Abstract
Understanding immunity to vaccinia virus (VACV) is important for the development of safer vaccines for smallpox- and poxvirus-vectored recombinant vaccines. VACV is also emerging as an outstanding model for studying CD8+ T cell immunodominance because of the large number of CD8+ T cell epitopes known for this virus in both mice and humans. In this study, we characterize the CD8+ T cell response in vaccinated BALB/c mice by a genome-wide mapping approach. Responses to each of 54 newly identified H-2d-restricted T cell epitopes could be detected after i.p. and dermal vaccination routes. Analysis of these new epitopes in the context of those already known for VACV in mice and humans revealed two important findings. First, CD8+ T cell epitopes are not randomly distributed across the VACV proteome, with some proteins being poorly or nonimmunogenic, while others are immunoprevalent, being frequently recognized across diverse MHC haplotypes. Second, some proteins constituted the major targets of the immune response by a specific haplotype as they recruited the majority of the specific CD8+ T cells but these proteins did not correspond to the immunoprevalent Ags. Thus, we found a dissociation between immunoprevalence and immunodominance, implying that different sets of rules govern these two phenomena. Together, these findings have clear implications for the design of CD8+ T cell subunit vaccines and in particular raise the exciting prospect of being able to choose subunits without reference to MHC restriction.
Vaccinia virus (VACV)3 is almost unique in that infections of known timing and similar pathogenesis can be achieved both in humans and mice. Moreover, in the last 5e years, the specificity of CD8+ T cell responses to VACV has gone from being completely unexplored to arguably the best documented, with epitopes mapped for many MHC restriction elements across these two species (1–15). This gives us the opportunity to ask basic questions about the specificity of CD8+ T cell responses generated by a complex viral pathogen and obtain answers that are broadly applicable to multiple host species. One such question is whether epitope hierarchy is entirely a property of the immunogenicity of individual epitopes or whether some proteins as a whole might be more immunogenic and therefore better sources of epitopes than others.
To address this question, herein we define immunoprevalent Ags or epitopes as those recognized more frequently, while major Ags or epitopes are the ones recognized most vigorously by the immune response. Major and minor epitopes or proteins document epitope or Ag hierarchy that generally takes place during an immune response. Maillere and coworkers (16) introduced the concept of an immunoprevalent epitope in their studies of HLA-DP4-restricted epitopes derived from HIV, hepatitis C virus, and the MAGE-A gene family in the human system (16–18). Immunoprevalence could result from recognition of epitopes restricted by distinct MHC types, but could also arise because certain protein Ags are recognized in the context of many MHC haplotypes. Conversely, Ag hierarchy as immunodominance could result from properties intrinsic to the epitope(s) recognized, such as MHC binding, TCR repertoire, or processing efficiency. It could also result from features intrinsic to the protein Ag as a whole, such as, for example, its level and pattern of expression. It remains to be determined whether immunoprevalent proteins are also the key contributors of the most major epitopes.
For small viruses that have been intensely studied, such as HIV and hepatitis C virus in humans and influenza and lymphocytic choriomeningitis viruses in mice, immunoprevalence will be of less relevance and also harder to discern. The limited coding capacity of these viral genomes, the need for epitopes to conform to MHC-binding motifs, and the ability of some of these viruses to mutate and escape CD8+ T cells, all mean that it is most likely that properties of epitopes rather than proteins will dictate all aspects of immunogenicity. However, for bigger pathogens, the immune system faces a far larger choice of potential epitopes and the properties of proteins may become more important in determining immunogenicity for CD8+ T cells.
Some suggestions that immunoprevalent proteins exist come from the literature relating to CD8+ T cell responses to human herpesviruses. EBV Ags can be placed in an immunodominance hierarchy for CD8+ T cell responses in acute infection with the earliest expressed proteins ranking most highly (19). In contrast, a single protein expressed late during infection, namely pp65, is considered to be dominant in CD8+ T cell responses to CMV infections (20, 21), and late gene products are thought to be most immunogenic after HSV infection (22, 23). When considering this data, it is important to remember that despite the large amount of work represented by these studies, they remain based on relatively few epitopes. In addition, herpesviruses establish lifelong infections characterized by latency and frequent reactivation, and the impact of repeated restimulation of CD8+ T cells must inevitably shape dominance hierarchies.
To date, nearly 200 CD8+ T cell epitopes have been described for VACV in the context of five mouse MHC alleles and seven human MHC supertypes. Of these, the most exhaustive analysis has been in C57BL/6 mice (1); for BALB/c mice, the next most commonly used strain, only three epitopes have been described (14). In this study, we extend our database of VACV epitopes by mapping >50 new specificities in BALB/c mice. We also show that the same set of epitopes are identified irrespective of whether a dermal route or i.p. route of immunization is used. Finally, using a combined set of epitope data from mice and humans, we present an analysis that shows that some proteins are immunoprevalent, being rich sources of epitopes restricted by multiple MHC alleles; however these proteins cannot be predicted from knowledge of epitope immunodominance hierarchies.
Materials and Methods
MHC binding predictions for H-2Kd and H-2Ld
Each predicted open reading frame (ORF) of the vaccinia virus Western Reserve (WR) strain (24) was scanned for peptide sequences of nine residues that are predicted to have a high-affinity binding capacity for the MHC class I molecules H-2Kd or H-2Ld. The prediction algorithm for H-2Kd peptides was generated based on a set of 196 peptides with previously determined binding affinity to Kd molecules and binding affinities of a combinatorial peptide library reported by Udaka et al. (25). These two data sets were used to generate a single stabilized matrix method scoring matrix following exactly the approach previously described (26). For the Ld predictions, the combinatorial peptide library alone was used, since too few individual peptides with measured binding affinities for Ld were available to reliably improve prediction quality above the prediction based solely on combinatorial peptide library.
Peptides and MHC-binding assays
Peptides used in initial screening experiments were synthesized as crude material by Pepscan Systems. MHC purification and quantitative assays to measure the binding affinity of peptides to purified H-2Kd, Ld, and Dd molecules were performed essentially as previously described (1, 4, 27). Briefly, 0.1–1 nM of a known MHC-binding radiolabeled peptide, along with varying amounts of unlabeled test peptide, were coincubated at room temperature with 1 μM to 1 nM of purified MHC in the presence of 1–3 μM human β2-microglubulin (Scripps Laboratories) and a mixture of protease inhibitors. After a 2-day incubation, binding of the radiolabeled peptide to the corresponding MHC class I molecule was determined by capturing MHC-peptide complexes on Greiner Lumitrac 600 microplates (Greiner Bio-one) coated with either the SF1-1.1.1 (anti-H-2Kd), 34-5-8s (anti-H-2Dd),or 28-14-8s (anti-H-2Ld) Ab and measuring bound cpm using the TopCount microscintillation counter (Packard Instrument). The concentration of unlabeled peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Peptides were typically tested at six different concentrations covering a 100,000-fold dose range and in three or more independent assays. Under the conditions used, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of KD values.
Viruses
The WR strain of VACV was obtained from B. Moss (National Institute of Allergy and Infectious Diseases, Bethesda, MD).
Mice
Specific pathogen-free BALB/c mice were obtained from The Jackson Laboratory. Mice were used between 6 and 12 wk of age, following National Institutes of Health guidelines and Institutional Animal Care and Use Committee-approved animal protocols.
Infection and immunizations
In all experiments, BALB/c mice were infected i.p or by dermal scarification, with 0.1–2 × 106 PFU of VACV-WR in PBS. After 7 days, the mice were sacrificed and the splenocytes were used for ex vivo mouse IFN-γ ELISPOT measurement or ICCS assay as described below.
Stimulator cells and cell lines
All cells, including P815 (provided by J. Yewdell, National Institute of Allergy and Infectious Diseases) were grown in RPMI 1640 medium containing 25 nM HEPES, 4 mM L-glutamine, 5 × 10−5 M 2-ME, 0.5 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids, 100 μg/ml streptomycin, and 100 U/ml penicillin (all Invitrogen) and 10% FBS (Gemini Bio-Products). For IFN-γ ELISPOT) assays, LPS-stimulated B lymphoblasts obtained by cultivating splenocytes in the presence of 8.5 μg/ml LPS and 7 μg/ml dextran sulfate (Sigma-Aldrich) for 3 days at 37°C were used as stimulator cells. P815 cells were used as stimulator cells for intracellular cytokine staining (ICCS) assays.
Ex vivo ELISPOT assay
ELISPOT assays were performed as previously described (28). Briefly, 2 × 106 syngeneic LPS blasts were either peptide pulsed (10 μg/ml) or incubated with VACV-WR (multiplicity of infection (MOI), 5 or 10) for 2 h and washed once before use as stimulators. In brief, 4 × 105 splenocytes were then cocultured with 1 × 104 syngeneic LPS blasts in flat-bottom 96-well nitrocellulose plates (Immobilon-P membrane; Millipore) precoated with anti-IFN-γ mAb (2 μg/ml; BD Pharmingen). After a 20-h incubation at 37°C, plates were washed with PBS/0.05% Tween 20 and wells were incubated with biotinylated anti-IFN-γ mAb (1 μg/ml; BD Pharmingen) for 3 h at 37°C. After additional washing, spots were developed by sequential incubation with Vectastain ABC peroxidase (Vector Laboratories) and 3-amino-9-ethylcarbazole solution (Sigma-Aldrich) and counted by computer-assisted image analysis (Zeiss KS ELISPOT Reader). Each assay was performed in triplicate and the experimental values were expressed as the mean net spots per 106 unfractionated splenocytes ± SEM for each peptide. Responses against medium only and 1% DMSO (corresponding to the concentration of DMSO in a pool of 10 peptides, each at 10 μg/ml) were measured to establish background values that were subtracted from the experimental values. To determine the level of statistical significance, Student’s t test was performed in which p ≤ 0.05 using the mean of triplicate values of the response against relevant peptides vs the response against irrelevant control peptides was considered significant. The net number of spots per 106 effector cells was calculated as ((number of spots against relevant peptide) − (number of spots against DMSO control)) × ((1 × 106)/(number of effector cells/well)).
Ex vivo ICCS assay
Ex vivo ICCS assays were performed as previously described, with minor modifications (1). Splenocytes from VACV-WR-infected mice were used as effectors and were incubated with P815 cells pulsed with peptides at 0.1 μg/ml in a 96-well plate or P815 cells were infected with VACV-WR for a total of 4.5 h. To this end, 5 × 106 P815 were infected with VACV (MOI, 5–10) in 200 μl of PBS at 37°C for 30 min in a 15-ml Falcon tube with occasional shaking. After this initial incubation, 9 ml of R10 medium was added and the incubation continued for an additional 4 h before use in T cell assays. Briefly, 1–2 × 106 splenocytes were then cultured with 2 × 105–1 × 106 P815 cells in a well of a 96-well plate in the presence of brefeldin A (10 μg/ml). The cells were cultured overnight, not exceeding 12 h before staining according to the protocol of the BD Biosciences Fix/Perm Solution Kit using anti-CD8-PerCP and anti-IFN-γ-FITC Abs (all BD Pharmingen). The cells were analyzed using a FACSCalibur (BD Biosciences) and analyzed with FlowJo software (Tree Star) by gating lymphocytes on forward scatter × side scatter followed by gating on CD8+ T cells to identify the IFN-γ-producing CD8+ T cells. Background values were determined from samples pulsed with DMSO only (no peptide) and subtracted from the experimental values. At least three independent experiments were performed for each peptide pool. A peptide pool was considered positive if the average of the individual experiments was at least >1 SD above the background.
Statistical tests
All standard statistical tests were performed using Microsoft Excel and the VassarStats web site (http://faculty.vassar.edu/lowry/VassarStats.html).
Statistical simulations of the epitope/ORF distribution
If a set of epitopes is equally distributed in the set of ORFs, the number of epitopes per ORF would follow the multinomial distribution. This is generated by randomly assigning each epitope to one ORF. To calculate p values for the hypothesis “some ORFs are prevalently recognized” against the null hypothesis of equally distributed epitopes, we performed Monte Carlo simulations implemented as python scripts. The first observation was that only 103 of 206 ORFs contain one or more of the 197 epitopes. For each epitope, one of 206 ORFs is randomly assigned. Once all epitopes are assigned, the number of ORFs that have one or more epitopes is counted. Repeating this 10,000 times, the number N of simulations in which 103 or less ORFs contain all epitopes is counted, which gives the p value N/10,000. The same approach is used to calculate p values for observing ORFs recognized by different MHC specificities.
Results
Bioinformatic predictions of H-2/Kd-, Dd-, and Ld-binding VACV peptides
To map CD8+ T cell epitopes in BALB/c mice, a bioinformatic approach similar to the one validated during the comprehensive mapping in C57BL/6 mice was used (1). All predicted ORFs contained within the VACV-WR sequence (14, 24) were screened for peptides predicted to bind with high affinity to H-2d molecules. For the H-2Dd molecule, all peptides matching its strict and infrequently occurring binding motif were selected. Fourteen 9-mer and eight 10-mer peptides in VACV matched the Dd motif of *GP*****[FIL] and *GP******[FIL], respectively. For the Kd and Ld molecules, quantitative binding predictions were made based on in vitro-binding data of individual peptides and combinatorial peptide libraries (25) to the respective MHC allele (see Materials and Methods and Ref. 26).
All 9-mer peptides derived from the putative VACV proteome were ranked by their predicted ability to bind H-2Kd or Ld molecules, and the top 0.5% (n = 292) of peptides randomly distributed throughout the VACV proteome were selected for each allele. The 0.5% cutoff was based on an analysis of the accuracy of prediction performance in the H-2b study (1), which indicated that 33 (67%) of 49 epitopes were included in the top 0.5% scoring peptides of the preferred peptide length for the restricting allele. Also, all of the three previously identified epitopes (14), KYGRLFNEI (A52R75–83), SPYAAGYDL (F2L26–34), and VGPSNSPTF (E3L140–148), restricted by Kd, Ld, and Dd respectively, met these scoring or motif selection criteria.
Identification of H-2d-restricted epitopes recognized following i.p. immunization
For the three MHC molecules, a total of 606 peptides (292 Kd 9-mer, 292 Ld 9-mer, 8 Dd 10-mer, and 14 Dd 9-mer) were analyzed for antigenicity in IFN-γ ELISPOT assays using splenocytes obtained 7 days after i.p. infection of BALB/c mice with VACV-WR. Peptides were initially tested in 28 pools, each consisting of ~22 peptides (supplemental Table Ia).4 These experiments were repeated two to three times and for each experiment spleens from four to five animals were pooled. Peptide pools generating >20 spot-forming cells (SFC)/106 cells, a stimulation index >1.4 and a p < 0.05 in a standard t test in at least two consecutive experiments were considered positive. Responses ranged from 20 to ≥500 SFC/106 cells. Screening of Kd 9-mer, Dd 9-mer, Dd 10-mer, and Ld 9-mer peptide pools identified 11 positive pools (data not shown). None of the pools elicited a positive response in naive mice (data not shown).
To identify the individual peptides responsible for the antigenic activity, positive pools were deconvoluted by testing each peptide individually for its capacity to stimulate IFN-γ production in CD8+ T cells. Deconvolution experiments were repeated two to three times for each positive pool. Peptides eliciting a response of >40 SFC/106 cells, stimulation index of >2, and a p < 0.05 were considered positive. A total of 54 H-2d-restricted T cell epitopes (39 Kd 9-mer, 2 Dd 9-mer, 1 Dd 10-mer, and 12 Ld 9-mer) were identified by ELISPOT assays (supplementary Table Ib). It is noteworthy that all three epitopes identified previously using the expression library approach (14) were also identified in the present analyses. Four epitopes, KYGRLFNEI (A52R75–83), SPYAAGYDL (F2L26–34), VGPSNSPTF (E3L140–148), and GFIRSLQTI (C6L74–82), elicited responses greater than 750 SFC/106 cells. Three of these had been previously identified (14), while a fourth (C6L74–83) is a novel epitope identified in the current study. Use of these peptides in ICCS assays (data not shown) further confirmed the major dominance of these four epitopes and the preeminence of SPYAAGYDL (F2L26–34) (14).
The same epitopes are identified in mice vaccinated by dermal and i.p. routes
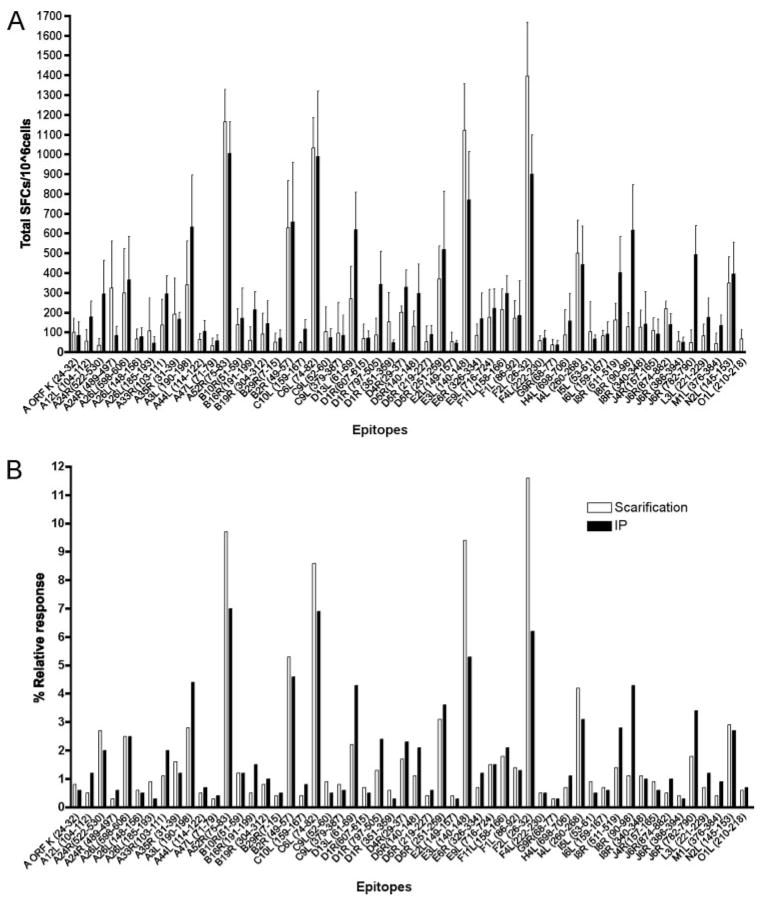
VACV epitope mapping in humans has largely been done after volunteers were vaccinated by the traditional scarification route. In contrast, most studies in mice have used i.p. injection. Because it has been suggested that the route of infection may affect immunodominance hierarchies (14, 15), we wanted to test whether it also affected the total breadth of epitopes recognized by CD8+ T cells. To this end, we tested the same set of 606 peptides used to analyze the responses to i.p. injection for recognition by T cells elicited following intradermal scarification and compared the responses observed following the two administration routes. As illustrated in Fig. 1 and supplementary Table Ib, the same 54 epitopes were identified following immunization by either route and no additional epitopes were identified. Thus, the route of administration did not qualitatively alter the breadth of the response to VACV infection.
Figure 1.
Immune response to VACV-WR; scarification vs i.p. immunization. Average responses of immunodominant epitopes, measured by IFN-γ expression in ELSIPOT assays, following VACV-WR immunization either by scarification (□) or i.p. infection (▪) are shown (A). Column bars of the percent relative responses to each identified VACV-WR-derived epitopes in either route of administration are also shown (B).
In quantitative terms, the overall response observed following scarification was somewhat lower than following i.p. immunization. When plotting the data as a percentage of the relative response (Fig. 1 and supplementary Table Ib), some epitopes appeared to be more vigorously recognized following one or the other immunization regimen, but these variations were never >2-fold. Strikingly, the major immunodominant epitopes, A52R75–83, F2L26–34, E3L140–148, and C6L74–82, have a much higher relative contribution to the overall immune response to VACV-WR after scarification compared with i.p. infection. When the percentage of CD8+ T cells involved in the response to each of these four immunodominant epitopes are added, we found that they represent ~25% of the total response after i.p. immunization, while accounting for ~40% of the total response after VACV-WR scarification in ELISPOT assays (Fig. 1 and supplementary Table Ib).
Further characterization of the responses to VACV observed in BALB/c mice
The 54 VACV epitopes were tested for their binding capacity to purified Kd, Db, and Ld molecules in vitro (Table I). As expected, significant binding to the relevant restriction element was demonstrated for all epitopes. Overall 90.7% (49 of 54) of the epitopes identified bound their predicted restricting MHC allele with high 500 nM). In 72.2% (39 of 54) of or intermediate affinity (IC50 ≤ the cases, they bound to their predicted MHC restriction molecule with a very high affinity (≤20 nM). Notably, the four major immunodominant epitopes bound their restricting MHC with even higher affinity (≤1.4 nM), reminiscent of what was observed in the H-2b mouse model of infection with the five major immunodominant epitopes (1), suggesting that binding affinity has some value to differentiate between major and minor immunodominant epitopes. Indeed, major immunodominant epitopes have highbinding affinity, and the data show that high affinity is necessary for immunodominance (4 peptides), but high affinity is not sufficient, since additional 11 high-affinity peptides are not immunodominant. Five of the 54 epitopes bound the predicted MHC restriction molecule with relatively weak affinities, with IC50 in the 500–7000 nM range. Taken together, these binding data support the MHC restriction assigned on the basis of our predictions.
Table I.
Summary of epitope characteristics
| Binding Affinity (IC50 nM)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ag Name (peptide position) | VACV-WR Name (peptide position) | Peptide Sequence | Restriction Size | Prediction Ranking | % of Top Scoring Peptides | ELISPOT Assay (SFC/106 cells) | Kd | Ld | Dd |
| NCa | VACVWR148 (598–606) | KYQQDRDTL | Kd, 9-mer | 21 | 0.0 | 365.0 | 6.4 | ND | 53,919 |
| NC | VACWR016 (52–60) | KCENHRSVI | Kd, 9-mer | 282 | 0.5 | 73.3 | 6,995 | ND | — |
| A3L (190–198) | VACVWR122 (190–198) | IYSPSNHHI | Kd, 9-mer | 159 | 0.3 | 632.5 | 1.9 | ND | 17,173 |
| A12L (104–112) | VACVWR131 (104–112) | SYNAMDGQI | Kd, 9-mer | 5 | 0.0 | 177.9 | 1.1 | ND | — |
| A24R (522–530) | VACVWR144 (522–530) | SYFETGFPI | Kd, 9-mer | 43 | 0.1 | 294.6 | 1.1 | ND | 8,768 |
| A24R (489–497) | VACVWR144 (489–497) | SVLSSITNI | Kd, 9-mer | 291 | 0.5 | 85.0 | 76 | ND | 41,894 |
| A33R (103–111) | VACVWR156 (103–111) | LYYQGSCYI | Kd, 9-mer | 38 | 0.1 | 295.0 | 2.8 | ND | — |
| A44L (114–122) | VACVWR170 (114–122) | IYTSSMEAI | Kd, 9-mer | 105 | 0.2 | 106.0 | 2.3 | ND | 35,870 |
| A47L (71–79) | VACVWR173 (71–79) | KIIQKSSSI | Kd, 9-mer | 243 | 0.4 | 56.7 | 191 | ND | — |
| A52R (75–83) | VACVWR178 (75–83) | KYGRLFNEI | Kd, 9-mer | 20 | 0.0 | 1,005.8 | 1.2 | ND | 18,914 |
| B2R (49–57) | VACVWR184 (49–57) | KYMWCYSQV | Kd, 9-mer | 186 | 0.3 | 658.8 | 22 | ND | — |
| B10R (51–59) | VACVWR192 (51–59) | IYKDKQWSI | Kd, 9-mer | 271 | 0.5 | 170.8 | 4,350 | ND | — |
| B16R (191–199) | VACVWR197 (191–199) | YYTCVLEYI | Kd, 9-mer | 283 | 0.5 | 215.2 | 1.3 | ND | 5,767 |
| B19R (304–312) | VACVWR200 (304–312) | SVLTSRGGI | Kd, 9-mer | 270 | 0.5 | 144.0 | 591 | ND | — |
| B22R/B28R/C22L (7–15) | VACVWR004/215 (7–15) | SYILFLSCI | Kd, 9-mer | 22 | 0.0 | 70.0 | 68 | ND | — |
| C6L (74–82) | VACVWR022 (74–82) | GFIRSLQTI | Kd, 9-mer | 258 | 0.4 | 989.4 | 1.4 | ND | 1,153 |
| C10L (159–167) | VACVWR010/209 (159–167) | SYNRSMTTI | Kd, 9-mer | 1 | 0.0 | 115.8 | 1.7 | ND | 34,745 |
| D1R (607–615) | VACVWR106 (607–615) | KYFYGEIAL | Kd, 9-mer | 26 | 0.0 | 70.4 | 36 | ND | — |
| D1R (797–505) | VACVWR106 (797–505) | KFINGASTM | Kd, 9-mer | 28 | 0.0 | 343.3 | 2.2 | ND | — |
| D1R (351–359) | VACVWR106 (351–359) | KYEGPFTTT | Kd, 9-mer | 292 | 0.5 | 47.9 | 2240 | ND | — |
| D4R (29–37) | VACVWR109 (29–37) | FYNEVASWL | Kd, 9-mer | 106 | 0.2 | 330.6 | 1.1 | ND | — |
| D5R (219–227) | VACVWR110 (219–227) | YYFSLQQRL | Kd, 9-mer | 190 | 0.3 | 89.2 | 2.3 | ND | 1,173 |
| D5R (251–259) | VACVWR110 (251–259) | SKIFINSII | Kd, 9-mer | 259 | 0.4 | 518.1 | 87 | ND | 11,247 |
| D5R (140–148) | VACVWR110 (140–148) | TYTTMDTLI | Kd, 9-mer | 91 | 0.2 | 296.8 | 9.4 | ND | 1,368 |
| D13L (61–69) | VACVWR118 (61–69) | QYITALNHL | Kd, 9-mer | 281 | 0.5 | 619.2 | 1.8 | ND | — |
| E2L (149–157) | VACVWR058 (149–157) | SYQFSENEI | Kd, 9-mer | 3 | 0.0 | 45.4 | 1.8 | ND | — |
| E6R (326–334) | VACVWR062 (326–334) | NFINGNTFI | Kd, 9-mer | 167 | 0.3 | 170.4 | 2.1 | ND | 24,184 |
| F1L (86–92) | VACVWR040 (86–92) | LPTKTRSYI | Kd, 9-mer | 268 | 0.5 | 186.0 | 890 | ND | 18,539 |
| F4L (222–230) | VACVWR043 (222–230) | SIITDAVSI | Kd, 9-mer | 290 | 0.5 | 70.0 | 273 | ND | — |
| F11L (158–166) | VACVWR050 (158–166) | LYIPGTSVI | Kd, 9-mer | 2 | 0.0 | 298.3 | 2.3 | ND | — |
| H4L (698–706) | VACVWR102 (698–706) | KYYYSNSYL | Kd, 9-mer | 10 | 0.0 | 157.9 | 1.4 | ND | — |
| I4L (260–268) | VACVWR073 (260–268) | SYISGTNGI | Kd, 9-mer | 16 | 0.0 | 443.8 | 1.4 | ND | — |
| I5L (53–61) | VCAVWR074 (53–61) | KYIGLFIYI | Kd, 9-mer | 11 | 0.0 | 67.1 | 157 | ND | — |
| I8R (511–519) | VACVWR077 (511–519) | SYMKSIQRI | Kd, 9-mer | 13 | 0.0 | 402.1 | 1.1 | ND | — |
| I8R (90–98) | VACVWR077 (90–98) | QYIYSEHTI | Kd, 9-mer | 31 | 0.1 | 617.5 | 2.5 | ND | 21,314 |
| J6R (386–394) | VACVWR098 (386–394) | RYNVIASSI | Kd, 9-mer | 32 | 0.1 | 50.4 | 1.1 | ND | — |
| J6R (874–882) | VACVWR098 (874–882) | KYFFTVSNI | Kd, 9-mer | 7 | 0.0 | 140.0 | 9.4 | ND | 13,876 |
| J6R (782–790) | VACVWR098 (782–790) | KYAANYTKI | Kd, 9-mer | 86 | 0.1 | 492.8 | 1.1 | ND | — |
| N2L (145–153) | VACVWR029 (145–153) | KYKPVYSYV | Kd, 9-mer | 168 | 0.3 | 394.6 | 1.3 | ND | — |
| NC | VACVWR148 (148–156) | IPAALIILL | Ld, 9-mer | 3 | 0.0 | 77.1 | 60 | 55 | 18,926 |
| AORF K (24–32) | A ORF K (24–32) | IPMFNKGHF | Ld, 9-mer | 2 | 0.0 | 85.0 | 45,474 | 78 | — |
| A26L (185–193) | VACVWR149 (185–193) | SPMYLWFNV | Ld, 9-mer | 32 | 0.1 | 45.2 | 17,159 | 11 | 13,672 |
| C9L (379–387) | VACVWR019 (379–387) | MPLFPTFSM | Ld, 9-mer | 8 | 0.0 | 84.0 | 35,852 | 18 | — |
| E9L (716–724) | VACVWR065 (716–724) | NPLSNPFYM | Ld, 9-mer | 16 | 0.0 | 222.3 | 11,740 | 1.6 | 13,144 |
| F2L (26–32) | VACVWR041 (26–32) | SPYAAGYDL | Ld, 9-mer | 14 | 0.0 | 900.6 | 27,686 | 0.38 | 32,218 |
| I6L (159–167) | VACVWR075 (159–167) | IPMSIISFF | Ld, 9-mer | 5 | 0.0 | 90.4 | 9,587 | 2.3 | 8,059 |
| I8R (340–348) | VACVWR077 (340–348) | LPNPAFIHI | Ld, 9-mer | 15 | 0.0 | 140.6 | 16,890 | 1.0 | 27,134 |
| J4R (157–165) | VACVWR096 (157–165) | IPLKIVRFF | Ld, 9-mer | 13 | 0.0 | 92.5 | 40,825 | 20 | 39,317 |
| L3L (221–229) | VACVWR090 (221–229) | LPQSCYLNF | Ld, 9-mer | 4 | 0.0 | 175.0 | 42,762 | 7.4 | 6,537 |
| M1L (376–384) | VACVWR030 (376–384) | IPFIAYFVL | Ld, 9-mer | 7 | 0.0 | 135.0 | 63,859 | 1.5 | 48,588 |
| O1L (210–218) | VACVWR068 (210–218) | YPMDNVINF | Ld, 9-mer | 6 | 0.0 | 100.4 | 4,255 | 10 | 59,978 |
| A35R (31–39) | VACVWR158 (31–39) | IGPYIIGNI | Dd, 9-mer | ND | ND | 167.7 | 61,401 | ND | 0.55 |
| E3L (140–148) | VACVWR059 (140–148) | VGPSNSPTF | Dd, 9-mer | ND | ND | 771.0 | — | ND | 0.93 |
| G9R (68–77) | VACVWR087 (68–77) | TGPGGLSALL | Dd, 10-mer | ND | ND | 37.3 | — | ND | 8.6 |
Dashes indicate IC50 > 70000 nM
NC, No classification; ND, no data available.
The 54 epitopes detected by the present study are derived from a total of 43 different Ags. The characteristics of each of the 43 recognized Ags are summarized in Table II. Ag names as defined by both the Poxvirus Bioinformatics Resource Center (http://www.poxvirus.org) (VACV-WR protein name) and by McCraith and coworkers (24) are listed. For C57BL/6 mice, we had found a statistically significant preference of recognition of early Ags by CD8+ T cells (1). In the present study, of the 39 Ags with known or predicted expression kinetics, 24 (61.5%) are early/early-late Ags and 15 (38.4%) are expressed as late stage Ags (Table II). Compared with the frequency of these expression categories in the entire vaccinia ORFs (97 early/early-late and 80 late), this also represents an increased recognition of early Ags, but does not by itself reach statistical significance (p = 0.13, one-tailed Fisher’s exact test).
Table II.
Structural characteristics of recognized Ags
| Copenhagen Name | VACV-WR Name | Protein length (aa) | Time of Expression | Function | Life Cycle |
|---|---|---|---|---|---|
| NC | 16 | 77 | Unknown | Gene fragment, ankyrin-like 77-kDa cowpox host range | Unknown |
| NC | 148 | 725 | Late | gene fragment, cowpox A-type inclusion protein | Structural protein |
| A ORF K | A ORF K | 70 | Unknown | Unknown | Unknown |
| A3L | 122 | 644 | Late | p4b precursor of core protein 4b | Structural protein |
| A12L | 131 | 192 | Late | Core protein | Structural protein |
| A26L | 149 | 500 | Late | Gene fragment, cowpox A-type inclusion protein | Structural protein |
| A24R | 144 | 1164 | Early/late | DNA-dependent RNA polymerase subunit rpo132 | Viral regulation |
| A33R | 156 | 185 | Early/late | EEV membrane phosphoglycoprotein | Structural protein |
| A35R | 158 | 176 | Early | Unknown | Unknown |
| A44L | 170 | 346 | Early | Hydroxysteroid dehydrogenase | Viral regulation |
| A47L | 173 | 252 | Early | Unknown | Unknown |
| A52R | 178 | 190 | Late | Toll/L-1R | Virulence factor |
| B2R | 184 | 219 | Early | Unknown | Unknown |
| B10R | 192 | 166 | Unknown | Unknown | Unknown |
| B16R | 197 | 326 | Late | IL-1β inhibitor | Virulence factor |
| B19R | 200 | 351 | Early | IFN-α/β-receptor-like secreted glycoprotein | Virulence factor |
| B22R/B28R/C22L | 4/215 | 122 | Unknown | TNF-γ receptor-like | Virulence factor |
| C9L | 19 | 635 | Early | Ankyrin-like protein | Unknown |
| C6L | 22 | 151 | Early | Unknown | Unknown |
| C10L | 10/209 | 331 | Early | Unknown | Virulence factor |
| D1R | 106 | 844 | Early | Large subunit of mRNA capping enzyme | Viral regulation |
| D4R | 109 | 218 | Early | Uracil-DNA glycosylase | Viral regulation |
| D5R | 110 | 785 | Early/late | NTPase interacts with A20R | Viral regulation |
| D13L | 118 | 551 | Late | Rifampicin target | Structural protein |
| E2L | 58 | 737 | Early | Unknown | Unknown |
| E3L | 59 | 190 | Early | dsRNA-binding protein | Virulence factor |
| E6R | 62 | 567 | Late | Unknown | Unknown |
| E9L | 65 | 1006 | Early | DNA polymerase | Viral regulation |
| F1L | 40 | 226 | Early | Inhibits apoptosis | Virulence factor |
| F2L | 41 | 147 | Early | dUTPase | Viral regulation |
| F4L | 43 | 319 | Early | Ribonucleotide reductase small subunit | Viral regulation |
| F11L | 50 | 348 | Early | Unknown | Unknown |
| G9R | 87 | 340 | Late | Myristylprotein | Viral regulation |
| H4L | 102 | 795 | Late | RAP94 | Viral regulation |
| I4L | 73 | 771 | Early | Ribonucleotide reductase large subunit | Viral regulation |
| I5L | 74 | 79 | Late | IMV protein VP13 | Structural, protein |
| I6L | 75 | 382 | Late | Unknown | Unknown |
| I8R | 77 | 676 | Early | RNA-helicase, DExH-NPH-II | Viral regulation |
| J4R | 96 | 185 | Late | DNA-dependent RNA polymerase subunit rpo22 | Viral regulation |
| J6R | 98 | 1286 | Early/late | DNA-dependent RNA polymerase subunit rpo147 | Viral regulation |
| L3L | 90 | 350 | Late | Unknown | Viral regulation |
| M1L | 30 | 472 | Early | Ankyrin-like protein | Unknown |
| N2L | 29 | 175 | Early | α-amanitin target | Unknown |
| O1L | 68 | 666 | Late | Unknown | Unknown |
N.C. = no classification
In terms of functional category, information for only 29 of the 43 VACV-WR-derived Ags is available. Of these 29 Ags, it was noted that 7 ORFs (24%) encode virulence factors, 6 (20.7%) encode structural proteins, and 16 (55%) encode genome regulation proteins (Table II). These percentages do not differ significantly from the incidence of these functional categories in the total proteome in which 28 ORFs (22.7%) with known function encode virulence factors, 38 (30.8%) encode structural proteins, and 57 (44.8%) encode genome regulation proteins (Fisher’s exact test with Freeman-Halton extension). These results are in accordance with our previously published data describing epitopes identified in humans and mice (1–3).
CD8+ T cell epitopes are not randomly distributed across the VACV proteome
Multiple H-2d-restricted epitopes were identified from certain Ags (C9L, J6R, I8R, VACVWR148, A24R, D1R, and D5R). Combining the data from the present study with previously identified epitopes results in a set of 197 epitopes restricted by five murine (H-2) alleles and seven human (HLA) supertypes. With these data available, two observations were made that suggested that epitopes are not distributed evenly across the VACV proteome. First, only 103 of the total of 206 unique genes present in VACV encoded CD8+ T cell epitopes and, second, six VACV genes were found to encode five or more discrete epitopes each. Most strikingly, the gene D1R was found to encode 10 epitopes recognized in the context of seven restriction elements. We then used statistical simulations to test whether these observations could have been due to chance. The finding that a subset of 103 of 206 unique Ags contains all 197 epitopes is highly statistically significant (p = 0.0004, see Materials and Methods). Furthermore, the maximum number of different epitopes likely to reside within a single Ag by chance alone is five and only a single Ag would be expected to be recognized this frequently. Finally, within the 103 recognized Ags, the recognition of an individual Ag in the context of as many as seven different MHC specificities is statistically significant (p = 0.011, see Materials and Methods). Altogether, these data and analyses suggest that recognized Ags share properties that distinguish them from nonrecognized Ags and that there is a subset of VACV proteins that is frequently immunogenic in the context of multiple MHC alleles. We refer to these Ags as immunoprevalent.
The identity and predictive value of immunoprevalent proteins from VACV
When examining the 72 proteins recognized in mice, 13 of these elicit CD8+ T cells in the context of both H-2b (C57BL/6) and H-2d (BALB/c). The genes for these are A3L, VACVWR148, A47L, B16R, B2R, D13L, D1R, E9L, F1L, J4R, J6R, M1L, and N2L. If these genuinely represent immunoprevalent Ags, we would predict that a high number of this subset of proteins is also recognized in the context of HLA supertypes. This was found to be true, with 8 of the 13 Ags identified as being immunoprevalent in mice also containing HLA-restricted epitopes. This is statistically significant because only 14.6% (32) of the 218 VACV ORFs are recognized by HLA class I molecules (p = 0.00007; one-tailed Fisher’s exact test). The identity of the genes encoding these proteins was VACVWR148, A3L, A47L, D1R, E9L, J6R, M1L, and N2L. It is noteworthy that five of these Ags (VACVWR148, A3L, A47L, D1R, and J6R) each encode two or more epitopes, restricted by at least two different MHC molecules in at least one model of infection (Table III and Refs. 1–4, 6, 9, 11, and 15). The D1R Ag particularly stands out, being recognized more than once in the context of two MHC restriction elements (H-2Kd and HLA-B7) and broadly in the context of four HLA supertypes (29) (A01, A03, B07, and B44) and three murine MHC alleles (H-2Kb, Db, and Kd). A total list of all VACV proteins containing at least three different epitopes restricted by at least three distinct MHC alleles, irrespective of species included D1R (10), D5R (8), J6R (6), A47L (5), A3L (5), B8R (5), E9L (3), VACVWR148 (3), M1L (3), and N2L (3) (Table III).
Table III.
VACV-derived immunoprevalent Ags
| Ag Name | VACV-WR Name | Peptide Sequence | Restriction | Ref. |
|---|---|---|---|---|
| — | 148 | NLWNGIVPT | HLA-A*0201 | Snyder et al. (11) |
| — | 148 | YLYTEYFLFL | HLA-A*0201 | Oseroff et al. (2) |
| — | 148 | SIYQYVRL | H-2Kb | Moutaftsi et al. (1) |
| — | 148 | KYQQDRDTL | H-2Kd | |
| — | 148 | IPAALIILL | H-2Ld | |
| A3L | 122 | DEVASTHDW | HLA-B*4403 | Jing et al. (6) |
| A3L | 122 | YEFRKVKSY | HLA-B*4403 | Jing et al. (6) |
| A3L | 122 | KSYNYMLL | H-2Kb | Moutaftsi et al. (1) |
| A3L | 122 | YSPSNHHIL | H-2Db | Moutaftsi et al. (1) |
| A3L | 122 | IYSPSNHHI | H-2Kd | |
| A47L | 173 | AFEFINSLLK | HLA-A*1101; H-2b | Pasquetto et al. (3) |
| A47L | 173 | LLYAHINAL | HLA-A*0201 | Terajima et al. (9) |
| A47L | 173 | AAFEFINSL | H-2Kb | Tscharke et al. (14); Moutaftsi et al. (1) |
| A47L | 173 | TMMINPFMI | H-2Db | Moutaftsi et al. (1) |
| A47L | 173 | AHINALEY | H-2Db | Mathew et al. (30) |
| A47L | 173 | KIIQKSSSI | H-2Kd | |
| B8R | 190 | DMCDIYLLY | HLA-A*0101; -A*2601; -A*2902 | Oseroff et al. (2) |
| B8R | 190 | FGDSKEPVPY | HLA-A*0101; -*2601; -A*2902 | Oseroff et al. (2) |
| B8R | 190 | FLSMLNLYKY | HLA-A*0101; -A*2902 | Oseroff et al. (2) |
| B8R | 190 | TEYDDHINL | HLA-B*4001 | Oseroff et al. (2) |
| B8R | 190 | TSYKFESV | H-2Kb | Tscharke et al. (15); Moutaftsi et al. (1) |
| D1R | 106 | FTIDFKLKY | HLA-A*2601; -A*2902 | Oseroff et al. (2) |
| D1R | 106 | KTKNFTIDFK | HLA-A*0301 | Oseroff et al. (2) |
| D1R | 106 | HPRHYATVM | HLA-B*0702 | Oseroff et al. (2) |
| D1R | 106 | RPSTRNFFEL | HLA-B*0702 | Pasquetto et al. (3, 30) |
| D1R | 106 | EERHIFLDY | HLA-B*4403 | Jing et al. (6) |
| D1R | 106 | LGYIIRYPV | H-2Kb | Moutaftsi et al. (1) |
| D1R | 106 | SMYCSKTFL | H-2Db | Moutaftsi et al. (1) |
| D1R | 106 | KYEGPFTTT | H-2Kd | |
| D1R | 106 | KYFYGEIAL | H-2 Kd | |
| D1R | 106 | KFINGASTM | H-2Kd | |
| D5R | 110 | EEIPDFAFY | HLA-B*4403 | Jing et al. (6) |
| D5R | 110 | LENGAIRIY | HLA-B*4403 | Jing et al. (6) |
| D5R | 110 | RYRFAFLYLL | HLA-A*2402 | Oseroff et al. (2) |
| D5R | 110 | VWINNSWKF | HLA-A*2402; - A*2301 | Oseroff et al. (2) |
| D5R | 110 | YLLVKWYRK | HLA-A*3303 | Oseroff et al. (2) |
| D5R | 110 | SKIFINSII | H-2Kd | |
| D5R | 110 | TYTTMDTLI | H-2Kd | |
| D5R | 110 | YYFSLQQRL | H-2Kd | |
| E9L | 65 | FLNISWFYI | HLA-A*0201 | Oseroff et al. (2) |
| E9L | 65 | RMNSNQVCI | H-2Db | Moutaftsi et al. (1) |
| E9L | 65 | NPLSNPFYM | H-2Ld | |
| J6R | 98 | NQVKFYFNK | HLA-A*0301 | Oseroff et al. (2) |
| J6R | 98 | MPAYIRNTL | HLA-B*0702 | Oseroff et al. (2) |
| J6R | 98 | INFEFVCL | H-2Kb | Moutaftsi et al. (1) |
| J6R | 98 | KYAANYTKI | H-2Kd | |
| J6R | 98 | KYFFTVSNI | H-2Kd | |
| J6R | 98 | RYNVIASSI | H-2Kd | |
| M1L | 30 | IIIPFIAYFV | HLA-A*0201 | Pasquetto et al. (3) |
| M1L | 30 | TSNVITDQTV | H-2Db | Moutaftsi et al. (1) |
We then examined the structure and function of these 10 immunoprevalent proteins. They ranged in size from 175 to 1286 residues in length. Two are structural proteins, four are regulatory proteins involved in transcription or DNA replication, one is an immunomodulator, and three are of unknown function. On the basis of published data, five of these Ags are expressed early during infection, two are expressed at early and late times, two are expressed late, and one remains unknown. These patterns mirror what is observed for all proteins containing CD8+ epitopes and there was no obvious property that distinguished the immunoprevalent proteins.
Immunoprevalent proteins are not the main source of major dominant epitopes
The identification of immunoprevalent proteins allowed us to ask whether these are also the main source of the major immunodominant epitopes. In this study, we define major immunodominant epitopes as peptides that trigger >5% of the total CD8+ T cell response to VACV. Of the 10 immunoprevalent Ags found to contain three or more discrete epitopes, only one (B8R) contains a major dominant epitope in C57BL/6 mice. Furthermore, all eight identified major dominant epitopes occur in different proteins and none of these proteins are immunogenic in both strains of mice. When defined as above, there is clearly no statistical association between epitope hierarchy and immunoprevalence on the whole protein level (p = 0.57; Fisher’s exact test). Thus, these observations confirm, as has been long known in the field that epitope hierarchy is largely determined by features of individual epitopes and not linked to the properties of the source protein.
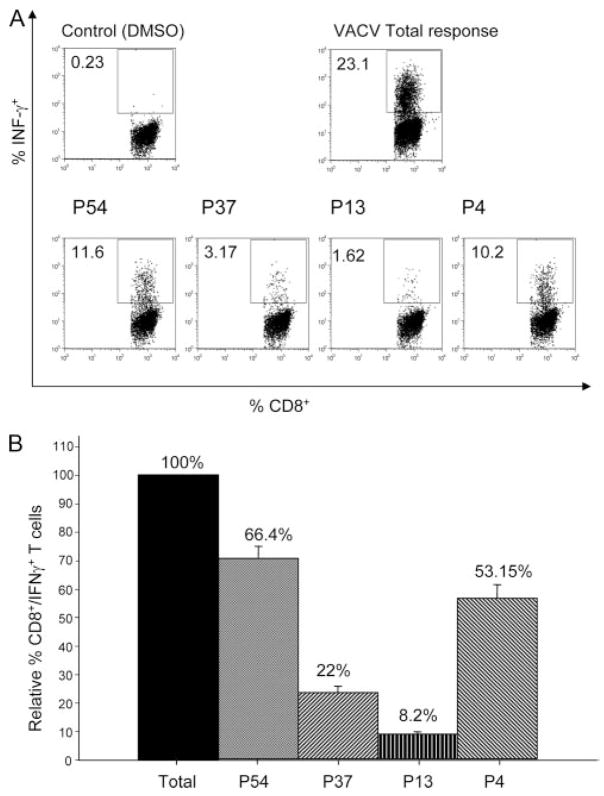
To explore this further experimentally, we determined the fraction of the total VACV-specific CD8+ T cell response that could be accounted for by major immunodominant epitopes compared with that attributable to epitopes from immunoprevalent proteins. Splenocytes from i.p.-immunized BALB/c mice were used in ICCS assays and stimulated with VACV-infected cells to determine the size of the total virus-specific response. The responses against four different peptide pools were also measured: 1) a pool of the 4 major immunodominant epitopes, 2) a pool of the 13 H-2d-restricted epitopes contained within immunoprevalent Ags that also contained epitopes restricted by H-2b and HLA alleles, 3) a pool of the 37 epitopes contained within the nonmajor and nonimmunoprevalent Ags, and 4) a pool of all 54 H-2d-restricted epitopes identified (Fig. 2). Fig. 2A depicts representative results, while Fig. 2B represents the overall average response of up to 16 individual measurements.
Figure 2.
CD8+ T cell responses induced by immunodominant vs immunoprevalent epitopes. Representative dot plots from IFN-γ-producing VACV-WR-specific CD8+ T cell following stimulation with P815 cells pulsed with peptide pools or infected with VACV-WR (MOI = 10) are shown (A). Column bars of the relative percentage compared with the total VACV-WR-specific CD8+ T cell response (represented by total activated CD8+ T cells after VACV infection) are also shown (B). The bars represent average values of up to 16 independent experiments; error bars, SEM. P4 represents a pool of the four H-2d major immunodominant epitopes. P13 represents a pool of the 13 H-2d-restricted epitopes contained in the 8 immunoprevalent Ags also restricted by H-2b and various HLA molecules. P37 represents the pool of epitopes identified not including P13 and P4. P54 represents the pool of all 54 identified epitopes. The total response elicited by VACV-WR represented by the total activated CD8+ T cells after infection is also shown.
The average size of the VACV-specific CD8+ T cell response was 20.13% of splenic CD8+ T cells and 66.4% of this could be accounted for by the pool of 54 known epitopes (P54). The four major immunodominant epitopes (P4) accounted for 53% of the total anti-VACV-WR response and 80% of the anti-P54 response; but in comparison the pool of peptides from immunoprevalent proteins (P13) represented only 8.2% of the total response to VACV-WR and 12% of the anti-P54 response. These data suggest that although immunodominance at the epitope and whole protein level are linked, immunodominance hierarchy at neither of these levels accurately reflects immunoprevalence, as we have defined it, at the protein level.
Discussion
In the present study, we have probed the epitope repertoire recognized in the H-2d haplotype following VACV-WR immunization by two different immunization routes. The epitope repertoire identified is composed of at least 54 epitopes, including 51 novel determinants, restricted by Kd, Ld, and Dd molecules, and 3 previously identified epitopes. These epitopes are derived from 43 different Ags representing various functional and temporal categories. This diversity echoes that observed in previous work performed in humans, HLA-transgenic mice, and C57BL/6 mice (1–3). Immunogenic proteins were heavily biased toward early expression as also found previously. With regard to functional category, viral regulation genes appear to be the most preferred sources of epitopes (n = 15). Thus, in this respect, the present study confirms and extends to a different mouse system the findings that CD8+ responses against a complex virus such as VACV-WR are very broad and diverse.
The goal of the present study was to identify the majority, but not necessarily the totality, of the H-2d responses observed using two different routes of administration. A retrospective analysis of the epitope mapping study in the H-2b model system indicated that two-thirds of the epitopes were included in the top 0.5% of peptides ranked by predicted MHC-binding affinity (1). Accordingly, in the present study, we chose to select the top 0.5% of scoring peptides for use in T cell assays. Consistently with these estimates, the epitopes identified account for approximately two-thirds of the total response. The missing one-third are maybe covered by a handful of immunodominant epitopes or by a very large number of minor epitopes. Either possibility might affect the conclusions about the relative impact of immunoprevalence vs immunodominance; therefore, our results should be interpreted with this caveat in mind.
Verification that T cells derived from animals immunized with epitopes identified by screening infected animals with predicted peptide pools recognize infected target cells has been addressed in two different studies from our group (30, 31). Overall, 14 (93.3%) of 15 of the lines raised by peptide immunizations recognized infected target cells. Indeed, screening samples from infected animals or individuals is a relatively common strategy used for the purpose of epitope identification. In the present study, whether all epitopes identified are actually produced in infected cells or whether some of the minor epitopes might be actually cross-reactive with major ones has also not been specifically addressed.
If epitope mapping data from mice and humans are to be combined for analyses, it is helpful if some of the variables that may lead to systematically different results can be eliminated. One of these variables is route of administration, with humans being vaccinated by dermal scarification and mice typically by i.p. injection. When we compared the range of CD8+ T cell epitopes that can be mapped in mice immunized by i.p. injection with those following dermal scarification, we found complete overlap. This suggests that route is not an important variable in determining the epitopes that are identified, at least using our predictive approach. However, the data here confirm previous observations that dermal immunization is associated with more pronounced immunodominance (15). Strikingly, the major immunodominant epitopes A52R75–83, F2L26–34, E3L140–148, and C6L74–82 have a significantly higher relative contribution to the overall immune response to VACV-WR after scarification compared with i.p. infection. Although they represent ~25% of the total response after i.p. immunization, these major immunodominant epitopes account for 40% of the total response after VACV-WR scarification. Although the biological relevance of a change in hierarchy is not known, it should be explored, even if the total number of epitopes recognized remains the same, because it has implications for the development of diagnostic tools and possibly vaccines.
We identified a total of 44 VACV protein Ags recognized by H-2d-restricted responses, 13 of which were also recognized in C57BL/6 mice. Of these 13 proteins, 8 (62%) have been shown to be immunogenic in humans vaccinated with the Dryvax vaccine or in HLA-transgenic mice immunized with VACV-WR. In contrast, although the number of epitopes mapped now approaches the total number of predicted genes in VACV, only around one-half of VACV genes have been found to encode CD8+ T cell epitopes. The absence of epitopes in such a large number of potential Ags and the presence of up to 10 epitopes in a single Ag, restricted by no less than seven MHC alleles (or supertypes), are significant deviations from what would be expected by chance alone (see Results, CD8+ T cell epitopes are not randomly distributed across the VACV proteome). This suggests that regardless of the host some proteins are more likely to contain CD8+ T cell epitopes than others and we refer to the frequently recognized proteins as being immunoprevalent.
Interestingly, with the exception of B8R, the immunoprevalent Ags do not contain major dominant epitopes and although they are frequently recognized across diverse MHC types, they are not as a group immunodominant. The reason for the dissociation between immunodominance and immunoprevalence is unclear. Indeed, these sets of proteins cannot be distinguished on the basis of biological function or time of expression. In all, protein length is likely an important factor, since E9L and J6R of these proteins are of relatively large size (>1000 residues). However, it should also be noted that relatively small proteins such as A47L, B8R, and N2L appear to be immunoprevalent.
Immunodominance is likely to be a function of epitopes rather than whole proteins and, if so, will be determined largely by factors such as affinity for MHC, efficiency of processing, and the diversity and/or frequency of cognate TCRs in the naive repertoire. Conversely, immunoprevalence appears to be MHC and epitope independent and is likely to reflect only properties of the whole protein, such as abundance, cellular localization, and kinetics of production, defined as the early vs late expression pattern and decay. Clearly, some of these factors, for example, high expression levels, would be expected to favor both immunodominance and immunoprevalence, but our data suggest that the sets of rules that underlie these phenomena are different.
This conclusion is novel and has substantial practical significance for the selection of Ags intended for diagnostic purposes or subunit vaccines. Selection of Ags on the basis of immunodominance in a given MHC background might lead to uneven diagnostic or vaccine performance, with a relatively large number of Ags being needed for good coverage of an outbred population. In contrast, an alternative approach based on selection of the most immunoprevalent Ags might elicit or detect CD8+ T cells in a much broader range of individuals. Finally, if immunoprevalent proteins can be predicted, this in turn will aid epitope discovery for large pathogens, where it might not always be cost effective to screen genome-wide libraries of synthetic peptides.
In conclusion, we present here not only the identity of >50 new CD8+ T cell epitopes for VACV, but evidence that some VACV proteins are immunoprevalent, being recognized frequently irrespective of MHC restriction. In addition, we show that immunoprevalence is a property of whole proteins and is distinct from immunodominance. These findings are important in their own right, but identifying the rules that underlie immunoprevalence will be of even more value to the development of vaccines and diagnostic tools for large pathogens.
Supplementary Material
Acknowledgments
We thank Kelly Riddle-Hilde for administrative assistance and Lori Giancola, Matt Maybeno, and Jean Glenn for outstanding technical help.
Footnotes
This work was supported by the National Institutes of Health, through Contract HHSN266200400024C, and R01 Grants AI-56268 (to A.S.) and AI067401 (to D.C.T.). D.C.T. is supported by Australian National Health and Medical Research Council Career Development Award 418108.
Abbreviations used in this paper: VACV, vaccinia virus; ORF, open reading frame; MOI, multiplicity of infection; WR, Western Reserve; SFC, spot-forming cell; ICCS, intracellular cytokine staining.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine TCD8+ -cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 2.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci USA. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, Oseroff C, Tscharke DC, Irvine K, Bennink JR, et al. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J Immunol. 2005;175:5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- 4.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, Brander C, Peters B, Grey H, Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 5.Smith CL, Dunbar PR, Mirza F, Palmowski MJ, Shepherd D, Gilbert SC, Coulie P, Schneider J, Hoffman E, Hawkins R, et al. Recombinant modified vaccinia Ankara primes functionally activated CTL specific for a melanoma tumor antigen epitope in melanoma patients with a high risk of disease recurrence. Int J Cancer. 2005;113:259–266. doi: 10.1002/ijc.20569. [DOI] [PubMed] [Google Scholar]
- 6.Jing L, Chong TM, McClurkan CL, Huang J, Story BT, Koelle DM. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J Immunol. 2005;175:7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang J, Murtadha M, Schnell M, Eisenlohr LC, Hooper J, Flomenberg P. Human T-cell responses to vaccinia virus envelope proteins. J Virol. 2006;80:10010–10020. doi: 10.1128/JVI.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terajima M, Cruz J, Raines G, Kilpatrick ED, Kennedy JS, Rothman AL, Ennis FA. Quantitation of CD8+ T cell responses to newly identified HLA-A*0201-restricted T cell epitopes conserved among vaccinia and variola (smallpox) viruses. J Exp Med. 2003;197:927–932. doi: 10.1084/jem.20022222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terajima M, Cruz J, Leporati AM, Demkowicz WE, Jr, Kennedy JS, Ennis FA. Identification of vaccinia CD8+ T-cell epitopes conserved among vaccinia and variola viruses restricted by common MHC class I molecules, HLA-A2, or HLA-B7. Hum Immunol. 2006;67:512–520. doi: 10.1016/j.humimm.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Drexler I, Staib C, Kastenmuller W, Stevanovic S, Schmidt B, Lemonnier FA, Rammensee HG, Busch DH, Bernhard H, Erfle V, Sutter G. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc Natl Acad Sci USA. 2003;100:217–222. doi: 10.1073/pnas.262668999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder JT, I, Belyakov M, Dzutsev A, Lemonnier F, Berzofsky JA. Protection against lethal vaccinia virus challenge in HLA-A2 transgenic mice by immunization with a single CD8+ T-cell peptide epitope of vaccinia and variola viruses. J Virol. 2004;78:7052–7060. doi: 10.1128/JVI.78.13.7052-7060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson KL, I, Ovsyannikova G, Madden BJ, Poland GA, Muddiman DC. Accurate mass precursor ion data and tandem mass spectrometry identify a class I human leukocyte antigen A*0201-presented peptide originating from vaccinia virus. J Am Soc Mass Spectrom. 2005;16:1812–1817. doi: 10.1016/j.jasms.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Dong Y, Denny TN. HLA-A2-restricted human CD8+ cytotoxic T lymphocyte responses to a novel epitope in vaccinia virus that is conserved among orthopox viruses. J Infect Dis. 2006;194:168–175. doi: 10.1086/505224. [DOI] [PubMed] [Google Scholar]
- 14.Tscharke DC, Woo WP, Sakala IG, Sidney J, Sette A, Moss DJ, Bennink JR, Karupiah G, Yewdell JW. Poxvirus CD8+ T-cell determinants and cross-reactivity in BALB/c mice. J Virol. 2006;80:6318–6323. doi: 10.1128/JVI.00427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SM, Williams S, Sidney J, Sette A, Bennink JR, Yewdell JW. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen WM, Pouvelle-Moratille S, Wang XF, Farci S, Munier G, Charron D, Menez A, Busson M, Maillere B. Scanning the HIV genome for CD4+ T cell epitopes restricted to HLA-DP4, the most prevalent HLA class II molecule. J Immunol. 2006;176:5401–5408. doi: 10.4049/jimmunol.176.9.5401. [DOI] [PubMed] [Google Scholar]
- 17.Wang XF, Cohen WM, Castelli FA, Almunia C, Lethe B, Pouvelle-Moratille S, Munier G, Charron D, Menez A, Zarour HM, et al. Selective identification of HLA-DP4 binding T cell epitopes encoded by the MAGE-A gene family. Cancer Immunol Immunother. 2007;56:807–818. doi: 10.1007/s00262-006-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castelli FA, Leleu M, Pouvelle-Moratille S, Farci S, Zarour HM, Andrieu M, Auriault C, Menez A, Georges B, Maillere B. Differential capacity of T cell priming in naive donors of promiscuous CD4+ T cell epitopes of HCV NS3 and core proteins. Eur J Immunol. 2007;37:1513–1523. doi: 10.1002/eji.200636783. [DOI] [PubMed] [Google Scholar]
- 19.Pudney VA, Leese AM, Rickinson AB, Hislop AD. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J Exp Med. 2005;201:349–360. doi: 10.1084/jem.20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riddell SR, Rabin M, Geballe AP, Britt WJ, Greenberg PD. Class I MHC-restricted cytotoxic T lymphocyte recognition of cells infected with human cytomegalovirus does not require endogenous viral gene expression. J Immunol. 1991;146:2795–2804. [PubMed] [Google Scholar]
- 21.Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, Sissons JG. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koelle DM, Liu Z, McClurkan CL, Cevallos RC, Vieira J, Hosken NA, Meseda CA, Snow DC, Wald A, Corey L. Immunodominance among herpes simplex virus-specific CD8 T cells expressing a tissue-specific homing receptor. Proc Natl Acad Sci USA. 2003;100:12899–12904. doi: 10.1073/pnas.2131705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tigges MA, Koelle D, Hartog K, Sekulovich RE, Corey L, Burke RL. Human CD8+ herpes simplex virus-specific cytotoxic T-lymphocyte clones recognize diverse virion protein antigens. J Virol. 1992;66:1622–1634. doi: 10.1128/jvi.66.3.1622-1634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udaka K, Wiesmuller KH, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- 26.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tangri S, Ishioka GY, Huang X, Sidney J, Southwood S, Fikes J, Sette A. Structural features of peptide analogs of human histocompatibility leukocyte antigen class I epitopes that are more potent and immunogenic than wild-type peptide. J Exp Med. 2001;194:833–846. doi: 10.1084/jem.194.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitiello A, Yuan L, Chesnut RW, Sidney J, Southwood S, Farness P, Jackson MR, Peterson PA, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 29.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 30.Mathew A, Terajima M, West K, Green S, Rothman AL, Ennis FA, Kennedy JS. Identification of murine poxvirus-specific CD8+ CTL epitopes with distinct functional profiles. J Immunol. 2005;174:2212–2219. doi: 10.4049/jimmunol.174.4.2212. [DOI] [PubMed] [Google Scholar]
- 31.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, Brander C, Peters B, Grey H, Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.