Abstract
Sphingolipids are present in membranes of all eukaryotic cells. Bioactive sphingolipids also function as signaling molecules that regulate cellular processes such as proliferation, migration, and apoptosis. Human cytomegalovirus (HCMV) exploits a variety of cellular signaling pathways to promote its own replication. However, whether HCMV modulates lipid signaling pathways is an essentially unexplored area of research in virus-host cell interactions. In this study, we examined the accumulation of the bioactive sphingolipids and the enzymes responsible for the biosynthesis and degradation of these lipids. HCMV infection results in increased accumulation and activity of sphingosine kinase (SphK), the enzyme that generates sphingosine 1-phosphate (S1P) and dihydrosphingosine 1-phosphate (dhS1P). We also utilized a mass spectrometry approach to generate a sphingolipidomic profile of HCMV-infected cells. We show that HCMV infection results in increased levels of dhS1P and ceramide at 24 h, suggesting an enhancement of de novo sphingolipid synthesis. Subsequently dihydrosphingosine and dhS1P decrease at 48 h consistent with attenuation of de novo sphingolipid synthesis. Finally, we present evidence that de novo sphingolipid synthesis and sphingosine kinase activity directly impact virus gene expression and virus growth. Together, these findings demonstrate that host cell sphingolipids are dynamically regulated upon infection with a herpes virus in a manner that impacts virus replication.
Human cytomegalovirus (HCMV)3 is a β-herpes virus that is endemic in the human population, and in healthy adults infection with this virus is relatively benign. The dramatic exceptions in which HCMV can cause serious diseases are in congenitally and perinatally infected infants, and in immunocompromised individuals or immunosuppressed transplant recipients (1). In recent years, some studies have suggested a potential link between HCMV and several chronic diseases including atherosclerosis, coronary restenosis, and possibly cancer (1-5). The key aspects of host cell-virus interactions responsible for HCMV persistence and pathogenesis are poorly understood. In addition to advancing fundamental aspects of virus and host cell biology, more detailed knowledge of this virus-host cell interface may also reveal rational points of intervention that could be exploited for the treatment of HCMV-associated diseases.
Many cell processes succumb to regulation by viral gene products, including cell communication systems. For example, HCMV can attenuate or block autocrine and paracrine signaling pathways that culminate in the activation of antiviral cellular defenses (6-12). Many cell signaling pathways are also activated by virus infection. Binding of the HCMV glycoprotein B (gB) to the epidermal growth factor receptor and the αvβ3 integrin coreceptor during virus binding and entry (13, 14) results in activation of phosphoinositide 3-kinase and phospholipase Cγ pathways promoting capsid transport to the nucleus (14-16).
Very early interactions of the virus with cellular receptors also results in activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) and p38 mitogen-activated protein kinases (MAPK), as well as cyclooxygenase-2, which are implicated in promoting expression of immediate early genes (17-20). Expression of viral genes, especially immediate early genes, also stimulates a second wave of MAPK activation, arachidonic acid release, and activation of E2Fs, cyclins, and p53 (17, 19, 21, 22).
However, nothing is known about whether HCMV modulates host cell signaling by sphingolipids. Sphingolipids are present in membranes of all eukaryotic cells and, in recent years, have been shown to function as signaling molecules (reviewed in Ref. 23). Most work has focused on the bioactive sphingolipids sphingosine 1-phosphate (S1P) and ceramide. Whereas S1P tends to promote cell survival and proliferation, ceramide has opposing effects often leading to cessation of cell proliferation and apoptosis (24).
S1P signals both intracellularly and through a family of G protein-coupled receptors termed S1PR1-5 to regulate diverse biological processes (25-27). These include increased cell growth and survival (24, 28), both positive and negative regulation of cell migration (29), vascular integrity (30), and angiogenesis (31, 32). Moreover, S1P impacts several key aspects of immunity including lymphocyte trafficking and chemotaxis, mast cell activity, and the activation and cytokine secretion profiles of T lymphocytes and dendritic cells (reviewed in Refs. 33-35). Dihydrosphingosine 1-phosphate (dhS1P), also known as sphinganine 1-phosphate, lacks the trans double bond at the 4-position. dhS1P is an S1P receptor agonist and can therefore elicit many of the same biological processes as S1P (36, 37), although overall much less is known about dhS1P compared with S1P.
S1P is produced from the phosphorylation of sphingosine, which is created by the deacylation of ceramide within the sphingolipid degradatory pathway, by sphingosine kinases (SphK). Two SphK isoforms have been cloned, termed SphK1 and SphK2 (38-40). dhS1P is produced from the phosphorylation of dihydrosphingosine, which is an intermediate of de novo sphingolipid synthesis, predominantly by the activity of SphK1 but not SphK2 (41). SphK1 is activated by a variety of signals including growth factors (42), immunoglobulin receptors (43, 44), and various G protein-coupled receptors (45), and has been shown to enhance cell proliferation, survival (46), transformation (47), and tumor malignancy (48). The regulation and function of SphK2 is less well understood. In some cellular contexts SphK2 is anti-proliferative and pro-apoptotic (49), whereas in others it may not affect growth and survival (50), or may even promote cell proliferation (48).
Because sphingolipid signaling impacts several fundamental physiological processes, we hypothesized that sphingolipid metabolism is regulated upon infection with viruses. Our results indicate that HCMV infection resulted in the dynamic regulation of both synthetic and degradative enzymes involved in sphingolipid metabolism at the transcriptional level. Additionally, SphK1 transcript abundance, protein levels, and activity were all elevated by HCMV infection. Analysis of the sphingolipid profile of HCMV-infected cells revealed an increase in the amount of dhS1P and ceramides at 24 h, coinciding with the up-regulation SphK1, and consistent with a stimulation of de novo sphingolipid synthesis. Interference with the de novo pathway led to reduced levels of virus replication. Later in infection, there was a decrease in dhS1P at 48 h that does not correlate with an increase in activity of degradative enzymes. Furthermore, small interfering RNA (siRNA) knockdown of SphK1 results in reduced expression of the immediate early gene product, IE1, whereas SphK1 overexpression results in increased accumulation of IE1. Finally, treatment of cells with a drug inhibitor of SphK1 activity causes a delay in virus growth. To our knowledge, this is the first report of regulation of sphingolipid metabolism upon infection by a member of the Herpesviridae family.
EXPERIMENTAL PROCEDURES
Cell Culture—U-373 MG glioma cell lines were obtained from ATCC. U-251 MG cells were provided by Dr. Allan Yates. Glioma cell lines were maintained in Eagle's minimum essential medium containing 10% fetal bovine serum, non-essential amino acids, and sodium pyruvate (all media from Mediatech, Herndon, VA). MRC-5 human primary fibroblasts were obtained from ATCC and maintained in modified Eagle's medium supplemented with 10% fetal calf serum, 1.7 mm sodium bicarbonate, 1.4 mm sodium chloride, essential and nonessential amino acids, vitamins, and sodium pyruvate at manufacturer-recommended concentrations (Sigma). Human foreskin-derived fibroblasts (HFF cells) were obtained from ATCC and maintained in Dulbecco's modified Eagle's media supplemented with 10% fetal bovine serum. Telomerase-immortalized human foreskin fibroblasts, HFF-TEL, were the kind gift of Tom Shenk and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Human umbilical vein endothelial cells were isolated from vessels as previously described (51) and propagated in endothelial cell growth medium consisting of M-199 (Invitrogen) supplemented with 20% fetal bovine serum (Hyclone, Logan, UT), 22.5 μg/ml bovine brain extract (BioWhitakker, Inc., Walkersville MD), 12 units/ml sodium heparin (Sigma), and 20 mm HEPES buffer. All growth surfaces for endothelial cells were pretreated with human fibronectin (25 μg/ml, Upstate Biotechnology, Lake Placid, NY). Cells were passed weekly by brief trypsin digestion at a ratio of 1:4, and used in experiments at passage 5-7. Cells were maintained at 37 °C in 95% air, 5% CO2.
Viruses—HCMV strain AD169 was purchased from ATCC and were propagated in MRC-5 cells. Virus titers were determined in MRC-5 cells by standard plaque assay (52). CMV strain VHL/E, originally isolated from duodenal biopsy material from a bone marrow transplant recipient (53), was propagated in human umbilical vein endothelial cells as detailed elsewhere (54) to preserve its natural endothelial cytopathogenicity. For UV inactivation of virus, virus stocks were subjected to 100 μ/J/cm2 UV irradiation in a Hoefer UVC 500 UV cross-linker for 5 min. The efficacy of UV treatment and optimal exposure time was measured by exposing virus to UV for increasing intervals of times. MRC-5 cells were then exposed to virus subjected to various irradiation times, and IE1 protein accumulation was measured in cell lysates harvested 16 h after infection by SDS-PAGE (see below) and immunoblot analysis (data not shown).
Antibodies and Drug Inhibitors—Rabbit polyclonal antibodies to SphK1 were made to a peptide (RNHARELVRSEELGRWD) near the N terminus representing residues 57-73 and affinity purified from serum by Quality Controlled Biochemicals (Hopkinton, MA). Antibody to serine palmitoyltransferase 1 (SPT) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antibody to the HCMV IE1 proteins were purchased from the Rumbaugh-Goodwin Institute. Antibodies to the HCMV pp150, UL44, and UL69 proteins were the gift of Bill Britt. Anti-glyceraldehyde phosphate dehydrogenase (GAPDH) was purchased from Chemicon. The SphK inhibitor, 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole HCl, (Calbiochem) was used at 1 μg/ml. HFF cells were exposed to SphK inhibitor or vehicle only (dimethyl sulfoxide) immediately after the virus inoculation period (0.5 pfu/cell). Combined cells and supernatant were assayed for virus by standard titration on MRC-5 cells at the times indicated under “Results.”
Sphingosine Kinase Assay—Sphingosine kinase activity was measured by phosphorylation of sphingosine with [γ-32P]ATP and thin layer chromatography as described (48).
S1P Degrading Enzyme Assay—S1P degrading activity due to SGPPs and SGPL was measured in lysates from mock- and HCMV-infected cells as described (55) with minor modifications. Briefly, cells were lysed by sonication in Buffer A (100 mm HEPES, pH 7.5, 10 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each leupeptin and aprotinin) and lysates were centrifuged to remove unbroken cells. Equal amounts of protein from lysates were incubated with 10 μm [32P]S1P (100,000 cpm/sample) for 1 h at 37 °C. Lipids were extracted and resolved by TLC as for the SphK assay. Radioactive spots were scraped from TLC plates and quantitated by liquid scintillation counting. Activity was calculated as nanomole of S1P degraded per min/mg of protein in comparison to blank samples, without cell lysates, run in parallel.
RNA Interference—Predesigned siRNA oligonucleotides to serine palmitoyltransferase 1 (SPT1), siRNA (ID code 11681), SPHK1 (ID code 1181), and random control siRNA (Silencer Negative control siRNA number 2, catalog number 4613) were purchased from Ambion. The negative control siRNA has no significant homology to known human, mouse, or rat genes. The sequence of the pre-designed siRNAs are as follows: SPHK1, sense 5′-GGCUGAAAUCUCCUUCACGtt-3′, antisense 5′-CGUGAAGGAGAUUUCAGCCtc-3′; SPT1, sense 5′-GCCACAAAACUGUGGUGAAtt-3′, antisense 5′-UUCACCACAGUUUUGUGGCtt-3′. The siRNA oligonucleotides were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 4 h medium was changed to normal growth medium and cells were incubated for the times indicated under “Results.”
SphK1 Expression Constructs and Nucleofection—The open reading frame of human SPHK1 was amplified from HEK293 cell RNA by reverse transcriptase-PCR using the following primers: 5′-GAGGAATTCTGGATCCAGCGGGCGGCCCC-3′; and 5′-GAGCTCGAGTCATAAGGGCTCTTCTGGCGGTGG-3′, containing EcoRI and XhoI linkers. The resulting PCR product was cloned into pcDNA3 (Invitrogen) in-frame with an N-terminal c-myc tag. The dominant negative G82D SPHK1 was generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions to change codon 82 from GGG to GAC. Mutagenic primers were as follows: 5′-GGTCATGTCTGGAGACGACCTGATGCACGAGG-3′ and 5′-CCTCGTGCATCAGGTCGTCTCCAGACATGACC-3′. Identity of clones was verified by sequencing MRC-5. For nucleofections, 1.5 × 106 MRC-5 cells were resuspended in 100 μl of basic nucleofector solution for primary fibroblasts with 3 μg of plasmid DNA and cells were nucleofected using the U23 program in the nucleofector device (Amaxa Biosystems).
Preparation of Cell Lysates and Immunoblotting—Cells grown in 6-well plates were solubilized at the times indicated under “Results” after exposure to the AD169 strain of virus with or without treatment with interfering RNAs. Cells were rinsed in phosphate-buffered saline and solubilized in lysis buffer containing 1% Triton X-100, 50 mm Tris, 150 mm NaCl, and 1% (v/v) protease inhibitor mixture (Sigma). After sonication, lysates were incubated at 4 °C for 30 min, and insoluble material was pelleted by centrifugation. Equivalent amounts of protein from each lysate were separated by electrophoresis in SDS-PAGE and transferred to nitrocellulose sheets (Amersham Biosciences). Sheets were reacted with primary antibodies indicated under “Results,” and subsequently to horseradish peroxidase-conjugated secondary antibody (Santa Cruz). Bound antibodies were visualized using a chemiluminescent detection system (ECL, Amersham Biosciences) and exposure to film. For SphK overexpression, cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, with a construct containing the human SPHK1 open reading frame cloned in-frame with a N-terminal c-MYC epitope tag as described above.
Real Time Quantitative PCR—Real time PCR analysis was performed as previously described (48). Briefly, total RNA was extracted from cultured cells using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, followed by treatment with DNaseI (Ambion, Austin, TX) for 20 min at 37 °C. cDNA was created using the Superscript II First Strand Synthesis System (Invitrogen) according to manufacturer's instructions. PCR were setup using TaqMan® Universal PCR Master Mix (Applied Biosystems, Foster City, CA). Real time PCR analysis was performed using Applied Biosystems ABI PRISM® 7700 Sequence Detection System for 40 cycles. Data were obtained using Sequence Detection System 1.7a software and exported to Microsoft Excel worksheets for analysis. SPHK1, SPHK2, S1P phosphohydrolase 1 and 2 (SGPP1 and SGPP2), and S1P lyase (SGPL1) expression levels were normalized to mock-infected HFF SPHK1 expression at 12 h. S1P receptor (S1PR1-3) expression levels were normalized to mock-infected HFF S1PR1 expression. Expression was determined relative to 18S rRNA as an internal control using the ΔΔCt method as described (48, 56).
Primer sequences of Applied Biosystems Assays-on-Demand™ gene expression assays are proprietary, however, RefSeq accession numbers for genes, location of binding sites for fluorescent probes, and amplicons, ∼50-150 base pairs surrounding the probe location, are provided as follows: S1PR1 (catalog number HS00173499 m1) based on RefSeq NM 001400. Probe was located at base 83 in the exon 1/exon 2 boundary. S1PR2 (catalog number HS00244677 s1) based on RefSeq NM005226, probe location was 968. S1PR3 (catalog number Hs00245464 s1) based on RefSeq NM004230, probe location was 623; SPHK1 (catalog number Hs00184211 m1) based on RefSeq NM021972, probe location was exon 5/6 boundary, base 802; SPHK2 (catalog number Hs00219999 m1) based on RefSeq NM020126, probe location was the exon 3/4 boundary, base 875; SGPP1 (catalog number Hs00229266_m1) based on RefSeq NM030791, probe location at was the exon 1/2 boundary, base 782; SGPP2 (catalog number Hs00544786_m1) based on RefSeq NM152386, probe location was the exon 4/5 boundary, base 649; SGPL1 (catalog number Hs00187407_m1) based on RefSeq NM003901, probe location was the exon 8/9 boundary, base 915.
Sphingolipid Quantitation—Quantitation of sphingolipids in mock- and CMV-infected cells was performed using liquid chromatography tandem mass spectrometry (LC MS/MS) as previously described (57) using the internal standard mixture from Avanti Polar Lipids (Alabaster, AL).
RESULTS
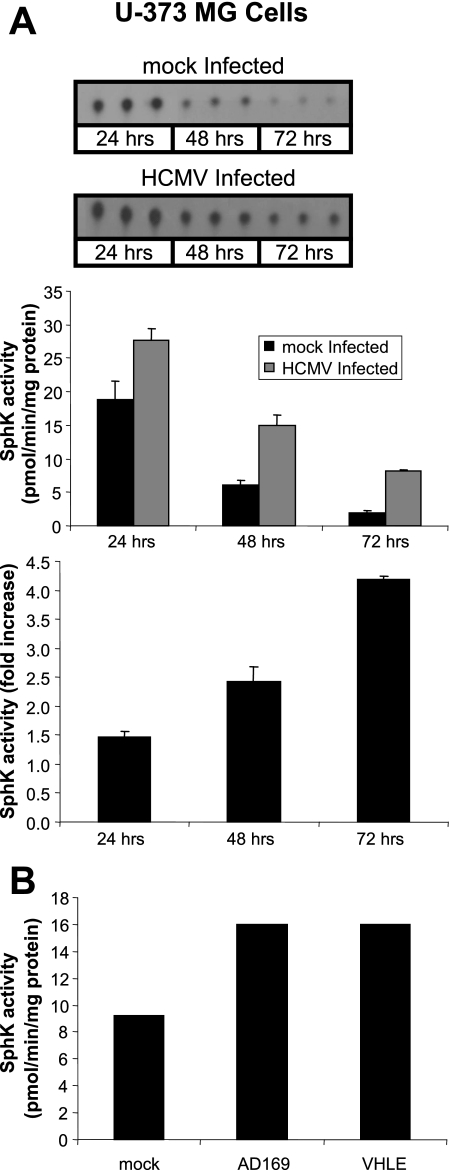
SphK Activity Is Elevated in HCMV-infected Cells—We first sought to investigate the effect of HCMV infection on SphK activity. U-373 MG glioma cells were exposed to 5 pfu/cell of the AD169 strain of HCMV and harvested 24, 48, and 72 h after infection. Cells were solubilized and cell lysates were then assayed for SphK activity as described under “Experimental Procedures.” As shown in the top panel of Fig. 1A, higher levels of S1P were generated in in vitro kinase reactions from infected cell lysates relative to uninfected cell lysates. Quantitation of the S1P spots detected by thin layer chromatography was translated into SphK activity (middle panel, Fig. 1A) allowing us to compare the increase in activity in infected cells relative to uninfected cells (bottom panel, Fig. 1A). Higher levels of SphK activity were observed in HCMV-infected U-373 MG cells at all time points and a notable increase in SphK activity was readily observable at 24 h after infection. We typically observe a decrease in SphK activity with respect to time in culture (48) as was the case in this experiment. Although the overall SphK activity decreased over the 72-h time period in both infected and uninfected cells, the relative SphK activity in infected cells continued to increase over the course of infection such that by 72 h, it was nearly 5-fold higher than that observed in uninfected cells. To ascertain if the increased SphK activity is specific to the laboratory-adapted AD169 strain of HCMV, we compared SphK activity in cell lysates derived from U-373 MG cells infected with AD169 or a clinical isolate strain, VHL/E, at 48 h after infection. SphK activity was higher in both the AD169 and the VHL/E-infected cells when compared with mock-infected U-373 MG cells (Fig. 1B). From these experiments we conclude that HCMV infection of U-373 MG cells results in increased SphK activity, and that maximal induction occurs 48 to 72 h after infection.
FIGURE 1.
Elevated SphK activity in HCMV-infected U-373 MG cells. A, time course of SphK activity of U-373 MG cells infected with HCMV. Shown in the top panel are film images of sphingosine kinase assays analyzed by TLC. Cell lysates from mock-infected U-373 MG cells, or cells exposed to 5 pfu/cell of the AD169 strain of HCMV were analyzed for SphK activity as described under “Experimental Procedures.” Quantitation of S1P spots in the top panel were translated into SphK activity (middle panel). Data are mean ± S.D. of three independent determinations. The -fold increase of SphK activity of mock-infected to virus-infected cells is shown in the bottom panel. B, SphK activity of U-373 MG cells exposed to 1 pfu/cell of the AD169 or VHLE strain of CMV.
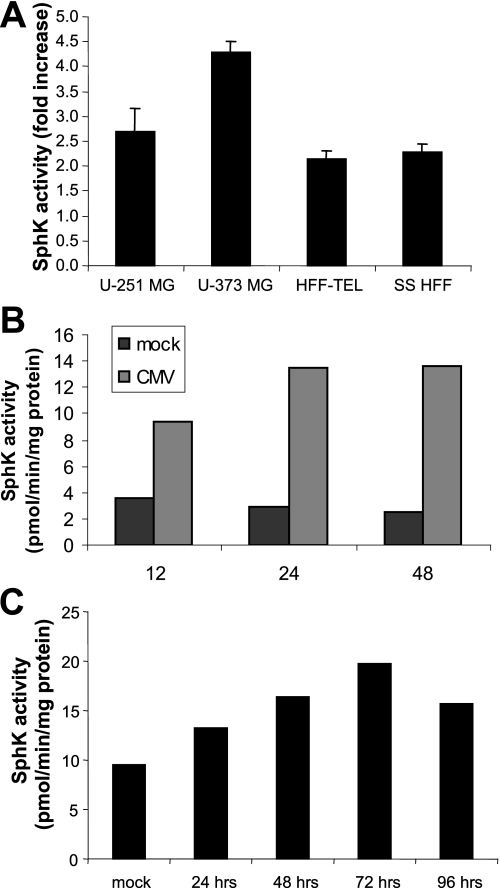
We next investigated whether this phenomenon was specific to HCMV infections of U-373 MG cells or if this was a general characteristic of HCMV infections in other glioma cells as well as fibroblasts. As shown in Fig. 2A, we compared SphK activity in U-373 MG cells to another glioma line, U-251 MG, and to immortalized human fibroblasts (HFF-TEL) as well as primary fibroblasts serum starved for 48 h. Cells were mock-infected or exposed to 5 pfu/cell of the AD169 strain of HCMV. At 48 h after infection, cells were harvested and analyzed for SphK activity. We observed a 4- and 2.5-fold increase in SphK activity in U-373 MG and U-251 cells, respectively. HFF-TEL cells and serum-starved HFF cells exhibited over a 2-fold increase under the same conditions. We also examined SphK regulation in another primary human fibroblast cell line, MRC-5. As show in Fig. 2B, infection of serum-starved MRC-5 cells with HCMV caused a nearly 3-fold increase in SphK activity at 12 h and more than 4-fold increase at 24 and 48 h.
FIGURE 2.
Induction of SphK1 activity is independent of cell type. A, -fold increase of SphK activity in HCMV-infected (5 pfu/cell) U-251 MG, U-373 MG, HFF telomerase immortalized cells (HFF-TEL), and primary HFF cells that were serum starved (SS HFF) for 48 h prior to infection. Data represent the -fold difference between HCMV-infected cells and mock-infected cells for each cell type and are given as mean ± S.D. of three independent determinations. B, time course of SphK activity in serum-starved MRC-5 primary human fibroblast cells infected with HCMV (2 pfu/cell). Data are given as mean ± S.D. of three independent determinations. C, time course of SphK activity in HCMV-infected endothelial cells (1 pfu/cell of the VHL/E strain of HCMV). Cell lysates from mock-infected or HCMV-infected cells were analyzed for SphK activity as described under “Experimental Procedures.”
We next measured SphK activity in endothelial cells (human umbilical vein endothelial cells), which are also known to be productively infected with clinical isolates of HCMV. Cells were left uninfected or exposed to 1 pfu/cell of the VHL/E strain of HCMV. Cells were harvested and analyzed over a 96-h time period. As shown in Fig. 2C, SphK activity was elevated in infected endothelial cells at all tested time points when compared with mock-infected cells. The maximal increase in SphK activity (over 2-fold) was observed at 72 h post-infection. We concluded from this series of experiments that activation of SphK is a common aspect of HCMV infection in primary, immortalized, and transformed cell types.
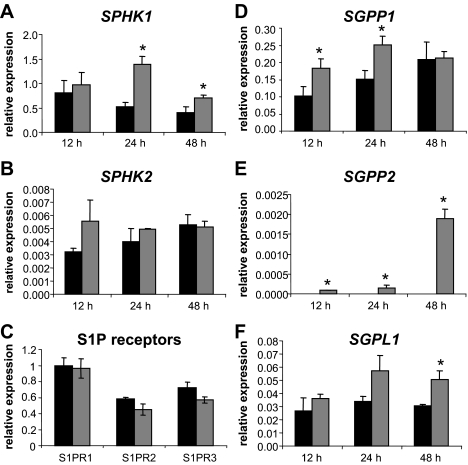
HCMV Infection Alters the Abundance of mRNA Transcripts for Key Enzymes Involved in Sphingolipid Metabolism—As discussed above, both SphK1 and SphK2 phosphorylate sphingosine to produce S1P, whereas predominantly SphK1 phosphorylates dihydrosphingosine to produce dhS1P. These bioactive lipids can then be converted back to sphingosine or dihydrosphingosine through the activity of SGPP1 and SGPP2 or irreversibly degraded by SGPL (Ref. 58 and Fig. 12A). In light of the changes in SphK activity in HCMV-infected cells, we wanted to investigate how virus infection impacts the levels of mRNA transcripts coding for SphK and other important sphingolipid metabolic enzymes. To do this we utilized real time quantitative PCR to measure the transcript levels of the aforementioned enzymes, as well as the S1P receptors, S1PR1-3. In this experiment HFF cells were left uninfected or exposed to 1 pfu/cell of the AD169 strain of HCMV for 12, 24, or 48 h in triplicate and then harvested for RNA extraction. mRNA was quantitated by real time PCR and the mRNA levels of all the enzymes were normalized to mock-infected HFF SphK1 levels at 12 h. S1P receptor (S1PR1-3) mRNA levels were normalized to mock-infected HFF S1PR1 levels. As shown in Fig. 3A, the levels of SPHK1 mRNA were significantly elevated in HCMV-infected cells at 24 and 48 h post-infection (although most prominently at 24 h) when compared with mock-infected cells. However, the levels of SPHK2 mRNA were not significantly elevated in HCMV-infected cells at any time point (Fig. 3B). It should be noted that the overall levels of SPHK2 transcript abundance was ∼100 times lower than that of SPHK1 in all tested samples. The S1P receptors, measured at 24 h post-infection, showed no significant difference in transcript abundance in HCMV-infected cells when compared with mock-infected cells (Fig. 3C).
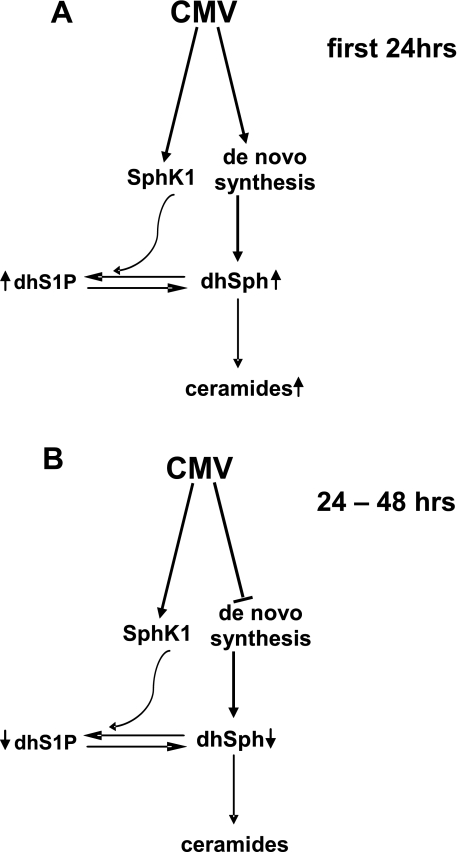
FIGURE 12.
Diagram depicting effects of CMV infection on sphingolipid metabolism. The early phase of infection (A) results in enhanced de novo sphingolipid synthesis as shown by increases in dhS1P and ceramides, whereas in the later phase (B) sphingolipid biosynthesis is diminished.
FIGURE 3.
Quantitation of transcripts specifying enzymes that regulate sphingolipid metabolism. HFF cells were mock-infected or exposed to 1 pfu/cell of the AD169 strain of HCMV for 12, 24, or 48 h and then harvested for RNA extraction. Relative transcript levels were analyzed by real time PCR. Transcript levels in A, B, and D-F are normalized to 18S rRNA levels and are all relative to SPHK1 transcript levels in mock-infected HFF cells harvested at the 12-h time point. S1P receptor mRNA levels are normalized to mock-infected S1RP1 levels. The black bars correspond to values derived from mock-infected cells and the gray bars correspond to values derived from HCMV-infected cells for: A, SPHK1; B, SPHK2; C, S1P receptors (S1PR1-3) at 24 h post-infection; D, SGPP1; E, SGPP2; and F, SGPL. p values of less than 0.05 are indicated with an asterisk. Three independent experiments provided similar results.
Interestingly, HCMV infection also resulted in increased transcript abundance of degradative enzymes SGPP1, SGPP2, and SGPL. Although, the levels of SGPP1 mRNA were significantly elevated in HCMV-infected cells at both the 12- and 24-h time points (Fig. 3D), we found that SGPP1 transcript levels were variable in replicate experiments (data not shown). SGPP2 mRNA levels could not be detected in mock-infected cells but were detected in HCMV-infected cells. All measured SGPP2 mRNA levels were significantly elevated in HCMV-infected cells at all time points when compared with mock-infected cells. Accumulation of SGPP2 mRNA was maximal at 48 h after infection, at which time levels were over 10-fold higher than that measured at 24 h after infection (Fig. 3E). However, the overall abundance of SGPP2 transcripts were much lower than the SGPP1 and SGPL transcripts. SGPL levels were very similar between mock and HCMV-infected cells at the 12-h time point, but were nearly 2-fold higher in HCMV-infected cells at 24 and 48 h; although, only the difference at the 48-h time point was statistically significant (Fig. 3F). We found the same pattern of increased mRNA levels for the degradative enzymes in infected MRC-5 fibroblasts (data not shown). Taken together these results show that HCMV infection results in the dynamic regulation of transcript accumulation for both synthetic (SphK1) and degradative (SGPP2 and SGPL) enzymes in the sphingolipid pathway.
HCMV Infection Alters the Abundance of Sphingolipids in a Time-dependent Manner—Altogether these studies show that virus infection leads to an increase in transcript levels and activity of SphK1 within the first 48 h after infection, at which time there is an increase in the transcript levels of enzymes involved in the degradation of bioactive sphingolipids. Based on these findings, we predicted that S1P and/or dhS1P should be elevated at early times after infection but levels would wane beginning at 48 h after infection. To test this prediction, we employed mass spectrometry to directly measure accumulation of bioactive sphingolipids in infected cells including Sph, dhSph, S1P, dhS1P, ceramide, and dihydroceramide. To examine the amounts of these sphingolipids in the context of HCMV infection, HFF cells were serum starved for 48 h and then mock-infected or exposed to 1 pfu/cell of the AD169 strain of HCMV for 24 or 48 h in triplicate. Cells were then harvested for the extraction of sphingolipids that were quantified by LC MS/MS. As shown in Fig. 4, A-D, when comparing levels of sphingoid bases and their 1-phosphate derivatives in HCMV-infected cells to mock-infected cells at 24 h, only the amount of dhS1P was significantly different, as dhS1P levels were over 2-fold higher in HCMV-infected cells (Fig. 4D). However, at the 48-h time point the levels of all measured sphingoid bases were significantly lower in HCMV-infected cells compared with mock-infected cells (Fig. 4, A-D).
FIGURE 4.
The sphingolipid profile of cells is altered by virus infection. HFF cells were serum starved for 48 h and then left uninfected or exposed to 1 pfu/cell of the AD169 strain of HCMV for 24 or 48 h in triplicate. Cells were then harvested, samples were spiked with an internal standard mixture, and sphingolipids were extracted for quantitation by tandem liquid chromatography/mass spectrometry. The data are given as the amount of each sphingolipid in picomoles per milligram of total protein and represent the mean ± S.D. of three independent determinations. A, quantitation of sphingosine (Sph); B, dihydrosphingosine (dhSph); C, sphingosine-1-phosphate (S1P), and D, dihydrosphingosine 1-phosphate (dhS1P) at 24 and 48 h of mock or HCMV infection. E, quantitation of total ceramide; and F, total dihydroceramide at 24 and 48 h of mock or HCMV infection. An asterisk indicates a p value of less than 0.05.
We also measured the levels of total ceramide and total dihydroceramide in mock- and HCMV-infected cells at 24 and 48 h. In HCMV-infected cells at 24 h both amounts of total ceramide (Fig. 4E) and total dihydroceramide (Fig. 4F) were elevated when compared with mock-infected cells at 24 h, however, only the difference in total ceramide levels at this time point was statistically significant. There was no significant difference in the levels of either total ceramide or total dihydroceramide when comparing HCMV-infected cells to mock-infected cells at 48 h. We also observed a similar trend of moderately increased glucosylceramide and sphingomyelin at 24 h, although these differences did not reach statistical significance (data not shown). Further measurements of different ceramide and dihydroceramide species, containing fatty acids of various chain lengths and degrees of saturation, did not reveal a significant alteration in the proportions of various species in response to HCMV infection (data not shown). These experiments show that a marked increase in the levels of dhS1P, total ceramide, and possibly dihydroceramide, are observed within 24 h of infection, whereas a significant decrease in the levels of Sph, dhSph, S1P, and dhS1P are observed at 48 h after infection. The transient increase in dihydrosphingolipids suggests that de novo sphingolipid synthesis is activated by HCMV during the first 24 h of infection, and is decreased following this time.
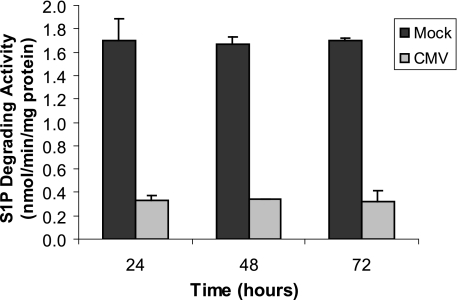
To determine whether increases in S1P degrading enzymes SGPP1, SGPP2, and SGPL contribute to the decrease in S1P and dhS1P at later times, we also measured S1P degrading activity in CMV-infected cells at 24, 48, and 72 h after infection. As shown in Fig. 5, S1P degrading activity is decreased in CMV-infected cells at all times. Thus, it is likely that the decrease in S1P and dhS1P at later times of infection is due to decreased de novo sphingolipid synthesis, in agreement with the lower levels of dihydrosphingosine, which only occurs as an intermediate in the synthetic pathway.
FIGURE 5.
HCMV infection decreases activity of S1P degrading enzymes. MRC-5 cells were mock-infected or exposed to 2 pfu/cell of the AD169 strain of HCMV for 24, 48, or 72 h in triplicate. Cells were then harvested and activity degrading S1P was measured as described under “Experimental Procedures.” Results are mean ± S.D. Three independent experiments provided similar results.
To test the hypothesis that stimulation of de novo sphingolipid synthesis at early times after infection impacts the virus life cycle, we measured virus growth in cells with reduced levels of SPT1 enzyme. SPT catalyzes the condensation of L-serine with a fatty acid acyl-CoA to generate 3-ketodihydrosphingosine. This is the first step in the de novo sphingolipid biosynthesis pathway. To target this step in the pathway, MRC-5 fibroblast cells were transfected with increasing amounts of siRNA specific to SPT or a control siRNA and SPT levels were measured 48 h after transfection. As shown in Fig. 6, we obtained a knockdown of 27, 53, and 54% of SPT protein levels using 1, 2, and 4 μg of siRNA, respectively. Next, we repeated this experiment in triplicate (using 2 μg/well of siRNA) and at 24 h after transfection, cells were exposed to 1 pfu/cell of the AD169 strain of HCMV. At 0, 48, and 96 h after infection, cells and supernatants (2 ml/well) were harvested and frozen. After sonication, infectious virus was measured by plaque assay in MRC-5 cells. The data are mean ± S.D. of three independent determinations of the number of counted plaques per ml within each experimental group. The -fold increase in virus titer over a 96-h period was 400-fold lower in cells treated with SPT siRNA relative to cells treated with a control siRNA (Table 1). From this we conclude that interference with SPT activity negatively impacts virus growth. Taken together with the sphingolipid profile analyses, we conclude that stimulation of de novo sphingolipid biosynthesis at early times after infection acts to promote virus replication in fibroblast cells.
FIGURE 6.
Knockdown of SPT by siRNA. Film image of immunoblot for SPT in cells treated with control or SPT-specific siRNA. MRC-5 cells were transfected with the indicated amount of random sequence, negative control siRNA, or siRNA specific to SPT. At 48 h after transfection cells were solubilized and proteins were separated by SDS-PAGE. Proteins were transferred to nitrocellulose sheets and reacted with antibody to SPT1. Blots were reprobed for GAPDH as a loading control. Integrated optical density (IOD) measurements were made for SPT and GAPDH bands and ratios are shown in the bottom panel.
TABLE 1.
Virus growth in cells with reduced SPT levels
|
Hours after infection |
siRNA treatment |
|
|---|---|---|
| Control | SPT | |
| pfu/ml ± S.D. | ||
| 0 | 4.00 × 102 ± 33 | 3.42 × 102 ± 56 |
| 48 | 1.45 × 103 ± 453 | 1.36 × 103 ± 405 |
| 96 | 2.56 × 105 ± 37,504 | 8.52 × 104 ± 35,360a |
| -Fold increase in virus titer | 639 | 240 |
p value <0.005 relative to control siRNA at 96 hr by Student's t test.
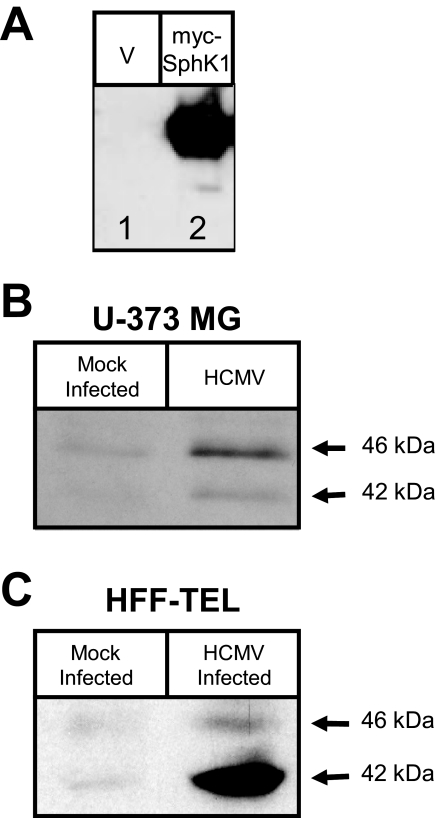
HCMV Infection Results in Increased Abundance of SphK1—Both the real time quantitation of transcripts and the increase in cell-associated dhS1P, but not S1P, directly implicates a role for SphK1 rather than SphK2 in infected cells. The increased SphK1 activity could be due to activation of SphK expressed at basal levels, and/or to an increase in accumulation of SphK protein. To begin to address the nature of SphK1 activation in infected cells, we generated an antibody specific to human SphK1 (Fig. 7A). We found that this antibody recognized a protein of the correct size (45 kDa) in lysates of Chinese hamster ovary-K1 cells transfected with a MYC-tagged SPHK1 gene (lane 2), but not lysates of cells transfected with vector alone (lane 1). This antibody also recognizes two bands of ∼42 and 46 kDa representing endogenous SphK1 protein expressed in U-373 MG cells and fibroblasts (Fig. 7, B and C). The two bands recognized by this antibody likely correspond to two of the three subtypes of SphK1 (1a, 1b, and 1c) that differ at the N termini (59). We used this antibody to compare total cellular levels of SphK1 in lysates from infected and uninfected cells. U-373 MG and HFF-TEL cells were mock-infected or were exposed to 5 pfu/cell of the AD169 strain of HCMV, and accumulation of SphK1 protein was measured at 48 h after infection. We observed an increased accumulation of SphK1 protein levels in HCMV-infected glioblastoma and fibroblast cells (Fig. 7, B and C). Although this does not preclude a role for signal-regulated activation of SphK1 activity, we conclude that the increased SphK activity is at least in part accounted for by increased accumulation of SphK1 protein in HCMV-infected cells.
FIGURE 7.
SphK1 protein levels are elevated upon infection. A, film image of an immunoblot of SphK1 using an antibody that was generated against human SphK1. Cell lysates generated from Chinese hamster ovary-K1 cells transfected with a myc-tagged SphK1 gene (lane 2) or transfected with vector alone (lane 1) were subjected to electrophoresis in an SDS-polyacrylamide gel. Proteins were transferred to a nitrocellulose sheet and probed with rabbit anti-SphK1 to detect the levels SphK1 at ∼45 kDa. B, film image of an immunoblot of SphK1 in U-373 MG cells infected with HCMV. U-373 MG cells were mock-infected or exposed to 5 pfu/cell of the AD169 strain of HCMV for 48 h. SphK1 levels were analyzed by immunoblot as described above. C, HFF-TEL cells were either mock-infected or exposed to 5 pfu/cell of the AD169 strain of HCMV for 48 h. SphK1 levels were analyzed by immunoblot as described above.
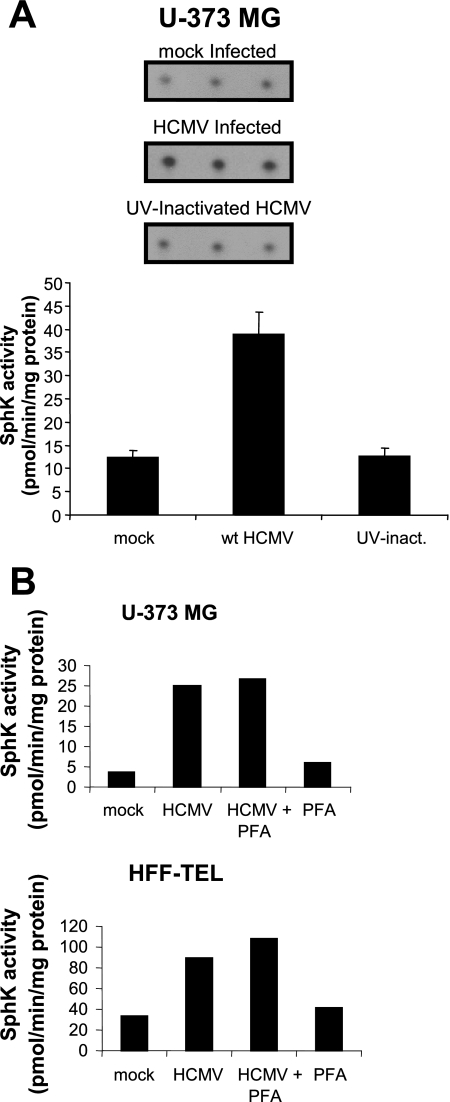
HCMV-mediated Induction of SphK Activity Occurs After Virus Entry but Prior to Viral DNA Replication—We next sought to characterize the stage of the virus life cycle associated with increased SphK activity. U-373 MG cells were mock-infected, exposed to 10 pfu/cell of the AD169 strain of HCMV, or exposed to the equivalent amount of UV-inactivated HCMV and harvested 48 h later for SphK activity. SphK activity in cells exposed to UV-inactivated virus was nearly identical to levels in uninfected cells, whereas 3-fold higher levels of SphK activity were observed in cells infected with the AD169 strain of HCMV (Fig. 8A). This finding suggests that elevated SphK activity cannot be accounted for by activation of signaling pathways caused by virus binding to cell surface receptors.
FIGURE 8.
HCMV-induced increase in SphK activity does not occur with UV-inactivated HCMV and is not affected by phosphonoformic acid (PFA). A, film image of TLC analysis of SphK1 assay (top). U-373 MG cells were mock-infected, exposed to 10 pfu/cell of HCMV, or exposed to 10 pfu/cell of UV-inactivated HCMV and analyzed for SphK activity 48 h after exposure to virus as described under “Experimental Procedures.” S1P spots were quantitated and translated into SphK1 activity (bottom). Data are mean ± S.D. of three independent determinations. B, SphK activity of U-373 MG cells and HFF-TEL cells infected with HCMV under conditions where viral DNA replication is inhibited. U-373 MG cells (top) and HFF telomerase-immortalized cells (bottom) were mock-infected or exposed to 10 pfu/cell of the AD169 strain of HCMV. Cells were cultured in the regular media or media supplemented with phosphonoformic acid. At 48 h after infection cell lysates were then analyzed for SphK activity as described under “Experimental Procedures.”
We next measured SphK activity in mock- and HCMV-infected cells exposed to 1 mm phosphonoformic acid, a drug inhibitor of viral DNA replication. As shown in Fig. 8B, blockade of viral DNA synthesis and subsequently, expression of late genes dependent on DNA synthesis, had no impact on the HCMV-mediated induction of SphK activity. SphK activity in mock-infected U-373 MG cells and cells treated with phosphonoformic acid were below 10 pmol/min/mg of protein. In contrast, SphK activity levels were ∼25 pmol/min/mg of protein in HCMV-infected U-373 MG cells both with and without phosphonoformic acid treatment. The identical pattern of SphK activity was observed in fibroblasts, although the overall magnitude of basal and induced SphK activity was greater in these cells relative to the U-373 MG cells. From these studies we conclude that induction of SphK activity in HCMV-infected cells occurs at a step of the virus life cycle subsequent to virion binding to surface receptors but prior to onset of viral DNA replication.
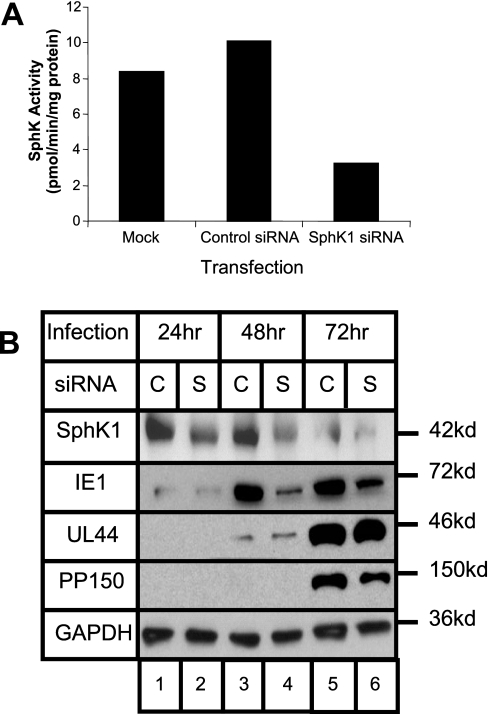
SphK1 Expression Regulates Accumulation of HCMV Immediate Early Gene Product IE1—The emerging picture from these studies is that early events in HCMV infection activate the de novo pathway of sphingolipid synthesis and especially the activity of SphK1. To test whether activation of SphK1 impacts the virus life cycle, we used RNA interference-mediated silencing to block accumulation of SphK1, and then measured accumulation of virus gene products in productively infected cells. In this experiment, primary HFF were transfected with silencing (si)RNA oligonucleotides specific for SPHK1 and subsequently fed with serum-free Dulbecco's modified Eagle's medium. We have previously shown that this siRNA oligonucleotide has no effect on expression of SphK2 (48). After 72 h of culture under serum-free conditions, cells were then harvested for a sphingosine kinase assay. As shown in Fig. 9A, siRNA treatment diminished SphK activity nearly 3-fold as measured by in vitro kinase assays relative to untreated cells and to cells treated with a control siRNA that does not target human genes. We next measured viral gene product accumulation after SphK1 knockdown in infected cells. Fibroblasts grown in 6-well plates were transfected with control siRNA, siRNA specific for SphK1, or were left untreated. After transfection, the cells were serum starved for 48 h and then exposed to 0.5 pfu/cell of AD169. Cells were then harvested 24, 48, and 72 h after infection. Protein lysates were separated by SDS-PAGE, transferred to nitrocellulose, and reacted with antibodies specific for SPHK1 or viral proteins. The SPHK1-specific siRNA led to a reduction in accumulation of SphK1 protein (Fig. 9B, lanes 2, 4, and 6). Viral genes are expressed in a regulated temporal cascade divided into immediate early, early, or late gene classes and we examined representative viral gene products of each of these classes. We observed a specific reduction in the accumulation of the immediate early, IE1 protein (second panel). No decrease was observed for the early protein UL44 at 48 or 72 h after infection and possibly a slight decrease in the late protein pp150 was seen at 72 h post-infection. Virus growth was not affected by this level of SphK1 knockdown when measured at 80 h after infection (data not shown).
FIGURE 9.
SphK knockdown with siRNA reduces accumulation of HCMV immediate early protein. A, SphK activity is suppressed by siRNA specific for SPHK1. HFF cells were left untransfected, transfected with control siRNA or SPHK1 siRNA, and then serum starved for 72 h. The cells lysates were then analyzed for SphK activity as described under “Experimental Procedures.” B, film image of immunoblot for SphK1, GAPDH, and viral proteins. HFF cells were transfected with control siRNA (C) or SphK1 siRNA (S) and subsequently serum starved for 48 h. Then cells were exposed to HCMV at 0.5 pfu/cell for 24, 48, or 72 h prior to harvesting cell lysates. Proteins were subjected to electrophoresis in an SDS-polyacrylamide gel and transferred to a nitrocellulose sheet. Immunoblots were performed as described under “Experimental Procedures” using antibodies to SphK1 and viral proteins. To control for loading of lysates, blots were also reacted to antibodies to cellular GAPDH proteins.
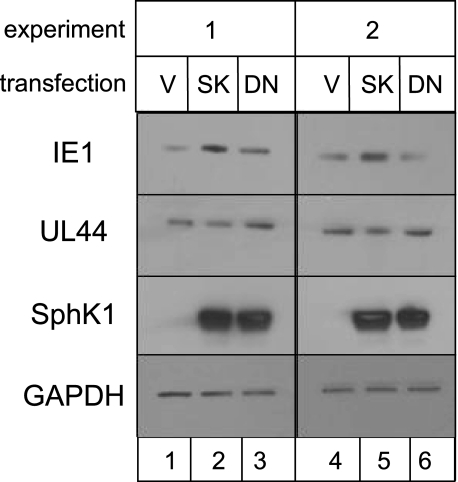
To further investigate the relationship between SphK1 activity and viral IE1 protein levels, fibroblasts were nucleofected with a plasmid harboring the SPHK1 gene, a dominant negative form of SPHK1 (DN-SPHK1), which blocks signal-induced but not basal SphK1 activity, or vector alone. For this experiment we used MRC-5 fibroblasts rather than HFF as MRC-5 provided much higher transfection efficiency, >95% (data not shown). As shown in Fig. 10, plasmid-expressed SphK1 and DN-SphK1 but not endogenous SphK1 was readily detected after a 3-s exposure (compares lanes 2-3 to lane 1 and lanes 5-6 to lane 4). Overexpression of SphK1 correlated with increased IE1 accumulation at 48 h after infection (lanes 2 and 5), whereas expression of the DN-SphK1 did not affect IE1 levels (lanes 3 and 6). UL44 protein accumulation was not substantially changed by overexpression of either SphK1 or DN-SphK1. It is important to note that decreased SphK1 activity mediated by treatment of cells with siRNA is not equivalent to overexpression of DN-SphK1. Whereas SphK1-specific siRNA treatment reduces basal and signal-inducible SphK1 activity, the DN-SphK1 functions to block signal-inducible SphK1 activity but has no effect on the basal SphK1 activity (60). Therefore, we conclude from this series of experiments that accumulation of HCMV IE1 protein can be regulated by basal activity of cellular SphK1.
FIGURE 10.
SphK overexpression increases accumulation of HCMV immediate early protein. Film image of immunoblot for SphK1 and viral proteins. MRC-5 cells were nucleofected with vector alone (V) or expression constructs harboring gene sequences for SPHK1 (SK) or a signal-activated dominant negative form of SPHK1 (DN) as described under “Experimental Procedures.” At 48 h after nucleofection, cells were exposed to 0.5 pfu/cell of HCMV. Cells were solubilized at 48 h after infection. Proteins were subjected to electrophoresis in an SDS-polyacrylamide gel and transferred to a nitrocellulose sheet. Immunoblots were performed as described under “Experimental Procedures” using antibodies to SphK1 and viral proteins (IE1 and UL44). To control for loading, blots were also reacted to antibodies to cellular GAPDH proteins.
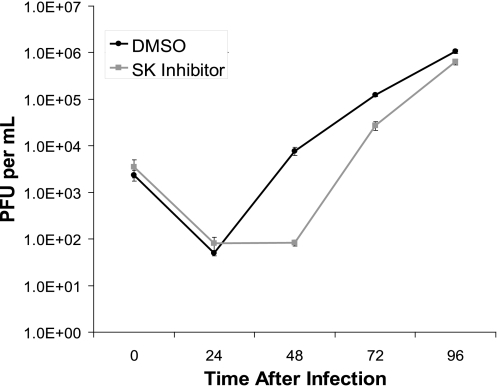
Finally, we measured virus growth in the presence of a drug inhibitor of SphK1 by single step growth curve analysis. After the infection period, HFF cells were exposed to the SK inhibitor or vehicle only and virus growth was measured over a 96-h time period (Fig. 11). Exposure to the SK inhibitor resulted in a 94-fold reduction in infectious progeny virus at 48 h after infection, a 4.6-fold reduction at 72 h after infection, and a 1.7-fold reduction at 96 h after infection. The diminished antiviral activity of this drug over time may be due to pharmacokinetic factors or the ability of viral factors that accumulate late in infection to overcome or compensate for the antiviral activity. In either case, this finding suggests that SphK1 activity is required for optimal virus growth. Interestingly, we did not observe a reduction in virus growth in cells in which SphK1 levels were knocked down by siRNA treatment (not shown). Because siRNA treatment targets SPHK1 transcripts, residual SphK1 protein levels in siRNA-treated cells may be sufficient to optimize virus replication. Although to our knowledge no off-target activities have been reported for this inhibitor, it also remains possible that the reduced accumulation of infectious progeny virus is attributable to activities of this drug unrelated to inhibition of SphK1. In summary, these findings suggest that SphK1 also plays a role in optimizing production of infectious progeny virus in infected cells.
FIGURE 11.
SphK inhibition decreases virus growth. Single step growth curve analysis of HCMV in cells exposed to a drug inhibitor of SphK. HFF cells were serum starved for 48 h and then exposed to 0.5 pfu/cell of AD169. Immediately following the 1-h inoculation period (time 0), cells were treated with 1 μg/ml SphK inhibitor (gray line) or DMSO (black line). Cells and supernatants (2 ml/well) were harvested at intervals of 24 h ranging from 0 to 96 h, sonicated, and infectious virus was measured by plaque assay in MRC-5 cells. The data are mean ± S.D. of three independent determinations of the number of counted plaques/ml within each experimental group.
DISCUSSION
Sphingolipids are important constituents of viral and cellular membranes and act as signaling mediators that regulate several fundamental physiological processes. Herein we provide evidence that cellular sphingolipids and the enzymes that regulate their accumulation are modulated during infection with a herpes virus. The major findings of this study are as follows. 1) HCMV infection leads to an increase in the accumulation of dhS1P and ceramides within the first 24 h of infection, consistent with a stimulation of the de novo pathway of sphingolipid metabolism. 2) Interference with the first step in the de novo pathway of sphingolipid synthesis mediated by SPT results in a reduction in virus growth. 3) By 48 h after infection, levels of S1P, dhS1P, and their precursors are diminished. 4) Protein levels and activity of SphK1, the key enzyme in the generation of S1P and dhS1P, are elevated in HCMV-infected cells. 5) Although the transcripts for cellular enzymes involved in the degradation of S1P and dhS1P are elevated during infection, S1P degradative activity is diminished in infected cells. 6) Sustained accumulation of the IE1 protein, the product of the UL123 gene, is diminished under conditions where SphK1 protein levels were reduced by siRNA treatment and enhanced when SphK1 was overexpressed in infected cells. 7) An SphK1 drug inhibitor exhibits antiviral activity against HCMV in primary human fibroblasts.
Earlier studies suggest that alteration of sphingolipid levels may have profound consequences on the life cycle of herpes viruses. In Niemann-Pick disease cells, which are deficient in sphingomyelinase, production of infectious herpes simplex virus is dramatically reduced (61). Also, addition of exogenous ceramides or stimulation of ceramide synthesis by treatment of cells with sphingomyelinase have been shown to reduce biosynthesis of HCMV glycoprotein B, although treatment of cells with C6-ceramide led to increased accumulation of glycoprotein B (62).
In the present study, we show for the first time that HCMV infection causes accumulation of SphK1 protein and increased activity of SphK1. Induction was observed after infection with laboratory-adapted and clinical isolates in primary fibroblasts and endothelial cells, immortalized fibroblasts, and in two distinct glioma cell lines. Together, these data suggest that induction of SphK1 activity is a common consequence of HCMV infection regardless of the virus strain or cell type.
We have not identified the precise trigger that causes up-regulation and/or activation of SphK1, but our studies show that exposure of cells to UV-inactivated virus is not sufficient to induce SphK1 activity. We also found that inhibition of viral DNA synthesis and consequent expression of strict late genes did not interfere with up-regulation of SphK1 activity. We conclude that one or more immediate early or early gene products is required to elicit the increased accumulation and activity of this enzyme.
SphK1 catalyzes the phosphorylation of sphingosine and dihydrosphingosine to produce S1P and dhS1P, respectively (38). S1P can function extracellularly through a group of cell surface G protein-coupled receptors (63). Moreover, S1P produced within cells can act in an autocrine or paracrine manner through its G protein-coupled receptors (64-66). dhS1P has been shown to be an S1P receptor agonist (36) and can presumably act in much the same way. Our results show a preferential increase in dhS1P rather than S1P in HCMV-infected cells. Extensive literature has emerged documenting the biological activities of S1P, whereas fewer studies have focused on elucidating the biological effects dhS1P. One recent study showed that dhS1P but not S1P mediated SphK-induced up-regulation of matrix metalloproteinase 1 gene expression in a manner consistent with G protein-coupled receptor activity (67). However, the extent of overlap and divergence in the biological activities of these two very similar sphingolipids have yet to be defined.
Our findings demonstrate that increased SphK1 activity promotes sustained accumulation of the IE1 gene product. IE1 is a multifunctional protein and the major viral transcriptional transactivator (68). Viral mutants harboring a deletion of the UL123 gene are viable but exhibit severe growth defects at low multiplicities of infection (69, 70).
Although these studies do not exclude the possibility that SphK1 has activity toward another substrate in virus-infected cells, a likely explanation for this result is that SphK1 activity stimulates formation of dhS1P and activation of S1P receptors. Subsequent transduction of the signals regulated by these receptors may act to promote IE1 biosynthesis or stability.
dhS1P released in this scenario could also function in a paracrine fashion via its G protein-coupled receptors to increase the survival and proliferation of target cells to maintain an ideal environment for infection, and may also affect the extracellular matrix. Evidence in support of a pro-survival activity of sphingolipid signaling comes from studies showing that the respiratory syncytial virus infection of lung epithelial cells results in increased levels of dhS1P (41) and SphK1-dependent activation of two survival pathways mediated by ERK and Akt (71). Interestingly, HCMV encodes several G protein-coupled receptors (72), one of which, US28, has been shown to signal through the Gi pathway (73), which is utilized by all 5 S1P receptors. It seems possible that HCMV has employed two ways of activating Gi-coupled receptors. By encoding these receptors HCMV can activate related pathways within an infected cell, and by boosting the activity of SphK and subsequently dhS1P levels, the virus may facilitate the priming of these pathways in infected or neighboring cells.
It is interesting to note that the increased dhS1P does not come at the expense of its precursor dihydrosphingosine or later derivatives ceramide and dihydroceramide, which are actually increased at 24 h. These data suggest that HCMV causes an up-regulation of de novo sphingolipid synthesis occurring at the endoplasmic reticulum. SPT catalyzes the first step in the de novo pathway. We found that virus replication is reduced in cells in which SPT levels are knocked down by siRNA treatment. These findings provide evidence for the possibility that stimulation of sphingolipid synthesis is a strategy the virus employs to promote its replication. These findings also substantially extend an earlier study linking HCMV infection with glycosphingolipid synthesis (74). Our results also indicate that at 48 h post-infection there is a significant decrease in the levels of sphingosine, dihydrosphingosine, S1P, and dhS1P. The decrease in all these lipids, especially dihydrosphingosine, which only occurs as an intermediate in the de novo pathway of sphingolipid synthesis, suggests that this synthetic pathway is inhibited at this later stage of infection. This is supported by the observation that, although there is a slight increase in mRNA for S1P degrading enzymes at later times of infection, S1P degrading activity was decreased at this stage, whereas SphK activity remained elevated. Thus, the decrease in S1P, dhS1P, Sph, and dhSph likely occurs due to decreased sphingolipid synthesis 48 h after infection, rather than an increase in enzyme activities degrading these lipids. A schematic depicting a summary of our data illustrating how HCMV infection interfaces with this pathway is presented in Fig. 12.
The decrease in sphingolipid synthesis at this stage of infection may represent an attempt of the cell to combat virus infection by lowering the levels of these sphingolipids. Alternatively, this may also be regulated by viral gene products to prevent signaling through S1P receptors at this time.
In summary, our findings demonstrate that sphingolipid metabolism is dynamically regulated in cells infected with a herpes virus. These findings warrant further studies to ascertain if and how virus infection impacts the activities of dhS1P through S1P receptor-regulated cellular processes such as cell survival, proliferation, and motility. Also, considering the important role of S1P receptor-mediated regulation of lymphocyte trafficking and immune responses, further studies to investigate how HCMV and other viruses interface with this signaling system may ultimately shed light on virus replication, spread, and pathogenesis.
Acknowledgments
We thank Dr. William J. Britt for the kind gift of antibodies, Dr. Tom Shenk for the gift of the HFF-TEL cells, and Cathy Jackson for the real time PCR work.
This work was supported, in whole or in part, by National Institutes of Health NINDS Grants R01 NS41517 (to J. R. V. B.), AI51411 (to J. T.), and GM069338 (Lipid MAPS Consortium) (to A. H. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HCMV, human cytomegalovirus; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; S1P, sphingosine 1-phosphate; dhS1P, dihydrosphingosine 1-phosphate; SphK, sphingosine kinase; HFF, human foreskin-derived fibroblasts; HFF-TEL, telomerase-immortalized human foreskin fibroblasts; GAPDH, glyceraldehyde phosphate dehydrogenase; siRNA, small interfering RNA; SGPP, S1P phosphohydrolase; SGPL, S1P lyase; LC MS/MS, liquid chromatography tandem mass spectrometry; Sph, sphingosine; dhSph, dihydrosphingosine; IE, immediate early; DN, dominant negative; pfu, plaque forming unit; SPT1, serine palmitoyltransferase 1.
References
- 1.Cobbs, C. S., Harkins, L., Samanta, M., Gillespie, G. Y., Bharara, S., King, P. H., Nabors, L. B., Cobbs, C. G., and Britt, W. J. (2002) Cancer Res. 623347 -3350 [PubMed] [Google Scholar]
- 2.Horvath, R., Cerny, J., Benedik, J., Jr., Hokl, J., Jelinkova, I., and Benedik, J. (2000) J. Clin. Virol. 16 17-24 [DOI] [PubMed] [Google Scholar]
- 3.Speir, E., Modali, R., Huang, E. S., Leon, M. B., Shawl, F., Finkel, T., and Epstein, S. E. (1994) Science 265391 -394 [DOI] [PubMed] [Google Scholar]
- 4.Harkins, L., Volk, A. L., Samanta, M., Mikolaenko, I., Britt, W. J., Bland, K. I., and Cobbs, C. S. (2002) Lancet 3601557 -1563 [DOI] [PubMed] [Google Scholar]
- 5.Samanta, M., Harkins, L., Klemm, K., Britt, W. J., and Cobbs, C. S. (2003) J. Urol. 170998 -1002 [DOI] [PubMed] [Google Scholar]
- 6.Boyle, K. A., Pietropaolo, R. L., and Compton, T. (1999) Mol. Cell. Biol. 193607 -3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne, E. P., Wing, B., Coleman, D., and Shenk, T. (2001) J. Virol. 7512319 -12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller, D. M., Rahill, B. M., Boss, J. M., Lairmore, M. D., Durbin, J. E., Waldman, J. W., and Sedmak, D. D. (1998) J. Exp. Med. 187675 -683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller, D. M., Zhang, Y., Rahill, B. M., Kazor, K., Rofagha, S., Eckel, J. J., and Sedmak, D. D. (2000) Transplantation 69687 -690 [DOI] [PubMed] [Google Scholar]
- 10.Miller, D. M., Zhang, Y., Rahill, B. M., Waldman, W. J., and Sedmak, D. D. (1999) J. Immunol. 1626107 -6113 [PubMed] [Google Scholar]
- 11.Navarro, L., Mowen, K., Rodems, S., Weaver, B., Reich, N., Spector, D., and David, M. (1998) Mol. Cell. Biol. 183796 -3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Preston, C. M., Harman, A. N., and Nicholl, M. J. (2001) J. Virol. 758909 -8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, X., Huong, S. M., Chiu, M. L., Raab-Traub, N., and Huang, E. S. (2003) Nature 424456 -461 [DOI] [PubMed] [Google Scholar]
- 14.Wang, X., Huang, D. Y., Huong, S. M., and Huang, E. S. (2005) Nat. Med. 11 515-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, N. L., Lewis, J. C., and Kilpatrick, B. A. (1986) Eur. J. Cell Biol. 41 304-312 [PubMed] [Google Scholar]
- 16.Ogawa-Goto, K., Tanaka, K., Gibson, W., Moriishi, E., Miura, Y., Kurata, T., Irie, S., and Sata, T. (2003) J. Virol. 778541 -8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodems, S. M., and Spector, D. H. (1998) J. Virol. 729173 -9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, R. A., Ma, X. L., Yurochko, A. D., and Huang, E. S. (2001) J. Gen. Virol. 82 493-497 [DOI] [PubMed] [Google Scholar]
- 19.Johnson, R. A., Huong, S. M., and Huang, E. S. (2000) J. Virol. 741158 -1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, H., Cong, J. P., Yu, D., Bresnahan, W. A., and Shenk, T. E. (2002) Proc. Natl. Acad. Sci. U. S. A. 993932 -3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nokta, M. A., Hassan, M. I., Loesch, K., and Pollard, R. B. (1996) J. Clin. Investig. 972635 -2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jault, F. M., Jault, J. M., Ruchti, F., Fortunato, E. A., Clark, C., Corbeil, J., Richman, D. D., and Spector, D. H. (1995) J. Virol. 696697 -6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiegel, S., and Milstien, S. (2003) Nat. Rev. Mol. Cell Biol. 4397 -407 [DOI] [PubMed] [Google Scholar]
- 24.Maceyka, M., Payne, S. G., Milstien, S., and Spiegel, S. (2002) Biochim. Biophys. Acta 1585193 -201 [DOI] [PubMed] [Google Scholar]
- 25.Spiegel, S., and Milstien, S. (2003) Biochem. Soc. Trans. 311216 -1219 [DOI] [PubMed] [Google Scholar]
- 26.Anliker, B., and Chun, J. (2004) Semin. Cell Dev. Biol. 15457 -465 [DOI] [PubMed] [Google Scholar]
- 27.Young, N., and Van Brocklyn, J. R. (2006) Scientific World J. 6946 -966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuvillier, O., Pirianov, G., Kleuser, B., Vanek, P. G., Coso, O. A., Gutkind, S., and Spiegel, S. (1996) Nature 381800 -803 [DOI] [PubMed] [Google Scholar]
- 29.Taha, T. A., Argraves, K. M., and Obeid, L. M. (2004) Biochim. Biophys. Acta 1682 48-55 [DOI] [PubMed] [Google Scholar]
- 30.McVerry, B. J., and Garcia, J. G. (2005) Cell. Signal. 17131 -139 [DOI] [PubMed] [Google Scholar]
- 31.Lee, M. J., Thangada, S., Claffey, K. P., Ancellin, N., Liu, C. H., Kluk, M., Volpi, M., Sha'afi, R. I., and Hla, T. (1999) Cell 99301 -312 [DOI] [PubMed] [Google Scholar]
- 32.English, D., Welch, Z., Kovala, A. T., Harvey, K., Volpert, O. V., Brindley, D. N., and Garcia, J. G. (2000) FASEB J. 142255 -2265 [DOI] [PubMed] [Google Scholar]
- 33.Payne, S. G., Milstien, S., Barbour, S. E., and Spiegel, S. (2004) Semin. Cell Dev. Biol. 15 521-527 [DOI] [PubMed] [Google Scholar]
- 34.Cyster, J. G. (2005) Annu. Rev. Immunol. 23127 -159 [DOI] [PubMed] [Google Scholar]
- 35.Rosen, H., and Goetzl, E. J. (2005) Nat. Rev. Immunol. 5560 -570 [DOI] [PubMed] [Google Scholar]
- 36.Tamama, K., Kon, J., Sato, K., Tomura, H., Kuwabara, A., Kimura, T., Kanda, T., Ohta, H., Ui, M., Kobayashi, I., and Okajima, F. (2001) Biochem. J. 353139 -146 [PMC free article] [PubMed] [Google Scholar]
- 37.Van Brocklyn, J. R., Lee, M. J., Menzeleev, R., Olivera, A., Edsall, L., Cuvillier, O., Thomas, D. M., Coopman, P. J., Thangada, S., Hla, T., and Spiegel, S. (1998) J. Cell Biol. 142229 -240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohama, T., Olivera, A., Edsall, L., Nagiec, M. M., Dickson, R., and Spiegel, S. (1998) J. Biol. Chem. 27323722 -23728 [DOI] [PubMed] [Google Scholar]
- 39.Liu, H., Sugiura, M., Nava, V. E., Edsall, L. C., Kono, K., Poulton, S., Milstien, S., Kohama, T., and Spiegel, S. (2000) J. Biol. Chem. 27519513 -19520 [DOI] [PubMed] [Google Scholar]
- 40.Pitson, S. M., D'Andrea, R. J., Vandeleur, L., Moretti, P. A., Xia, P., Gamble, J. R., Vadas, M. A., and Wattenberg, B. W. (2000) Biochem. J. 350429 -441 [PMC free article] [PubMed] [Google Scholar]
- 41.Berdyshev, E. V., Gorshkova, I. A., Usatyuk, P., Zhao, Y., Saatian, B., Hubbard, W., and Natarajan, V. (2006) Cell. Signal. 181779 -1792 [DOI] [PubMed] [Google Scholar]
- 42.Olivera, A., and Spiegel, S. (1993) Nature 365557 -560 [DOI] [PubMed] [Google Scholar]
- 43.Choi, O. H., Kim, J.-H., and Kinet, J.-P. (1996) Nature 380634 -636 [DOI] [PubMed] [Google Scholar]
- 44.Melendez, A., Floto, R. A., Gillooly, D. J., Harnett, M. M., and Allen, J. M. (1998) J. Biol. Chem. 2739393 -9402 [DOI] [PubMed] [Google Scholar]
- 45.Meyer zu Heringdorf, D., Lass, H., Alemany, R., Laser, K. T., Neumann, E., Zhang, C., Schmidt, M., Rauen, U., Jakobs, K. H., and van Koppen, C. J. (1998) EMBO J. 172830 -2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olivera, A., Kohama, T., Edsall, L., Nava, V., Cuvillier, O., Poulton, S., and Spiegel, S. (1999) J. Cell Biol. 147545 -558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia, P., Gamble, J. R., Wang, L., Pitson, S. M., Moretti, P. A., Wattenberg, B. W., D'Andrea, R. J., and Vadas, M. A. (2000) Curr. Biol. 101527 -1530 [DOI] [PubMed] [Google Scholar]
- 48.Van Brocklyn, J. R., Jackson, C. A., Pearl, D. K., Kotur, M. S., Snyder, P. J., and Prior, T. W. (2005) J. Neuropathol. Exp. Neurol. 64695 -705 [DOI] [PubMed] [Google Scholar]
- 49.Maceyka, M., Sankala, H., Hait, N. C., Le Stunff, H., Liu, H., Toman, R., Collier, C., Zhang, M., Satin, L., Merrill, A. H., Jr., Milstien, S., and Spiegel, S. (2005) J. Biol. Chem. 28037118 -37129 [DOI] [PubMed] [Google Scholar]
- 50.Okada, T., Ding, G., Sonoda, H., Kajimoto, T., Haga, Y., Khosrowbeygi, A., Gao, S., Miwa, N., Jahangeer, S., and Nakamura, S. I. (2005) J. Biol. Chem. 28036318 -36325 [DOI] [PubMed] [Google Scholar]
- 51.Sedmak, D. D., Roberts, W. H., Stephens, R. E., Buesching, W. J., Morgan, L. A., Davis, D. H., and Waldman, W. J. (1990) Transplantation 49458 -462 [DOI] [PubMed] [Google Scholar]
- 52.Wentworth, B. B., and French, L. (1970) Proc. Soc. Exp. Biol. Med. 135253 -258 [DOI] [PubMed] [Google Scholar]
- 53.Waldman, W. J., Roberts, W. H., Davis, D. H., Williams, M. V., Sedmak, D. D., and Stephens, R. E. (1991) Arch. Virol. 117143 -164 [DOI] [PubMed] [Google Scholar]
- 54.Waldman, W. J., Adams, P. W., Orosz, C. G., and Sedmak, D. D. (1992) Transplantation 54 887-896 [DOI] [PubMed] [Google Scholar]
- 55.Maceyka, M., Milstien, S., and Spiegel, S. (2007) Methods Enzymol. 434243 -256 [DOI] [PubMed] [Google Scholar]
- 56.Livak, K. J., and Schmittgen, T. D. (2001) Methods (Amst.) 25402 -408 [DOI] [PubMed] [Google Scholar]
- 57.Merrill, A. H., Jr., Sullards, M. C., Allegood, J. C., Kelly, S., and Wang, E. (2005) Methods (Amst.) 36 207-224 [DOI] [PubMed] [Google Scholar]
- 58.Oskouian, B., and Saba, J. D. (2004) Semin. Cell Dev. Biol. 15529 -540 [DOI] [PubMed] [Google Scholar]
- 59.Venkataraman, K., Thangada, S., Michaud, J., Oo, M. L., Ai, Y., Lee, Y. M., Wu, M., Parikh, N., Khan, F., Proia, R. L., and Hla, T. (2006) Biochem. J. 397461 -471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pitson, S. M., Moretti, P. A., Zebol, J. R., Xia, P., Gamble, J. R., Vadas, M. A., D'Andrea, R. J., and Wattenberg, B. W. (2000) J. Biol. Chem. 27533945 -33950 [DOI] [PubMed] [Google Scholar]
- 61.Steinhart, W. L., Busch, J. S., Oettgen, J. P., and Howland, J. L. (1984) Intervirology 21 70-76 [DOI] [PubMed] [Google Scholar]
- 62.Allan-Yorke, J., Record, M., de Preval, C., Davrinche, C., and Davignon, J. L. (1998) J. Virol. 722316 -2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hla, T. (2003) Pharmacol. Res. 47 401-407 [DOI] [PubMed] [Google Scholar]
- 64.Hobson, J. P., Rosenfeldt, H. M., Barak, L. S., Olivera, A., Poulton, S., Caron, M. G., Milstien, S., and Spiegel, S. (2001) Science 2911800 -1803 [DOI] [PubMed] [Google Scholar]
- 65.Jolly, P. S., Bektas, M., Olivera, A., Gonzalez-Espinosa, C., Proia, R. L., Rivera, J., Milstien, S., and Spiegel, S. (2004) J. Exp. Med. 199959 -970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goparaju, S. K., Jolly, P. S., Watterson, K. R., Bektas, M., Alvarez, S., Sarkar, S., Mel, L., Ishii, I., Chun, J., Milstien, S., and Spiegel, S. (2005) Mol. Cell. Biol. 254237 -4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bu, S., Yamanaka, M., Pei, H., Bielawska, A., Bielawski, J., Hannun, Y. A., Obeid, L., and Trojanowska, M. (2006) FASEB J. 20184 -186 [DOI] [PubMed] [Google Scholar]
- 68.Stenberg, R. M. (1996) Intervirology 39343 -349 [DOI] [PubMed] [Google Scholar]
- 69.Mocarski, E. S., Kemble, G. W., Lyle, J. M., and Greaves, R. F. (1996) Proc. Natl. Acad. Sci. U. S. A. 9311321 -11326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greaves, R. F., and Mocarski, E. S. (1998) J. Virol. 72366 -379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monick, M. M., Cameron, K., Powers, L. S., Butler, N. S., McCoy, D., Mallampalli, R. K., and Hunninghake, G. W. (2004) Am. J. Respir. Cell Mol. Biol. 30 844-852 [DOI] [PubMed] [Google Scholar]
- 72.Stropes, M. P., and Miller, W. E. (2004) Biochem. Cell Biol. 82636 -642 [DOI] [PubMed] [Google Scholar]
- 73.Billstrom, M. A., Johnson, G. L., Avdi, N. J., and Worthen, G. S. (1998) J. Virol. 725535 -5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Radsak, K., and Wiegandt, H. (1984) Virology 138300 -309 [DOI] [PubMed] [Google Scholar]