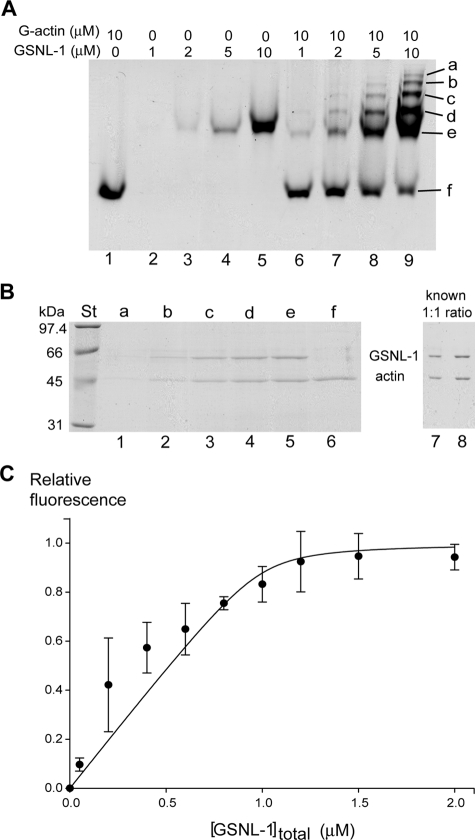
GSNL-1 Binds to G-actin—To clarify whether GSNL-1 binds to G-actin, we first tested their binding by nondenaturing gel electrophoresis. Only a single band was detected with G-actin alone (10 μm) (Fig. 8A, lane 1) or GSNL-1 alone (1, 2, 5, or 10 μm) (Fig. 8A, lanes 2–5). At 1 μm GSNL-1, the band of GSNL-1 was very faint (Fig. 8A, lane 2). However, when 1 μm GSNL-1 was incubated with G-actin, a more intense band than GSNL-1 alone appeared nearly at the same mobility as the band of GSNL-1 (Fig. 8A, lane 6, band e), suggesting that a complex of GSNL-1 and G-actin migrated similarly to GSNL-1 alone. As the concentrations of GSNL-1 were increased, band e was also increased and were consistently more intense than the bands of GSNL-1 alone (Fig. 8A, compare lanes 6–8 with lanes 2–4, respectively). At the same time, the band of actin became less intense as GSNL-1 was increased (Fig. 8A, lanes 6–9, band f), indicating an increase in the GSNL-1-actin complex and a decrease in free G-actin. Moreover, four additional bands with slower mobility incrementally appeared (Fig. 8A, lane 9, bands a–d).
FIGURE 8.
GSNL-1 binds to G-actin. A, interaction between GSNL-1 and G-actin was examined by nondenaturing gel electrophoresis. 10 μm G-actin alone (lane 1), GSNL-1 alone (1, 2, 5, and 10 μm) (lanes 2–5), and 10 μm G-actin with GSNL-1 (1, 2, 5, and 10 μm) (lanes 6–9) were incubated for 30 min at room temperature, and the samples were analyzed by nondenaturing gel electrophoresis. Six major bands (a–f) were further analyzed by SDS-PAGE in B. B, the proteins from the bands a–f detected in the mixture of 10 μm GSNL-1 and 10 μm G-actin in nondenaturing electrophoresis (A, lane 9) were extracted and separated on SDS-PAGE (lanes 1–6). The band intensities were compared with a known 1:1 mixture (1 and 2 μl) of GSNL-1 (1 μm) and G-actin (1 μm) (lanes 7 and 8). Molecular weight standards (lane St) are shown on the left. C, binding of GSNL-1 to G-actin was monitored by changes in the fluorescence of pyrene. 1 μm G-actin (20% pyrene-labeled) was incubated with varying concentrations of GSNL-1 at room temperature for 30 min. Relative fluorescence values were plotted as a function of total GSNL-1 concentrations and fitted to an equation as described under “Experimental Procedures” to calculate a Kd value for GSNL-1 interaction with monomeric actin. Data are the average ± S.D. of three experiments.
To determine protein compositions in these bands, proteins from bands a–f were extracted and examined by SDS-PAGE (Fig. 8B). Band f contained only actin, as expected (Fig. 8B, lane 6). Interestingly, bands a–e contained both actin and GSNL-1 with the band of GSNL-1 slightly more intense than the band of actin (Fig. 8B, lanes 1–5). This is similar to the pattern of a known mixture of GSNL-1 and actin in a 1:1 molar ratio (Fig. 8B, lanes 7 and 8). If GSNL-1 binds to G-actin in a 1:2 molar ratio as does gelsolin, the band of actin should be more intense. It is not clear as to why the GSNL-1-actin complex is resolved in five different bands. One possibility is that the GSNL-1:actin complex forms oligomers. We attempted to analyze the GSNL-1:actin complex by gel filtration, but the resolution was not high enough to resolve the complex possibly due to interactions between the proteins and the gel matrix under low ionic conditions.3 At least we did not detect a high molecular weight protein complex by gel filtration. The other possibility is that these complexes have conformational differences that affect the electrophoretic mobility. These results strongly suggest that GSNL-1 binds to G-actin in a 1:1 molar ratio.
The interaction between GSNL-1 and G-actin was also examined in solution. During examination of the effect of GSNL-1 on actin polymerization using pyrene fluorescence, we noticed that incubation of GSNL-1 and pyrene-labeled G-actin increased the fluorescence without inducing polymerization (Fig. 7). Therefore, we interpreted that the change in the pyrene fluorescence is due to a conformational change upon binding of GSNL-1 to G-actin. Incubation of pyrene-labeled G-actin (20% labeled) with various concentrations of GSNL-1 increased the fluorescence in a saturable manner (Fig. 8C). A dissociation constant (Kd) for the binding of GSNL-1 to G-actin was estimated to be 0.02 μm. Taken together, these results show that GSNL-1 binds to G-actin in a 1:1 molar ratio with high affinity.
DISCUSSION
In this study we biochemically characterized C. elegans GSNL-1, a novel gelsolin-like protein with four G domains, and found both similarities and differences as compared with gelsolin that has six G domains. Despite the fact that GSNL-1 has an unconventional number of G domains, we predicted that GSNL-1 has an ability to fragment actin filaments, because GSNL-1 has similar sequences to gelsolin in a region of G1 and a linker between G1 and G2 that is crucial for severing (Fig. 1B) (37). Indeed, we found that GSNL-1 severs actin filaments in a calcium-dependent manner, although the activity was weaker than that of gelsolin (Figs. 2, 3, and 6). In addition, GSNL-1 capped the barbed end of actin filaments and preferentially severed ADP-actin to ADP-Pi actin in a similar manner to gelsolin (Figs. 2B, 4, and 5) (39). In contrast, GSNL-1 had different properties from gelsolin in that it remains bound to the side of F-actin (Figs. 3 and 6) and does not nucleate actin polymerization (Fig. 7) (43). Thus, these results demonstrate that GSNL-1 is a novel member of the gelsolin family of actin severing proteins.
What might explain the differences between GSNL-1 and gelsolin? Gelsolin has six G domains and contains three major actin binding sites. Monomeric actin binding sites are located in G1 and G4, and the F-actin binding site is in G2 (17). The N-terminal half (G1-G3) of gelsolin alone can sever actin filaments in contrast to the C-terminal half (G4-G6) that lacks severing activity (47). However, G4-G6 is proposed to enhance the severing activity of the full-length molecule in a cooperative manner (48). Filament severing by gelsolin is a stepwise process (17). In the absence of calcium, the G2 F-actin binding site is masked by the C-terminal latch of gelsolin. Upon calcium binding, the F-actin binding site in G2 is exposed and allowed to interact with F-actin. In the following steps, G1 and G4 bind to actin subdomains 1 and 3, resulting in dissociation of the longitudinal actin-actin contacts and subsequent capping of the barbed end. Thus, G4 also significantly contributes to the strong severing activity of gelsolin. Interestingly, our sequence alignment (Fig. 1B) suggests that GSNL-1 lacks two G domains that are equivalent to G4 and G5 of gelsolin. Therefore, GSNL-1 may sever actin filaments by G1-G3 without contribution of the C-terminal G domain. This may explain why GSNL-1 has weaker severing activity than gelsolin.
Why does GSNL-1 remain bound to F-actin? One possibility might be that the C-terminal G domain of GSNL-1 possesses an F-actin binding site that may compete with filament severing by G1-G3. GSNL-1 binds to F-actin until binding is saturated at ∼1 GSNL-1 molecule per 5 actin monomers with a KD value of 0.47 μm (Fig. 6C). However, our microscopic analysis demonstrated that GSNL-1 severs F-actin at nanomolar concentrations. At low GSNL-1 concentrations, F-actin binding of the C-terminal G domain is not significant, so G1-G3 can simply sever filaments. However, at high GSNL-1 concentrations, the C-terminal G domain remains associated with F-actin and may compete with G1-G3 of other GSNL-1 molecules for binding to F-actin. Previous studies on gelsolin support this idea. It has been shown that removal of G1 dramatically reduces the severing activity of gelsolin (49). Furthermore, Fujita et al. (50) reported that N-terminal-truncated gelsolin (G2-G6) inhibits the severing activity of full-length gelsolin by remaining bound to F-actin with high affinity. At this point it is not clear whether the severing mechanism of GSNL-1 resembles that of gelsolin. Thus, dissection of functional domains of GSNL-1 and structural analysis of GSNL-1 are required to understand how GSNL-1 severs actin filaments.
The gelsolin family proteins nucleate actin polymerization. Surprisingly, our data showed that GSNL-1 does not nucleate actin polymerization (Fig. 7), although it binds to G-actin with high affinity (Kd 0.02 μm) forming a 1:1 complex (Fig. 8). To our knowledge, GSNL-1 is the first member of the gelsolin family lacking the ability to induce nucleation. Two actin monomers bind to G1 and G4 of gelsolin, and this ternary complex nucleates actin polymerization (51–53). Therefore, it is possible that the 1:1 complex of GSNL-1 and G-actin is not sufficient to act as a nucleus. However, previous studies demonstrated that G2-G6 of gelsolin is sufficient for nucleation (49) and that G6 of gelsolin is essential for nucleation (54). Thus, GSNL-1 may bind to actin monomer in a different manner from gelsolin. Furthermore, Dictyostelium severin (55), which has three G domains, and Lilium ABP29, which has two G domains (23), can nucleate polymerization. Thus, different gelsolin-related proteins have diverse effects on actin polymerization because of different modes of interaction with G-actin. The high affinity binding of GSNL-1 to G-actin may modulate actin dynamics by competing with nucleation activities of other actin-binding proteins.
To date, biological function of C. elegans GSNL-1 is unknown. mRNA of GSNL-1 is enriched in body wall muscle (25), and the promoter of gsnl-1 is specifically active in body wall muscle (56). These data strongly suggest that GSNL-1 is involved in regulation of actin filament dynamics in body wall muscle. Nonetheless, RNA interference of gsnl-1 resulted in no detectable phenotype (57). One possibility is that GSNL-1 is functionally redundant with two other gelsolin-related genes, viln-1 (a villin-like protein) and fli-1 (a Flightless-1 homolog). Mutations of fli-1 cause disorganization of myofibrils in body wall muscle (15), but the phenotype appears mild as compared with severe phenotypes in unc-60B (ADF/cofilin) (6, 7) and unc-78 (AIP1) mutants (8, 9). To determine the functions of gelsolin-related genes in C. elegans, multiple genes will have to be deleted or knocked down by RNA interference to analyze a phenotype. Alternatively, GSNL-1 may have a distinct function in actin remodeling from ADF/cofilin and AIP1. Organization of actin filaments in body wall muscle is disturbed in a calcium-dependent manner in mutant backgrounds of dys-1 (dystrophin) (58) and mup-2/tnt-1 (troponin T) (59). Thus, GSNL-1 may be involved in these calcium-dependent alterations in the actin filament organization. Further genetic and cell biological analyses of gelsolin-related proteins in C. elegans should reveal distinct and redundant functions of gelsolin-related proteins and a functional relationship with ADF/cofilin and AIP1.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AR48615. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ADF, actin depolymerizing factor; Bicine, N,N-bis(2-hydroxyethyl)glycine; GSNL-1, gelsolin-like protein-1; Cc, critical concentration.
T. Klaavuniemi, S. Yamashiro, and S. Ono, unpublished data.
References
- 1.Ono, S. (2007) Int. Rev. Cytol. 258 1-82 [DOI] [PubMed] [Google Scholar]
- 2.Southwick, F. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6936-6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono, S., Minami, N., Abe, H., and Obinata, T. (1994) J. Biol. Chem. 269 15280-15286 [PubMed] [Google Scholar]
- 4.Vartiainen, M. K., Mustonen, T., Mattila, P. K., Ojala, P. J., Thesleff, I., Partanen, J., and Lappalainen, P. (2002) Mol. Biol. Cell 13 183-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal, P. B., Greenleaf, R. S., Tomczak, K. K., Lehtokari, V. L., Wallgren-Pettersson, C., Wallefeld, W., Laing, N. G., Darras, B. T., Maciver, S. K., Dormitzer, P. R., and Beggs, A. H. (2007) Am. J. Hum. Genet. 80 162-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono, S., Baillie, D. L., and Benian, G. M. (1999) J. Cell Biol. 145 491-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono, K., Parast, M., Alberico, C., Benian, G. M., and Ono, S. (2003) J. Cell Sci. 116 2073-2085 [DOI] [PubMed] [Google Scholar]
- 8.Ono, S. (2001) J. Cell Biol. 152 1313-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohri, K., Ono, K., Yu, R., Yamashiro, S., and Ono, S. (2006) Mol. Biol. Cell 17 2190-2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dissmann, E., and Hinssen, H. (1994) Eur. J. Cell Biol. 63 336-344 [PubMed] [Google Scholar]
- 11.Yin, H. L., Albrecht, J. H., and Fattoum, A. (1981) J. Cell Biol. 91 901-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtsuka, Y., Nakae, H., Abe, H., and Obinata, T. (1994) Zool. Sci. 11 407-412 [Google Scholar]
- 13.Ohtsuka, Y., Nakae, H., Abe, H., and Obinata, T. (1998) Biochim. Biophys. Acta 1383 219-231 [DOI] [PubMed] [Google Scholar]
- 14.Campbell, H. D., Schimansky, T., Claudianos, C., Ozsarac, N., Kasprzak, A. B., Cotsell, J. N., Young, I. G., de Couet, H. G., and Miklos, G. L. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 11386-11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., Xia, D., Fang, B., and Zhang, H. (2007) Genetics 177 847-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miklos, G. L., and De Couet, H. G. (1990) J. Neurogenet. 6 133-151 [DOI] [PubMed] [Google Scholar]
- 17.McGough, A. M., Staiger, C. J., Min, J. K., and Simonetti, K. D. (2003) FEBS Lett. 552 75-81 [DOI] [PubMed] [Google Scholar]
- 18.Silacci, P., Mazzolai, L., Gauci, C., Stergiopulos, N., Yin, H. L., and Hayoz, D. (2004) Cell. Mol. Life Sci. 61 2614-2623 [DOI] [PubMed] [Google Scholar]
- 19.Sun, H. Q., Yamamoto, M., Mejillano, M., and Yin, H. L. (1999) J. Biol. Chem. 274 33179-33182 [DOI] [PubMed] [Google Scholar]
- 20.Yin, H. L., Janmey, P. A., and Schleicher, M. (1990) FEBS Lett. 264 78-80 [DOI] [PubMed] [Google Scholar]
- 21.Gloss, A., Rivero, F., Khaire, N., Muller, R., Loomis, W. F., Schleicher, M., and Noegel, A. A. (2003) Mol. Biol. Cell 14 2716-2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamoto, S., Suzuki, T., Aki, T., Katsutani, T., Tsuboi, S., Shigeta, S., and Ono, K. (2002) FEBS Lett. 516 234-238 [DOI] [PubMed] [Google Scholar]
- 23.Xiang, Y., Huang, X., Wang, T., Zhang, Y., Liu, Q., Hussey, P. J., and Ren, H. (2007) Plant Cell 19 1930-1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goshima, M., Kariya, K., Yamawaki-Kataoka, Y., Okada, T., Shibatohge, M., Shima, F., Fujimoto, E., and Kataoka, T. (1999) Biochem. Biophys. Res. Commun. 257 111-116 [DOI] [PubMed] [Google Scholar]
- 25.Fox, R. M., Watson, J. D., Von Stetina, S. E., McDermott, J., Brodigan, T. M., Fukushige, T., Krause, M., and Miller, D. M., 3rd (2007) Genome Biology 8 R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardee, J. D., and Spudich, J. A. (1982) Methods Enzymol. 85 164-181 [DOI] [PubMed] [Google Scholar]
- 27.Kouyama, T., and Mihashi, K. (1981) Eur. J. Biochem. 114 33-38 [PubMed] [Google Scholar]
- 28.Kurokawa, H., Fujii, W., Ohmi, K., Sakurai, T., and Nonomura, Y. (1990) Biochem. Biophys. Res. Commun. 168 451-457 [DOI] [PubMed] [Google Scholar]
- 29.Ono, S., and Benian, G. M. (1998) J. Biol. Chem. 273 3778-3783 [DOI] [PubMed] [Google Scholar]
- 30.Soeno, Y., Abe, H., Kimura, S., Maruyama, K., and Obinata, T. (1998) J. Muscle Res. Cell Motil. 19 639-646 [DOI] [PubMed] [Google Scholar]
- 31.Ichetovkin, I., Han, J., Pang, K. M., Knecht, D. A., and Condeelis, J. S. (2000) Cell Motil. Cytoskeleton 45 293-306 [DOI] [PubMed] [Google Scholar]
- 32.Ono, S., Mohri, K., and Ono, K. (2004) J. Biol. Chem. 279 14207-14212 [DOI] [PubMed] [Google Scholar]
- 33.Yamashiro, S., Gimona, M., and Ono, S. (2007) J. Cell Sci. 120 3022-3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safer, D. (1989) Anal. Biochem. 178 32-37 [DOI] [PubMed] [Google Scholar]
- 35.Carlier, M. F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G. X., Hong, Y., Chua, N. H., and Pantaloni, D. (1997) J. Cell Biol. 136 1307-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007) Bioinformatics 23 2947-2948 [DOI] [PubMed] [Google Scholar]
- 37.Southwick, F. S. (1995) J. Biol. Chem. 270 45-48 [DOI] [PubMed] [Google Scholar]
- 38.Burtnick, L. D., Koepf, E. K., Grimes, J., Jones, E. Y., Stuart, D. I., McLaughlin, P. J., and Robinson, R. C. (1997) Cell 90 661-670 [DOI] [PubMed] [Google Scholar]
- 39.Allen, P. G., Laham, L. E., Way, M., and Janmey, P. A. (1996) J. Biol. Chem. 271 4665-4670 [DOI] [PubMed] [Google Scholar]
- 40.Carlier, M. F., and Pantaloni, D. (1988) J. Biol. Chem. 263 817-825 [PubMed] [Google Scholar]
- 41.Yamashiro, S., Mohri, K., and Ono, S. (2005) Biochemistry 44 14238-14247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris, H. E., and Weeds, A. G. (1984) FEBS Lett. 177 184-188 [DOI] [PubMed] [Google Scholar]
- 43.Yin, H. L., Hartwig, J. H., Maruyama, K., and Stossel, T. P. (1981) J. Biol. Chem. 256 9693-9697 [PubMed] [Google Scholar]
- 44.Pollard, T. D. (1986) J. Cell Biol. 103 2747-2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casella, J. F., Craig, S. W., Maack, D. J., and Brown, A. E. (1987) J. Cell Biol. 105 371-379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casella, J. F., Maack, D. J., and Lin, S. (1986) J. Biol. Chem. 261 10915-10921 [PubMed] [Google Scholar]
- 47.Bryan, J., and Hwo, S. (1986) J. Cell Biol. 102 1439-1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selden, L. A., Kinosian, H. J., Newman, J., Lincoln, B., Hurwitz, C., Gershman, L. C., and Estes, J. E. (1998) Biophys. J. 75 3092-3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Way, M., Gooch, J., Pope, B., and Weeds, A. G. (1989) J. Cell Biol. 109 593-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujita, H., Allen, P. G., Janmey, P. A., Azuma, T., Kwiatkowski, D. J., Stossel, T. P., Furu-uchi, K., and Kuzumaki, N. (1997) Eur. J. Biochem. 248 834-839 [DOI] [PubMed] [Google Scholar]
- 51.Ditsch, A., and Wegner, A. (1994) Eur. J. Biochem. 224 223-227 [DOI] [PubMed] [Google Scholar]
- 52.Doi, Y., and Frieden, C. (1984) J. Biol. Chem. 259 11868-11875 [PubMed] [Google Scholar]
- 53.Porte, F., and Harricane, M. C. (1986) Eur. J. Biochem. 154 87-93 [DOI] [PubMed] [Google Scholar]
- 54.Kwiatkowski, D. J., Janmey, P. A., and Yin, H. L. (1989) J. Cell Biol. 108 1717-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown, S. S., Yamamoto, K., and Spudich, J. A. (1982) J. Cell Biol. 93 205-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunt-Newbury, R., Viveiros, R., Johnsen, R., Mah, A., Anastas, D., Fang, L., Halfnight, E., Lee, D., Lin, J., Lorch, A., McKay, S., Okada, H. M., Pan, J., Schulz, A. K., Tu, D., Wong, K., Zhao, Z., Alexeyenko, A., Burglin, T., Sonnhammer, E., Schnabel, R., Jones, S. J., Marra, M. A., Baillie, D. L., and Moerman, D. G. (2007) PLoS Biol. 5 e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., Welchman, D. P., Zipperlen, P., and Ahringer, J. (2003) Nature 421 231-237 [DOI] [PubMed] [Google Scholar]
- 58.Mariol, M. C., and Segalat, L. (2001) Curr. Biol. 11 1691-1694 [DOI] [PubMed] [Google Scholar]
- 59.McArdle, K., Allen, T. S., and Bucher, E. A. (1998) J. Cell Biol. 143 1201-1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5857-5864 [DOI] [PMC free article] [PubMed] [Google Scholar]