Abstract
Thymidylate synthase (TS) catalyzes the reductive methylation of deoxyuridine monophosphate (dUMP) using methylene tetrahydrofolate (CH2THF) as cofactor, the glutamate tail of which forms a water-mediated hydrogen-bond with an invariant lysine residue of this enzyme. To understand the role of this interaction, we studied the K48Q mutant of Escherichia coli TS using structural and biophysical methods. The kcat of the K48Q mutant was 430 fold lower than wild-type TS in activity, while the the Km for the (R)-stereoisomer of CH2THF was 300 µM, about 30 fold larger than Km from the wild-type TS. Affinity constants were determined using isothermal titration calorimetry, which showed that binding was reduced by one order of magnitude for folate-like TS inhibitors, such as propargyl-dideaza folate (PDDF) or compounds that distort the TS active site like BW1843U89 (U89). The crystal structure of the K48Q-dUMP complex revealed that dUMP binding is not impaired in the mutamt, and that U89 in a ternary complex of K48Q-nucleotide-U89 was bound in the active site with subtle differences relative to comparable wild type complexes. PDDF failed to form ternary complexes with K48Q and dUMP. Thermodynamic data correlated with the structural determinations, since PDDF binding was dominated by enthalpic effects while U89 had an important entropic component. In conclusion, K48 is critical for catalysis since it leads to a productive CH2THF binding, while mutation at this residue does not affect much the binding of inhibitors that do not make contact with this group.
Keywords: thymidylate synthase, protein crystallography, isothermal titration calorimetry, tryptophan fluorescence, mutant K48Q
1. Introduction
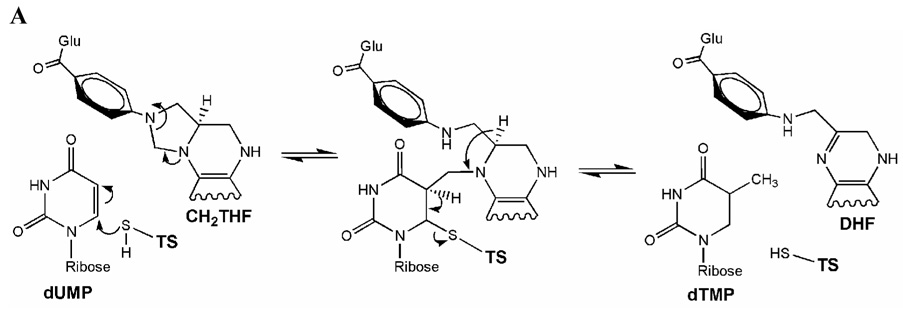
The search for antiproliferative drugs based on enzyme targets has led to drugs such as methotrexate, which inhibits dihydrofolate reductase (Huennekens, 1994; Kimura et al., 2004). A second related enzyme involved in folate metabolism is serine hydroxymethyltransferase, which has been designated also as a drug target (Appaji Rao, Talwar & Savithri, 2000). Thymidylate synthase (TS, E.C. 2.1.1.45) is the third enzyme in the methylene-tetrahydrofolate cycle that catalyzes the reductive methylation of 2’-deoxyuridine-5’-monophosphate (dUMP) to 2’- deoxythymidine-5’-monophosphate (dTMP) using 5,10-methylene-5,6,7,8-tetrahydrofolate (CH2THF) as cofactor (Fig. 1A) (Carreras & Santi, 1995). In many organisms, TS is the only de novo source for dTMP required for DNA synthesis (Finer-Moore, Santi & Stroud, 2003). The wealth of information on the structure, function and mechanism of TS has led to the design of substrate analogs to inhibit human TS, since impairing this enzyme inhibits the growth of proliferating cells including those that cause cancer. Human TS pharmacogenomics has been focused to 5’ and 3’ untranslated polymorphisms, tandem repeats, and also in mutations in the TS coding region (Barbour, Berger & Berger, 1990; Tong et al., 1998). Furthermore, TS stands as an important model to understand structure-function relationships and as a paradigm for structure based drug design against various bacterial and proliferative diseases (Stout et al., 1999).
Fig. 1.
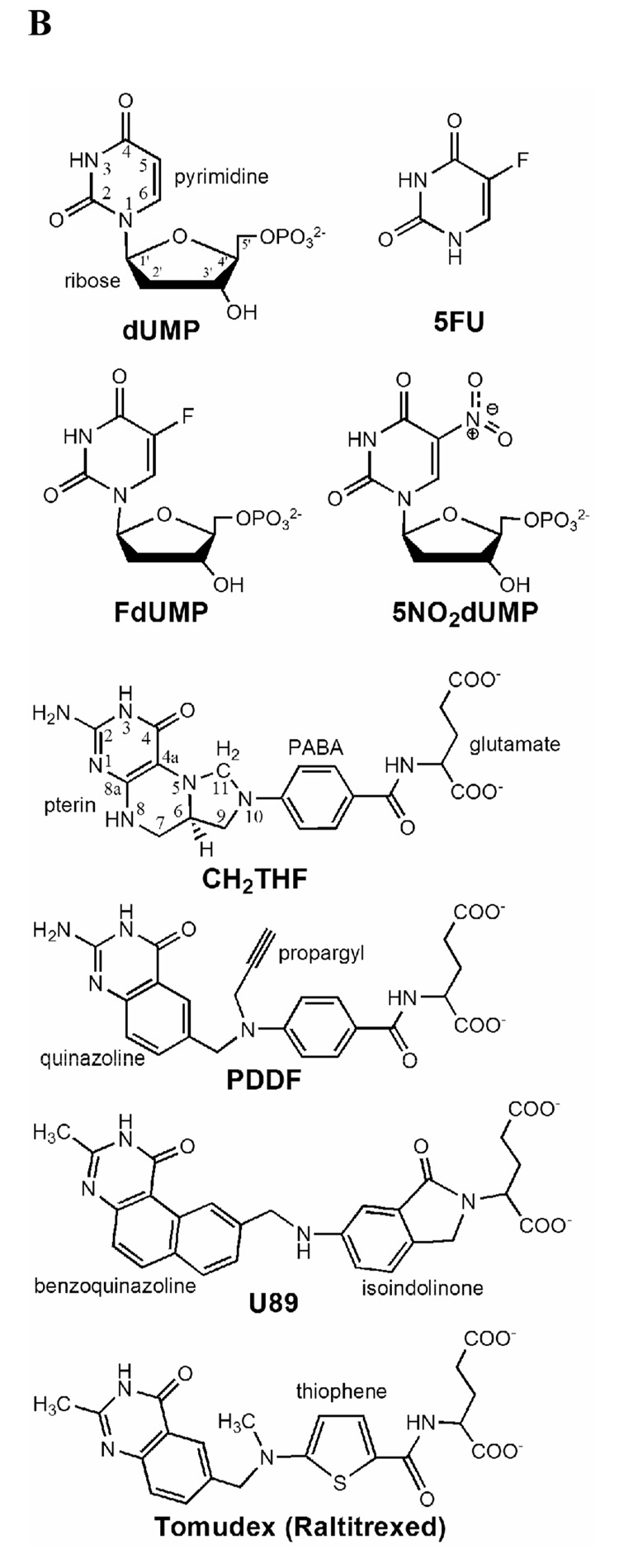
Mode of action of thymidylate synthase. (A) Enzymatic mechanism after Carreras & Santi (1995). (B) Chemical structure of substrate, cofactor and inhibitors.
Several nucleotide and folate analogs have been developed as anticancer drugs. 5-fluorouracil is widely used as in cancer chemotherapy as it is converted to 5-fluoro-dUMP (FdUMP), which is a suicide-inhibitor of TS (Fig. 1B) (Danenberg, Langenbach & Heidelberger, 1974; Santi, McHenry & Sommer, 1974).
The search for anti-folate analogues of TS lead to 10-propargyl-5,8-dideazafolate (PDDF, Fig. 1B), which bound to TS in a similar manner as the natural cofactor and like the cofactor induced a conformational change that promoted active site closure (Matthews, Appelt, Oatley & Xuong, 1990; Montfort et al., 1990). Another antifolate, BW1843U89 (U89) is structurally similar to PDDF (Fig. 1B), but has an extra ring attached to the quinazoline group that makes it a larger molecule than PDDF. When bound to TS, U89 produces an active site distortion and binds in a new position (Stout & Stroud, 1996; Weichsel & Montfort, 1995). But despite the latter effect it still induced a conformational change that promoted closure of the C-terminus and a reduction in size of the active site cavity (Montfort et al., 1990). PDDF and U89 eventually lead to the development of drugs such as Tomudex (raltitrexed), an antifolate targeted to TS that is used against colon cancer (Chu, Callender, Farrell & Schmitz, 2003).
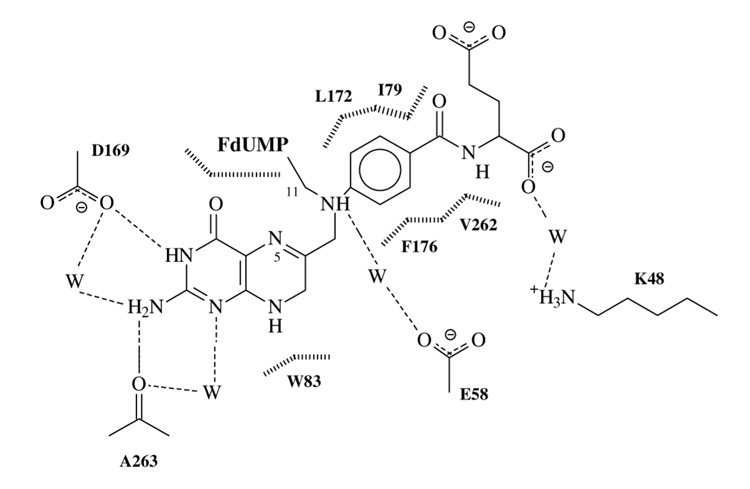
In general, conserved or invariant residues are critical for function, and their mutation result in reduced catalytic activity (Finer-Moore, Santi & Stroud, 2003). One such residue is K481, which is an invariant non-catalytic residue that stabilizes the negative charge of the glutamate-end of CH2THF by a water-mediated H-bond (Maley, Maley & Baugh, 1982; Montfort et al., 1990) (Fig. 2). This interaction was observed in the crystal structure of the wild-type (WT) ternary covalent complex of TS with FdUMP and CH2THF (Hyatt, Maley & Montfort, 1997) indicating that K48 is essential for the binding of CH2THF and its analogues (Kamb, Finer-Moore, Calvert & Stroud, 1992). The CH2THF γ-glutamate is critical for folate processing, since it is polyglutamylated by folylpoly-gamma-glutamate synthetase. This enzyme is key for folate metabolism since it also processes drugs such as methotrexate, another long-standing antineoplasic drug, which targets dihydrofolate reductase (Huennekens, 1994).
Fig. 2.
Schematic drawing of the interactions between the cofactor CH2THF and wild-type TS in the crystal structure of the ternary complex with FdUMP. Water molecules are represented by W, hydrogen-bonds are represented with dashed lines and van der Waals contacts with vertical broken lines. Note the water-mediated hydrogen-bond between the glutamate tail of cofactor and the lysine residue at position 48 of wild-type TS.
In studies with the bacteriophage T4 TS, the conservative mutation K48R decreased kcat and increased Km for CH2THF by two orders of magnitude compared to WT (LaPat-Polasko, Maley & Maley, 1990). Our hypothesis is that the K48Q mutation in TS impairs the binding of antifolates structurally similar to the natural cofactor (and cognate folates), such as PDDF. However, U89 has not been used to characterize mutant TSs such as K48Q and since the U89-glutamate group is remote from the K48 site (Weichsel & Montfort, 1995), it may help to understand the role of this residue.
In this work, we tested the functional role of K48, an invariant non-catalytic residue in the vicinity of the active site of TS, by means of protein crystallography, isothermal titration calorimetry (ITC), protein fluorescence and circular dichroism. We also determined the crystal structures of the K48Q TS mutant in the presence of various ligands to evaluate the effect of the mutation on folate binding, ligand-induced conformational change and the role of the ordered water molecule in contact with K48.
2. Materials and methods
2.1 Protein isolation and characterization
Wild-type TS and its K48Q mutant were prepared from an overexpressing strain of E. coli as previously described (Maley, Pedersen-Lane & Changchien, 1995). Enzyme activity was measured using the spectrophotometric assay (Wahba & Friedkin, 1962) with modifications (Maley, Pedersen-Lane & Changchien, 1995). After purification and precipitation with ammonium sulfate, WT and K48Q TS were frozen at −80 °C.
2.2 Protein crystallization
For crystal growth, the enzyme was extensively dialyzed against 20 mM potassium phosphate, pH 7.0, 0.1 M EDTA and 5 mM DTT. All crystallization experiments were done using the hanging drop method at room temperature. For each complex, crystallization conditions were searched using a screening grid with 20 mM potassium phosphate buffer, 4 mM DTT and increments in ammonium sulfate from 2.05 to 2.6 M (in 0.05 M increments) and pH from 7.5 to 8.0 (in 0.1 pH increments) (Montfort et al., 1990). Crystals appeared after a few days in different crystalline forms. The complexes of mutant TS with dUMP crystallized in the cubic form. Binary and ternary complexes with 5NO2dUMP crystallized in the hexagonal form. The final protein concentration was 3 mg/ml and the concentration of the compounds used to form complexes in the crystallization drops was 3 mM each.
2.3 Structure determinations
A single crystal was used and mounted in a quartz capillary to obtain diffraction data for each complex on an Enraf-Nonius area detector attached to a Cuα X-ray source and a FAST area detector at room temperature. Data reduction was done with MADNES (Messerschmidt & Pflugrath, 1987), PROCOR (Kabsch, 1988), and CCP4 (CCP4, 1994). Electron density maps were calculated using Refmac from CCP4 or CNS 1.2 (Brunger et al., 1998). All crystal forms were isomorphous with the corresponding WT complexes, and they were used as starting models for refinement with CNS 1.2 (Brunger et al., 1998). Rebuilding was done with O (Jones, Zou, Cowan & Kjelgard, 1991) and structure comparisons with Coot (Emsley & Cowtan, 2004). Final statistics for data collection and model refinement are shown in Table 1. Figures were drawn with CCP4mg (Potterton et al., 2004) and Pymol 1.0 (DeLano, 2002).
Table 1.
Crystallographic data for the E. coli TS K48Q mutant structures.
| K48Q-dUMP | K48Q-5NO2dUMP | K48Q-5NO2dUMP-U89 | |
|---|---|---|---|
| PDB accession code | 2VET | 3B5B | 2VF0 |
| Unit Cell | |||
| Space group | I213 | P63 | P63 |
| a (Å) | 132.9 | 127.4 | 127.2 |
| c (Å) | 67.8 | 67.9 | |
| Data Collection | |||
| Resolution(Å) | 15-2.2 | 25.5-2.7 | 34-3.0 |
| Unique reflections | 18,173 | 16,931 | 12,094 |
| Multiplicity | 4.4 | 4.8 | 4.7 |
| Completeness (%)1 | 91.5(76.6) | 97.4(92.7) | 95.1(89.3) |
| I/σ(I) | 4.8/1.5 | 7.2/1.7 | 3.9/1.3 |
| Rsym2 | 0.09 | 0.09/0.40 | 0.17/0.5 |
| Refinement | |||
| Rcrys3 (working set / test set) | 0.185(0.24) | 0.193(0.30) | 0.180(0.28) |
| Rfree (working set / test set) | 0.205(0.24) | 0.234(0.35) | 0.239(0.35) |
| Root mean square deviations | |||
| Bond lengths | 0.010 Å | 0.007 Å | 0.009 Å |
| Bond angles | 1.8 ° | 1.4 ° | 1.4 ° |
| Dihedral angles | 25.5 ° | 24.7 ° | 23.3 ° |
| Improper angles | 1.22 ° | 0.74 ° | 0.9° |
| Standard error from Luzzati plot | 0.24 Å | 0.3 Å | 0.29 Å |
Total (outer shell).
Rsym = (Σh|Ih - 〈I〉)| / (ΣhIh).
Rcrys = (Σh|Fobs - Fcalc|) / (ΣhFobs), where the working and test sets are randomly selected.
2.4 Near UV Circular dichroism (CD)
Ellipticities were measured from 240 to 400 nm at room temperature using a Jasco J-720 (Jasco Inc., Easton, MD) spectropolarimeter, essentially as previously described (Galivan, Maley & Maley, 1975; Weichsel, Montfort, Ciesla & Maley, 1995). Difference spectra were obtained by subtracting the ellipticities of TS (WT or mutant) to the TS-nucleotide-folate solution complex using a program provided by Jasco.
2.5 Isothermal titration calorimetry
For ITC, the enzymes were dialyzed against 20 mM potassium phosphate buffer, pH 7.5 plus 10 % ethylene glycol and 5 mM DTT. However, DTT was omitted in the final buffer of dialysis because of its interference with the calorimetric determinations. ITC measurements were performed using a VP-ITC calorimeter (MicroCal, Inc.). Calorimetric titrations were assessed at 25 °C with a previously equilibrated binary complex of 1 mg/mL of TS (WT or K48Q) and a ten fold molar excess of dUMP in the sample cell. The stirring speed was kept constant at 394 rpm and the binding reaction was monitored in duplicate against the addition of 8 µl aliquots of antifolate to the binary complex in the cell. The antifolate (PDDF or U89) concentration in the syringe was 0.333 mM in 20 mM potassium phosphate buffer plus dUMP in order to avoid nucleotide dilution. Microcalorimetry titration experiments were continued until a three fold excess of antifolate was reached. The binding reactions from ITC were analyzed using the Microcal Origin software supplied with the calorimeter, averaging duplicate titration data for calculation of binding constants and thermodynamical parameters.
2.5 Tryptophan fluorescence
Trp fluorescence was used to observe the antifolate binding regarding to the Trp residues at the active site (W80 and W83) (Anderson, O’Neil, DeLano & Stroud, 1999; Felder, Dunlap, Dix & Spencer, 2002; Lovelace, Gibson & Lebioda, 2007; Sharma & Kisliuk, 1975). Samples were prepared by dialysis as described above for ITC. Trp fluorescence was monitored using a PTI QM-2003 fluorometer with a Xenon lamp as a light source (Photon Technology International). The excitation wavelength was 295 nm and emission data were collected from 300 to 500 nm with accumulation of two readings. Measurements of Trp fluorescence were performed with a solution of 1 µM of TS (WT or K48Q) containing 10 µM of dUMP in potassium phosphate buffer, pH 7.5, plus 10 % ethylene glycol. Trp fluorescence data were collected in the absence of and in the presence of a three fold excess of antifolate, as the final ITC titration condition.
3. Results
3.1 Catalysis by K48Q
The K48Q mutant of TS was impaired both in catalysis and ligand binding. The kcat was found to be 0.016 s−1, which is 430 fold less than that for the WT enzyme, while the Km for dUMP was 50 µM, 10 fold higher than the WT. The Km for the (R)-stereoisomer of CH2THF was 300 µM, about 30 fold larger than for the WT.
3.2 Ligand selection
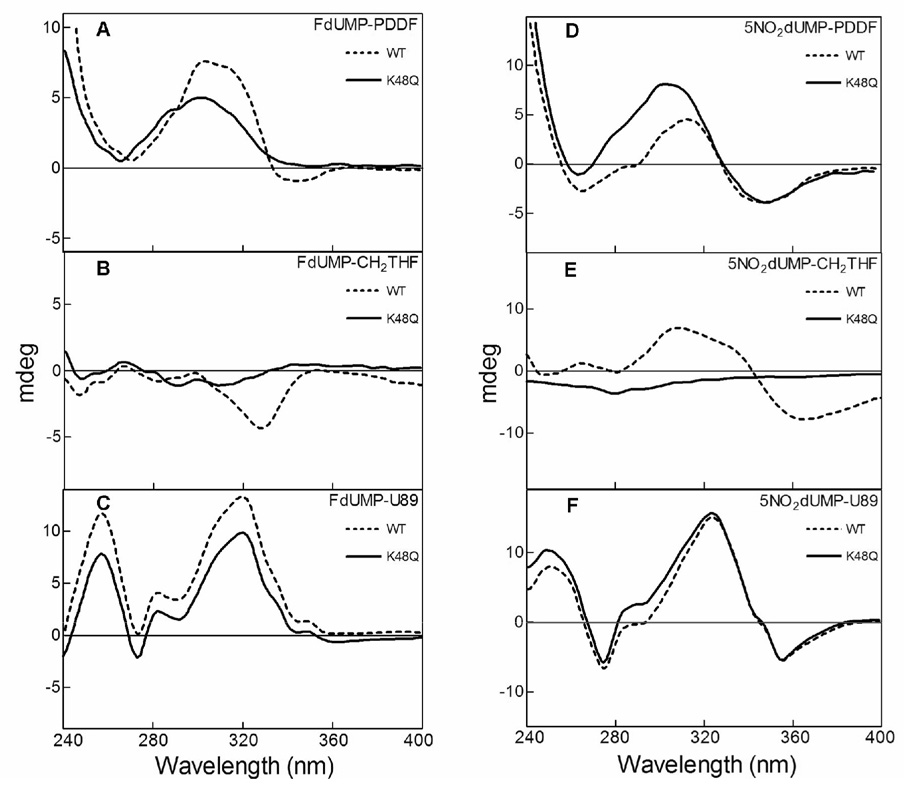
Kinetic data indicated that the mutation of K48 to a glutamine was deleterious to the enzyme’s function and may not produce a stable ternary complex with the K48Q mutant. We used near-UV circular dichroism (240–400 nm) as a screening method (Danenberg, Langenbach & Heidelberger, 1974), so that changes in the difference spectra would be indicative of productive binding as in our previous work with the binding of purine and pyrimidine nucleotides to the TS active site (Galivan, Maley & Maley, 1975; Weichsel, Montfort, Ciesla & Maley, 1995) (Fig. 3). In this manner, it was determined which combination of ligands produce a ternary complex similar to that of WT. Two nucleotides known to stabilize a ternary complex were tested: FdUMP and 5NO2dUMP; and three folates: PDDF, (R)-CH2THF and U89. A combination of ligands that produced spectra with K48Q that was very similar to those with the WT was 5NO2dUMP and U89 (Fig. 3F), and these compounds were used to crystallize a K48Q ternary complex. Note that although all three folates are glutamylated, U89 has an isoindolinone instead of a PABA ring compared to CH2THF and PDDF (Fig. 1B).
Fig. 3.
Near-UV circular dichroism difference spectra for wild-type (dashed lines) and K48Q mutant (continuous lines) TS in presence of nucleotide and folate analogs. Difference spectra pattern were calculated by substracting the ellipticities in millidegrees of the protein-nucleotide-folate solution to the protein-nucleotide. The nucleotide analogs tested were FdUMP (left column) and 5NO2dUMP (right column). The folates evaluated were PDDF (A and B), CH2THF (C and D) and U89 (E and F).
3.3 Nucleotide binding to the K48Q mutant
From the nature of K48Q mutant, nucleotide binding was predicted not to be affected, and to test this hypothesis, we determined the crystal structure of the K48Q-dUMP binary complex. The final model was refined to a resolution of 2.2 Å (Table 1, PDB 2VET). After a three-dimensional superposition with the binary complex of WT E. coli TS with dUMP (Stout, Sage & Stroud, 1998), the RMS deviation of the Cα backbone between the models was 0.15 Å. The nucleotide was well ordered in the crystal structure although there is a displacement in the position of the deoxyribose ring by approximately 0.3 Å compared to its position in the WT structure. Such movement did not affect the H-bonding with the enzyme and only the H-bond distances of the nucleotide-ribose hydroxyl with H207 and Y209 are increased by 0.2 Å. We also found a displacement of the catalytic C146 (0.3 Å) in parallel direction of the uridine ring and observe a double conformation in this residue.
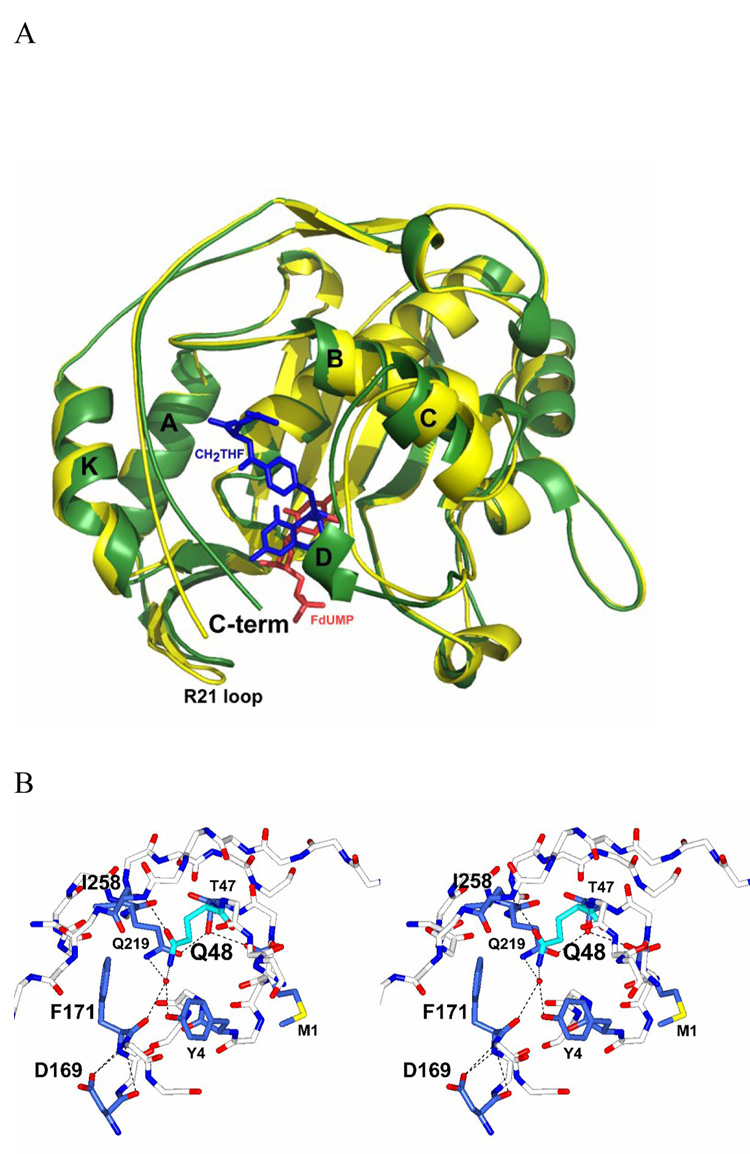
We found that Q48 disrupts the H-bond network that exists in the WT binary complex. Since in the latter structure, K48 contacts the main chain of I258 through a water molecule, while Y4 and Q219 are also H-bonded through a water molecule (Hyatt, Maley & Montfort, 1997). However, in the K48Q mutant binary complex, the contact with the F171 carbonyl is lost and the side-chain carbonyl of Q48 now contacts the main chain amide of I258 (Fig. 4B). Y4 and Q219 are H-bonded by a water molecule in contact with the amino group of the Q48 side-chain. The loss of contact with F171 does not produce any significant shift in the connecting secondary structure elements and confirms that K48 does not participate in the nucleotide binding process.
Fig. 4.
Ligand induced TS conformational change, (A) Ribbon diagram of the apo TS (PDB 1TJS) in yellow and the ternary complex TS-FdUMP-CH2THF (PDB 1TSN) in green. FdUMP is represented in red and the folate in blue. The ligand-induced conformational change is defined by a C-terminus movement inward towards the active site by about 5 Å. The loop between helix A and B moves about 1 Å, allowing R21 to contact the nucleotide phosphate and the I264 C-terminal carboxylate. Secondary structure elements are named after Montfort et al, (1990). (B) Stereo drawing of the mutation at the K48Q site on the crystal structure bound to dUMP. The glutamine residue at position 48 is colored in cyan bonds and the residues implicated in the hydrogen-bond network with Q48 are highlighted in dark blue bonds. The water molecule in contact with Q48 is colored in red and hydrogen-bonds are represented with dashed lines. The final model was refined to a resolution of 2.2 Å.
3.4 Non-folate dependent active site closure in K48Q
To investigate whether the barrier for closing the active site was larger in the K48Q mutant, we determined the crystal structure of the mutant with the nucleotide analog 5NO2dUMP, a strong competitive inhibitor of TS with a Ki of 27 nM (Wataya, Matsuda & Santi, 1980) which forms a covalent adduct with TS and induces closure of the active site in the absence of folate (Arendall, 2001).
This structure essentially confirms that the mutant enzyme adopts a closed conformation, and that 5NO2dUMP forms a covalent bond with the catalytic residue C146 (Fig. 5A). The density for the nucleotide analog was good in both active sites, since the whole dimer was contained in the asymmetric unit. The C-terminus was also ordered with the R21 H-bonded to the C-terminal carbonyl (I264) and establishes the presence of the nucleotide-phosphate in both active sites (Fig. 5B). All other contacts with the enzyme were found as in the WT complex with 5NO2dUMP. However, two water molecules that form H-bonds between the nitro group and to E58 and H147 were missing in the mutant structure. The marginal differences in the backbone of the mutant structure are probably due to the disruption of the H-bond network with Y4, Q219 and I258. In conclusion, K48Q did not interfere with the nucleophilic attack of 5NO2-dUMP on the catalytic C146 nor did it prevent the ligand-induced conformational change.
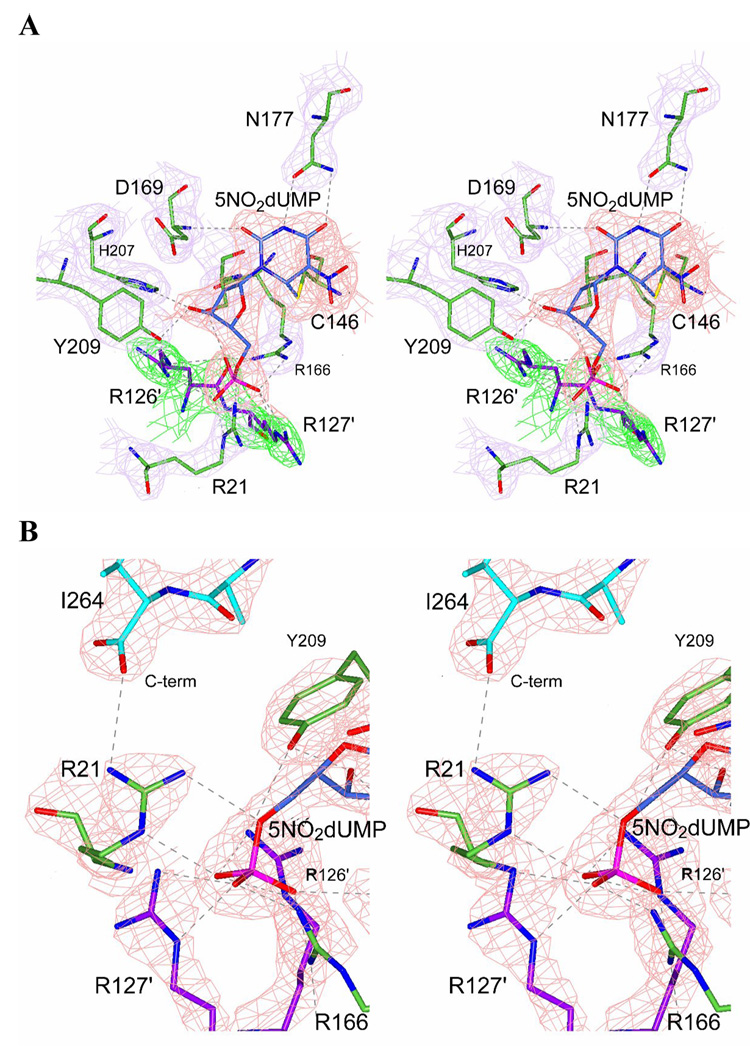
Fig. 5.
Structure of the covalent binary complex K48Q-5NO2dUMP. (A) Stereo drawing with electron density showing contacts with the active site and 5NO2dUMP and the covalent bond between C6 of nucleotide and C146 of the K48Q mutant TS. Electron density map for the nucleotide is colored in red and for the residues of the protein in magenta. R126′ and R127′ (green electron density map) designate to the residues of the other chain in the homodimer mutant TS. (B) Stereo drawing with electron density showing the hydrogen-bond of R21 with the carboxyl-terminus of the K48Q mutant TS, indicating active site closure. R21 is colored in green bonds and I264 (C-terminus) in cyan bonds. Hydrogen-bonds are represented by dashed lines.
3.5 Folate binding for the K48Q mutant
In view of earlier studies (Maley, Maley & Baugh, 1982), the binding of CH2THF to the K48Q mutant was predicted to be impaired, and consistent with this hypothesis, the Km was increased by 30 fold compared to WT (see above). Since the natural substrate CH2THF will react with dUMP in the titration experiments, the antifolates PDDF and U89 were used for these equilibrium experiments (Fig. 6).
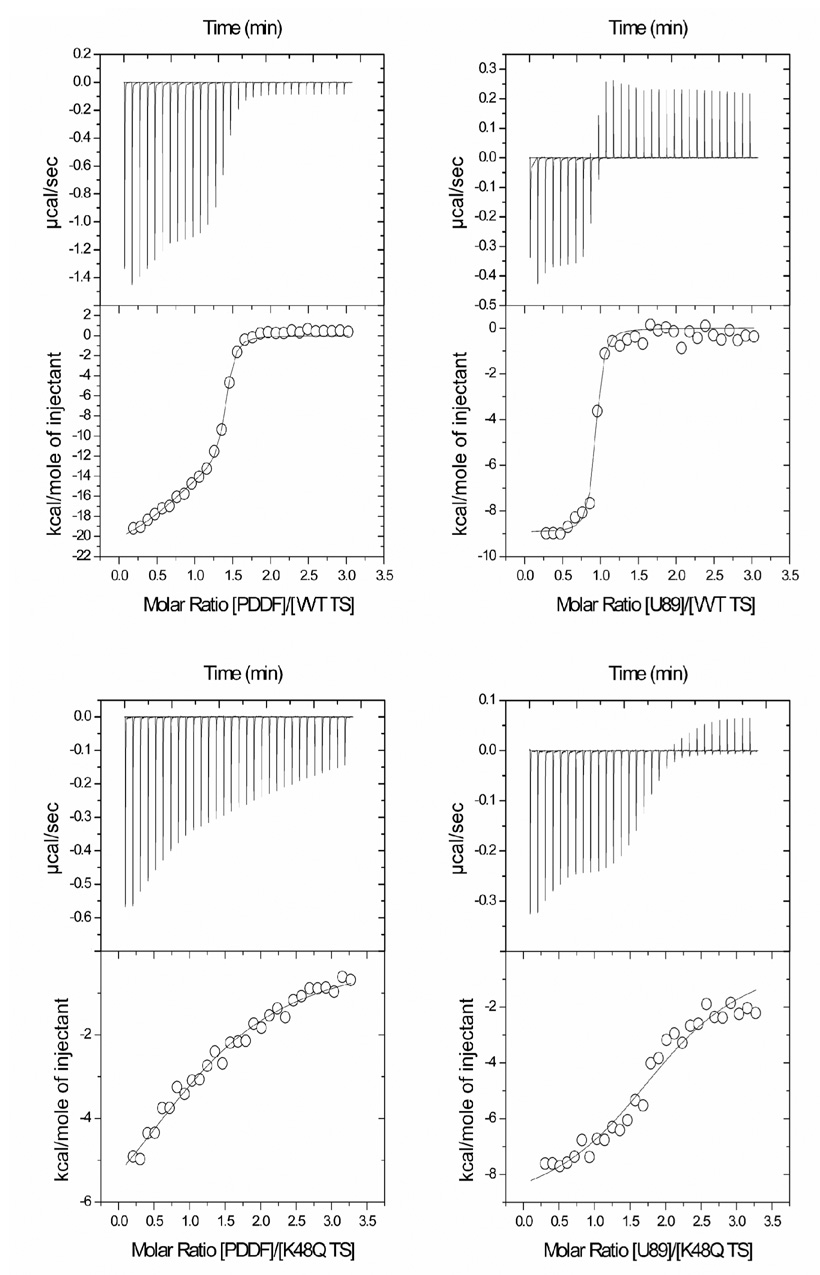
Fig. 6.
Isothermal titration calorimetry of antifolate binding to wild-type and K48Q mutant TS in the presence of dUMP. In the left, titration with PDDF for the wild-type (A) and the K48Q mutant (C). In the right titration with U89 for the wild-type (B) and the K48Q mutant (D). Titration experiments were done in potassium phosphate buffer (20 mM, pH 7.5), ethylene glycol (10%) and dUMP (166.6 µM) at 25 °C. Protein concentration in the cell was 1 mg/mL. The concentration of antifolate in the syringe was 0.333 mM.
U89 was more tightly bound compared to PDDF for WT TS, and the constants (Table 2) are similar to previous reports (Dev et al., 1994). We also found that only one active site is occupied by the folate, which speaks to the asymmetry of the TS active site. These are the first ITC folate-titration experiments on TS, and since they do not rely on indirect binding measurements, they are a reference for future work in understanding antifolate binding.
Table 2.
Thermodynamic data for the binding of antifolates to E. coli TS WT and K48Q determined by ITC.
| Ligand | ΔH | TΔS | ΔG | Kd (µM) | n | |
|---|---|---|---|---|---|---|
| WT-dUMP | U89 | −8.9 | 1.00 | −9.94 | 0.05 | 0.9 |
| K48Q-dUMP | U89 | −9.4 | −2.17 | −7.22 | 5.00 | 2.0 |
| WT-dUMP | PDDF | −19.9 | −11.06 | −8.82 | 0.33 | 1.2 |
| K48Q-dUMP | PDDF | −21.6 | −15.59 | −5.97 | 42.70 | 1.3 |
Values of ΔH, TΔS and ΔG are in kcal/mol
The enthalpic and entropic contributions provided by ITC are considerably different for the binding of the antifolates. This confirms differences in the mode of binding between U89 and PDDF, and it is also suported by the crystallographic evidence. For the binding of U89 to the WT TS, there is a favorable contribution from both the enthalpic and entropic components, with −8.9 and 1 kcal mol−1, respectively. This suggests that the binding mode of U89 is dominated by hydrophobic interactions, possibly by releasing relatively ordered water molecules from the ligand binding interface as seen by differential scanning calorimetry (Chen, Davis & Maley, 1996).
In contrast, PDDF binding to WT TS is driven by a strong enthalpic contribution (−19.9 kcal mol−1), derived from extensive H-bonds with the active site of WT TS and by water-mediated H-bonds. Also, this increase in the system order will have enthalpy-entropy compensation, as usually seen for many other ligand-protein interactions (Cooper, Johnson, Lakey & Nollmann, 2001). For the K48Q mutant, the enthalpic and entropic contributions for the binding of both antifolates are similar to those for the WT TS (see Table 2). However, the contribution from the enthalpy change for K48Q is increased while the entropy change is decreased in a major manner.
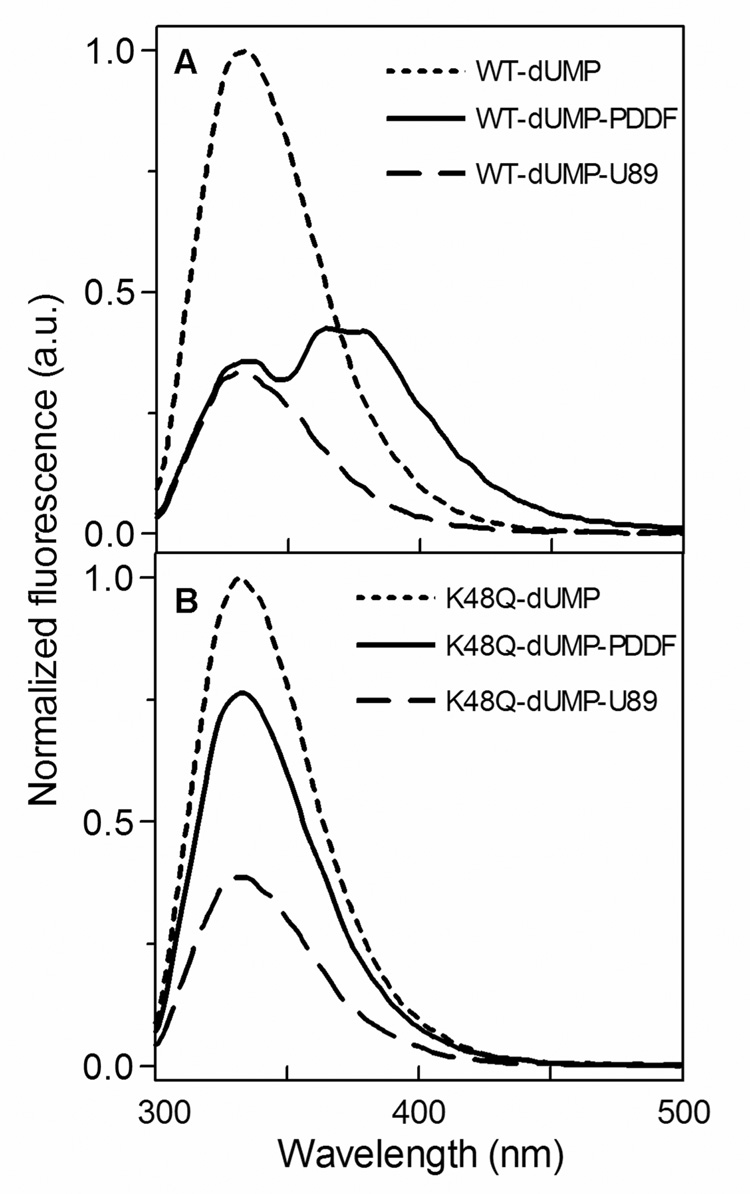
Trp fluorescence was used to examine the binding of antifolates to the WT and K48Q mutant active sites of the E. coli TS (Fig. 7). During PDDF titration, Trp fluorescence at 332 nm was quenched related to the binding of PDDF, but an emission at 379 nm appeared, suggesting a fluorescence resonance energy transfer (FRET) between the WT TS and PDDF. A similar red shift in maximum emission during binding was reported in the literature for TS from Lactobacillus casei (Sharma & Kisliuk, 1975) and Pneumocystis carinii (Anderson, O’Neil, DeLano & Stroud, 1999). Trp fluorescence experiments indicate that the K48Q mutant has a lower affinity for both antifolates, consistent with the ITC data.
Fig. 7.
Tryptophan fluorescence of wild-type (A) and K48Q mutant (B) TS with various ligands. The experiments were carried out in potassium phosphate buffer (20 mM, pH 7.5), ethylene glycol (10%) and dUMP (10 µM). Protein concentration in the cell was 1 µM and data were collected in the absence of antifolate and 3 µM of PDDF or U89.
3.6 Crystal structure of an antifolate-K48Q mutant complex
To further understand folate binding and to obtain ternary closed mutant complexes, we tried co-crystallization experiments of K48Q-dUMP-PDDF and K48Q-FdUMP-CH2THF, but failed to obtain diffracting crystals. Using the ligand selection strategy described above (Fig. 3), neither CH2THF-FdUMP nor PDDF-dUMP in complex with K48Q gave an ellipticity pattern similar to the corresponding WT complex. Only the ternary complex K48Q-5NO2dUMP-U89 had a near-UV CD pattern similar to the WT complex. Using the pH and ammonium sulfate screening previously described, we obtained hexagonal crystals belonging to the P63 space group, which diffracted to 3 Å (PDB 2VF0).
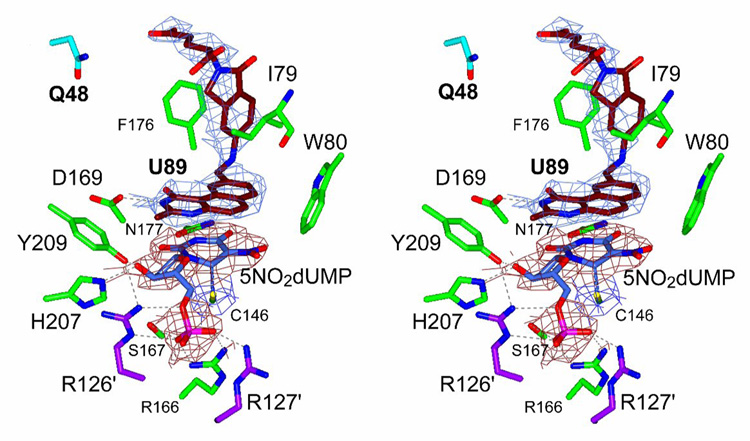
Even at the low resolution obtained, we could model the ligands within the K48Q active site and the enzyme structure appeared in a closed conformation (Fig. 8). A strong covalent bond existed between the active site C146 and position C6 of the pyrimidine ring of 5NO2dUMP. The ordering of the loop containing R21 and the shift of the C-terminus towards the active site were the main features of the structure. The closure of the active site was assessed from the H-bonds between the 3’-hydroxyl of 5NO2dUMP and Y209 and H207, and from the contacts of R21 and the C-terminus with the ligands (Montfort et al., 1990). We detected two conformations of U89 in the K48Q active site, which differed from that described in the WT enzyme complex (Stout & Stroud, 1996; Weichsel & Montfort, 1995). The isoindolinone carboxyl group is oriented about 1 Å away from the observed position in WT TS and residue S54 maintained an H-bond with the isoindolinone carboxyl. We resolved different conformations with respect to the WT enzyme, which showed a displacement of the main chain in both subunits of about 0.5 Å.
Fig. 8.
Stereo drawing of the active site of the crystal structure of the ternary complex of K48Q mutant TS with 5NO2dUMP and U89. Electron density is showed for nucleotide (red) and for antifolate (blue). Q48 is drawn with cyan bonds and other residues in the active site are colored in green. R126′ and R127′ from the other chain are colored in magenta. A covalent bond is present between C6 of nucleotide and C146 of TS. Hydrogen-bonds are represented with dashed lines.
4. Discussion
The structural analyses of the E. coli TS K48Q mutant complexes confirmed the critical role of K48 for folate binding. The crystallographic results presented here show that the early steps in the reaction mechanism, namely nucleotide binding and covalent adduct formation, are not impaired by the mutation. Nonetheless, there is a decrease in kcat (~430 fold) and an increase in Km for CH2THF (~30 fold).
From this view of ligand binding, K48 functions as an electrostatic anchor that positions the glutamate tail of folate in the active site (Maley, Maley & Baugh, 1982; Matthews, Appelt, Oatley & Xuong, 1990). Although CH2THF and some antifolates are polyglutamylated by folylpolyglutamate synthase, it is the nucleotide-dependent folate binding that is consistent with the enzyme mechanism (Spencer, Villafranca & Appleman, 1997). Also, the polyglutamylated folates interact with K48 via ordered water molecules (Kamb, Finer-Moore, Calvert & Stroud, 1992).
Active site closure is thought to be favored by the Van der Waal contacts with the folate PABA more than the H-bonds made by the C-terminus, as seen in the closed conformation of TS binary complex with PDDF (Kamb, Finer-Moore & Stroud, 1992) and ternary complexes (Montfort et al., 1990; Stout, Sage & Stroud, 1998). In support of this thesis, the ITC and Trp fluorescence data indicate that binding of U89 to K48Q is only slightly affected by the mutation.
However, for PDDF it appears that the PABA ring of the folate is not enough to support productive conformation, since near-UV CD pattern were not similar to their WT counterparts. Interestingly, K48 is not essential for the ligand-induced conformational change to occur, since K48Q was able to stabilize a covalent adduct and the closed conformation when crystallized with 5NO2dUMP.
The altered mode of binding of U89 appears not to require the water-mediated K48 contact with the glutamate tail. These shifts enable the larger isoindolinone group of U89 to be accommodated in the active site, where the smaller PABA ring of the substrate CH2THF binds normally. As a consequence of these shifts, the glutamate tail from U89 is about 7 Å away from the position of K48. We found a closed conformation in the ternary complex of K48Q with 5NO2dUMP and U89, as expected.
Trp fluorescence quenching of both WT and K48Q in the presence of PDDF suggests that the antifolate is located in the active site. However, the fact that FRET is only present when PDDF binds to the WT enzyme indicates that a specific orientation is required between the ligand and the residues in the active site that is not present in K48Q. The Trp fluorescence data clearly shows that the K48Q mutation affects the mode of binding of PDDF to the active site, and reflects the role of Trp residues at the active site in folate binding (Fritz, Liu, Finer-Moore & Stroud, 2002; Hong, Haddad, Maley, Jensen & Kohen, 2006; Kealey, Eckstein & Santi, 1995). This thesis may be extrapolated to the natural folate CH2THF, which also contains a PABA-ring and as a result it may be stated that this mutation precludes productive folate binding and may be the cause for the two-order loss of catalytic activity found for the K48Q mutant TS.
The combination of biophysical techniques such as calorimetric titrations and protein fluorescence was useful in correlating ligand binding and ligand-induced conformational change in E. coli TS in solution when compared to the x-ray structure. The K48Q mutation in TS disrupts the H-bond network that exists in the active site, which as a result impairs cofactor binding. K48 does not participate in the nucleotide binding process and the nucleophilic attack on the catalytic C146 nor does it prevent the ligand-induced conformational change. The altered mode of binding of U89 makes it unnecessary for the glutamate of this inhibitor to interact electrostatically with K48 and leads to a ternary complex in the closed conformation. In this work, we demonstrate that the K48Q mutation in E. coli TS severely impairs the binding of classical folates such as CH2THF or PDDF, relative to the WT enzyme. Also, the mutation of this critical amino acid residue does not have the same effect on a modified antifolate (U89) whose mode of binding is altered. One aspect of studying mutation in invariant residues of TS is that a drug resistant TS mutant can be developed for ex-vivo gene therapy to prevent inmunosuppresion in bone marrow (Banerjee & Bertino, 2002; Banerjee et al., 2002). Such potential applications will certainly benefit from understanding the structure and function of a model system such as that found in the case of bacterial TS.
Acknowledgements
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACYT) México D.F. grant 48991-Z (RRS), by American Cancer Society grant RPG-93-041-04-CDD (WRM); Arizona Disease Control Research Commissions Grant 1-208A (WRM); National Cancer Institutes grant CA44355 (FM), the State Committee for Scientific Research Grant No. 4 P05F 030 11p02 (WRM). We thank Secretaría de Educación Pública (México) for grant PIFI 2004-26-09 for the purchase of the isothermal titration calorimeter. We thank Drs. Sue Roberts and Andrzej Weichsel (Univ. of Arizona) for critical reading and expert assistance. A. Arvizu-Flores received a graduate scholarship from CONACYT and R. Arreola received a postdoctoral fellowship from UNAM (Universidad Nacional Autónoma de México). A. Arvizu-Flores designed and performed research, analyzed data and wrote the paper, R. Sugich-Miranda performed research and analyzed data, R. Arreola analyzed data and wrote the paper, K. Garcia-Orozco performed research, E. Velazquez-Contreras designed research and contributed analytical tools, F. Maley designed research, performed research, contributed new reagents and wrote the paper, W. Montfort designed research and analyzed data, R. Sotelo-Mundo designed and performed research, analyzed data and wrote the paper.
Abbreviations and Textual Footnotes
- TS
Thymidylate synthase
- CH2THF
5,10-methylene-5,6,7,8-tetrahydrofolate
- dUMP
2′-deoxyuridine-5′-monophosphate
- dTMP
2’-deoxythymidine-5’-monophosphate
- FdUMP
5-fluoro-dUMP
- PDDF
10-propargyl-5,8-dideazafolate
- DTT
dithiotreithol
- PABA
para-aminobezoic acid
- 5NO2dUMP
5-nitro-dUMP
- ITC
isothermal titration calorimetry
- U89
(S)-2-(5(((1,2-dihydro-3-methyl-1-oxobenzo[f]quinazolin-9-yl)methyl)amino)1-oxo-2-isoindolinyl) glutaric acid
- CD
circular dichroism
- Trp fluorescence
tryptophan fluorescence
- FRET
fluorescence resonance energy transfer
- H-bond
hydrogen bond
Footnotes
K48 is referred to the E. coli sequence numbering and is equivalent to K77 in the human TS amino acid sequence.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson AC, O'Neil RH, DeLano WL, Stroud RM. The structural mechanism for half-the-sites reactivity in an enzyme, thymidylate synthase, involves a relay of changes between subunits. Biochemistry. 1999;38:13829–13836. doi: 10.1021/bi991610i. [DOI] [PubMed] [Google Scholar]
- Appaji Rao N, Talwar R, Savithri HS. Molecular organization, catalytic mechanism and function of serine hydroxymethyltransferase -- a potential target for cancer chemotherapy. International Journal of Biochemistry & Cell Biology. 2000;32:405–416. doi: 10.1016/s1357-2725(99)00126-0. [DOI] [PubMed] [Google Scholar]
- Arendall WBI. Doctoral Dissertation. Department of Biochemistry. Tucson AZ: The University of Arizona; 2001. X-ray structures of novel intermediates in the thymidylate synthase models for chemical mechanism and conformational change; p. 148. [Google Scholar]
- Banerjee D, Bertino JR. Myeloprotection with drug-resistance genes. Lancet Oncology. 2002;3:154–158. doi: 10.1016/s1470-2045(02)00678-2. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR. Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochimica et Biophysica Acta. 2002;1587:164–173. doi: 10.1016/s0925-4439(02)00079-0. [DOI] [PubMed] [Google Scholar]
- Barbour KW, Berger SH, Berger FG. Single amino acid substitution defines a naturally occurring genetic variant of human thymidylate synthase. Molecular Pharmacology. 1990;37:515–518. [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica D Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carreras CW, Santi DV. The Catalytic Mechanism and Structure of Thymidylate Synthase. Annual Review of Biochemistry. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallographica D Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen CH, Davis RA, Maley F. Thermodynamic stabilization of nucleotide binding to thymidylate synthase by a potent benzoquinazoline folate analogue inhibitor. Biochemistry. 1996;35:8786–8793. doi: 10.1021/bi952953t. [DOI] [PubMed] [Google Scholar]
- Chu E, Callender MA, Farrell MP, Schmitz JC. Cancer Chemotherapy and Pharmacology. Vol. 52. 2003. Thymidylate synthase inhibitors as anticancer agents: from bench to bedside; pp. S80–S89. [DOI] [PubMed] [Google Scholar]
- Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophysical Chemistry. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- Danenberg PV, Langenbach RJ, Heidelberger C. Structures of reversible and irreversible complexes of thymidylate synthetase and fluorinated pyrimidine nucleotides. Biochemistry. 1974;13:926–933. doi: 10.1021/bi00702a016. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Dev IK, Dallas WS, Ferone R, Hanlon M, McKee DD, Yates BB. Mode of binding of folate analogs to thymidylate synthase. Evidence for two asymmetric but interactive substrate binding sites. Journal of Biological Chemistry. 1994;269:1873–1882. [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica D Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Felder T, Dunlap RB, Dix D, Spencer T. Differences in natural ligand and fluoropyrimidine binding to human thymidylate synthase identified by transient-state spectroscopic and continuous variation methods. Biochimica et Biophysica Acta. 2002;1597:149–156. doi: 10.1016/s0167-4838(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme thymidylate synthase mutagenesis function and structure. biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Liu L, Finer-Moore JS, Stroud RM. Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access. Biochemistry. 2002;41:7021–7029. doi: 10.1021/bi012108c. [DOI] [PubMed] [Google Scholar]
- Galivan JH, Maley GF, Maley F. The effect of substrate analogs on the circular dichroic spectra of tlymidylate synthase from Lactobacillus casei. Biochemistry. 1975;14:3338–3344. doi: 10.1021/bi00686a008. [DOI] [PubMed] [Google Scholar]
- Hong B, Haddad M, Maley F, Jensen JH, Kohen A. Hydride transfer versus hydrogen radical transfer in thymidylate synthase. Journal of the American Chemical Society. 2006;128:5636–5637. doi: 10.1021/ja060196o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huennekens FM. The methotrexate story: a paradigm for development of cancer chemotherapeutic agents. Advances in Enzyme Regulation. 1994;34:397–419. doi: 10.1016/0065-2571(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Hyatt DC, Maley F, Montfort WR. Use of Strain in a Stereospecific Catalytic Mechanism: Crystal Structures of Escherichia coli Thymidylate Synthase Bound to FdUMP and Methylenetetrahydrofolate. Biochemistry. 1997;36:4585–4594. doi: 10.1021/bi962936j. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjelgard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallographica A Foundations of Crystallography. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Evaluation of Single-Crystal X-ray Diffraction Data from a Position-Sensitive Detector. J. Appl. Crystallogr. 1988;21:916–934. [Google Scholar]
- Kamb A, Finer-Moore JS, Calvert AH, Stroud RM. Structural Basis for Recognition of Polyglutamyl Folates by Thymidylate Synthase. Biochemistry. 1992;31:9883–9890. doi: 10.1021/bi00156a005. [DOI] [PubMed] [Google Scholar]
- Kamb A, Finer-Moore JS, Stroud RM. Cofactor Triggers the Coformational Change in Thymidylate Synthas: Implications for an Ordered Binding Mechanism. Biochemistry. 1992;31:12876–12884. doi: 10.1021/bi00166a024. [DOI] [PubMed] [Google Scholar]
- Kealey JT, Eckstein J, Santi DV. Role of the conserved tryptophan 82 of Lactobacillus casei thymidylate synthase. Chemistry & biology. 1995;2:609–614. doi: 10.1016/1074-5521(95)90125-6. [DOI] [PubMed] [Google Scholar]
- Kimura E, Nishimura K, Sakata K, Oga S, Kashiwagi K, Igarashi K. Methotrexate differentially affects growth of suspension and adherent cells. International Journal of Biochemistry & Cell Biology. 2004;36:814–825. doi: 10.1016/j.biocel.2003.09.002. [DOI] [PubMed] [Google Scholar]
- LaPat-Polasko L, Maley GF, Maley F. Properties of bacteriophage T4 thymidylate synthase following mutagenic changes in the active site and folate binding region. Biochemistry. 1990;29:9561–9572. doi: 10.1021/bi00493a010. [DOI] [PubMed] [Google Scholar]
- Lovelace LL, Gibson LM, Lebioda L. Cooperative inhibition of human thymidylate synthase by mixtures of active site binding and allosteric inhibitors. Biochemistry. 2007;46:2823–2830. doi: 10.1021/bi061309j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley F, Pedersen-Lane J, Changchien L. Complete Restoration of Activity to Inactive Mutants of Escherichia coli Thymidylate Synthase: Evidence that E. coli Thymidylate Synthase is a Half-the-Sites Activity Enzyme. Biochemistry. 1995;34:1469–1474. doi: 10.1021/bi00005a001. [DOI] [PubMed] [Google Scholar]
- Maley GF, Maley F, Baugh CM. Studies on identifying the folylpolyglutamate binding sites of Lactobacillus casei thymidylate synthetase. Archives of Biochemistry and Biophysics. 1982;216:551–558. doi: 10.1016/0003-9861(82)90244-2. [DOI] [PubMed] [Google Scholar]
- Matthews DA, Appelt K, Oatley SJ, Xuong NH. Crystal Structure of Escherichia coli Thymidylate Synthase Containing Bound 5-fluoro-2'-deoxyuridylate and 10-Propargyl-5,8-dideazafolate. Journal of Molecular Biology. 1990;214:923–936. doi: 10.1016/0022-2836(90)90346-N. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A, Pflugrath JW. Crystal Orientation and X-ray Pattern Prediction Routines for Area-Detector Diffractometer Systems in Macromolecular Crystallography. Journal of Applied Crystallography. 1987;20:306–315. [Google Scholar]
- Montfort WR, Perry KM, Fauman EB, Finer-Moore JS, Maley GF, Hardy L, Maley F, Stroud RM. Structure, Multiple Site Binding, and Segmental Accommodation in Thymidylate Synthase on Binding dUMP and an Anti-Folate. Biochemistry. 1990;29:6964–6977. doi: 10.1021/bi00482a004. [DOI] [PubMed] [Google Scholar]
- Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallographica D Biological Crystallography. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Santi DV, McHenry CS, Sommer H. Mechanism of interaction of thymidylate synthase with 5-fluorodexoyuridylate. Biochemistry. 1974;13:471–481. doi: 10.1021/bi00700a012. [DOI] [PubMed] [Google Scholar]
- Sharma RK, Kisliuk RL. Quenching of thymidylate synthetase fluorescence by substrate analogs. Biochemical and Biophysical Research Communications. 1975;64:648–655. doi: 10.1016/0006-291x(75)90370-8. [DOI] [PubMed] [Google Scholar]
- Spencer HT, Villafranca JE, Appleman JR. Kinetic Scheme for Thymidylate Synthase from Escherichia coli: Determination from Measurments of Ligand Binding Primary and Secondary Isotope Effects and Pre-Steady-State Catalysis. Biochemistry. 1997;36:4212–4222. doi: 10.1021/bi961794q. [DOI] [PubMed] [Google Scholar]
- Stout TJ, Sage CR, Stroud RM. The additivity of substrate fragments in enzyme-ligand binding. Structure. 1998;6:839–848. doi: 10.1016/S0969-2126(98)00086-0. [DOI] [PubMed] [Google Scholar]
- Stout TJ, Stroud RM. The complex of the anti-cancer therapeutic BW1843U89 with thymidylate synthase at 2.0 Å resolution implications for a new mode of inhibition. Structure. 1996;4:67–77. doi: 10.1016/s0969-2126(96)00010-x. [DOI] [PubMed] [Google Scholar]
- Stout TJ, Tondi D, Rinaldi M, Barlocco D, Pecorari P, Santi DV, Kuntz ID, Stroud RM, Shoichet BK, Costi MP. Structure-based design of inhibitors specific for bacterial thymidylate synthase. Biochemistry. 1999;38:1607–1617. doi: 10.1021/bi9815896. [DOI] [PubMed] [Google Scholar]
- Tong Y, Liu-Chen X, Ercikan-Abali EA, Zhao SC, Banerjee D, Maley F, Bertino JR. Probing the folate-binding site of human thymidylate synthase by site- directed mutagenesis. Generation of mutants that confer resistance to raltitrexed, Thymitaq, and BW1843U89. Journal of Biological Chemistry. 1998;273:31209–31214. doi: 10.1074/jbc.273.47.31209. [DOI] [PubMed] [Google Scholar]
- Wahba AJ, Friedkin M. Direct spectrophotometric evidence for oxidation of tetrahydrofolate during enzymatic synthesis of thymidylate. Journal of Biological Chemistry. 1962;237:3794–3801. [PubMed] [Google Scholar]
- Wataya Y, Matsuda A, Santi DV. Interaction of Thymidylate Synthetase with 5 Nitro-2′-deoxyuridylate. Journal of Biological Chemistry. 1980;255:5538–5544. [PubMed] [Google Scholar]
- Weichsel A, Montfort WR. Ligand-Induced Distortion of an Active Site in Thymidylate Synthase Upon Binding Anticancer Drug 1843U89. Nature Structural Biology. 1995;2:1095–1101. doi: 10.1038/nsb1295-1095. [DOI] [PubMed] [Google Scholar]
- Weichsel W, Montfort WR, Ciesla J, Maley F. Promotion of Purine Nucleotide Binding to Thymidylate Synthase by a Potent Folate Analogue Inhibitor 1843U89. Proceedings of the National Acadamy of Sciences USA. 1995;92:3493–3497. doi: 10.1073/pnas.92.8.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]