Abstract
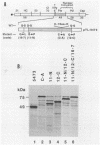
The genome of tobacco etch virus contains a single open reading frame with the potential to encode a 346-kilodalton (kDa) polyprotein. The large polyprotein is cleaved at several positions by a tobacco etch virus genome-encoded, 49-kDa proteinase. The locations of the 49-kDa proteinase-mediated cleavage sites flanking the 71-kDa cytoplasmic pinwheel inclusion protein, 6-kDa protein, 49-kDa proteinase, and 58-kDa putative polymerase have been determined by using cell-free expression, proteolytic processing, and site-directed mutagenesis systems. Each of these sites is characterized by the conserved sequence motif Glu-Xaa-Xaa-Tyr-Xaa-Gln-Ser or Gly (in which cleavage occurs after the Gln residue). The amino acid residue (Gln) predicted to occupy the -1 position relative to the scissile bond has been substituted, by mutagenesis of cloned cDNA, at each of four cleavage sites. The altered sites were not cleaved by the 49-kDa proteinase. A series of synthetic polyproteins that contained the 49-kDa proteinase linked to adjoining proteins via defective cleavage sites were expressed, and their proteolytic activities were analyzed. As part of a polyprotein, the proteinase was found to exhibit cis (intramolecular) and trans (intermolecular) activity.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison R. F., Sorenson J. C., Kelly M. E., Armstrong F. B., Dougherty W. G. Sequence determination of the capsid protein gene and flanking regions of tobacco etch virus: Evidence for synthesis and processing of a polyprotein in potyvirus genome expression. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3969–3972. doi: 10.1073/pnas.82.12.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J. C., Dougherty W. G. Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. J Virol. 1987 Aug;61(8):2540–2548. doi: 10.1128/jvi.61.8.2540-2548.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorssers L., van der Krol S., van der Meer J., van Kammen A., Zabel P. Purification of cowpea mosaic virus RNA replication complex: Identification of a virus-encoded 110,000-dalton polypeptide responsible for RNA chain elongation. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1951–1955. doi: 10.1073/pnas.81.7.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari V., Siegel A., Rozek C., Timberlake W. E. The RNA of tobacco etch virus contains poly(A). Virology. 1979 Jan 30;92(2):568–571. doi: 10.1016/0042-6822(79)90159-4. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsen G. R., Anderson C. W., Dorner A. J., Semler B. L., Wimmer E. Cleavage sites within the poliovirus capsid protein precursors. J Virol. 1982 Jan;41(1):340–344. doi: 10.1128/jvi.41.1.340-344.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler B. L., Hanecak R., Anderson C. W., Wimmer E. Cleavage sites in the polypeptide precursors of poliovirus protein P2-X. Virology. 1981 Oct 30;114(2):589–594. doi: 10.1016/0042-6822(81)90242-7. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Wellink J., Rezelman G., Goldbach R., Beyreuther K. Determination of the proteolytic processing sites in the polyprotein encoded by the bottom-component RNA of cowpea mosaic virus. J Virol. 1986 Jul;59(1):50–58. doi: 10.1128/jvi.59.1.50-58.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Semler B. L. In vitro molecular genetics as a tool for determining the differential cleavage specificities of the poliovirus 3C proteinase. Nucleic Acids Res. 1987 Mar 11;15(5):2069–2088. doi: 10.1093/nar/15.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel P., Moerman M., Lomonossoff G., Shanks M., Beyreuther K. Cowpea mosaic virus VPg: sequencing of radiochemically modified protein allows mapping of the gene on B RNA. EMBO J. 1984 Jul;3(7):1629–1634. doi: 10.1002/j.1460-2075.1984.tb02021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]