Abstract
Secretory phospholipase A2 group IIA (sPLA2-IIA) plays an important role in the pathogenesis of inflammatory diseases. Catalytic activity of this enzyme that generates arachidonic acid is a major target for development of anti-inflammatory agents. Independent of its catalytic activity, sPLA2-IIA induces pro-inflammatory signals in a receptor-mediated mechanism (e.g. through the M-type receptor). However, the M-type receptor is species-specific: sPLA2-IIA binds to the M-type receptor in rodents and rabbits, but not in human. Thus sPLA2-IIA receptors in human have not been established. Here we demonstrated that sPLA2-IIA bound to integrin αvβ3 at a high affinity (KD = 2 × 10-7 m). We identified amino acid residues in sPLA2-IIA (Arg-74 and Arg-100) that are critical for integrin binding using docking simulation and mutagenesis. The integrin-binding site did not include the catalytic center or the M-type receptor-binding site. sPLA2-IIA also bound to α4β1. We showed that sPLA2-IIA competed with VCAM-1 for binding to α4β1, and bound to a site close to those for VCAM-1 and CS-1 in the α4 subunit. Wild type and the catalytically inactive H47Q mutant of sPLA2-IIA induced cell proliferation and ERK1/2 activation in monocytic cells, but the integrin binding-defective R74E/R100E mutant did not. This indicates that integrin binding is required, but catalytic activity is not required, for sPLA2-IIA-induced proliferative signaling. These results suggest that integrins αvβ3 and α4β1 may serve as receptors for sPLA2-IIA and mediate pro-inflammatory action of sPLA2-IIA, and that integrin-sPLA2-IIA interaction is a novel therapeutic target.
The phospholipase A2 (PLA2)2 family is a group of intracellular and secreted enzymes that hydrolyzes the sn-2 ester bond in the glyceroacyl phospholipids present in lipoproteins and cell membranes to form nonesterified fatty acids and lysophospholipids. These products act as intracellular second messengers or are further metabolized into potent mediators of a broad range of cellular processes, including inflammation, apoptosis, and atherogenesis (1). The mammalian secretory PLA2 isoforms are comprised of the groups named IB, IIA, IIC, IID, IIE, IIF, V, X, and XII (2, 3). All secretory PLA2 isoforms have in common a Ca2+-dependent catalytic mechanism, a low molecular mass (13–16 kDa), several disulfide bridges, and a well-conserved overall three-dimensional structure (2, 4, 5). Secretory PLA2 type IIA (sPLA2-IIA) was first isolated and purified from rheumatoid synovial fluid. sPLA2-IIA is an acute phase reactant and is found in markedly increased plasma concentrations in diseases that involve systemic inflammation such as sepsis, rheumatoid arthritis, and cardiovascular disease (up to 1000-fold and >1 μg/ml). Inflammatory cytokines such as IL-6, TNF-α, and IL-1β induce synthesis and release of sPLA2-IIA in arterial smooth muscle cells and hepatocytes, which are the major sources of the plasma sPLA2-IIA in these systemic inflammatory conditions (6, 7). In addition to being a pro-inflammatory protein, sPLA2-IIA expression is elevated in neoplastic prostatic tissue (8) and dysregulation of sPLA2-IIA may play a role in prostatic carcinogenesis (9), and is a potential therapeutic target in prostate cancer (10).
Notably some biological effects associated with sPLA2-IIA are independent of its catalytic function (11). Catalytically inactive sPLA2-IIA mutants retained the ability to enhance cyclooxygenase-2 expression in connective tissue mast cells (11). Also inactivation of sPLA2-IIA by bromophenacyl bromide did not affect the ability of sPLA2-IIA to induce secretion of β-glucuronidase, IL-6, and IL-8 from human eosinophils (12). It has thus been proposed that sPLA2-IIA action is mediated through interaction with specific receptors. Indeed the enzyme binds to a high affinity receptor of 180 kDa present on rabbit skeletal muscle (13). This so-called M (muscle)-type receptor belongs to the superfamily of C-type lectins and mediates some of the physiological effects of mammalian sPLA2-IIA, and binding of sPLA2-IIA to this receptor induces internalization of sPLA2-IIA (14). However, the interaction between sPLA2-IIA and the M-type receptor is species-specific, and human sPLA2-IIA binds to the human or mouse M-type receptor very weakly (15). Thus, sPLA2-IIA receptors in human have not been established. Mammalian sPLA2-IIAs bind to heparan sulfate proteoglycans like glypican-1 (16) and decorin in apoptotic human T cells (17). The binding of sPLA2-IIA to heparan sulfate proteoglycans has been implicated in the release of arachidonic acid from apoptotic T cells (18), but it is unclear whether this process plays a role in other situations.
Integrins are a family of cell adhesion receptors that recognize ECM ligands and cell surface ligands (19). Integrins are transmembrane αβ heterodimers, and at least 18 α and 8 β subunits are known (20). Integrins transduce signals to the cell upon ligand binding (19). In this study, we investigated whether integrins are involved in the pro-inflammatory functions of sPLA2-IIA. Here we demonstrate that sPLA2-IIA bound to integrins and induced proliferative signals in an integrin-dependent manner. We first showed that sPLA2-IIA specifically bound to integrin αvβ3 at a high affinity in several different assays, and localized the integrin-binding site in sPLA2-IIA using docking simulation and mutagenesis. The integrin-binding site did not include the catalytic center or the M-type receptor-binding site. We obtained evidence that sPLA2-IIA also bound to α4β1 and competed with vascular cell adhesion molecule (VCAM)-1 for binding to α4β1. Wt and the catalytically inactive mutant of sPLA2-IIA induced cell proliferation, but an integrin-binding defective mutant did not induce cell proliferation in cells that express αvβ3 and/or α4β1. This indicates that integrin binding is required, but catalytic activity is not required, for sPLA2-IIA-induced cell proliferation. sPLA2-IIA induced cell proliferation of monocytic U937 cells (αvβ3+/α4β1+) and induced ERK1/2 activation in an integrin-dependent manner. These results suggest that integrins αvβ3 and α4β1 may serve as receptors for sPLA2-IIA and mediate pro-inflammatory action of sPLA2-IIA in human. Thus integrin-sPLA2-IIA interaction is a novel therapeutic target in inflammation.
EXPERIMENTAL PROCEDURES
Materials—Recombinant soluble αvβ3 was synthesized as previously described (21). CHO cells expressing recombinant αvβ3 (designated β3-CHO cells) (22), Wt or mutant α4 (23), and K562 human erythroleukemia cells that express human α4 (α4-K562) (24) have been described. K562 cells that express human αvβ3(αvβ3-K562) (25) were provided by Eric Brown (UCSF). Human β3 was stably expressed in the CHO pgs745 mutant cells (26) deficient in xylosyltransferase, an essential enzyme in proteoglycan synthesis, as described (22).
Synthesis of sPLA2-IIA—A cDNA fragment encoding sPLA2-IIA was amplified with sPLA2-IIA cDNA (ATCC) as a template using synthetic oligonucleotide primers 5′-GAAGATCTAATTTGGTGAATTTCCAC-3′ and 5′-GGAATTCTCAGCAACGAGGGGTGCTCCC-3′ by PCR. After digestion with BglII and EcoRI, the cDNA fragment was subcloned into the BamHI/EcoRI sites of PET28a/Amp vector. We generated the PET28a/amp vector by replacing the kanamycin-resistant gene of PET28a with the ampicillin-resistant gene of PET21a. We generated sPLA2-IIA as an insoluble protein in bacteria BL21 and purified it by Ni-NTA affinity chromatography under denatured conditions and refolded following the protocols (“Isolation of proteins from inclusion bodies” available from the Björkman laboratory). Briefly, purified proteins in 8 m urea were diluted into refolding buffer (100 mm Tris-HCl, pH 8.0, 400 mm l-Arg, 2 mm EDTA, 0.5 mm oxidized glutathione, 5 mm reduced glutathione, and protease inhibitors) and kept for 8 h at 4 °C, and then concentrated by ultrafiltration. The refolded sPLA2-IIA was more than 90% homogeneous in SDS-PAGE. We performed site-directed mutagenesis by QuikChange method (27). The presence of mutations was confirmed by DNA sequencing. To remove endotoxin, we washed the Ni-NTA resin with 1% Triton X-114 before eluting the bound protein. We confirmed that the sPLA2-IIA (Wt and mutants) had no detectable endotoxin as tested by the Limulus amebocyte lysate assay (Fisher Scientific).
Binding of Soluble αvβ3 to Immobilized sPLA2-IIA—sPLA2-IIA was immobilized to wells of 96-well microtiter plates and the remaining protein-binding sites were blocked by BSA as described (28). Soluble recombinant αvβ3 at 5 μg/ml in Hepes-Tyrodes buffer supplemented with 1 mm MnCl2 was added to the well and incubated for 2 h at room temperature. Bound αvβ3 was measured using anti-integrin β3 (mAb AV-10) followed by horseradish peroxidase-conjugated goat anti-mouse IgG and peroxidase substrates.
Binding of FITC-labeled sPLA2-IIA to Integrins on the Cell Surface—sPLA2-IIA was labeled with FITC using fluorescein labeling kit (Pierce) according to the manufacturer's instructions. Cells were harvested with 3.5 mm EDTA in phosphate-buffered saline. Cells were double-labeled with (a) FITC-labeled sPLA2-IIA (10 μg/ml in the presence of 10 mm Mg2+ at room temperature for 30 min) and (b) with non-blocking anti-human integrin β3 subunit mAb AV10 and PE (phycoerythrin)-conjugated secondary antibody. Bound FITC (FL1) and PE (FL2) were quantified in flow cytometry.
Surface Plasmon Resonance Study—Recombinant soluble integrin αvβ3 was immobilized to Biacore Sensor chip CM5 (Biacore, Piscataway, NJ) by the standard amine coupling method. 2-fold serially diluted sPLA2-IIA and its mutants R74E/R100E (ranging from 2 nm to 500 pm) and H47Q (ranging from 4 to 1 nm) in running buffer HBS-P buffer containing 1 mm of Mn2+ were injected for 3 min at the flow rate of 50 μl/min. Then the sensor chip was washed with the running buffer alone at the same flow rate for another 5 min (the dissociation phase). Two consecutive 1-minute injections of 0.5 m, pH 8 EDTA solution at the same flow rate were used to regenerate the chip for another cycle of injection. The resonance unit elicited from the reference flow cell was subtracted from the resonance unit elicited from the integrin flow cell to eliminate the nonspecific protein-flow cell interaction and the bulk refractive index effect. The recorded binding curves were fitted with the 1:1 binding with drifting baseline model by using the Biaevaluation Version 4.1.
Cell Proliferation and MAP Kinase Activation—K562 cells and human monocytic lymphoma U937 cells were maintained in RPMI1640 medium supplemented with 10% fetal calf serum. Cells were plated in 96-well plates (1 × 104 cells/well), and serum-starved for 48 h at 37 °C in 5% CO2 atmosphere. Cells were then treated with or without sPLA2-IIA in medium without serum for 48 h. Cell proliferation was determined by MTS assays using the Aqueous Cell Proliferation Assay kit (Promega). For MAP kinase activation assays, cells were serum-starved in RPMI1640 medium supplemented with 0.4% fetal calf serum for 24 h, and stimulated with Wt and mutant sPLA2-IIA (0.5 μg/ml) for 10 min at 37 °C. ERK1/2 activation was measured as described (28).
Other Methods—We performed docking simulation of interaction between sPLA2-IIA and integrin αvβ3 using the AutoDock3 as previously described (28). Adhesion assays were performed as described previously (29). mAb 7E3 was used at 10 μg/ml. We assayed PLA2 activity by arachidonoyl-Thio-PC hydrolysis (Cayman Chemicals, Ann Arbor, MI) as described (30).
RESULTS
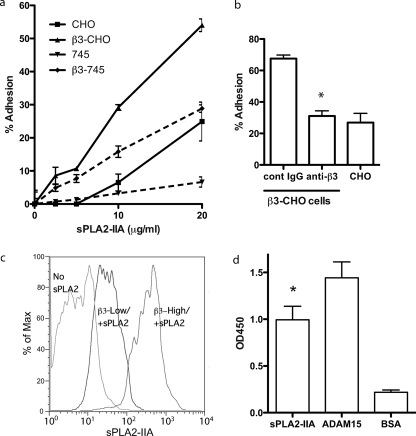
sPLA2-IIA Bound to Integrin αvβ3—We have previously shown that CHO cells that express human β3 (designated β3-CHO) bound to several different ligands to αvβ3 (e.g. angiostatin (31), cardiotoxin (32), ADAM15 (22), and FGF1 (28)) while mock-transfected CHO cells did not. Thus β3-CHO cells are useful tools to demonstrate the specific binding of ligands to αvβ3. Because sPLA2-IIA binds to proteoglycans we also used the proteoglycan-deficient variants of CHO cells (pgs745) (26) that express human β3 (designated as β3–745 cells). These cells express αvβ3 as a hamster αv/human β3 hybrid. We found that β3-CHO and β3–745 cells adhered to immobilized sPLA2-IIA at a much higher level than mock-transfected CHO or 745 cells (Fig. 1a). Consistent with the previous report that proteoglycans support binding of positively charged sPLA2-IIA to the cell surface (34), mock-CHO cells adhered to sPLA2-IIA better than mock-transfected 745 cells. These results suggest that the difference in adhesion between β3-CHO and CHO or between β3–745 and 745 reflects the integrin αvβ3-mediated adhesion to sPLA2-IIA. We found that mAb against human integrin β3 subunit (mAb 7E3) effectively reduced the adhesion of β3-CHO cells to the background level (from 67 to about 30%) (Fig. 1b), indicating that the adhesion is specific to αvβ3. These results indicate that αvβ3 mediated cell adhesion to sPLA2-IIA, that proteoglycans partly supported cell adhesion to sPLA2-IIA, consistent with a previous report (18).
FIGURE 1.
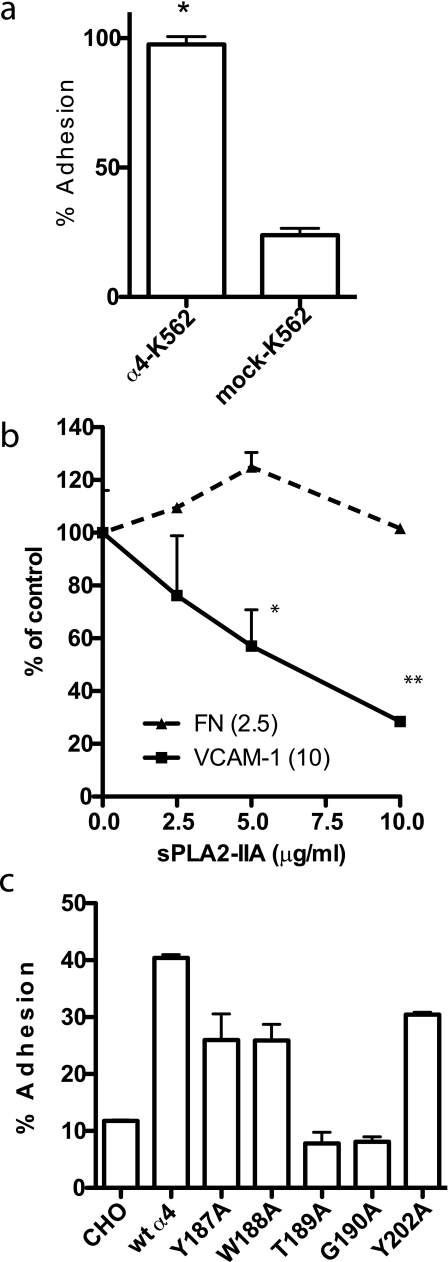
sPLA2-IIA binding to integrin αvβ3. a, cell adhesion to sPLA2-IIA in an αvβ3- and proteoglycan-dependent manner. Wells of a 96-well microtiter plate were coated with sPLA2-IIA at the indicated coating concentrations. The remaining protein-binding sites were blocked with BSA. CHO cells expressing recombinant αvβ3(β3-CHO), mock-transfected CHO cells, proteoglycan-deficient CHO cell variant (pgs745) expressing recombinant αvβ3 (β3–745), and mock-transfected pgs745 cells (105 cells per well in 100 μl of Tyrodes-HEPES with 1 mm MgCl2) were added to the wells. After incubating for 1 h at 37 °C, unbound cells were removed by gentle rinsing, and bound cells were quantified using endogenous phosphatase activity (33). Data are shown as means ± S.E. of triplicate experiments. b, effect of anti-β3 mAb 7E3 on adhesion of β3-CHO cells to immobilized sPLA2-IIA. Adhesion assays were done as described in a. mAb 7E3 (specific to human β3 subunit, function blocking) or purified mouse IgG as a negative control was added to the medium during adhesion assays at 10 μg/ml. *, p < 0.05 between control IgG and anti-β3 (7E3) by Student's t test. c, binding of FITC-labeled sPLA2-IIA to αvβ3 on the cell surface. β3-CHO cells (about 50% are positive in αvβ3 expression) were harvested with 3.5 mm EDTA in phosphate-buffered saline. Cells were double-stained with (i) FITC-labeled sPLA2-IIA (10 μg/ml in the presence of 10 mm Mg2+ at room temperature for 30 min) and (ii) with non-blocking anti-human integrin β3 subunit mAb AV10 and PE (phycoerythrin)-conjugated secondary antibody. Bound FITC (FL1) and PE (FL2) were quantified in flow cytometry. FITC binding to the PE-positive population (αvβ3-high) and the PE–negative population (αvβ3-low) is shown. d, binding of recombinant soluble αvβ3 to immobilized sPLA2-IIA. Soluble αvβ3 was incubated with sPLA2-IIA, the ADAM15 disintegrin domain (a positive control) (22), and BSA, which were immobilized to wells of a 96-well microtiter plate (20 μg/ml coating concentration). Remaining protein-binding sites were blocked with BSA. Bound αvβ3 was detected using anti-β3 mAb (AV10) and peroxidase-conjugated anti-mouse IgG antibody. Bound peroxidase activity was measured. Data are shown as means ± S.E. of triplicate experiments. *, p < 0.05 between sPLA2-IIA and BSA by Student's t test.
Next we tested if soluble sPLA2-IIA binds to cell surface αvβ3. We first stained the β3-CHO cells for β3 expression with anti-β3 mAb and PE-labeled secondary antibody, and then incubated with FITC-labeled sPLA2-IIA. We selected cell populations that express high or low levels of β3 and tested if sPLA2-IIA binding is related to the levels of β3 expression in flow cytometry. We found that FITC-sPLA2-IIA bound at much higher levels to cells expressing high level αvβ3(β3-high) than to cells expressing little αvβ3(β3-low) (Fig. 1c), indicating the significant contribution of αvβ3 in sPLA2-IIA binding. The low level binding of sPLA2-IIA to αvβ3-low cells probably represents contribution of proteoglycans and other receptors.
We next demonstrated that recombinant soluble αvβ3 bound to immobilized sPLA2-IIA in ELISA-type assays (Fig. 1d). Soluble αvβ3 bound to the disintegrin domain of ADAM15, a known αvβ3 ligand (22) (as a positive control), but did not significantly bound to BSA (as a negative control). These results indicate that αvβ3 directly binds to sPLA2-IIA. We showed that soluble sPLA2-IIA bound to immobilized αvβ3 in surface plasmon resonance studies at a high affinity (see below).
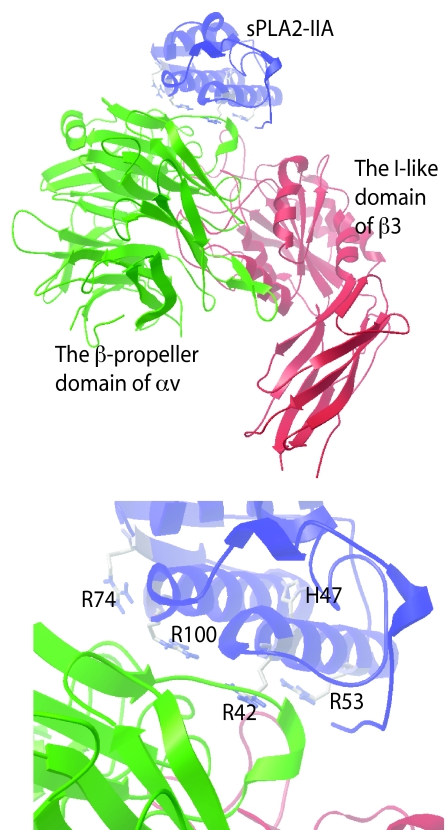
Docking Simulation of Interaction between Integrin and sPLA2-IIA—To determine how sPLA2-IIA binds to integrin αvβ3, we performed docking simulation by using AutoDock 3. AutoDock is a set of docking tools widely used for predicting the conformation of small ligands bound to receptors (35–37), and the methods has been used to predict protein-protein complex poses (38). We performed 50 dockings, each one starting with a random initial position and orientation of sPLA2-IIA (PDB code 1DCY1 and 1AYP) with respect to the headpiece of αvβ3 (PDB code 1L5G). The results were clustered together by positional RMSD (0.5 Å) into families of similar poses. 24 of the 50 docking poses clustered well with the lowest docking energy (cluster 1) with a docking energy -26.1 kcal/mol with 1DCY1 and -25.5 kcal/mol with 1AYP. These results predict that the docking pose of cluster 1 represents the most probable stable sPLA2-IIA pose upon binding to αvβ3 (Fig. 2a)). While the poses obtained by using two structures are slightly different, the integrin-binding sites are overlapping. This model predicts that the αvβ3 integrin-binding interface of sPLA2-IIA with integrin αvβ3 does not include the catalytic center of sPLA2-IIA (e.g. His-47). The interface on the αvβ3 side contains several αv (green) or β3 (red) residues that have been shown to be critical for ligand binding by mutagenesis and crystallographic studies (39–41). Thus the predicted docking model is consistent with the previous biochemical studies of integrin-ligand interaction.
FIGURE 2.
Docking simulation of αvβ3-sPLA-IIA interaction. a, a model of αvβ3-sPLA2-IIA interaction from cluster 1, in which 24 of 50 docking poses clustered with the lowest docking energy (-25.5 kcal/mol) within 0.5 Å RMSD. b, several amino acid residues within the predicted integrin binding interface of sPLA2-IIA.
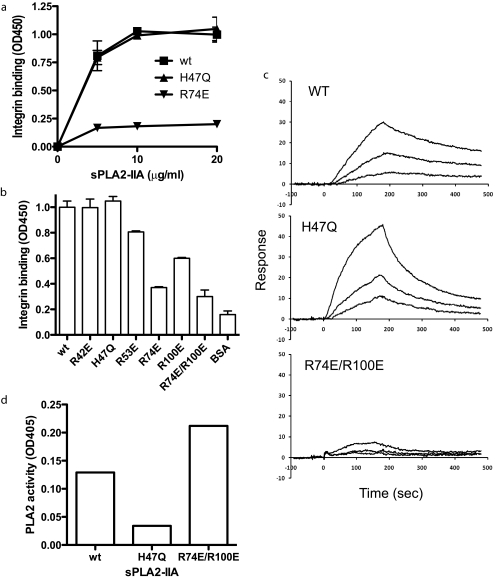
Mutagenesis Study of the Predicted Integrin-binding Interface of sPLA2-IIA—To test if the docking model is correct, we introduced several mutations within the predicted interface of sPLA2-IIA with integrin αvβ3. Positively charged amino acids at the predicted interface common to 1AYP and 1DCY (Arg-42, Arg-53, Arg-74, and Arg-100) were mutated to Glu (charge reversal mutagenesis) (Fig. 2b). We found that the R74E and R100E mutations in sPLA2-IIA reduced the binding to soluble αvβ3, while the R42E and R53E mutations had little or no effect on integrin binding (Fig. 3, a and b). We generated the catalytically inactive mutant of sPLA2-IIA by mutating His-47 to Gln (the H47Q mutation) as a control. The H47Q mutation did not affect the binding to soluble αvβ3. These results are consistent with the prediction by docking simulation. The combined R74E/R100E mutation effectively reduced the binding of sPLA2-IIA to soluble αvβ3 (Fig. 3b), and was used for further analysis of the role of integrins in sPLA2-IIA signaling.
FIGURE 3.
Localization of the integrin-binding site of sPLA2-IIA. a, binding of sPLA2-IIA mutants to soluble αvβ3. Based on the docking model we introduced mutations in the integrin-binding interface of sPLA2-IIA. The mutant proteins were tested for binding to soluble integrin αvβ3. The low level background binding to BSA was subtracted. b, summary of the mutagenesis study of sPLA2-IIA-integrin interaction. The binding of soluble αvβ3 to immobilized Wt and mutant sPLA2-IIA was determined at the saturating conditions (20 μg/ml coating concentrations). The H47Q mutation is located in the catalytic center of the enzyme. c, the surface plasmon resonance study of sPLA2-IIA binding to αvβ3. Soluble integrin αvβ3 was immobilized to a sensor chip, and the binding of Wt sPLA2-IIA, and the H47Q and R74E/R100E mutants (concentrations at 2, 1, and 0.5 nm) was analyzed. KD was calculated as 2.11 × 10-7 m for Wt sPLA2-IIA, 4.47 × 10-8 m for H47Q, and 1.08 × 10-6 m for R74E/R100E. d, effect of sPLA2-IIA mutations on PLA2 activity. PLA2 activity was measured as described under “Experimental Procedures.” A similar result was obtained from another independent experiment.
We determined binding kinetics of Wt and mutant sPLA2-IIA to αvβ3 using surface plasmon resonance (SPR) (Fig. 3c). We immobilized recombinant soluble αvβ3 to a sensor chip and monitored the association and dissociation of Wt or mutant sPLA2-IIA in solution with αvβ3 on the chip. Wt and H47Q sPLA2-IIA showed high affinity to αvβ3(KD = 2.11 × 10-7 m and 4.47 × 10-8 m, respectively) and the R74E/R100E mutant showed much lower affinity (KD = 1.08 × 10-6 m). This is consistent with the results obtained by ELISA-type binding assays.
PLA2 activity was measured to confirm that the integrin binding-defective mutation did not affect catalytic activity (Fig. 3d). The data suggest that the H47Q mutation reduced PLA2 activity (while its integrin binding was not affected). In contrast, the R74E/R100E mutation did not affect PLA2 activity (while its integrin binding was suppressed).
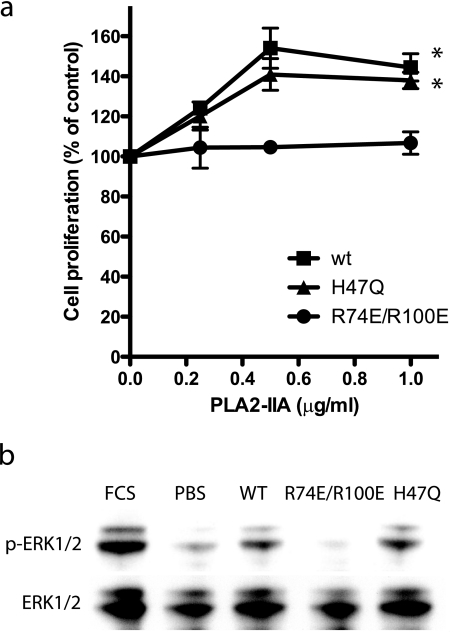
sPLA2-IIA Induced Proliferation of Monocytic Cells in an Integrin-dependent Manner—It has been reported that sPLA2-IIA induced proliferation of LNCap prostate cancer cells in a dose-dependent manner (10) and induced resistance to apoptosis in baby hamster kidney (BHK) cells (42). To address if sPLA2-IIA-integrin interaction is involved in monocyte proliferation, we tested if the integrin binding-defective or catalytically inactive mutations affects sPLA2-IIA's ability to induce proliferative signals in monocytic U937 cells. Notably, we found that Wt sPLA2-IIA and H47Q induced robust proliferation of U937 cells, but R74E/R100E did not (Fig. 4a). These results suggest that integrin binding to sPLA2-IIA plays a critical role in sPLA2-IIA-induced cell proliferation, but catalytic activity is not important in this process. Consistent with this observation, Wt sPLA2-IIA and H47Q induced, but R74E/R100E did not induce, ERK1/2 activation in U937 cells (Fig. 4b). While it has been reported that U937 cells express αvβ3 (43), 7E3 did not block adhesion of U937 cells to sPLA2-IIA (not shown), suggesting that other receptors are involved in the binding of sPLA2 to U937 cells. We hypothesized that integrin α4β1, a major integrin in U937 cells (44), may be involved in sPLA2-IIA signaling in U937 cells.
FIGURE 4.
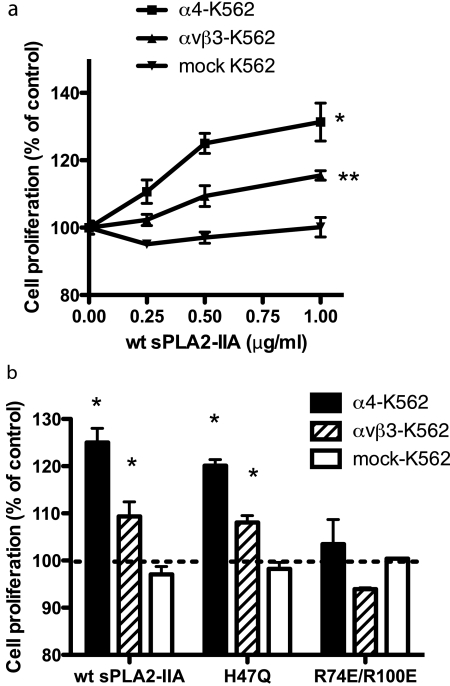
sPLA2-IIA-induced proliferation of U937 human monocytic lymphoma cells in an-integrin-dependent manner. a, effect of sPLA2-IIA mutants on cell proliferation. U937 cells (αvβ3+, α4β1+) were plated in wells of 96-well plates (10,000 cells/well), and serum-starved for 48 h. After treatment with Wt or mutant sPLA2-IIA for 48 h, we measured cell proliferation by MTS assays. p < 0.0001 between Wt and R74E/R100E by 2-way analysis of variance. b, Wt and catalytically inactive H47Q mutant of sPLA2-IIA induced ERK1/2, but integrin binding-defective R74E/R100E mutant did not. U937 cells were serum-starved for 24 h, and stimulated with Wt and mutant sPLA2-IIA (0.5 μg/ml) for 10 min at 37 °C. Cell lysates were analyzed by Western blotting with anti-phospho ERK1/2 or anti-ERK1/2 antibodies. The blot is representative of three independent experiments.
We tested this hypothesis using K562 cells that express recombinant α4β1(α4-K562 cells, α4β1+/α5β1+). α4-K562 cell adhered to immobilized Wt sPLA2-IIA better than mock-transfected K562 cells (α5β1+) (Fig. 5a), suggesting that α4β1 interacts with sPLA2-IIA. We obtained similar results with CHO cells that express human α4(α4-CHO) and mock-transfected CHO cells (Fig. 5c), suggesting that the observation is not cell-specific. However, a small molecular weight α4β1 ligand (LLP2A) (45) or anti-α4 mAb SG73 or P4C2 (23) did not block α4β1 binding to sPLA2-IIA in transfected K562 cells (data not shown). To confirm that sPLA2-IIA binds to α4β1, we tested if sPLA2-IIA competes with known α4β1-specific ligand such as VCAM-1 for binding to α4β1. We found that sPLA2-IIA suppressed α4β1-mediated adhesion of U937 cells to VCAM-1 (Fig. 5b), whereas sPLA2-IIA did not suppress cell adhesion to α5β1-specific ligand fibronectin domains 8–11. This suggests that sPLA2-IIA competed with VCAM-1 for binding to α4β1.
FIGURE 5.
sPLA2-IIA binding to integrin α4β1. a, cells expressing recombinant α4β1 adhered to Wt sPLA2-IIA better that cells expressing other recombinant integrins. We used transfected K562 cells clonally express different human integrins for adhesion assays. Low background adhesion to BSA was subtracted. Data are shown as means ± S.E. of triplicate experiments. b, sPLA2-IIA blocked adhesion of U937 cells to VCAM-1, but did not block adhesion of U937 cells to the cell-binding domain of fibronectin. Wells of 96-well microtiter plate were coated with VCAM-1 and FN-GST, and incubated with U937 cells (1 × 104 cells/plate) in the presence of the increasing concentrations of sPLA2-IIA for 1 h at 37 °C. Adherent cells were quantified using endogenous phosphatase after gently rinsing the well to remove unbound cells. Data are shown as means ± S.E. of triplicate experiments. *, p = 0.0101 and **, p < 0.0001 by Student's t test. c, amino acid sequence in the α4 subunit that is critical for VCAM-1 and CS-1 binding is also critical for sPLA2-IIA binding. CHO cells that clonally express Wt or mutant α4 (23) were used for adhesion assays. Data are shown as means ± S.E. of triplicate experiments. Similar results were obtained using K562 cells expressing α4 mutants (not shown).
To confirm that sPLA2-IIA binds to α4β1, we mapped the sPLA2-IIA-binding site in the α4 subunit. We previously identified several amino acid residues in the α4 subunit (e.g. Tyr-187 and Gly-190) that are critical for VCAM-1 and CS-1 binding (23) and for binding of LLP2A (45) to α4 by introducing point mutations in the α4 subunit. These amino acid residues are located within the ligand-binding site of integrins (46). We tested if these α4 mutations affect sPLA2-IIA binding to α4β1 using CHO cells that express Wt or mutant human α4 as a human α4/hamster β1 hybrid (23). We found that mutating Tyr-189 and Gly-190 of α4 to Ala blocked binding to sPLA2-IIA (Fig. 5c), suggesting that sPLA2-IIA-binding site in α4 is close to or overlaps with the VCMA-1 or CS-1 binding sites. We obtained similar results using K562 cells that express the α4 mutants (data not shown). These findings are consistent with the observation that sPLA2-IIA and VCAM-1 competed for binding to α4β1.
To test if αvβ3 and α4β1 individually mediate sPLA2-IIA-induced cell proliferation, we used K562 cells that overexpress αvβ3 or α4β1 (αvβ3- and α4-K562 cells, respectively). sPLA2-IIA induced proliferation of α4-K562 cells, and to a less extent, of αvβ3-K562 cells. sPLA2-IIA did not induce proliferation of mock-transfected K562 cells (Fig. 6a). Consistent with the results with U937 cells, H47Q induced proliferation of αvβ3- and α4-K562 cells, but R74E/R100E did not induce proliferation of αvβ3- or α4-K562 (Fig. 6b). These results suggest that sPLA2-IIA-induced proliferation of K562 cells required the binding of sPLA2-IIA to α4β1 or αvβ3, but did not require catalytic activity of sPLA2-IIA.
FIGURE 6.
sPLA2-induced proliferation of K562 cells in an integrin-dependent and catalytic activity-independent manner. a, Wt sPLA2-IIA enhanced proliferation of K562 cells that clonally express αvβ3 or α4β1, but not mock-K562 cells. Cells were serum-starved for 48 h and stimulated with sPLA2-IIA for 48 h. Cell proliferation was determined by MTS assays. Data are shown as means ± S.E. of triplicate experiments. α4- and αvβ3-K562 cells proliferated faster than mock-K562 cells. p < 0.0001 (α4-K562) and **, p = 0.0353 (αvβ3-K562) compared with mock K562 cells by 2-way analysis of variance. b, sPLA2-IIA-induced cell proliferation required integrin binding but did not require catalytic activity. Serum-starved cells were stimulated with Wt or mutant sPLA2-IIA (0.5 μg/ml) for 48 h. Data are shown as means ± S.E. of triplicate experiments. α4-K562 and αvβ3-K562 cells grew faster than mock K562 cells with Wt and H47Q sPLA2-IIA (*, p < 0.05 by Student's t test).
DISCUSSION
The present study establishes for the first time that human sPLA2-IIA specifically bound to integrin αvβ3 at a high affinity (KD 2 × 10-7 m). Using docking simulation and mutagenesis, we developed an integrin binding-defective mutation of sPLA2-IIA (the R74E/R100E mutation) that effectively reduced αvβ3 binding without affecting catalytic activity. In contrast the H47Q mutation destroyed catalytic activity, but did not reduce αvβ3 binding. SPR studies showed that the R74E/R100E mutation markedly reduced the binding affinity to αvβ3, but the H47Q mutant did not. These results are consistent with the prediction from the simulation, and that the integrin-binding site is distinct from the catalytic center or the M-type receptor-binding site, in which Gly-30 and Asp-49 of sPLA2-IIA are involved (47).
Integrin αvβ3 is a ubiquitous receptor that is expressed on a variety of cell types (48, 49). Consistent with its expression profile in vivo, αvβ3 plays a key role in the initiation or progression of several human diseases, including rheumatoid arthritis, cancer, and ocular diseases, and cardiovascular diseases (48, 49). Endothelial cells are primary targets in angiogenesis in chronic inflammation and cancer, and activated endothelial cells express high levels of αvβ3 (48). Macrophages represent a major mononuclear cell population in inflammation (50), and macrophages express high level αvβ3. Its expression is modulated by several cytokines (e.g. interleukin-4, tumor necrosis factor-α) and growth factors (e.g. platelet-derived growth factor, fibroblast growth factor). αvβ3 is consistently detected on the macrophages in early and advanced human atherosclerotic lesions, and its expression is up-regulated by atherogenic stimuli (oxidized low-density lipoprotein, macrophage colony-stimulating factor) in vitro (50). These reports suggest that sPLA2-IIA and αvβ3 co-exist in the inflammatory lesion and directly connect the pro-inflammatory action of sPLA2-IIA and αvβ3, the newly identified receptor of sPLA2-IIA.
We also presented evidence that α4β1 that is widely expressed in immunocompetent cells (19) mediated sPLA2-IIA binding using cells that express recombinant α4. Although mAbs or small molecular weight ligand tested against α4 did not significantly inhibit α4β1-sPLA2-IIA interaction, we showed that sPLA2-IIA competed with VCAM-1 for binding to α4β1. Also, amino acid residues within the ligand-binding site of α4 (Tyr-189 and Gly-190) that are critical, or close to the critical residues, for VCAM-1 and CS-1 binding were also critical for sPLA2-IIA binding. These findings suggest that sPLA2-IIA binds to α4β1 in a ligand-binding site common to those for other known α4β1 ligands.
We showed that Wt sPLA2-IIA and H47Q induced proliferation and ERK1/2 activation in U937 cells (αvβ3+, α4β1+), while R74E/R100E did not, suggesting that sPLA2-IIA induced proliferative signals of monocytic cells in an integrin-dependent manner. These observations directly connect the pro-inflammatory functions of sPLA2-IIA and integrins. Although relative contribution of α4β1 and αvβ3 in sPLA2-IIA-induced proliferative signals in U937 cells is unclear, we showed α4β1 and to a less extent αvβ3 can individually mediate cell proliferation using αvβ3- and α4-K562 cells. In both cases sPLA2-IIA induced cell proliferation in an integrin-dependent and catalytic activity-independent manner. Because K562 cells have very low proteoglycans (51), the effect of sPLA2 binding to proteoglycans is not important in this cell type.
It has been reported that specific inhibitors of sPLA2-IIA catalytic activity S-5920/LY315920Na and S-3013/LY333013 failed to demonstrate a significant therapeutic effect in rheumatoid arthritis (52) and asthma (53). The present results suggest that sPLA2-IIA-integrin interaction is a novel potential therapeutic target in inflammation. Because anti-α4 antibodies we used and a small molecular weight α4 ligand (LLP2A) did not block α4β1-sPLA2-IIA interaction, we were not able to directly test the role of sPLA2-IIA-α4β1 interaction in inflammation. Function-blocking antibodies against integrins (including α4β1) have been selected for inhibition of cell adhesion to large extracellular matrix proteins (such as fibronectin). Because sPLA2-IIA is a small ligand, it is not surprising that the antibodies against α4 did not effectively block sPLA2-IIA-α4β1 binding. We will need to develop anti-α4 antibodies that block sPLA2-IIA binding to α4β1. Also, the docking simulation predicted that sPLA2-IIA will not occupy the RGD motif-binding site between the α and β subunits. This will probably explain why LLP2A did not block α4β1-sPLA2-IIA interaction, because LLP2A is expected to bind to the LDV motif (which is related to RGD) binding site in α4β1 (45). It would be important to develop antagonists that effectively block this interaction to fully evaluate the significance of this interaction in future studies.
Acknowledgments
We thank Eric Brown for reagents. We have no conflicting financial interests.
This work was supported, in whole or in part, by National Institutes of Health Grant AG027350 (to Y. T.). This work was also supported by American Heart Association Grant-in-aid 0555102Y. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PLA2, phospholipase A2; IL, interleukin; TNF, tumor necrosis factor; Wt, wild type; BSA, bovine serum albumin; FITC, fluorescein isothiocyanate; RMSD, root mean square deviation; FN, fibronectin; VCAM, vascular cell adhesion molecule.
References
- 1.Tatulian, S. A. (2001) Biophys. J. 80 789-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Six, D. A., and Dennis, E. A. (2000) Biochim. Biophys. Acta 1488 1-19 [DOI] [PubMed] [Google Scholar]
- 3.Gelb, M. H., Valentin, E., Ghomashchi, F., Lazdunski, M., and Lambeau, G. (2000) J. Biol. Chem. 275 39823-39826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelb, M. H., Cho, W., and Wilton, D. C. (1999) Curr. Opin. Struct. Biol. 9 428-432 [DOI] [PubMed] [Google Scholar]
- 5.Valentin, E., and Lambeau, G. (2000) Biochim. Biophys. Acta 1488 59-70 [DOI] [PubMed] [Google Scholar]
- 6.Jaross, W., Eckey, R., and Menschikowski, M. (2002) Eur. J. Clin. Investig. 32 383-393 [DOI] [PubMed] [Google Scholar]
- 7.Niessen, H. W., Krijnen, P. A., Visser, C. A., Meijer, C. J., and Erik Hack, C. (2003) Cardiovasc. Res. 60 68-77 [DOI] [PubMed] [Google Scholar]
- 8.Jiang, J., Neubauer, B. L., Graff, J. R., Chedid, M., Thomas, J. E., Roehm, N. W., Zhang, S., Eckert, G. J., Koch, M. O., Eble, J. N., and Cheng, L. (2002) Am. J. Pathol. 160 667-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Q., Patel, M., Scott, K. F., Graham, G. G., Russell, P. J., and Sved, P. (2006) Cancer Lett. 240 9-16 [DOI] [PubMed] [Google Scholar]
- 10.Sved, P., Scott, K. F., McLeod, D., King, N. J., Singh, J., Tsatralis, T., Nikolov, B., Boulas, J., Nallan, L., Gelb, M. H., Sajinovic, M., Graham, G. G., Russell, P. J., and Dong, Q. (2004) Cancer Res. 64 6934-6940 [DOI] [PubMed] [Google Scholar]
- 11.Tada, K., Murakami, M., Kambe, T., and Kudo, I. (1998) J. Immunol. 161 5008-5015 [PubMed] [Google Scholar]
- 12.Triggiani, M., Granata, F., Balestrieri, B., Petraroli, A., Scalia, G., Del Vecchio, L., and Marone, G. (2003) J. Immunol. 170 3279-3288 [DOI] [PubMed] [Google Scholar]
- 13.Lambeau, G., Ancian, P., Barhanin, J., and Lazdunski, M. (1994) J. Biol. Chem. 269 1575-1578 [PubMed] [Google Scholar]
- 14.Nicolas, J. P., Lambeau, G., and Lazdunski, M. (1995) J. Biol. Chem. 270 28869-28873 [DOI] [PubMed] [Google Scholar]
- 15.Cupillard, L., Mulherkar, R., Gomez, N., Kadam, S., Valentin, E., Lazdunski, M., and Lambeau, G. (1999) J. Biol. Chem. 274 7043-7051 [DOI] [PubMed] [Google Scholar]
- 16.Murakami, M., Kambe, T., Shimbara, S., Yamamoto, S., Kuwata, H., and Kudo, I. (1999) J. Biol. Chem. 274 29927-29936 [DOI] [PubMed] [Google Scholar]
- 17.Sartipy, P., Johansen, B., Gasvik, K., and Hurt-Camejo, E. (2000) Circ. Res. 86 707-714 [DOI] [PubMed] [Google Scholar]
- 18.Boilard, E., Bourgoin, S. G., Bernatchez, C., Poubelle, P. E., and Surette, M. E. (2003) Faseb. J. 17 1068-1080 [DOI] [PubMed] [Google Scholar]
- 19.Hynes, R. O. (2002) Cell 110 673-687 [DOI] [PubMed] [Google Scholar]
- 20.Shimaoka, M., and Springer, T. A. (2003) Nat. Rev. Drug Discov. 2 703-716 [DOI] [PubMed] [Google Scholar]
- 21.Takagi, J., Erickson, H. P., and Springer, T. A. (2001) Nat. Struct. Biol. 8 412-416 [DOI] [PubMed] [Google Scholar]
- 22.Zhang, X. P., Kamata, T., Yokoyama, K., Puzon-McLaughlin, W., and Takada, Y. (1998) J. Biol. Chem. 273 7345-7350 [DOI] [PubMed] [Google Scholar]
- 23.Irie, A., Kamata, T., Puzon-McLaughlin, W., and Takada, Y. (1995) EMBO J. 14 5550-5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming, F. E., Graham, K. L., Taniguchi, K., Takada, Y., and Coulson, B. S. (2007) Arch. Virol. 152 1087-1101 [DOI] [PubMed] [Google Scholar]
- 25.Blystone, S., Graham, I., Lindberg, F., and Brown, E. (1994) J. Cell Biol. 127 1129-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esko, J. D., Stewart, T. E., and Taylor, W. H. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 3197-3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, W., and Malcolm, B. A. (1999) BioTechniques 26 680-682 [DOI] [PubMed] [Google Scholar]
- 28.Mori, S., Wu, C. Y., Yamaji, S., Saegusa, J., Shi, B., Ma, Z., Kuwabara, Y., Lam, K. S., Isseroff, R. R., Takada, Y. K., and Takada, Y. (2008) J. Biol. Chem. 283 18066-18075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eto, K., Huet, C., Tarui, T., Kupriyanov, S., Liu, H. Z., Puzon-McLaughlin, W., Zhang, X. P., Sheppard, D., Engvall, E., and Takada, Y. (2002) J. Biol. Chem. 277 17804-17810 [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, L. J., Hughes, L. L., Yu, L., and Dennis, E. A. (1994) Anal. Biochem. 217 25-32 [DOI] [PubMed] [Google Scholar]
- 31.Tarui, T., Miles, L. A., and Takada, Y. (2001) J. Biol. Chem. 276 39562-39568 [DOI] [PubMed] [Google Scholar]
- 32.Wu, P. L., Lee, S. C., Chuang, C. C., Mori, S., Akakura, N., Wu, W. G., and Takada, Y. (2006) J. Biol. Chem. 281 7937-7945 [DOI] [PubMed] [Google Scholar]
- 33.Prater, C. A., Plotkin, J., Jaye, D., and Frazier, W. A. (1991) J. Cell Biol. 112 1031-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuentes, L., Hernandez, M., Nieto, M. L., and Sanchez Crespo, M. (2002) FEBS Lett. 531 7-11 [DOI] [PubMed] [Google Scholar]
- 35.Goodsell, D. S., and Olson, A. J. (1990) Proteins 8 195-202 [DOI] [PubMed] [Google Scholar]
- 36.Morris, G. M., Goodsell, D. S., Halliday, R. S., Fig Huey, R., Hart, W. E., Belew, R. K., and Olson, A. J. (1998) J. Comp. Chem. 19 1639-1662 [Google Scholar]
- 37.Morris, G. M., Goodsell, D. S., Huey, R., and Olson, A. J. (1996) J. Comput. Aided Mol. Des. 10 293-304 [DOI] [PubMed] [Google Scholar]
- 38.Saphire, E. O., Parren, P. W., Pantophlet, R., Zwick, M. B., Morris, G. M., Rudd, P. M., Dwek, R. A., Stanfield, R. L., Burton, D. R., and Wilson, I. A. (2001) Science 293 1155-1159 [DOI] [PubMed] [Google Scholar]
- 39.Takagi, J., Isobe, T., Takada, Y., and Saito, Y. (1997) J. Biochem. 121 914-921 [DOI] [PubMed] [Google Scholar]
- 40.Humphries, J. D., Askari, J. A., Zhang, X. P., Takada, Y., Humphries, M. J., and Mould, A. P. (2000) J. Biol. Chem. 275 20337-20345 [DOI] [PubMed] [Google Scholar]
- 41.Xiong, J. P., Stehle, T., Zhang, R., Joachimiak, A., Frech, M., Goodman, S. L., and Arnaout, M. A. (2002) Science 296 151-155 [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y., Lemasters, J., and Herman, B. (1999) J. Biol. Chem. 274 27726-27733 [DOI] [PubMed] [Google Scholar]
- 43.Nath, D., Slocombe, P. M., Stephens, P. E., Warn, A., Hutchinson, G. R., Yamada, K. M., Docherty, A. J., and Murphy, G. (1999) J. Cell Sci. 112 579-587 [DOI] [PubMed] [Google Scholar]
- 44.Hemler, M. E., Huang, C., Takada, Y., Schwarz, L., Strominger, J. L., and Clabby, M. L. (1987) J. Biol. Chem. 262 11478-11485 [PubMed] [Google Scholar]
- 45.Peng, L., Liu, R., Marik, J., Wang, X., Takada, Y., and Lam, K. S. (2006) Nat. Chem. Biol. 2 381-389 [DOI] [PubMed] [Google Scholar]
- 46.Takada, Y., Ye, X., and Simon, S. (2007) Genome Biol. 8 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambeau, G., Ancian, P., Nicolas, J. P., Beiboer, S. H., Moinier, D., Verheij, H., and Lazdunski, M. (1995) J. Biol. Chem. 270 5534-5540 [DOI] [PubMed] [Google Scholar]
- 48.Eliceiri, B. P., and Cheresh, D. A. (1999) J. Clin. Investig. 103 1227-1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Byzova, T. V., Rabbani, R., D'Souza, S. E., and Plow, E. F. (1998) Thromb. Haemost. 80 726-734 [PubMed] [Google Scholar]
- 50.Antonov, A. S., Kolodgie, F. D., Munn, D. H., and Gerrity, R. G. (2004) Am. J. Pathol. 165 247-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, H. C., Derian, C. K., Andrade-Gordon, P., Hoekstra, W. J., McComsey, D. F., White, K. B., Poulter, B. L., Addo, M. F., Cheung, W. M., Damiano, B. P., Oksenberg, D., Reynolds, E. E., Pandey, A., Scarborough, R. M., and Maryanoff, B. E. (2001) J. Med. Chem. 44 1021-1024 [DOI] [PubMed] [Google Scholar]
- 52.Bradley, J. D., Dmitrienko, A. A., Kivitz, A. J., Gluck, O. S., Weaver, A. L., Wiesenhutter, C., Myers, S. L., and Sides, G. D. (2005) J. Rheumatol. 32 417-423 [PubMed] [Google Scholar]
- 53.Bowton, D. L., Dmitrienko, A. A., Israel, E., Zeiher, B. G., and Sides, G. D. (2005) J. Asthma 42 65-71 [DOI] [PubMed] [Google Scholar]