Abstract
Heterochromatin Protein 2 is a nonhistone chromosomal protein from Drosophila melanogaster that binds to HP1 and has been implicated in heterochromatin-induced gene silencing. Heretofore, HP1 has been the only known binding partner of HP2, a large protein devoid of sequence motifs other than a pair of AT-hooks. In an effort to identify proteins that interact with HP2 and assign functions to its various domains, nuclear proteins were fractionated under non-denaturing conditions. On separation of nuclear proteins, Nap-1, or Nucleosome assembly protein 1, has an overlapping elution profile with HP2 (assayed by Western blot) and has been identified by mass spectrometry in fractions with HP2. Upon probing fractions in which HP2 and Nap-1 are both present, we find that NURF, an ISWI-dependent chromatin remodeling complex, is also present. Results from coimmunoprecipitation experiments suggest that HP2 interacts with Nap-1 as well as with NURF; NURF appears to interact directly with both HP2 and Nap-1. Three distinct domains within HP2 mediate the interaction with NURF, allowing us to assign NURF binding domains in addition to the AT-hooks and HP1 binding domains already mapped in HP2. Mutations in Nap-1 are shown to suppress position effect variegation, suggesting that Nap-1 functions to help assemble chromatin into a closed form, as does HP2. Based on these interactions, we speculate that HP2 may cooperate with these factors in the remodeling of chromatin for silencing.
Heterochromatin Protein 2 (HP2) was originally identified based on its ability to bind to Heterochromatin Protein 1 (HP11), one of the best-characterized nonhistone chromosomal proteins, in a yeast two-hybrid assay (1). HP2 colocalizes with HP1 at the pericentric heterochromatin of Drosophila polytene chromosomes, coimmunoprecipitates with HP1 from a Drosophila embryo extract, and is recruited to ectopic sites upon mislocalization of HP1. Analysis of the structure of the gene coding for HP2, Su(var)2-HP2, reveals two isoforms, a consequence of alternative splicing. Both proteins are large; the larger isoform of HP2 (HP2-L) is 356 kDa and the smaller isoform of HP2 (HP2-S) is 175 kDa. Both proteins are devoid of recognizable sequence motifs, except for two AT-hooks that are present only in the larger isoform. [AT-hooks are small, 9-aa DNA binding motifs which preferentially interact with AT-rich DNA through the minor groove, typically found in high mobility group (HMG) proteins (2)]. Recently the domain in HP2 that binds to HP1 has been identified and found to be conserved in Drosophila (3).
Mutations in Su(var)2-HP2 act as dominant suppressors of position effect variegation (PEV) monitored by wm4, a chromosome rearrangement (1). PEV occurs when a gene that is normally found in a euchromatic region is transposed to a position within or in close proximity to heterochromatin; a variegating phenotype results as the gene is silenced in some of the cells in which it is normally expressed (4). When mutations are made within Su(var)2-HP2, the silent state is suppressed, resulting in increased expression of white (1). This implicates HP2 in initiation or spreading of the heterochromatic state, in parallel with HP1 (5).
We have used biochemical approaches to identify protein-binding partners of HP2 that may contribute to the regular array of nucleosomes that are commonly found in heterochromatin (6, 7). In addition to possible interactions with enzymes that generate appropriate histone modifications [such as SU(VAR)3-9 or another critical HMT], one might anticipate identifying proteins that can bind to nucleosomes and remodel them into a regular array.
The assembly of nucleosomes, the fundamental subunits of chromatin, is essential for proper genome function. The process of chromatin assembly begins with a tetramer of histones H3 and H4 being deposited onto the DNA by histone chaperones, followed by deposition of two heterodimers of H2A and H2B to yield a histone octamer around which 146 base pairs of DNA is wrapped. During chromatin assembly in S phase, there is random deposition of the preexisting as well as newly made histones onto the two daughter strands of DNA. In vivo, chromatin assembly appears to occur immediately following DNA replication (8, 9); chromatin assembly can also take place in the absence of replication, presumably in response to nucleosome displacement or disassembly, thought to occur during transcription (10).
When chromatin is reconstituted in vitro the nucleosomes are randomly distributed along the DNA molecules. However, in native chromatin, nucleosomes are distributed at approximately regular intervals. It appears that histone chaperones alone are insufficient to emulate the in vivo assembly of chromatin. Biochemical analysis has shown that multipeptide chromatin remodeling complexes can use the energy from ATP to alter nucleosome positioning and structure (for review see (11)). Three distinct families of complexes that remodel chromatin using the energy from ATP have been identified: SWI2/SNF2-like, ISWI-like, and Mi-2-like (for review see (12)). Some or all might play a role in heterochromatin formation, generating the regular nucleosome array observed.
Biochemical experiments have identified several negatively charged proteins and protein complexes that bind to histones and deposit them onto the DNA in an ATP-dependent manner. Chromatin Assembly Factor-1 (CAF-1; (13)), Antisilencing Function Protein 1 (ASF1; (14)), and Histone Regulatory A (HIRA; (15)) show a preference for the H3-H4 tetramer, whereas other histone chaperones, such as Nap-1, deposit histones H2A and H2B onto the DNA (16).
A few histone chaperones, such as CAF-1 and ASF1, have been directly implicated in assembly of heterochromatin, as have some proteins that are components of multiprotein chromatin remodeling complexes, such as Acf1. When CAF-1 is deleted in budding yeast, silencing at telomeres, mating type loci, and ribosomal DNA is impaired (17-21), suggesting a role for CAF-1 in heterochromatin assembly. This interpretation is supported by the finding that CAF-1 can be found associated with Heterochromatin Protein 1 (HP1α) in mammalian cells (22). In Drosophila, mutations in ASF1 or in acf1 result in suppression of PEV, indicating a role in heterochromatin-induced gene silencing (23). Acf1 is a subunit of the ACF (ATP-utilizing chromatin assembly and remodeling factor) complex, which also contains ISWI. Thus, components of the chromatin assembly machinery appear to contribute to the establishment of epigenetic chromatin states.
In this report, we have identified two novel binding partners of HP2, Nap-1 and the NURF complex. These factors have an overlapping elution profile with HP2 on fractionation of a nuclear extract using multiple conventional chromatography columns. Nap-1 is an H2A-H2B histone chaperone, as mentioned previously, and NURF (Nucleosome Remodeling Factor) is a remodeling complex in Drosophila that is composed of NURF301, ISWI, p55, and p38 (24). We find that HP2 antibodies can immunoprecipitate Nap-1 as well as all four members of the NURF complex, thus confirming an interaction with both. Nap-1 antibodies are also able to immunoprecipitate the NURF complex. There appear to be direct interactions between HP2 and NURF and between Nap-1 and NURF, but not between HP2 and Nap-1. Three distinct sites within HP2 mediate the interaction with NURF, allowing us to assign functional domains to HP2. Mutations in Nap-1 are found to result in suppression of PEV, further implicating Nap-1 in heterochromatin-induced gene silencing, a role already assigned to HP2. This data identifies novel factors that bind to HP2 and gives us hints of a possible role for HP2 in chromatin remodeling.
Experimental Procedures
Chromatography
100 g of frozen 6-18 hr Oregon R embryos are dechorionated and homogenized in 100 mls of buffer containing 50 mM HEPES pH 7.5, 60 mM KCl, 15 mM NaCl, 0.25 M sucrose, 1 mM EDTA, and 0.1 mM EGTA. Protease inhibitors are added to a final concentration of 1 mM PMSF, 0.01 mg/ml phenanthroline, aprotinin, leupeptin, and pepstatin, and 20 mM benzamidine HCl. The homogenate is filtered through Miracloth and nuclei recovered by centrifugation in Corex tubes at 7000 rpm for 15 min. in a Sorvall SS-34 rotor. The supernatant is then poured off, and the pellet is resuspended in the above buffer; the spin is repeated, and the supernatant is decanted. This wash procedure is repeated twice more using the above buffer without sucrose. The recovered nuclear pellets are then resuspended in extraction buffer containing 25 mM HEPES pH 8, 400 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.2% NP40 (HEPES-400, with the number indicating the concentration of NaCl). The nuclei are incubated in this buffer with the above protease inhibitors and a final concentration of 1mM DTT for 30 minutes on ice. The nuclei are then pelleted at 7000 rpm for 30 min. as above, and the supernatant recovered is used for column fractionation. Approximately 90 mg of total nuclear protein is diluted to 150 mM NaCl with HEPES-0. This sample is then loaded onto a 30 ml DEAE Sepharose Fast Flow column (GE Healthcare) that is equilibrated in HEPES-150 pH 8. The proteins are then step eluted in HEPES-150, HEPES-250, and HEPES-1M. The 250 mM NaCl eluate is collected in 10 ml fractions; fractions 3-9 are combined and spun down at 10,000 g for 15 minutes. (The fractions chosen for downstream applications are those containing HP2 as shown by Western blots. Western blots were originally performed after each fractionation step until a reproducible procedure was established.) The supernatant is then diluted with HEPES-0 to a final concentration of 125 mM NaCl. The following columns are used on an AKTA FPLC (GE Healthcare). The sample is loaded onto a 1 ml Mono Q column (GE Healthcare). A gradient is run from 125 mM NaCl to 600 mM NaCl at a flow rate of 1 ml/min. 1 ml fractions are collected and Mono Q fractions 8-14 are then combined and spun down in Centricon YM30 spin columns (Millipore) until the volume is less than or equal to 500 μl. The sample is then eluted from the column and spun down at 10,000 g for 15 min. The sample is then injected onto a Superose 6 column (GE Healthcare) equilibrated in HEPES-50 pH 8. 500 μl fractions are collected at a flow rate of 0.4 ml/min. Fractions 18-23 are then combined and centrifuged at 10,000 g for 15 min. This sample is then separated on a 1 ml Mono S column (GE Healthcare) equilibrated in HEPES-50. 1 ml fractions are collected at a flow rate of 1 ml/min. The Mono S fractions are then precipitated with trichloroacetic acid and frozen at –80° C until further examination. Those bands that peaked with HP2 on SDS-PAGE gels upon staining with SYPRO Ruby were excised and analyzed by mass spectrometry.
Western blotting
Proteins are loaded onto SDS-PAGE gels, size separated, and transferred to Immobilon-P (Millipore) membranes for 1 hr using 100 V at 4° C. Incubations with primary antibodies are done for 1-2 hrs at room temperature or 4° C overnight. Antibodies specific for HP2 were produced in chicken using a KLH-conjugated peptide, the first 14 amino acids of HP2, MEDIEYLDEYKDZ (Aves Lab), and used at 1:5000. Nap-1 antibodies obtained from Jim Kadonaga (UC San Diego) are used at 1:10,000; NURF301 antibodies (25), used to follow NURF301 after column chromatography, are used at 1:2500. FLAG M2 antibodies (Sigma Aldrich) are used at 1:10,000 to detect FLAG-tagged NURF301. ISWI antibodies obtained from Jim Kadonaga (UC San Diego) are used at 1:2500 and p55 antibodies obtained from Jessica Tyler (Univ. of Colorado Health Sciences Center) are used at 1:10,000. p38 antibodies are used at 1:2000 (26), and HP1 WA191 antibodies (produced in rabbits against a chromo domain peptide CYAVEKIIDRRVRKGKVEYYLKWKG) are used at 1:2500. Incubations with secondary antibodies are done at room temperature for 30-45 minutes at an approximately 1:25,000-100,000 dilution. Western detection is performed with ECL+ (GE Healthcare) or Immobilon Western (Millipore) according to the manufacturer's instructions.
Coimmunoprecipitations
Coimmunoprecipitation experiments using HP2 rabbit antibodies [prepared using the C-terminal domain of HP2 isolated in the original yeast two-hybrid screen (1) as immunogen] confirmed an interaction between HP2 and the NURF complex, as well as an interaction of HP2 with Nap-1. The source of HP2 in the immunoprecipitation experiments with the purified NURF complex is a transcription/translation reaction (Promega) product produced with the cDNA of the smaller isoform of HP2, described previously (3). The source of NURF is a FLAG affinity-purified complex described previously (25). 20-40 μl of a nonradioactive HP2-S transcription/translation reaction is added to 1.5-3 μl (62.5-125 ng) of the NURF complex, HEPES-150 (described above), and protease inhibitors. The mixture is incubated for 1 hr at 4° C while rocking. 10 μl of Bleed 7 rabbit HP2 antibody is then added, and the reaction is allowed to continue for 1 hr. The reaction mixture is then added to 2 mg of Protein A Sepharose CL-4B (GE Healthcare) that has been washed in binding buffer and incubated with a 2% BSA solution. The reaction is allowed to continue for 1 hr at 4° C, or overnight, while rocking. As a control for specificity, preimmune serum from the rabbits in which the HP2 antibody was made is used for comparison. The beads are collected, washed, resuspended in load dye, boiled, and the eluted proteins loaded onto SDS-PAGE gels. Western blotting is done as previously described. The antibody used is FLAG M2. The coimmunoprecipitation observed might be spurious if both proteins were binding independently to the same DNA molecule. To show that DNA is not the sole mediator of the interaction between HP2 and NURF, 100-fold excess of plasmid DNA was added to the immunoprecipitation reactions and the binding efficiency monitored. Similar experiments were done to confirm direct interactions between Nap-1 and NURF.
Coimmunoprecipitation of Nap-1 and HP2 as well as of NURF301 and HP2 was also observed using an embryo nuclear extract. In the case of Nap-1 and HP2, the DNA in the extract was digested with micrococcal nuclease to completion. HP2 preimmune serum and Nap-1 preimmune serum are used as controls as appropriate. Approximately 200-400 μg of salt-extracted nuclear protein, prepared as in the chromatography experiments, is added to HP2-specific antibodies, Nap-1-specific antibodies, or preimmune serum, along with protease inhibitors. This binding reaction is allowed to continue for approximately four hours. 2 mg of Protein A Sepharose CL-4B (GE Healthcare), washed two times with binding buffer and incubated with a 2% BSA solution, is then added, and the interaction is allowed to continue overnight at 4° C. The beads are treated as described above. Nap-1 binding is detected with Nap-1 antibodies, described previously. The secondary antibody used is a mouse anti-rabbit IgG directed against the light chain (Jackson Immunoresearch Laboratories). NURF301 is detected with rabbit NURF301 antibodies.
Coimmunoprecipitation experiments with HP2 and NURF301 and Nap-1 and NURF301 were also done using an Sf9 extract from cells in which the NURF301 subunit is overproduced. The experiment is done as described above for embryo extracts, except that NURF301 is detected with FLAG M2 antibody.
Immunoprecipitation assays for domain mapping were done using a protocol similar to the experiments demonstrating an interaction between HP2 and purified NURF. In this instance the HP2 peptides from the rabbit reticulocyte lysate system have a T7 tag, and T7 antibodies are used for immunoprecipitation. (The T7 tag is present at the N-terminal end of each HP2 peptide.) Other conditions and reagents are the same as above. A Distalless protein with a T7 tag is used as a control. The coprecipitation of NURF is detected by Western blot using the FLAG M2 antibody. The HP2 peptides have been described previously (3) except for those which include only the first through the fifth exons of HP2. In these cases, PCR products containing the T7 tag are produced as described in the aforementioned paper, and used as templates for transcription/translation.
Construction of Nap-1 single copy knockouts
The alleles used for assaying the suppressor activity of Nap-1 are targeted single-copy Nap-1SSA mutations (27). The SSA mutations were derived from tandem duplications of the Nap-1 gene using “ends-in targeting” of a double mutant Nap-1 gene. Each targeted Nap-1 duplicate of the original Nap-1KO1 flies contains four diagnostic restriction sites. XhoI and SalI sites are diagnostic for the targeted recombination reaction. HindIII and BclI sites, in addition to being diagnostic for recombination, interrupt the Nap-1 open reading frame. The starting SSA mutations and the molecular specifics of subsequent mismatch-repair reactions at the heteroduplex intermediate resulted in eleven different combinations of the markers. Knockout mutations Nap-1SSA2, Nap-1SSA3, and Nap-1SSA4 were established as stocks and used in this study. Nested PCR on genomic DNA from these flies was performed as published elsewhere (28). Diagnostic restriction enzyme digests of the PCR products with XhoI, HindIII, BclI, and SalI verified the structure of each Nap-1SSA allele.
Drosophila melanogaster stocks and crosses for Nap-1 PEV assays
Drosophila stocks are raised on cornmeal sucrose based medium (29). Crosses are carried out at 25° C and 70% relative humidity. To assess modification of variegation, males carrying various mutant Nap-1 chromosomes balanced over CyO are first crossed to SM6/nocsco virgin females. Male progeny carrying nocsco and the chromosome encoding the Nap-1 mutant to be tested are then crossed to virgins carrying a ywm4 chromosome, and the progeny compared. The nocsco chromosome is chosen as a standard because it contains no major enhancers or suppressors of variegation. Males from these crosses are then collected, aged to three to five days post-eclosion, and photographed. Pigment assays were performed by extracting pigment and measuring OD at 480 nm (30). Fifteen male fly heads were used in one preparation for pigment quantification. 4-5 replicates were done for each mutant allele.
Results
Nap-1 and NURF, as well as HP1, are present in protein fractions with HP2 upon fractionation under non-denaturing conditions
In an effort to find proteins that interact with HP2, conventional chromatography was performed under non-denaturing conditions; an embryo nuclear extract was run through a series of chromatographic separations and HP2 followed by Western blot. The fractionation procedure is illustrated in Figure 1A. On the fourth and final column, Mono S, fractions containing HP2-S and HP2-L were separated (fractions 4-7 and 11-12 respectively), as shown by Western blot (Figure 1B, top row).
FIGURE 1.
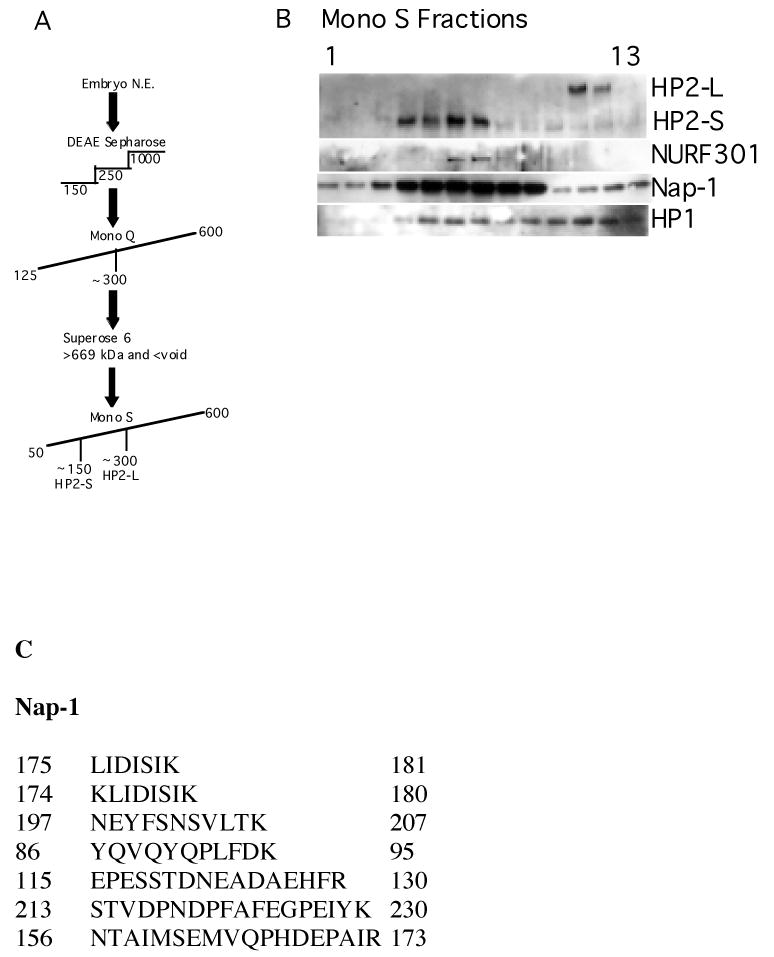

HP2 has an overlapping elution profile with NURF301 as well as Nap-1 and HP1.
FIGURE 1A. Flow chart of the chromatography protocol used to identify HP2 binding partners. See Materials and Methods for additional details.
FIGURE 1B. Elution profiles of components that are present in fractions with HP2. Mono S column fractions were probed with antibodies against HP2 as well as Nap-1 and NURF301. As can be seen in the top panel, HP2-S is eluting in fractions 4-7 while HP2-L is eluting in fractions 11 and 12. The NURF 301 blot in the next panel shows that NURF301 is present in fractions 6 and 7, which also contain HP2-S. Nap-1 is present in all of the fractions shown but is present to a greater extent with HP2-S. HP1, a known HP2-interactor, is present in fractions with both isoforms, as expected.
FIGURE 1C. Nap-1 peptides identified by mass spectrometry. Numbers at the left and right of each sequence indicate the first and the last amino acids identified, respectively.
FIGURE 1D. Complete amino acid sequence for Drosophila Nap-1. Residues shown in boldface represent amino acids identified by mass spectrometry.
Proteins in fractions from the Mono S column were separated and stained on SDS-PAGE gels, and those bands that peaked with HP2 were subsequently analyzed using mass spectrometry. MALDI-TOF and LC/MS mass spectrometry both identified Nap-1 as a possible candidate that interacts with HP2 (Figure 1C and 1D). Other polypeptides were also identified but were not confirmed in multiple protein preparations. Analysis of the Mono S column fractions by Western blot, using antibodies specific for Nap-1, determined that Nap-1 is present in protein fractions with HP2-S. Nap-1 is also present in fractions with HP2-L, but to a lesser extent (Figure 1B, first and third rows). HP2-S and Nap-1 were present in overlapping protein fractions throughout the chromatographic separation (data not shown). Since other proteins were present in these fractions with HP2 and Nap-1, as seen by total protein staining following gel electrophoresis, antibodies were obtained against proteins that are involved in chromatin remodeling, as is Nap-1, to check for their presence along with HP2 and Nap-1. After obtaining antibodies against ISWI, which was revealed to be present in fractions with HP2-S by Western analysis, antibodies against the unique component of NURF, NURF301, as well as against Acf1 were obtained, as these polypeptides are found in protein complexes that contain ISWI. As shown in the second row of Figure 1B, NURF301 elutes in overlapping fractions with HP2-S. Interestingly, the smaller isoform of HP2, HP2-S, is present in fractions with NURF301 while the larger isoform, HP2-L, is not, suggesting different functions for the two HP2 isoforms. Acf1 was not present in any of the fractions. These fractions were also analyzed using HP1 antibodies (Figure 1B, fourth row), since the association of HP1 and HP2 has been previously demonstrated (1, 3). HP1 is present in fractions containing HP2, as well as in fractions that do not contain HP2, not unexpected given the large number of HP1 binding partners. The elution profile of HP2 with Nap-1 and NURF led us to further investigate a possible interaction between HP2 and these proteins.
HP2 coimmunoprecipitates with Nap-1 as well as with all four components of NURF, and Nap-1 coimmunoprecipitates with NURF
We then asked whether HP2 and Nap-1 physically interact, using coimmunoprecipitation assays. As shown in Figure 2A, HP2-specific antibodies are able to pull down Nap-1 from an embryo extract, whereas the rabbit preimmune serum does not. This interaction is not DNA-mediated; upon complete digestion of the DNA in the nuclear extract with micrococcal nuclease, no change in binding is seen. Nap-1 antibodies have also been shown to immunoprecipitate both isoforms of HP2 from an embryo nuclear extract (data not shown). Experiments were also done to investigate a direct interaction between HP2 and Nap-1 by producing both of the proteins in a rabbit reticulocyte lysate system and performing coimmunoprecipitation experiments. A direct interaction was not observed.
FIGURE 2.
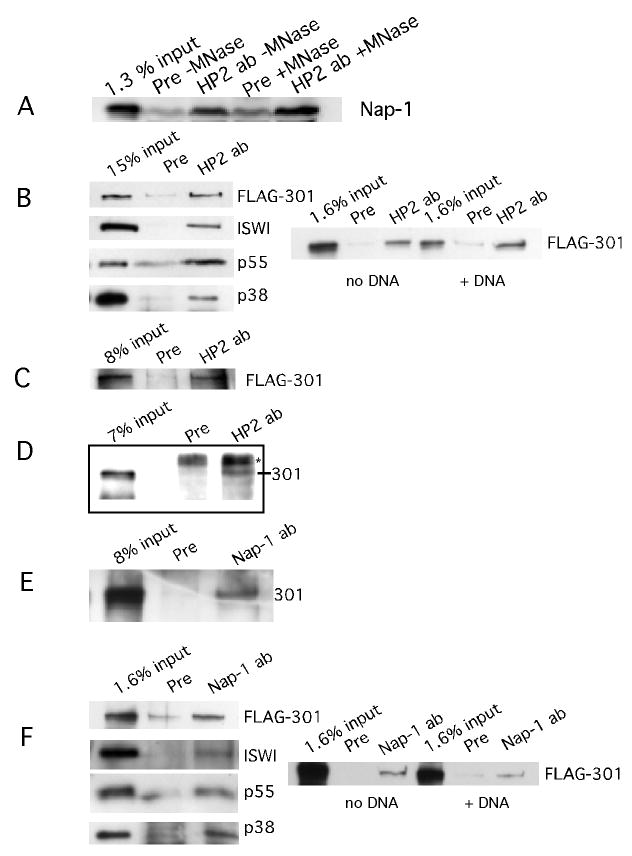
HP2 interacts with Nap-1 and NURF.
FIGURE 2A. HP2 interacts with Nap-1 in an embryo extract. HP2 antibodies or HP2 preimmune serum were added to Drosophila embryo extract and the immunoprecipitated material was tested for the presence of Nap-1 by western blot. The input is shown in the first lane; the following lanes indicate that in the presence of preimmune serum, no Nap-1 is pulled down while in the presence of HP2 antibodies, Nap-1 is immunoprecipitated. By comparison with the input sample, we estimate that 1-2% of the Nap-1 is immunoprecipitated. This percent is indicative of our immunoprecipitation efficiency throughout this paper. No loss of binding occurs between Nap-1 and HP2 upon digestion of the nuclear DNA to completion with micrococcal nuclease.
FIGURE 2B. HP2 interacts with the NURF complex; DNA does not mediate the interaction. HP2 rabbit antibodies were mixed with FLAG purified NURF complex plus HP2-S transcribed and translated in the rabbit reticulocyte lysate system. Preimmune serum was used as a control. As can be seen in the top panel, NURF301 is pulled down when HP2-specific antibodies are present, but not in the presence of the preimmune serum. The first lane indicates the input. The same is true for ISWI, p55, and p38, the other three components of NURF. To the right, it can be seen that upon the addition of 100-fold excess plasmid DNA to the binding reaction, HP2 antibodies are still able to pull down the NURF complex, demonstrating that the coprecipitation is not dependent on the binding of the individual proteins to DNA. The input into the reactions is shown; rabbit preimmune serum acts as a negative control.
FIGURE 2C. HP2 rabbit antibodies can pull down NURF301 from Sf9 cell extract. Sf9 extract from cells in which the NURF301 subunit was overproduced was incubated with HP2 antibodies, immunoprecipitated, the pellet run out on SDS-PAGE gels, and probed with antibodies against the FLAG tag. The first lane shows FLAG-tagged NURF301 in the load. HP2 antibodies pull down NURF301, whereas the preimmune serum does not, as shown in lanes 3 and 2.
FIGURE 2D. HP2 rabbit antibodies immunoprecipitate NURF from a Drosophila embryo extract. Embryo extract was prepared as in the chromatography experiments and immunoprecipitations performed as described for the Sf9 cell extract. The western blots were then probed with rabbit NURF301 antibodies. The first lane shows NURF 301 in the load and the following lanes show that NURF301 is precipitated with HP2 rabbit antibodies and not with HP2 preimmune serum. The * indicates a nonspecific product.
FIGURE 2E. Nap-1 antibodies can pull down NURF from an Sf9 extract in which NURF301 is overproduced. A preimmune serum was used as a control. The presence of NURF was assayed by probing the blot with FLAG M2 antibodies. In the first lane is the material added to the immunoprecipitation reactions. Lane 2 shows the results of the immunoprecipitation performed with preimmune serum. Lane 3 shows that Nap-1 antibodies can pull down NURF301, the unique subunit of NURF.
FIGURE 2F. Nap-1 interacts with the NURF complex; DNA does not mediate the interaction. Nap-1 antibodies were mixed with FLAG purified NURF complex plus Nap-1 transcribed and translated in the rabbit reticulocyte lysate system. Preimmune serum was used as a control. As can be seen in the top panel, NURF301 is pulled down when Nap-1-specific antibodies are present, but not in the presence of the preimmune serum. The first lane indicates the input. The same is true for ISWI, p55, and p38, the other three components of NURF. To the right, it can be seen that upon the addition of 100 fold excess plasmid DNA to the binding reaction, Nap-1 antibodies are still able to pull down the NURF complex. The input into the reactions is shown; rabbit preimmune serum acts as a negative control. Note that the right panel in Figure 2F was exposed longer than that in Figure 2B.
As seen in Figure 1A, HP2 has an overlapping elution profile with NURF301, the unique subunit of the NURF complex. We were thus interested in determining if HP2 interacts with the NURF complex. Purified NURF was obtained as described (Materials and methods) and mixed with HP2-S made in rabbit reticulocyte lysate, along with HP2 rabbit antibodies. HP2 antibodies are able to pull down all four components of the NURF complex, NURF301, ISWI, p55, and p38. The addition of 100-fold excess of plasmid DNA to the immunoprecipitation reaction does not abrogate binding, indicating that the interaction is not DNA-mediated (Figure 2B). None of the NURF products are precipitated in the presence of rabbit preimmune serum. For additional confirmation of the binding interaction, we determined that HP2 antibodies specifically pull down NURF301, a protein so far found only in the NURF complex, from both an Sf9 cell extract and an embryo extract (Figures 2C and 2D respectively). In Figure 2C, an Sf9 extract, from cells overexpressing a FLAG-tagged NURF301 subunit, was used for immunoprecipitation with HP2 antibodies. Western blots of the Sf9 extract confirm the presence of both isoforms of HP2 (data not shown). Upon producing HP2 and the individual subunits of the NURF complex in the rabbit reticulocyte lysate system, direct interactions were not found. The assembled NURF complex may be required for binding to HP2 and/or posttranslational modifications needed for binding may be lacking in this system.
Upon determining that HP2 interacts with Nap-1 and NURF, we were interested in determining whether Nap-1 and NURF interact with one another. Nap-1 antibodies as well as preimmune serum were tested for their ability to pull down the unique subunit of NURF, NURF301. Upon the addition of Nap-1 antibodies to the Sf9 extract, the NURF301 subunit is precipitated, whereas a preimmune serum does not pull down NURF301 (Figure 2E). Upon producing Nap-1 in the rabbit reticulocyte lysate system and performing coimmunoprecipitations as with HP2-S, we find that antibodies against Nap-1 pull down all four subunits of NURF (Figure 2F). DNA does not mediate this interaction since the addition of 100-fold excess of plasmid DNA to the binding reaction did not abrogate binding.
Three domains within HP2 mediate binding to the NURF complex
Upon determining that HP2 antibodies pull down all four subunits of the NURF complex in the presence of HP2, we were interested in determining which domain or domains of HP2 can mediate binding with the NURF complex. Western blots failed to detect HP1 in association with FLAG-purified NURF, (data not shown) suggesting a direct interaction with HP2.
The various HP2 test peptides used for mapping the interaction domain are shown in Figure 3A. Synthesis of appropriate-size products in the rabbit reticulocyte system was confirmed by gel electrophoresis. Upon incubating the test peptides that contain various domains of HP2 (in all cases tagged with a T7 epitope) with the purified NURF complex as described previously, three domains of HP2 appear able to mediate binding to NURF; these are indicated in Figure 3A. The NURF complex was followed using an antibody against the FLAG tag on the NURF 301 subunit. The data that narrowed down the potential interaction sites to these three domains is shown in Figures 3B, C, and D. In all cases, the far left column shows the FLAG-tagged NURF301 protein. The second column is a control immunoprecipitation using the Distalless protein, which contains the T7 tag, but does not interact with NURF, demonstrating the specificity of the interaction. In the third column, T7 antibodies, T7-tagged HP2 fragments, and NURF were added to the immunoprecipitation mixture and the NURF 301 subunit was pulled down, as shown by Western. In each immunoprecipitation, Distalless and HP2-S were used as negative and positive controls respectively to ensure that the assay was working properly. In Fig. 3B, the domain of interaction in the N-terminal portion of HP2 is described. As shown in the first row, a portion of HP2 that contains amino acids 1-1353 is able to mediate an interaction with NURF, whereas an HP2 test peptide that encompasses amino acids 1328-1900 cannot. In the second row data is presented that indicate that a peptide which contains amino acids 400-1353 does not mediate an interaction with NURF and it thus appears that the domain of interaction is somewhere within the first 400 amino acids of the protein. Upon testing peptides that contained portions of this N-terminal 400 amino acids, it can be seen in the third row of Figure 3B that a region within amino acids 229-276 of HP2 is necessary for mediating an interaction with NURF in vitro. As expected, binding also occurred for an HP2 peptide which contains the N-terminal domain up to amino acid 360.
FIGURE 3.
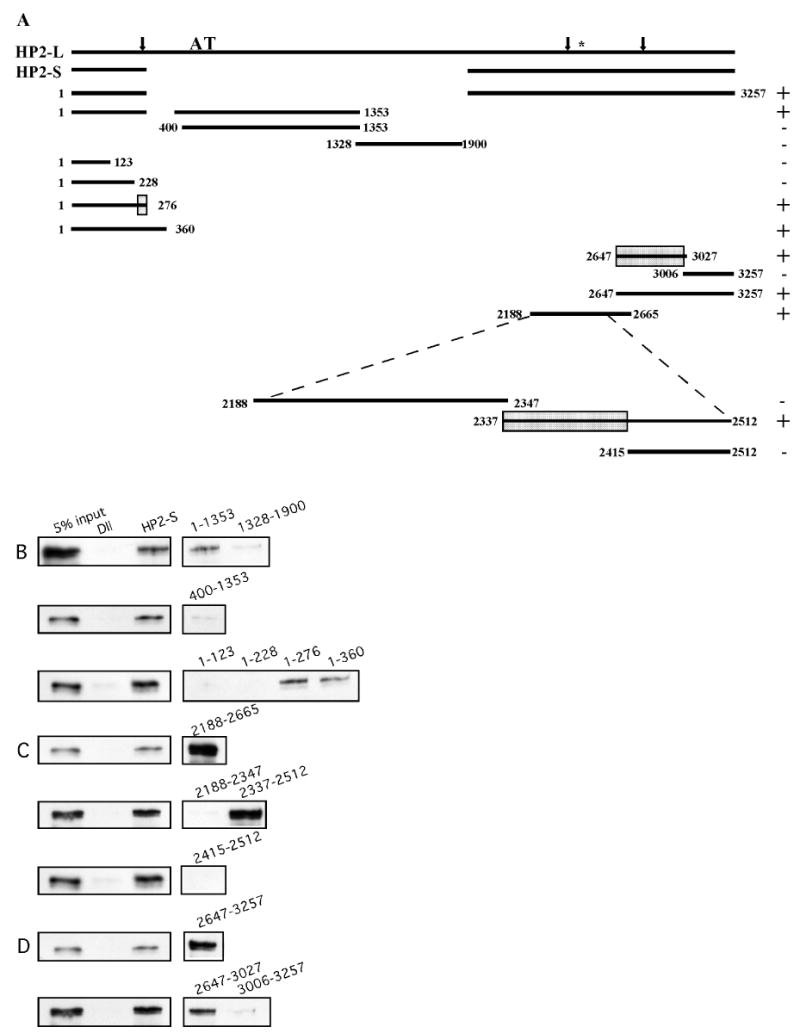
Three domains in HP2-S mediate binding to NURF. T7-tagged peptides of HP2 were produced in a rabbit reticulocyte lysate system and added to purified NURF complex along with T7 antibodies. The immunoprecipitate was then probed with FLAG M2 antibodies. The load is shown in each experiment as well as results of the Distalless immunoprecipitation (negative control) and an HP2-S immunoprecipitation (positive control).
FIGURE 3A. A schematic indicating the HP2 test peptides used to identify the NURF interacting domains. The structure of HP2-L and HP2-S is shown at the top of the schematic. The large AT letters show the location of the pair of AT-hooks. An asterisk indicates the HP1-interacting domain. Arrows indicated the domains of NURF interaction. Below the structure of HP2 are the HP2 test peptides used to determine the domain(s) of interaction with NURF. For each test peptide, the amino acids encompassed in the peptide are given. On the right side of the figure is a + or - sign indicating if the peptide binds to NURF. The minimal domains responsible for the interaction are boxed on the test peptide.
FIGURE 3B. A conserved region in the N-terminal domain of HP2 interacts with NURF. As can be seen in the first and second rows, a domain exclusive to HP2-L does not appear to mediate the interaction with NURF. The third row indicates that a region within amino acids 229-276 is necessary for the interaction.
FIGURE 3C. A domain N-terminal to the HP1 binding domain mediates an interaction with NURF. A test peptide containing amino acids 2188-2665 binds to NURF, as can be seen in the first row. A peptide that encompasses amino acids 2188-2347 does not bind to NURF whereas a peptide with amino acids 2337-2512 does, as seen in the second row. Since this peptide includes the HP1 binding domain, a 100 amino acid domain that has been found to be both necessary and sufficient for HP1 binding (3) was tested for interaction. This domain, as seen in the third row, does not mediate an interaction and thus the NURF interaction domain appears to be distinct, in a region containing amino acids 2337-2414.
FIGURE 3D. A third domain in the C-terminal half HP2 mediates an interaction with NURF. A test peptide containing amino acids 2647-3257 binds to NURF as can be seen in the first row. A peptide encompassing amino acids 2647-3027 does bind to NURF whereas a peptide containing amino acids 3006-3257 does not. Thus, a third interaction domain is found in the HP2 domain containing amino acids 2647-3006.
In Figure 3C, it can be seen that a region encompassing amino acids 2188-2665 is able to mediate an interaction with NURF. Upon further division of this domain, we find that an HP2 test peptide containing amino acids 2188-2347 is not able to bind to NURF whereas an HP2 test peptide containing amino acids 2337-2512 is able to mediate an interaction with NURF, as seen in the second row. Interestingly, the HP1 binding domain of HP2 described previously lies within the region encompassing amino acids 2337-2512 (3). As shown in the third row of Figure 3C, the domain of HP2 that interacts with HP1 (2415-2512) is not the domain that mediates the interaction with NURF. By deduction, it then appears that the interaction domain includes a region in amino acids 2337-2414.
In Figure 3D, a third NURF interaction domain is described in the C-terminal half of the protein. A test fragment that encompasses amino acids 2647-3257 is able to interact with NURF as well. Further division of this fragment shows that amino acids 2647-3027 contain the third interaction domain, since amino acids 3006-3257 do not interact with NURF. Thus, it appears that HP2 has three different domains that mediate an interaction with NURF. Peptides which contained regions unique to the larger isoform were tested to determine if there were any HP2/NURF interactions specific to HP2-L; none were observed. The results indicate that an interaction with HP1 is not required, but that HP2 interacts directly with the NURF complex.
Nap-1 is a dominant suppressor of position effect variegation
We were interested in evaluating whether mutations in Nap-1 would result in suppression of PEV; such a result would place Nap-1 into a group of fifty or so proteins in Drosophila, including HP1 and HP2, which function to create a heterochromatic structure in vivo.
To test the possibility that Nap-1 may be required to create a heterochromatic nucleosome array in vivo, we analyzed the impact of mutations in Nap-1 on heterochromatic silencing displayed by ywm4. In this line, an inversion on the X chromosome places the white gene next to the pericentric heterochromatin, resulting in a variegated appearance where the white gene is expressed in some cells in the eyes and not in others (31). In males that carry ywm4 and are heterozygous for a knockout of Nap-1 (Nap-1SSA/+), the white gene shows a loss of silencing as can be seen in Figure 4A. In siblings that do not contain the Nap-1 knockout (nocsco/+), there is more variegation (more silencing) in the fly eye as can be seen by the fly eye on the left. We have quantitatively assessed the amount of pigment in the fly eyes. As can be seen in Figure 4B, flies carrying any of the three Nap-1 mutant alleles that were tested using pigment assays have higher pigment levels (statistically significant p values) when compared to their siblings without the Nap-1 mutations. In a two-tailed t-test the p value for the SSA2 set is 0.017, for SSA3 is 0.006, and for SSA4 is .008. Thus, mutations in Nap-1 that prevent production of Nap-1 protein act as dominant suppressors of PEV. This finding suggests a function for Nap-1 in the spread of heterochromatin, a function shared by HP2, as mentioned previously. Previous reports have shown that mutations in nurf301 do not result in dominant suppression of PEV. Unfortunately, a recessive effect cannot be tested due to lethality.
FIGURE 4.
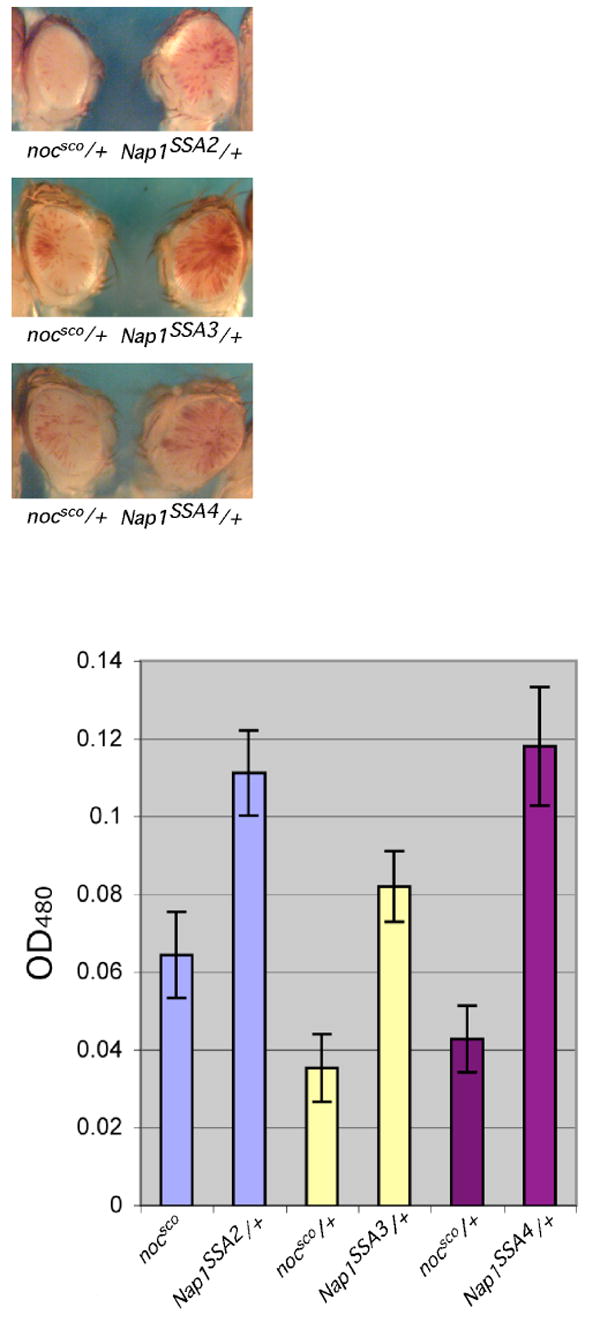
Heterozygous Nap-1 mutations result in suppression of variegation (loss of silencing) in ywm4.
FIGURE 4A. The white gene is derepressed in Nap-1 mutants. In males that carry ywm4 and are heterozygous for a knockout of Nap-1 (Nap-1SSA/+), the white gene is derepressed as can be seen from the extent of eye pigmentation in the flies on the right side of each panel. Siblings that do not contain the Nap-1 knockout (nocsco/+) show less expression of white in the fly eye as seen on the left side of each panel.
FIGURE 4B. Pigment values show an increase in white expression in response to mutations in Nap-1. 15 male heads were used for quantitative pigment assays upon aging for 3-5 days. 4-5 determinations are reported with the standard error indicated (thin line).
Discussion
To identify novel HP2-binding partners, we have fractionated nuclear proteins by column chromatography under non-denaturing conditions. Obtaining overlapping elution profiles of HP2-S and HP2-L with Nap-1, as well as HP2-S with NURF301, suggests that HP2 may have functional interactions with proteins that have histone chaperone activity (Nap-1) as well as those that are capable of remodeling chromatin (NURF) (Fig. 1B). Interestingly, HP2-S and HP2-L are eluting from the last column, Mono S, in different fractions, suggesting some separable functions; the elution profile indicates that HP2-L fractions do not contain NURF. The binding studies presented in Figure 3 indicate that either isoform of HP2 is potentially capable of interacting with NURF. Other proteins that are bound to HP2-L may affect its capacity to bind to NURF.
Based on the aforementioned elution profiles of HP2, Nap-1, and NURF, we performed coimmunoprecipitation experiments to explore these interactions. As can be seen in Fig. 2A, HP2 antibodies are able to pull down Nap-1 from an embryo nuclear extract. Given that HP2 and Nap-1 synthesized in the rabbit reticulocyte lysate system do not coimmunoprecipitate, it would appear that this interaction is indirect, as mentioned previously. Given the presence of a pair of AT-hooks in HP2 and Nap-1's role in chromatin remodeling, one could speculate that HP2 and Nap-1 are binding to DNA and not to one another, and that they are coimmunoprecipitated based on a DNA interaction. Data shown in Figure 2 (including complete digestion of DNA in the reaction mixture) indicates that DNA does not mediate this binding interaction. An alternative explanation could be that the interactions are impacted by post-translational modifications, and so were not revealed in the rabbit reticulocyte lysate system, or that another protein or proteins mediate this interaction, possibly the NURF complex.
Examining the HP2-NURF interaction, one finds that anti-HP2 antibodies pull down all four subunits of the NURF complex in vitro (Figure 2B). Since the four-component NURF complex was purified on the basis of the FLAG tag on the NURF301 subunit, it appears that HP2 has a direct interaction with at least one of the NURF components. Again, experiments in Figure 2B indicate that DNA is not mediating the interaction and since HP1 did not coprecipitate with NURF upon FLAG-based purification, it also does not mediate the binding reaction (data not shown). Anti-HP2 antibodies were able to pull down NURF301 from a Drosophila nuclear extract as well as an Sf9 extract from cells where NURF301 was overproduced. Results from this experiment suggest that HP2 may interact with NURF via NURF301. However, there remains the possibility that one or more of the other components of NURF are conserved in Sf9 cells and are mediating the interaction.
Coimmunoprecipitation experiments with Nap-1 antibodies precipitate all four subunits of the NURF complex in vitro as well as precipitating NURF301 from Sf9 cell extract where NURF301 is overproduced; this interaction appears to be direct (Figures 2E and 2F). We were able to eliminate the possibility that the interaction is the consequence of each protein binding independently to the small amount of DNA present; upon 100-fold addition of plasmid DNA to our binding reactions, the binding efficiency was only slightly decreased. One of the components of NURF, p55, has been found to be an H3/H4 chaperone. Hence, one possibility, given that an interaction exists between Nap-1, the H2A-H2B chaperone, and NURF, an ISWI dependent chromatin remodeling complex, is that the chaperone and remodeling complex are both present to construct the nucleosome and/or to remodel it.
It was of interest to us to determine the domains in HP2 that were responsible for these protein interactions. HP2 is devoid of any defined protein motifs except for the AT-hooks and the HP1-binding domain. Comparative analysis of HP2 in various Drosophila species has provided insight into the domains that have been conserved during the evolution of Drosophilidae (3). We were interested in determining if any of these conserved domains were sites of NURF binding, given that this interaction was found to be direct.
The NURF binding studies in Figure 3B show that there are three sites within HP2 mediating interactions with NURF. The first four exons of HP2 have been found to be highly conserved among the Drosophila species studied in our previous work (3). Binding studies indicate that a domain containing this region of HP2 is sufficient for NURF binding. Various regions in the C-terminal domain have also been found to be conserved and another domain mediating interaction is found within a polypeptide encompassing amino acids 2647-3006. The third domain is just N-terminal to the HP1 binding site characterized previously (3). This NURF-binding domain does not appear to be conserved among the Drosophila species studied thus far. It is possible that this domain is structurally conserved but does not have sequence conservation. Based on these studies, we are now able to assign additional functional domains to HP2.
It is to be noted that in vitro transcription/translation reactions vary in efficiency based on template size and other factors such as sequence composition. A large protein product that is barely detectable by Western, HP2-S, precipitates NURF effectively, while some peptides, such as that encompassing amino acids 3006-3257, which are highly expressed, do not. Thus, coprecipitation is not a function of the amount of product being made in the rabbit reticulocyte system but reflects specific sites of interaction.
What do these interactions mean? Previous work has implicated histone chaperones such as Nap-1 in the regulation of DNA accessibility. In in vitro experiments in yeast, Nap-1 can remove H2A-H2B dimers from folded nucleosomes and replace them with canonical or variant histones (32, 33). Thus, Nap-1 can disassemble as well as reassemble chromatin.
Nap-1 has been shown to interact genetically with acf1. Acf1 is a subunit of ACF, a complex that has been shown to catalyze an ATP-dependent assembly of nucleosomes in a very regular array in vitro. acf1 has previously been shown to be a recessive suppressor of PEV (34). Thus there is precedence for Nap-1 cooperating with other proteins in generating a regularly spaced nucleosomal array, characteristic of heterochromatin (6, 7).
In budding yeast, mutations in CAF-1, a histone chaperone complex mediating H3/H4 assembly, result in defects in transcriptional silencing at the telomeres and at the mating type loci (17-20). The gene encoding ASF1, an H3/H4 histone chaperone in Drosophila, is a suppressor of PEV when mutant, indicating that ASF1 contributes to the heterochromatic state (23). Thus, there is precedence for histone chaperones contributing to a condensed nucleosomal array as is found in heterochromatin. Our genetic analysis shows that Nap-1 has a similar role, contributing to the heterochromatic state (Fig. 4). Based on our findings that Nap-1 is a suppressor of PEV, it is not surprising that Nap-1 is found to bind to other proteins such as HP2 that promote heterochromatin formation.
The lack of Nap-1-specific antibodies that will stain Drosophila polytene chromosomes has prevented analysis to localize Nap-1 to specific chromosomal sites, such as heterochromatin. Nap-1 may be only transiently localized to the chromatin, and thus its presence undetectable by immunostaining of polytene chromosomes. Alternatively, it may be denatured or rendered inaccessible by the standard chromosome fixation protocol.
As mentioned previously, Nap-1 has been associated with generation of periodic nucleosome arrays, but this has not been reported for NURF. NURF has been shown to bind to factors that are involved in active transcription, such as GAGA factor and HSF (25). However, the same chromatin remodeling complex might well function to move nucleosomes into a more regular ordered array when bound to factors such as HP2, which contributes to initiation and/or spreading of heterochromatin in PEV (1). Recent data indeed indicates this. Whole genome expression analysis of mutants lacking the NURF-specific subunit NURF301 (35) found that approximately two-thirds of the affected genes showed decreased expression in the NURF mutant background; these all appeared to be ecdysone-responsive targets. In addition, a second group of genes in which expression increased in the NURF301 mutant background was observed, indicating that the NURF complex has dual functions. NURF can function both in gene activation and in gene repression.
Previous work has shown that reduction in nurf301 gene dosage has no dominant effect on suppression of PEV (36). Work in our laboratory has confirmed this observation. Interestingly, Acf1, a member of the ACF chromatin remodeling complex mentioned previously, does not have an impact on PEV in the heterozygous state, but is a recessive suppressor of PEV. The arrangement of various motifs in Acf1 is similar to the arrangement of the same motifs in NURF301 (37-41). In the case of Acf1, some of the homozygous mutants survive to adulthood and thus can be tested in the PEV assay (34). NURF301 may be a recessive suppressor of PEV, but since the homozygous mutants die before adulthood, this cannot be determined.
As with Nap-1, the lack of antibodies specific for NURF301 that are suitable for immunostaining has precluded experiments to determine if NURF301 colocalizes with HP2 at sites on Drosophila polytene chromosomes and/ or if it localizes to any sites of heterochromatin.
In this work, we have found two new HP2 binding factors, Nap-1 and NURF, in addition to the previously identified HP2 binding factor HP1. The functional consequences of these interactions remain to be determined. One would speculate that HP2 has a role in chromatin remodeling. Upon successful purification of purified HP2, it would be interesting to perform chromatin remodeling assays to monitor whether HP2 cooperates with Nap-1 and/or NURF to remodel chromatin into the heterochromatic state.
Acknowledgments
We thank the Elgin lab for critical review of the manuscript; Susanne Lankenau, Ming-Fei Lang, Tobias Jursch, and Zhu Jiayun for help with construction of the Nap-1 mutants; Jim Kadonaga (UCSD) for anti Nap-1 antibodies and anti-ISWI antibodies; Jessica Tyler (Univ. of Colorado Health Sciences Center) for anti-p55 antibodies, and the Protein and Nucleic Acid Chemistry Laboratory at Washington University for mass spectrometry analysis. We are very grateful to Craig Pikaard for use of the FPLC, Keith Earley for showing us how to use it, and Amy Caudy for advice on protein fractionation.
This work was supported by National Institutes of Health Grant GM068388 (to S. C. R. E.). G. E. S. was supported in part by National Institutes of Health Training Grant T32 GM07067. H. X. and C. W. are supported by the Intramural Research Program of the National Cancer Institute. Construction of the Nap-1 mutants by D-H. L. was supported in part by a Max-Planck grant to D-H. L. and Ming-Fei Lang.
Footnotes
The abbreviations used are: HP1, Heterochromatin Protein 1; HP2, Heterochromatin Protein 2; HP2-L, the larger isoform of HP2; HP2-S, the smaller isoform of HP2; PEV, Position Effect Variegation; Nap-1, Nucleosome Assembly Protein 1; NURF, Nucleosome Remodeling Factor; CAF-1, Chromatin Assembly Factor 1; ASF1, Antisilencing Function Protein 1; HIRA, Histone Regulatory A; ACF, ATP-utilizing Chromatin Assembly and Remodeling Factor
References
- 1.Shaffer CD, Stephens GE, Thompson BA, Funches L, Bernat JA, Craig CA, Elgin SC. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc Natl Acad Sci U S A. 2002;99:14332–7. doi: 10.1073/pnas.212458899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–82. [PubMed] [Google Scholar]
- 3.Stephens GE, Slawson EE, Craig CA, Elgin SC. Interaction of Heterochromatin Protein 2 with HP1 Defines a Novel HP1-Binding Domain. Biochemistry. 2005;44:13394–403. doi: 10.1021/bi051006+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 5.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–7. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–77. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 7.Sun FL, Cuaycong MH, Elgin SC. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol. 2001;21:2867–79. doi: 10.1128/MCB.21.8.2867-2879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKnight SL, Miller OL., Jr Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977;12:795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- 9.McKnight SL, Bustin M, Miller OL., Jr Electron microscopic analysis of chromosome metabolism in the Drosophila melanogaster embryo. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):741–54. doi: 10.1101/sqb.1978.042.01.075. [DOI] [PubMed] [Google Scholar]
- 10.Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. Embo J. 1988;7:2211–9. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varga-Weisz P. Chromatin remodeling factors and DNA replication. Prog Mol Subcell Biol. 2005;38:1–30. doi: 10.1007/3-540-27310-7_1. [DOI] [PubMed] [Google Scholar]
- 12.Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–52. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–14. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 14.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–60. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 15.Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 16.Ishimi Y, Kojima M, Yamada M, Hanaoka F. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur J Biochem. 1987;162:19–24. doi: 10.1111/j.1432-1033.1987.tb10535.x. [DOI] [PubMed] [Google Scholar]
- 17.Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–70. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–57. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 19.Monson EK, de Bruin D, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci U S A. 1997;94:13081–6. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–32. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–97. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–40. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 23.Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–6. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–20. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–43. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 26.Gdula DA, Sandaltzopoulos R, Tsukiyama T, Ossipow V, Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998;12:3206–16. doi: 10.1101/gad.12.20.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lankenau DH. Germline Double-Strand Break Repair and Gene Targeting in Drosophila: a Trajectory System throughout Evolution. In: Lankenau DH, editor. Genome Integrity: Facets and Perspectives. Vol. 1. Springer; Heidelberg: 2006. pp. 153–197. [Google Scholar]
- 28.Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau DH. Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics. 2003;163:611–23. doi: 10.1093/genetics/163.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer CD, Wuller JM, Elgin SC. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 1994;44:99–108. doi: 10.1016/s0091-679x(08)60908-5. [DOI] [PubMed] [Google Scholar]
- 30.Khesin RB, Leibovitch BA. Influence of deficiency of the histone gene-containing 38B-40 region on X-chromosome template activity and the white gene position effect variegation in Drosophila melanogaster. Mol Gen Genet. 1978;162:323–8. doi: 10.1007/BF00268858. [DOI] [PubMed] [Google Scholar]
- 31.Muller H. The remaking of chromosomes. Collecting Net. 1938;XIII:181–195. [Google Scholar]
- 32.Bruno M, Flaus A, Owen-Hughes T. Site-specific attachment of reporter compounds to recombinant histones. Methods Enzymol. 2004;375:211–28. doi: 10.1016/s0076-6879(03)75014-9. [DOI] [PubMed] [Google Scholar]
- 33.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–25. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 34.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–83. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–5. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–98. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–39. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci U S A. 2000;97:1038–43. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–90. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 40.Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. Embo J. 2000;19:3377–87. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. Embo J. 2001;20:3781–8. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]


