Abstract
The Ras-like small GTPase Rheb is an upstream activator of the mammalian target of rapamycin (mTOR). It has recently been shown that Rheb activates mTOR by binding to its endogenous inhibitor FKBP38 and preventing it from association with mTOR. The interaction of Rheb with FKBP38 is controlled by its guanine nucleotide binding states, which are responsive to growth factor and amino acid conditions. In this study, we show that Rheb interacts with FKBP38 through a section within its switch I region that is equivalent to the effector domain of other Ras-like small GTPases. We find that the ability for Rheb to interact with FKBP38 correlates with its activity for mTOR activation. Our findings suggest that FKBP38 is a bona fide effector of Rheb and that the ability to interact with FKBP38 is important for Rheb as an activator of mTOR.
Rheb is a Ras-related small GTPase that functions as an upstream activator of the mammalian target of rapamycin (mTOR)2 (1, 2). Like other members of the Ras superfamily of small GTPases, the activity of Rheb is regulated by its guanine nucleotide binding states. It is active upon GTP bound but becomes inactive following hydrolysis of the bound GTP to GDP (3). The nucleotide binding states of Rheb are controlled mainly by the TSC1-TSC2 complex, which acts as a GTPase-activating protein that stimulates GTP hydrolysis of Rheb and prevents its activation (4–7). A complex signaling network converges both growth factor and energy signals to the TSC1-TSC2 complex, altering its GTPase-activating protein activity and consequently the guanine nucleotide binding states of Rheb (8). Changes in amino acid conditions also regulate Rheb activity through mechanisms independent of the TSC1-TSC2 complex (9, 10). In response to those diverse signals, Rheb cycles between active GTP-bound and inactive GDP-bound states, acting as a molecular switch to control mTOR activity (8).
How Rheb activates mTOR has been a key focus of current studies in mTOR signaling. mTOR elicits its rapamycin-sensitive function in the context of a multiple protein complex termed the mTOR complex 1 (mTORC1). Upon activation, mTORC1 promotes protein synthesis by phosphorylating S6 kinase (S6K) at position Thr389 and 4E-BP1 at positions Thr37/Thr46, two factors involved in translation initiation (11). It has been recently shown that Rheb activates mTORC1 by antagonizing its endogenous inhibitor, FKBP38 (12, 13). Under growth factor deprivation or amino acid starvation condition, FKBP38 binds directly to mTOR and down-regulates mTORC1 activity. When amino acids and growth factors are present, Rheb interacts with FKBP38 and releases mTORC1 for activation. The interaction of Rheb with FKBP38 is guanine nucleotide-dependent. It binds strongly to FKBP38 in GTP-bound form and weakly in GDP bound form. This GTP-dependent binding suggests that FKBP38 is an effector of Rheb in the mTOR pathway, which represents the first known effector of Rheb that is regulated by guanine nucleotide (12).
FKBP38, also known as FKBP8, belongs to the peptidyl prolyl cis/trans-isomerase family of FK506-binding protein (FKBP) (14). It contains a domain, referred to as FKBP-C, that is highly similar to FKBP12, the receptor for the immunosuppressive drug rapamycin (15). It has been shown previously that the FKBP-C domain in FKBP38 is sufficient for its interaction with mTOR. Although the binding of FKBP12 to mTOR requires rapamycin, the binding of the FKBP-C domain of FKBP38 with mTOR is independent of the drug (12). In addition to the FKBP-C domain, FKBP38 also contains several other distinct domains, including a tetratricopeptide repeat (TPR) domain, a Ca2+/calmodulin (CaM) binding domain, and a transmembrane domain (TM) (16–19). The TM domain of FKBP38 is unique among all of the members of the FKBP protein family and is essential for targeting this protein to mitochondria, where it functions (20).
The identification of FKBP38 as a target of Rheb allows us to examine how Rheb, a unique member of the Ras superfamily, relays its signaling activity downstream. It is well established that Ras, in GTP-bound form, interacts with its effectors through a section located in the switch I region of the protein. This section, commonly referred to as the effector domain, is critical for the signaling activity of Ras (21, 22). Since the switch I region in Rheb is highly similar to that in Ras, it is possible that Rheb signals to its effector in the mTOR pathway through a mechanism similar to that of Ras. In this study, we investigated the role of the switch I region in Rheb for its interaction with FKBP38 and its ability to activate mTORC1.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids—Antibodies against Rheb, S6K, phospho-S6K (Thr389), 4E-BP1, and phospho-4E-BP1 (Thr37/Thr46) were purchased from Cell Signaling Inc. Anti-HA (12CA5) and c-Myc (9E10) antibodies were purchased from Roche Applied Science, and anti-FLAG M2 antibody was from Sigma. The pCMV-Myc-FKBP38 vector was constructed by PCR amplifying FKBP38 from pcDNA3–2×HA-FKBP38 (14) and cloning into pCMV-tag3 vector. The sources for pRK7-HA-S6K1 and Rheb constructs were described before (12). The FKBP52 gene was amplified by PCR from the FKBP52 cDNA clone (ID 3542330) obtained from OpenBiosystems and cloned into pcDNA3.1-Myc vector to create pcDNA3.1-Myc-FKBP52. pcDNA3.1-Myc-FKBP52TM was created by inserting the sequence for the transmembrane domain of FKBP38 at the 3′-end of FKBP52.
Generation of Rheb and FKBP38 Mutants—Plasmids encoding all mutant versions of Rheb were generated by site-directed mutagenesis using QuikChange kit from Stratagene. The wild type Rheb in pcDNA-FLAG-Rheb vector was used as the template (6). Primers were designed to contain the desired mutations, and the mutagenesis was performed following the manufacturer's instruction. Deletion mutants of FKBP38 were generated by PCR from wild type FKBP38 construct with suitable primers and cloned into pcDNA3.1-Myc vector to create Myc-tagged truncated FKBP38 expression vectors. All mutations were verified by sequencing.
Cell Culture and Transfection—HEK293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum from PAA Laboratories (Ontario, Canada). For amino acid starvation and repletion, HEK293 cells were incubated in Dulbecco's modified Eagle's medium free of amino acids for 1 h, amino acid stock mixture (50× the standard concentration) from Sigma was added to the culture to a final concentration of 1×. Cells were harvested and lysed after a 30-min incubation. For serum deprivation and restimulation, cells were shifted to Dulbecco's modified Eagle's medium containing 0.2% of fetal bovine serum for 16 h. Fetal bovine serum was then added back to the medium to a final concentration of 20%. Cells were harvested and lysed upon incubation for another 30 min. DNA transfection was performed using Lipofectamine™ 2000 following the manufacturer's instructions (Invitrogen).
Coimmunoprecipitation—HEK293 cells were co-transfected with vectors expressing FKBP38 and Rheb. After incubation in Dulbecco's modified Eagle's medium for 30 h, transfected cells were washed with cold PBS on ice and lysed in buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture (Roche Applied Science). Lysates (0.5 mg) were incubated with anti-Myc antibody (9E10)-linked Protein A-agarose beads for 3 h at 4 °C with agitation. Beads were washed four times with lysis buffer and once with 20 mm Tris-Cl, pH 7.4. Washed beads were resuspended in 60 μl of 2× SDS sample buffer and boiled for 5 min. Samples were subjected to SDS-PAGE followed by Western blotting.
Immunofluorescence—HeLa cells grown on glass coverslips were stained with 200 nm Mitotracker Red (Molecular Probes) for 20 min and fixed with 4% paraformaldehyde in PBS for 10 min. Fixed cells were permeabilized by incubating with 0.1% Triton X-100 in PBS for 10 min and blocked with 5% bovine serum albumin in PBS for 30 min. Cells were then incubated overnight at 4 °C with anti-Rheb antibody (1:100 dilution), followed by staining with Alexa Fluor 488-conjugated anti-rabbit IgG (Molecular Probes) (1:1000). Cells were washed and mounted with ProLong® Gold antifade reagent (Molecular Probes) and visualized by confocal microscopy.
All of the experiments in this study were repeated independently no fewer than three times. Representative data are shown in the figures.
RESULTS
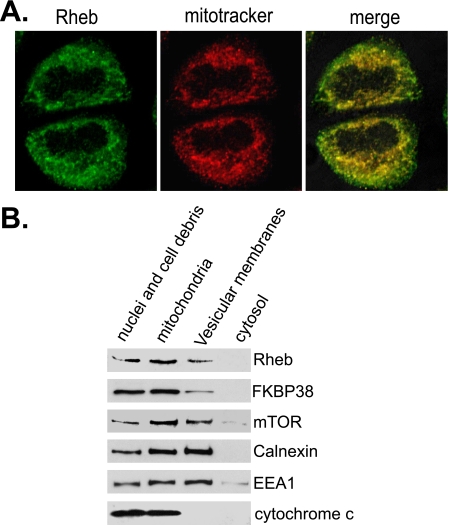
We have previously shown that Rheb physically interacts with FKBP38, which suggests a co-localization of both proteins. Since FKBP38 is found mainly on mitochondria (20), it is likely that Rheb resides on the same organelle. To test this notion, we examined intracellular localization of the endogenous Rheb using immunofluorescence microscopy. As shown in Fig. 1A, Rheb appeared to localize to some punctate structures, which were mostly overlapped with mitochondria that was marked by mitotracker. This observation indicates that Rheb is mainly localized to mitochondria. To further confirm the mitochondrial association of Rheb, we isolated the mitochondrion-enriched fraction from HEK293 cells by differential fractionation and analyzed the amount of Rheb associated with the fraction. As shown in Fig. 1B, we found that the majority of Rheb existed in the mitochondrial fraction that was marked by the presence of cytochrome c. Consistent with previous findings (20), FKBP38 was mostly found in the mitochondrion-enriched fraction. A small portion of Rheb and FKBP38 existed in the vesicular membrane fraction that was enriched in the endoplasmic reticulum and endosomal membranes. In addition, mTOR was also found to concentrate in the mitochondrial fraction, which is in accordance with previous observations that mTOR associates with mitochondria (23, 24). Altogether, these findings indicate that Rheb, FKBP38, and mTOR coexist on mitochondria.
FIGURE 1.
Rheb associates with mitochondria. A, HeLa cells were stained with mitotracker (red), followed by fixation and staining with anti-Rheb antibody in conjunction with Alexa Fluor-labeled secondary antibody (green), and imaged with confocal microscopy. B, lysates from HEK293 cells were fractionated by differential centrifugation to obtain fractions containing nuclei and cell debris, the mitochondrion-enriched membranes (mitochondria), vesicular membranes, and cytosol. The fractions were blotted for Rheb, FKBP38, and mTOR and markers for endoplasmic reticulum (calnexin), endosomal membranes (EEA1), and mitochondria (cytochrome c).
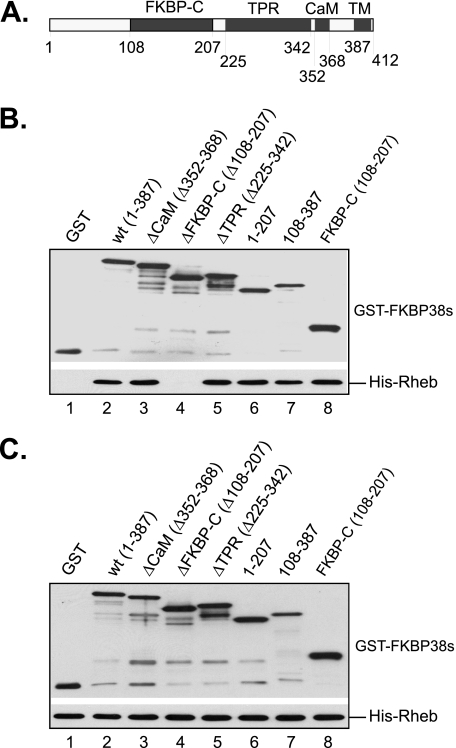
As an attempt to understand how Rheb affects the binding of FKBP38 with mTOR, we determined the regions in FKBP38 that mediate its interaction with Rheb. Since FKBP38 contains several distinct domains (Fig. 2A), we constructed GST-fused mutant proteins lacking each of the domains and expressed them in bacterial cells. To ensure their solubility, none of the mutant proteins contained the transmembrane domain. The interaction of the expressed mutant proteins with Rheb was examined using an in vitro binding assay. As shown in Fig. 2B, we found that a mutant FKBP38 protein lacking the FKBP-C domain was unable to interact with Rheb (lane 4), whereas those deleted for the CaM domain (lane 3), the tetratricopeptide repeat domain (lane 5), or the first 108 amino acids (lane 7) were. Furthermore, deletion mutants containing the first 207 amino acids (lane 6) or the FKBP-C domain alone (lane 8) were able to interact with Rheb. These findings suggest that Rheb interacts with FKBP38 by binding to its FKBP-C domain, the region that is highly similar to FKBP12. Similar results were obtained by co-immunoprecipitation using FKBP38 mutants expressed in cultured mammalian cells (Fig. S1).
FIGURE 2.
FKBP38 interacts with Rheb through its FKBP-C domain. GST alone or GST-fused deletion mutants of FKBP38 without the transmembrane domain were expressed in bacteria and purified. The recombinant proteins at a final concentration of 0.2 μm were incubated with the same amount of bacterially expressed and purified His-Rheb in 500 μl of PBS buffer containing 0.5% Triton X-100 on ice for 30 min. The reaction mixtures were precipitated with glutathione-conjugated Sepharose beads, and the amount of GST-FKBP38 and its truncated versions and that of co-purified His-Rheb were determined by Western blotting. A, a schematic presentation of FKBP38 structures. B, co-purified GST-FKBP38 mutants (top) and His-Rheb (bottom). C, input of GST-FKBP38 (top) and His-Rheb (bottom).
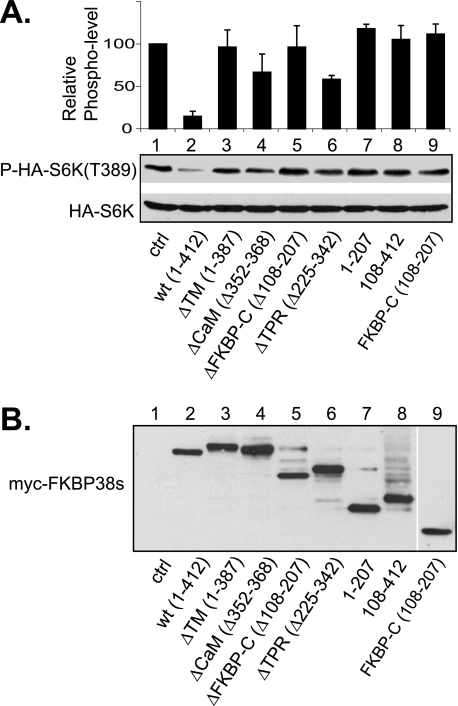
We next investigated the role for each of the unique domains in the function of FKBP38 as an mTORC1 inhibitor. Accordingly, we examined the serum-stimulated phosphorylation of S6K at Thr389, an mTORC1-dependent phosphorylation site, in cells that expressed the deletion mutants of FKBP38. In accordance with the previous finding (12), overexpression of wild type FKBP38 inhibited the serum-stimulated phosphorylation on S6K (Fig. 3A, compare lanes 1 and 2). In comparison, a mutant FKBP38 protein lacking the FKBP-C domain was largely ineffective in inhibition of the phosphorylation (lane 5), consistent with the fact that this domain is required for interaction with mTOR (6). Mutants without the transmembrane domain, including the regions of amino acids 1–387 (ΔTM; lane 3), amino acids 1–207 (lane 7), and amino acids 108–207 (FKBP-C; lane 9), also lost the inhibitory ability. Because these mutants were unable to associate with mitochondria (Fig. S3), this finding suggests that the mitochondrial localization is essential for the inhibitory activity of FKBP38. In addition, the region containing the N-terminal 108 amino acids appeared to be important for FKBP38 function, since a mutant protein lacking the region was incapable of inhibiting S6K phosphorylation (lane 8), despite being able to associate with mitochondria (Fig. S3). In contrast, mutants lacking the TPR domain retained some of the inhibitory activity (lane 6), suggesting that this domain is not essential for the inhibitory activity of FKBP38. Although a mutant deleted for the CaM domain exhibited a certain degree of inhibition, the activity is not statistically significant in comparison with that of the mutant lacking the transmembrane domain (lanes 3 and 4). Similar effects were observed for these mutant FKBP38 proteins in the amino acid-stimulated S6K phosphorylation at Thr389 (Fig. S2). These findings indicate three regions in FKBP38 are critical for its inhibitory activity toward mTORC1. These regions are the N-terminal 108 amino acids, the FKBP-C domain, and the transmembrane domain.
FIGURE 3.
Effect of FKBP38 deletion mutants on mTORC1 activity. Myc-tagged deletion mutants of FKBP38 were transfected together with HA-S6K into HEK293 cells. Transfected cells were incubated for 24 h. Cells were serum-starved (0.2%) for 16 h followed by readdition of serum (20%) and harvested after incubation for another 30 min. A, the presence of HA-S6K (bottom) and phosphorylation at Thr389 (top) were determined by Western blotting. The protein and phosphorylation levels were determined by densitometry. The ratio of phosphorylation versus protein level of each sample was compared with that of the control (ctrl; lane 1), and the relative values were shown. Data are the mean ± S.D. of values from three independent experiments. B, the expression levels of the mutant FKBP38 proteins in the lysates.
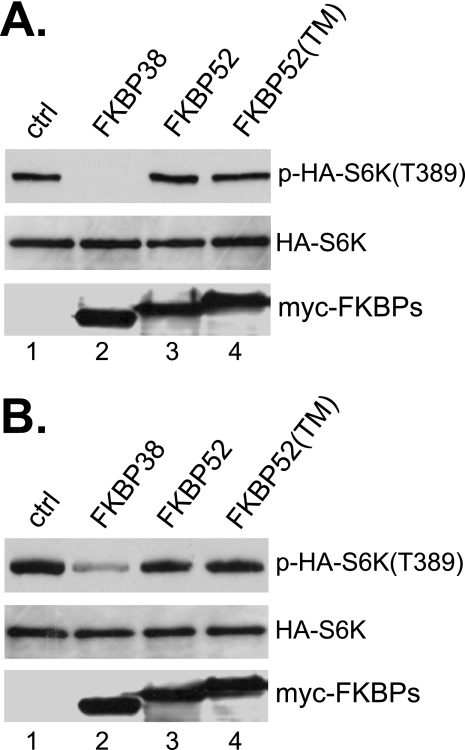
In the family of FK506-binding proteins, another member, FKBP52, shares similar structural features with FKBP38, including the FKBP-C domain, the TPR domain, and the CaM binding site (25). We thus tested whether FKBP52 possessed a similar inhibitory activity toward mTORC1. As shown in Fig. 4, although overexpression of FKBP38 prevented amino acid- or serum-stimulated phosphorylation of S6K at Thr389, that of FKBP52 failed to do so. The lack of inhibitory effect on mTORC1 does not appear to be caused by its inability to attach to membrane, since a hybrid version of the protein containing the transmembrane domain from FKBP38 remained incapable of mTORC1 inhibition. These data indicate that the FKBP38 is a unique member in the family of the FK506-binding proteins that possesses the ability to inhibit mTORC1 in the absence of rapamycin.
FIGURE 4.
Effect of FKBP52 on mTORC1 activity. HEK293 cells were transfected with HA-S6K together with control vector (ctrl; lane 1) or vector expressing Myc-tagged FKBP38 (lane 2), FKBP52 (lane 3), or FKBP52 attached at the 3′-end with the transmembrane domain sequence from FKBP38 (lane 4). Transfected cells were incubated for 24 h. A, cells were then starved for amino acids for 1 h, followed by the readdition of amino acids, and harvested after incubation for another 30 min. B, cells were deprived of serum (0.2%) for 16 h followed by serum readdition (20%) and harvested after incubation for another 30 min. The levels of HA-S6K (middle), phosphorylation at Thr389 (top), and expressed FKBP proteins (bottom) were determined by Western blotting.
The finding that Rheb interacts with FKBP38 and regulates its activity in a GTP-dependent manner suggests that FKBP38 is an effector of Rheb. As a small GTPase that is closely related to Ras, Rheb is expected to employ a similar mechanism to interact and activate its effectors. We thus determined whether the switch I region in Rheb is required for its interaction with FKBP38 and whether the ability to interact with FKBP38 is essential for Rheb signaling to mTORC1.
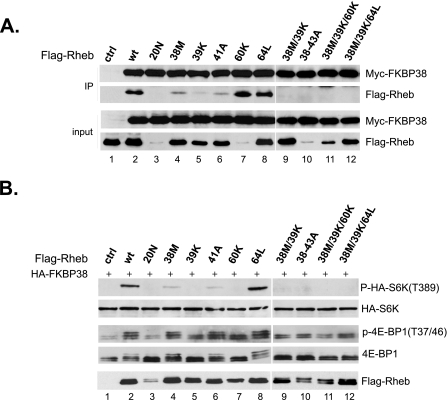
Previous studies have identified a few mutations within the switch I region of Rheb that impair its ability to activate mTORC1 but not that for nucleotide binding, including T38M, I39K, and N41A (26, 27). We first examined whether these mutations affect the binding of Rheb with FKBP38. Accordingly, we expressed Myc-tagged FKBP38 together with different FLAG-tagged Rheb mutants in HEK293 cells and examined their interaction by co-immunoprecipitation. As shown in Fig. 5A, we found that the interaction of Rheb with FKBP38 was partially disrupted by T38M and N41A mutations (lanes 4 and 6) and largely abolished by the I39K mutation (lane 5). A T38M/I39K combined mutation or alanine substitution of residues from position 38 to 43 (38–43A) completely abolished the binding (lanes 9 and 10). We next determined the ability of these Rheb mutants to antagonize the inhibitory effect of FKBP38 on mTORC1 signaling activity. Consistent with our previous findings, we found that overexpression of FKBP38 blocked amino acid-stimulated phosphorylation of S6K at Thr389 and of 4E-BP1 at Thr37/Thr46. However, a simultaneous expression of wild type Rheb abolished the inhibitory effect of FKBP38 (Fig. 5B, compare lanes 1 and 2). Although T38M and N41A mutants retained some residual activity for stimulating the S6K phosphorylation in the presence of FKBP38 (lanes 4 and 6), the T38M/I39K and 38–43A mutants were devoid of any activity (lanes 9 and 10). Thus, the ability of Rheb to bind with FKBP38 correlates with its ability to stimulate mTORC1 signaling. One exception was D60K, a dominant negative allele, which bound strongly to FKBP38 yet failed to stimulate the S6K phosphorylation (Fig. 5, A and B, lane 7). Despite this, a triple mutation, containing T38M, I39K, and D60K substitutions, was unable to interact with FKBP38, suggesting that the binding of D60K with FKBP38 still requires the switch I region (Fig. 5A, lane 11).
FIGURE 5.
The Rheb effector domain is required for its interaction with FKBP38. A, lysates (input) from cells expressing Myc-tagged FKBP38 and FLAG-tagged Rheb mutants were precipitated with Protein A-agarose beads cross-linked with anti-Myc antibody. Precipitates (IP) were immunoblotted for the presence of Myc-FKBP38 and FLAG-Rheb. The amount of input represents 5% of that used for immunoprecipitation. B, HEK293 cells were transfected with FLAG-tagged Rheb mutants together with HA-FKBP38 and HA-S6K. After incubation for 24 h, transfected cells were starved for amino acids for 1 h, followed by amino acid repletion. Cells were collected after 30 min and lysed. The levels of phosphorylation at Thr389 of HA-S6K (top), the HA-S6K protein (second panel), phosphorylation at Thr37/Thr46 of 4E-BP1 (third panel), the endogenous 4E-BP1 (fourth panel), and the expressed Rheb mutants (bottom panel) were determined by Western blotting. ctrl, control.
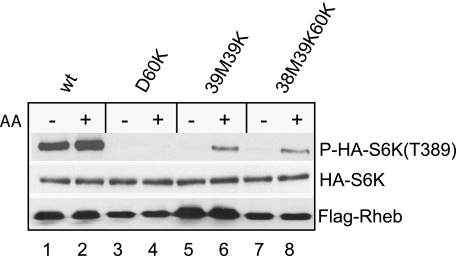
D60K was originally identified as a dominant negative allele, whose overproduction in cells blocks mTORC1-dependent S6K phosphorylation and renders it largely unresponsive to amino acids or growth factor stimulation (28). We have previously shown that D60K binds to and sequesters FKBP38 on mTOR in such a way that the binding between FKBP38 and mTOR is no longer sensitive to amino acids availability or growth factor stimulation (12). This observation raised a possibility that the dominance of D60K over the endogenous Rheb originated from its binding and sequestering FKBP38. To test this notion, we examined whether nullifying the ability of D60K to bind with FKBP38 eliminates its dominance. As shown in Fig. 6, we found that whereas the S6K phosphorylation at Thr389 was insensitive to amino acid availability in cells overexpressing D60K (lanes 3 and 4), it became responsive in cells overexpressing the triple T38M/I39K/D60K mutant (lanes 7 and 8), which was incapable of FKBP38 binding (Fig. 5A, lane 11). This finding suggests that the dominance of D60K is caused by sequestering its effector, FKBP38.
FIGURE 6.
The negative dominance of Rheb D60K mutant is caused by sequestering FKBP38. HEK293 cells were transfected with HA-S6K together with different Rheb mutants. After incubated for 24 h, transfected cells were starved for amino acids for 1 h followed by amino acid repletion. Cells were harvested and lysed after incubation for another 30 min. The levels of HA-S6K, phosphorylation at Thr389, and expressed Rheb were determined by Western blotting. wt, wild type.
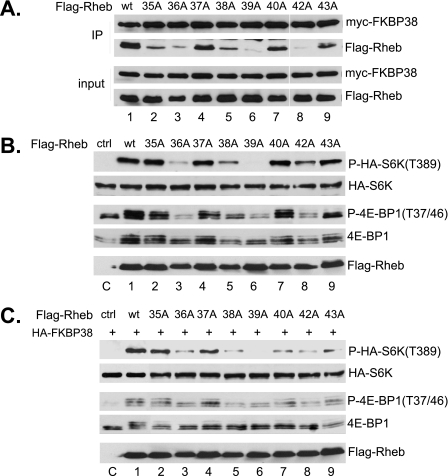
The region in Rheb that corresponds to the effector domain of Ras contains 9 residues, including amino acids 35–43. We examined the role for each of these residues in mediating its interaction with FKBP38 by substituting them individually with alanine. As shown in Fig. 7, we found that the substitutions at position 35, 40, or 43 partially impair the ability of the mutant Rheb to interact with FKBP38; that at position 38 modestly affected the interaction, whereas those at position 36, 39, or 42 largely abolished the interaction. We next determined the ability of the alanine substitution mutants to activate mTORC1 signaling in the absence of amino acids. We found that the mutants with largely impaired FKBP38 binding ability, including those with alanine substitution at positions 36, 38, 39, and 42, were ineffective in stimulating the mTORC1-dependent phosphorylation of S6K and 4E-BP1 (Fig. 7B). This finding suggests that the interaction with FKBP38 is important for Rheb to activate mTORC1. To further confirm the notion, we examined the ability of these mutants in antagonizing FKBP38 activity. We found that these mutants were incapable of reversing the inhibitory effect of FKBP38 on mTORC1 (Fig. 7C). Thus, the ability of Rheb to interact with FKBP38 correlates with its activity for mTORC1 stimulation.
FIGURE 7.
Analysis of the effector domain of Rheb. A, effect of individual residue in the effector domain of Rheb on its interaction of with FKBP38. Lysates (input) from HEK293 cells expressing Myc-tagged FKBP38 and FLAG-tagged Rheb mutants were precipitated with Protein A-agarose beads cross-linked with anti-Myc antibody. Precipitates (IP) were immunoblotted for the presence of Myc-FKBP38 and FLAG-Rheb. The amount of input represents 5% of that used for immunoprecipitation. B, stimulation of S6K (Thr389) phosphorylation by the alanine substitution mutants of Rheb under amino acid starvation conditions. HEK293 cells were transfected with Rheb mutants together with HA-S6K. After 24 h, cells were starved for amino acids for 1 h before being collected. The levels of HA-S6K (second panel), phosphorylation at Thr389 of HA-S6K (top), phosphorylation at Thr37/Thr46 of 4E-BP1 (third panel), the endogenous 4E-BP1 (fourth panel), and expressed Rheb (bottom panel) were determined by Western blotting. C, the ability of the Rheb mutants to antagonize the inhibitory effect of FKBP38. Rheb mutants were transfected together with FKBP38 into HEK293 cells. Upon incubation for 24 h, cells were starved for amino acids for 1 h, followed by the readdition of amino acids. Cells were harvested after incubation for another 30 min. The levels of HA-S6K (second panel), phosphorylation at Thr389 of HA-S6K (top), phosphorylation at Thr37/Thr46 of 4E-BP1 (third panel), the endogenous 4E-BP1 (fourth panel), and expressed Rheb (bottom) were determined by Western blotting. wt, wild type; ctrl, control.
DISCUSSION
Previous studies have shown that Rheb is localized in cells on some punctate particles that resemble endosomal compartments. These observations have led to the suggestion that Rheb associates with endosomal compartments (29, 30). However, our analysis of Rheb localization revealed that the majority of Rheb is co-localized with mitotracker (Fig. 1), suggesting that the protein is associated with mitochondria. Because the endosomal compartments and mitochondria are morphologically indistinguishable under a light microscope, it is possible that some of the Rheb-containing punctate particles observed earlier were actually mitochondria. The mitochondrial association of Rheb is also consistent with the fact that Rheb directly interacts with FKBP38, which has been shown previously to be a mitochondrial protein (20, 31). The target of FKBP38, mTOR, has been shown to localize to various compartments, including endosomal membrane, endoplasmic reticulum, Golgi, nucleus, and mitochondria (23, 24, 32–35). Our fractionation analysis indicates that at least a portion of mTOR is localized to mitochondria, where Rheb and FKBP38 reside. This result is supported by previous findings showing that mTOR associated with outer membrane of mitochondria (23). Given that mitochondria are the major metabolic organelle, co-localization of mTOR, Rheb, and FKBP38 on mitochondria is consistent with the role of mTORC1 in nutrient, energy, and oxygen sensing.
The FKBP-C domain of FKBP38 shares 53% sequence similarity with FKBP12 (16). We have previously shown that the FKBP-C domain in FKBP38 but not FKBP12 is able to interact with mTOR in the absence of rapamycin (12). Consistent with this result, we show in this study that the FKBP-C domain of FKBP38 is essential for its inhibitory activity toward mTORC1 signaling activity. However, the FKBP-C domain alone does not appear to be sufficient for mTORC1 inhibition. Deletion mutants of FKBP38 lacking the transmembrane domain or the N-terminal 108 amino acids are largely defective in mTORC1 inhibition, suggesting that these domains are also important for FKBP38 function. Since the transmembrane domain is needed for targeting FKBP38 to mitochondria (20), the requirement of this domain for FKBP38 function indicates that mitochondrial association is important for FKBP38 function. On the other hand, the region containing the N-terminal 108 amino acids of FKBP38 is not required for its association with mitochondria (Fig. S3) or for its interaction with Rheb (Fig. 2) and mTOR (12). Its role in FKBP38 function requires further study.
Interestingly, the FKBP-C domain of FKBP38 also mediates its binding with Rheb (Fig. 2), raising the possibility that Rheb may compete with mTOR for FKBP38 binding. However, the fact that the D60K mutant of Rheb is able to bind with FKBP38 without displacing mTOR from it suggests that the bindings of FKBP38 with Rheb and mTOR are not mutually exclusive (12). It is likely that Rheb and mTOR bind to different sides of the FKBP-C domain in such a way that Rheb is able to modulate FKBP38 allosterically and consequently affects its association with mTOR. Further analysis of the three-dimensional structure of the Rheb-FKBP38 and FKBP38-mTOR complexes will shed light on the structural bases for the interactions.
Upon GTP binding, small GTPases of Ras family interact with their effectors through the effector domain within the switch I region of the proteins. As such, the effector domain of a small GTPase plays a critical role in relaying its activity to downstream effectors (36). Previous studies have identified a few mutations within the switch I region, including T38M, I39K, and N41A, that affect the ability of Rheb to stimulate mTORC1 activity (26). In the present study, we find that these mutations in Rheb disrupt its interaction with FKBP38. These observations demonstrate a correlation between the ability to interact with FKBP38 and the ability to stimulate mTORC1 activity, which is consistent with the notion that Rheb activates mTORC1 by removing FKBP38 from mTOR (12). Our mutational analysis also identifies two other residues in the effector domain that are important for the interaction of Rheb with FKBP38, including Asp36 and Thr42. Alanine substitutions at these positions drastically reduce the ability of Rheb to interact with FKBP38 and to stimulate mTORC1. These results suggest that the interaction with FKBP38 is critical for Rheb to activate mTORC1, thus supporting a view that FKBP38 is a key factor that links Rheb activity to mTORC1. Importantly, the finding that Rheb interacts with FKBP38 through its effector domain in a GTP-dependent manner suggests that the signaling mechanism of Rheb is similar to that of other members of the Ras superfamily.
Supplementary Material
Acknowledgments
We thank Q. Wang and members of the Jiang laboratory for constructive discussions.
This work was supported, in whole by National Institutes of Health Grants GM068832 and CA129821 (to Y. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: mTOR, mammalian target of rapamycin; S6K, S6 kinase; FKBP, FK506-binding protein; CaM, calmodulin; TM, transmembrane; TPR, tetratricopeptide repeat; PBS, phosphate-buffered saline; GST, glutathione S-transferase.
References
- 1.Li, Y., Corradetti, M. N., Inoki, K., and Guan, K. L. (2004) Trends Biochem. Sci. 29 32-38 [DOI] [PubMed] [Google Scholar]
- 2.Manning, B. D., and Cantley, L. C. (2003) Trends Biochem. Sci. 28 573-576 [DOI] [PubMed] [Google Scholar]
- 3.Aspuria, P. J., and Tamanoi, F. (2004) Cell. Signal. 16 1105-1112 [DOI] [PubMed] [Google Scholar]
- 4.Garami, A., Zwartkruis, F. J., Nobukuni, T., Joaquin, M., Roccio, M., Stocker, H., Kozma, S. C., Hafen, E., Bos, J. L., and Thomas, G. (2003) Mol. Cell 11 1457-1466 [DOI] [PubMed] [Google Scholar]
- 5.Zhang, Y., Gao, X., Saucedo, L. J., Ru, B., Edgar, B. A., and Pan, D. (2003) Nat. Cell Biol. 5 578-581 [DOI] [PubMed] [Google Scholar]
- 6.Saucedo, L. J., Gao, X., Chiarelli, D. A., Li, L., Pan, D., and Edgar, B. A. (2003) Nat. Cell Biol. 5 566-571 [DOI] [PubMed] [Google Scholar]
- 7.Stocker, H., Radimerski, T., Schindelholz, B., Wittwer, F., Belawat, P., Daram, P., Breuer, S., Thomas, G., and Hafen, E. (2003) Nat. Cell Biol. 5 559-565 [DOI] [PubMed] [Google Scholar]
- 8.Avruch, J., Hara, K., Lin, Y., Liu, M., Long, X., Ortiz-Vega, S., and Yonezawa, K. (2006) Oncogene 25 6361-6372 [DOI] [PubMed] [Google Scholar]
- 9.Long, X., Ortiz-Vega, S., Lin, Y., and Avruch, J. (2005) J. Biol. Chem. 280 23433-23436 [DOI] [PubMed] [Google Scholar]
- 10.Nobukuni, T., Joaquin, M., Roccio, M., Dann, S. G., Kim, S. Y., Gulati, P., Byfield, M. P., Backer, J. M., Natt, F., Bos, J. L., Zwartkruis, F. J., and Thomas, G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14238-14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, D. E., and Hall, M. N. (2005) Curr. Opin. Cell Biol. 17 158-166 [DOI] [PubMed] [Google Scholar]
- 12.Bai, X., Ma, D., Liu, A., Shen, X., Wang, Q. J., Liu, Y., and Jiang, Y. (2007) Science 318 977-980 [DOI] [PubMed] [Google Scholar]
- 13.Proud, C. G. (2007) Science 318 926-927 [DOI] [PubMed] [Google Scholar]
- 14.Harrar, Y., Bellini, C., and Faure, J. D. (2001) Trends Plant Sci. 6 426-431 [DOI] [PubMed] [Google Scholar]
- 15.Fischer, G., Tradler, T., and Zarnt, T. (1998) FEBS Lett. 426 17-20 [DOI] [PubMed] [Google Scholar]
- 16.Maestre-Martinez, M., Edlich, F., Jarczowski, F., Weiwad, M., Fischer, G., and Lucke, C. (2006) J. Biomol. NMR 34 197-202 [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, J. V., Mitchelmore, C., Pedersen, K. M., Kjaerulff, K. M., Finsen, B., and Jensen, N. A. (2004) Genomics 83 181-192 [DOI] [PubMed] [Google Scholar]
- 18.Pedersen, K. M., Finsen, B., Celis, J. E., and Jensen, N. A. (1999) Electrophoresis 20 249-255 [DOI] [PubMed] [Google Scholar]
- 19.Wang, H. Q., Nakaya, Y., Du, Z., Yamane, T., Shirane, M., Kudo, T., Takeda, M., Takebayashi, K., Noda, Y., Nakayama, K. I., and Nishimura, M. (2005) Hum. Mol. Genet. 14 1889-1902 [DOI] [PubMed] [Google Scholar]
- 20.Shirane, M., and Nakayama, K. I. (2003) Nat. Cell Biol. 5 28-37 [DOI] [PubMed] [Google Scholar]
- 21.Marshall, C. J. (1996) Curr. Opin. Cell Biol. 8 197-204 [DOI] [PubMed] [Google Scholar]
- 22.Wittinghofer, A., and Nassar, N. (1996) Trends Biochem. Sci. 21 488-491 [DOI] [PubMed] [Google Scholar]
- 23.Desai, B. N., Myers, B. R., and Schreiber, S. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4319-4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schieke, S. M., Phillips, D., McCoy, J. P., Jr., Aponte, A. M., Shen, R. F., Balaban, R. S., and Finkel, T. (2006) J. Biol. Chem. 281 27643-27652 [DOI] [PubMed] [Google Scholar]
- 25.Davies, T. H., and Sanchez, E. R. (2005) Int. J. Biochem. Cell Biol. 37 42-47 [DOI] [PubMed] [Google Scholar]
- 26.Long, X., Lin, Y., Ortiz-Vega, S., Yonezawa, K., and Avruch, J. (2005) Curr. Biol. 15 702-713 [DOI] [PubMed] [Google Scholar]
- 27.Tee, A. R., Blenis, J., and Proud, C. G. (2005) FEBS Lett. 579 4763-4768 [DOI] [PubMed] [Google Scholar]
- 28.Tabancay, A. P., Jr., Gau, C. L., Machado, I. M., Uhlmann, E. J., Gutmann, D. H., Guo, L., and Tamanoi, F. (2003) J. Biol. Chem. 278 39921-39930 [DOI] [PubMed] [Google Scholar]
- 29.Buerger, C., DeVries, B., and Stambolic, V. (2006) Biochem. Biophys. Res. Commun. 344 869-880 [DOI] [PubMed] [Google Scholar]
- 30.Takahashi, K., Nakagawa, M., Young, S. G., and Yamanaka, S. (2005) J. Biol. Chem. 280 32768-32774 [DOI] [PubMed] [Google Scholar]
- 31.Portier, B. P., and Taglialatela, G. (2006) J. Biol. Chem. 281 40493-40502 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, X., Shu, L., Hosoi, H., Murti, K. G., and Houghton, P. J. (2002) J. Biol. Chem. 277 28127-28134 [DOI] [PubMed] [Google Scholar]
- 33.Withers, D. J., Ouwens, D. M., Nave, B. T., van der Zon, G. C., Alarcon, C. M., Cardenas, M. E., Heitman, J., Maassen, J. A., and Shepherd, P. R. (1997) Biochem. Biophys. Res. Commun. 241 704-709 [DOI] [PubMed] [Google Scholar]
- 34.Drenan, R. M., Liu, X., Bertram, P. G., and Zheng, X. F. (2004) J. Biol. Chem. 279 772-778 [DOI] [PubMed] [Google Scholar]
- 35.Kim, J. E., and Chen, J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14340-14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bos, J. L. (1997) Biochim. Biophys. Acta 1333 M19-31 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.