Abstract
In a screen for genes expressed in the Drosophila embryonic salivary gland, we identified a tryptophanyl-tRNA synthetase gene that maps to cytological position 85D (WRS-85D). WRS-85D expression is dependent on the homeotic gene Sex combs reduced (Scr). In the absence of Scr function, WRS-85D expression is lost in the salivary gland primordia; conversely, ectopic expression of Scr results in expression of WRS-85D in new locations. Despite the fact that WRS-85D is a housekeeping gene essential for protein synthesis, we detected both WRS-85D mRNA and protein at elevated levels in the developing salivary gland. WRS-85D is required for embryonic survival; embryos lacking the maternal contribution were unrecoverable, whereas larvae lacking the zygotic component died during the third instar larval stage. We showed that recombinant WRS-85D protein specifically charges tRNATrp, and WRS-85D is likely to be the only tryptophanyl-tRNA synthetase gene in Drosophila. We characterized the expression patterns of all 20 aminoacyl-tRNA synthetases and found that of the four aminoacyl-tRNA synthetase genes expressed at elevated levels in the salivary gland primordia, WRS-85D is expressed at the highest level throughout embryogenesis. We also discuss the potential noncanonical activities of tryptophanyl-tRNA synthetase in immune response and regulation of cell growth.
INTRODUCTION
How master regulators, such as those encoded by homeotic genes, determine the final structure and physiology of an organism is a long-standing scientific inquiry. We know that homeotic genes are expressed in limited domains along the anterior–posterior body axis where they control which structures develop (Duncan, 1987; Krumlauf et al., 1987; Kaufman et al., 1990; McGinnis and Krumlauf, 1992; Krumlauf, 1994). Mutations in homeotic genes cause structures within a particular segment or segments to be replaced by structures normally found elsewhere. Each homeotic gene encodes one or more related DNA-binding transcription factors (Levine and Hoey, 1988; Scott et al., 1989). Thus homeotic genes control cell fate by regulating transcription of downstream target genes. However, despite a concerted effort to identify these genes in the past several years, only a limited number have been found (Andrew and Scott, 1992; Biggin and McGinnis, 1997; Graba et al., 1997). To identify novel downstream target genes and to learn how homeotic genes control cell fate, we have focused on how Sex combs reduced (Scr), a Drosophila homeotic gene, controls the formation of a relatively simple organ, the larval salivary gland. Our approach has been to identify and characterize candidate target genes based solely on their SCR-dependent expression in the early salivary gland.
Salivary glands provide a simple developmental system to study how early acting regulatory molecules control the assembly of multicellular organs (Campos-Ortega and Hartenstein, 1997). In Drosophila, salivary glands start out as two ventrolateral plates of ∼100 cells each in the region of the presumptive posterior head, an area known as parasegment 2 (PS2).1 No additional cell divisions occur during salivary gland differentiation. Instead, the salivary glands increase in size simply by increasing the volume of individual cells. These cells invaginate and move dorsally and posteriorly, led by cells near the posterior–lateral edge of each plate, leading to the internalization of the salivary glands. By late embryogenesis, salivary gland cells reach the most posterior extent of their migration, reaching to the middle of the third thoracic segment. The salivary gland duct cells are the last to invaginate. These tube-forming cells, which contribute to both the lateral individual ducts and the central common duct, connect the secretory cells of the salivary glands to the larval mouth. The developing salivary gland thus provides a simple system to model cell growth, cell shape changes, cell migration, tube formation, and tissue-specific gene regulation.
Formation of the salivary gland is dependent on the homeotic gene Scr. In the absence of Scr function, salivary glands do not form, and when Scr is expressed everywhere in the embryo, salivary glands form in new locations (Panzer et al., 1992; Andrew et al., 1994; Isaac and Andrew, 1996). However, not every cell that expresses Scr becomes salivary gland. Other genes limit the recruitment of Scr-expressing cells to a salivary gland fate. The transforming growth factor-β signaling cascade limits salivary gland formation to the ventral–lateral regions of PS2 (Panzer et al., 1992; Isaac and Andrew, 1996; Andrew, 1998; Henderson et al., 1998), whereas the localized transcription factors TEASHIRT and ABDOMINAL-B block salivary gland formation in posterior segments when SCR is expressed everywhere (Andrew et al., 1994).
To identify genes expressed in the embryonic salivary gland, we screened several different enhancer trap stock collections (O’Kane and Gehring, 1987; Bellen et al., 1989; Bier et al., 1989; Grossniklaus et al., 1989; Wilson et al., 1989). Enhancer traps are created by the insertion of transposable elements containing the Escherichia coli lacZ gene fused to a relatively inactive promoter. Insertion of the transposon within or near enhancers for different genes often results in the expression of β-galactosidase (β-gal) in patterns that mirror the expression of those genes. This allows us to sample a large portion of the Drosophila genome for genes expressed in various tissues by assaying β-gal expression in different lines that each harbor a single transposable element. DNA flanking the transposon insertion site can be recovered using the E. coli origin of replication present in the enhancer trap. Mutations in selected genes can be made by mobilizing the transposon and selecting for imprecise excisions. Thus enhancer trap lines provide molecular as well as mutational access to genes selected solely on the basis of their expression within a particular tissue.
We identified 36 different lines in which β-gal is expressed in embryonic salivary glands using enhancer trap collections available through three different groups (M. Scott and M. Fuller; A. Spradling; and C. Goodman and G. Rubin). We cloned the gene corresponding to two independent insertions and have shown that it encodes the only Drosophila tryptophanyl-tRNA synthetase (WRS-85D). Furthermore, we have demonstrated that WRS-85D is essential for Drosophila development.
MATERIALS AND METHODS
Cloning and Molecular Characterization
DNA flanking the P-element inserts in lines l(3)03559 and l(3)04410 was obtained by plasmid rescue (Hamilton and Zinn, 1994) and used to isolate both genomic and cDNA WRS-85D clones (Zinn et al., 1988; Tamkun et al., 1991). Library screening, plasmid, phage, and genomic DNA preparations, subcloning, and labeling of radioactive probes were performed as described (Maniatis et al., 1989). The 2.6- and 1.6-kb cDNAs were subcloned into pGEM7Zf to yield plasmids pPS10.1 and 11.1, respectively. The developmental Northern blot was prepared as described (Henderson and Andrew, 1998). DNA sequencing was performed at the Johns Hopkins University Core DNA Analysis Facility and also as described (Isaac and Andrew, 1996). Sequence alignments were done using the CLUSTAL X (Higgins, 1993) and MacBoxshade programs (Baron, 1997, MacBoxshade; http://www.netaxs.com/∼jayfar/mops.html.).
Reduced Stringency Genomic Southern Blot
Plasmid pPS10.1 was digested with EcoRI, and the 660-bp fragment encoding the N-terminal 200 residues of WRS-85D was isolated and used to probe a blot of Oregon R (wild-type) Drosophila melanogaster genomic DNA under reduced stringency hybridization conditions (42°C, 5× SSC, 30% formamide) (Laird et al., 1969).
Polytene Chromosome In Situ Hybridizations
Polytene chromosome in situ hybridizations were done by the procedure of Pardue (1994) omitting the RNase treatment and acetylation steps and using the Vectastain kit (Vector Laboratories, Burlingame, CA) for HRP signal detection.
Antibodies, Embryo Staining, and Whole-Mount In Situ Hybridization
The β-gal mouse mAb was obtained from Promega (Madison, WI). The rat polyclonal antisera to WRS-85D was raised against an N-terminal 200-residue peptide. The PCR (Saiki et al., 1985) was used to amplify the most 5′ 600 bp of the ORF of WRS-85D using as template clone pPS10.1, which contains the 2.6-kb WRS-85D cDNA clone, with forward primer (5′-GGGGCTCGAGAATGGCGGACACCAAGGAG) and reverse primer (5′-TGCCCTTGACCTGATTGA). The resulting product was digested with XhoI and EcoRI and subcloned into the XhoI–EcoRI sites of pTrcHisB (Invitrogen, Carlsbad, CA) downstream of and in frame with the His6 tag. This construct, pPS12.2, was transformed into BL21(DE3) cells (Studier et al., 1990). A 500-ml culture of the pPS12.2-transformed cells was grown to an OD600 of 0.6, isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.1 mM, and the cells were grown for an additional 5.5 h to induce expression of the fusion protein. WRS-85D protein was purified from the induced cells by isolation of inclusion bodies (Rio et al., 1986) followed by Ni-NTA (Qiagen, Santa Clarita, CA) affinity chromatography (Invitrogen). Rat polyclonal antisera were raised against 1.3 mg of the Ni-NTA–purified protein (Covance Research Products, Denver, PA).
Embryo fixation and staining were performed as described, except Bouin’s solution (Sigma, St. Louis, MO) was used as fixative for WRS-85D immunostaining. Homozygous mutant embryos were identified by the absence of staining with α-β-gal, which stains the nonmutant embryos that carry a balancer chromosome marked with an ftz-lacZ insert on TM3. Immunostained embryos were mounted in methyl salicylate (Sigma).
Whole-mount in situ hybridization to detect embryonic mRNA accumulation was performed as described by Lehmann and Tautz (1994). Embryos were mounted in 70% glycerol to limit diffusion of the alkaline phosphatase reaction products.
Both immunostained embryos and embryos used for whole-mount in situ hybridization were visualized and photographed on a Zeiss (Thornwood, NY) Axiophot microscope using Normarski optics. Ektar print film (Eastman Kodak, Rochester, NY) was used for photography.
Cuticle Preparations
Cuticle preparations were as described in Andrew et al. (1994). Preparations were examined using both phase and dark-field optics and photographed with TMAX 100 print film (Kodak).
Fly Stocks, Excisional Mutagenesis, Lethal Phase Determination, and Generation of Germ Line Clones
The wild-type flies used in all experiments were Canton S or Oregon R. The P-element insertion alleles, l(3)03559 and l(3)04410, are described in FlyBase (http://www.flybase.org). Excisional mutagenesis to revert the lethality and to create additional alleles of WRS-85D was performed as described (Hamilton and Zinn, 1994). The lethal phase for WRS-85D zygotic loss of function was determined by collecting embryos and counting the number of balancer (Tubby) and nonbalancer (non-Tubby) animals at each developmental stage.
To obtain embryos missing both maternal and zygotic function of WRS-85D, we generated homozygous mutant germ line clones using the dominant female sterile P[ovoD1] flippase (FLP)–FRT recombination technique (Chou and Perrimon, 1996). Females with FRT82B and a WRS-85D loss-of-function allele on the third chromosome were crossed with males with FRT82B and P[ovoD1] on the third chromosome and one copy of hs-flippase (hsFLP) on the X chromosome. hsFLP-induced germ line clone-bearing females were crossed to males heterozygous for a loss-of-function mutation in WRS-85D, and embryos were collected.
Purification of WRS-85D for Aminoacylation Activity Assay
Clone pPS10.1 was digested with EcoRI, and the fragment from position 601-1527 of the cDNA, which contains the remainder of the WRS-85D ORF, was subcloned downstream of and in frame with the 5′ region of WRS-85D in pPS12.2. This construct, pPS17.6, was transformed into BL21(DE3). Expression of the fusion protein was induced as described above. The cells were resuspended in NBB (Invitrogen), sonicated, and centrifuged at 26,000 × g. The supernatant was then purified by Ni-NTA affinity chromatography (Invitrogen). The pooled fractions were dialyzed against 100 mM Tris, pH 8.0, 1 mM EDTA, 10% glycerol. Protein concentration was determined using the BCA assay (Pierce, Rockford, IL). An identical protocol was applied to BL21(DE3) cells transformed with the vector pTrcHisB to obtain mock-purified protein as a negative control.
Determination of Aminoacylation Activity
Twenty-three micrograms of purified WRS-85D protein were combined with 142 mM Tris, pH 8.0, 1.42 mM EDTA, 15 mM MgOAc, 0.05 mg/ml BSA, 0.1 mM [14C]l-tryptophan (54 mCi/mmol; DuPont NEN, Wilmington, DE), 7 mg/ml total tRNA from Brewer’s yeast (Boehringer Mannheim, Indianapolis, IN) and 4.2% glycerol in a 60-μl reaction volume (Bange et al., 1992). ATP was added to 8 mM to initiate the reaction, and the mixture was incubated at 30°C. Aliquots of 9 μl were removed at various time points, precipitated with 5% trichloroacetic acid (TCA)/0.5% Trp, spotted on GF/C filters (Whatman, Clifton, NJ), and washed with 5% TCA. Radioactivity retained on the filters was quantitated with a scintillation counter. Twenty micrograms of mock-purified protein from pTrcHisB-transformed cells was used in parallel experiments as a negative control, and 1.1 μl of reticulocyte lysate (Promega) were used in parallel experiments as a positive control. Each experiment was independently repeated three times. Experiments were also carried out in triplicate using 0.1 mM [14C][scsp]l-leucine to control for enzyme specificity.
RESULTS
Tryptophanyl-tRNA Synthetase (WRS) Is Expressed to High Levels in the Drosophila Salivary Gland
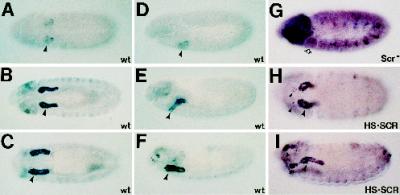
In an enhancer trap screen for genes expressed in the early Drosophila salivary gland, two independent insertions at cytological position 85D7,8 were identified, l(3)03559 and l(3)04410. Both lines presented very high levels of β-gal expression in the secretory portion of the salivary gland with additional low-level staining in a subset of cells in the peripheral nervous system (PNS) (Figure 1, A–F). The salivary gland expression of β-gal in these lines was SCR dependent. In Scr null embryos, we did not detect β-gal expression in the salivary gland primordia even when embryos were overdeveloped to show strong staining in the PNS (Figure 1G). In embryos that carried an HS-SCR transgene, a construct containing the Hsp70 enhancer fused to an Scr cDNA (Zeng et al., 1993), and were heat shocked, we observed ectopic β-gal expression in cells derived from PS0, PS1, and PS14 (Figure 1, H and I). Cells expressing ectopic β-gal are found at approximately the same dorsal–ventral position as salivary gland primordial cells in PS2 (Figure 1H). Furthermore, β-gal–expressing cells derived from PS1 often invaginate and remain attached to the salivary gland cells of PS2 (Figure 1I), suggesting that these cells have adopted a salivary gland fate.
Figure 1.
Expression of enhancer traps in WRS-85D is observed at high levels in the salivary gland and is under the control of the homeotic gene Scr. Embryos have been immunostained with an mAb to β-gal. Embryos in A–C, and H are ventral views; embryos in D–G and I are lateral views. Embryos in A–F show staining dependent on the P-element insertion in the stock l(3)03559 in a wild-type genetic background. In addition to the high-level expression of β-gal in the salivary gland, we also detected expression in the head sensory sensilla that will form the dorsal and ventral organs (F) (Campos-Ortega and Hartenstein, 1997). (G) Immunostaining from the P-element stock in an embryo missing Scr function (an Scr4 homozygote). Note the loss of expression in the salivary gland primordia even when the embryo is significantly overstained to demonstrate both head and PNS staining. (H and I) β-Gal staining in an l(3)03559 embryo where SCR is expressed everywhere under the control of an induced heat-shock promoter (HS·SCR). Note the ectopic β-gal expression in ventrolateral cells of PS1 in H and in PS0, PS1, and PS14 in I. An identical profile of β-gal expression is observed in embryos carrying the l(3)04410 insertion (our unpublished results).
To identify the gene corresponding to the enhancer trap lines, we used plasmid rescue to isolate genomic DNA flanking one side of the P-element insertion site. In situ hybridization to Drosophila polytene chromosomes localized the genomic DNA to the 85D7,8 region of chromosome 3, consistent with the position of the P-element insertions (our unpublished results). We sequenced the junction between the genomic DNA and the P-element and found that in both l(3)03559 and l(3)04410, the P-element had inserted into the identical position in the genome, 103 bp upstream of an ORF. Therefore, we consider l(3)03559 and l(3)04410 to be equivalent.
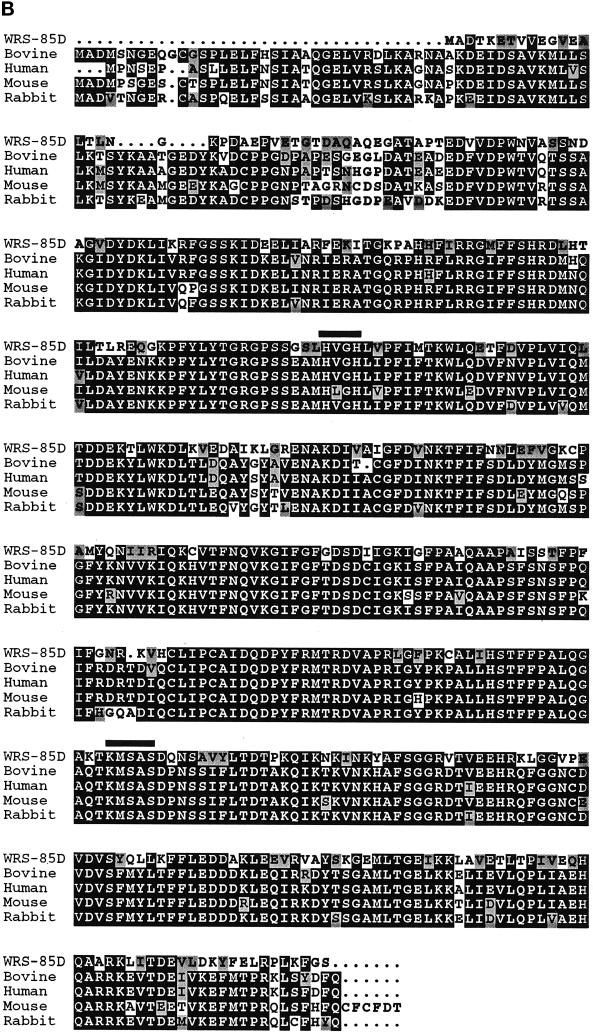
The plasmid rescue DNA was used to isolate several cDNAs ranging in size from 1.2 to 2.6 kb. The 2.6-kb cDNA clone was sequenced in its entirety. Conceptual translation of this cDNA revealed a 430-residue ORF with strong homology to mammalian tryptophanyl-tRNA synthetases (TrpRS/WRS) (BLAST at National Center for Biotechnology Information, Bethesda, MD; Altschul et al., 1997). WRS-85D and mammalian TrpRS are 53% identical and 63% similar (Figure 2B). Tryptophanyl-tRNA synthetases covalently link tryptophan to its cognate tRNA before protein translation. TrpRS is a class I aminoacyl-tRNA synthetase, whose members contain the “HVGH” and “KMSAS” signature sequences (Meinnel et al., 1995; Arnez and Moras, 1997). The HVGH and KMSAS motifs are conserved in WRS-85D, supporting its identification as a tryptophanyl-tRNA synthetase.
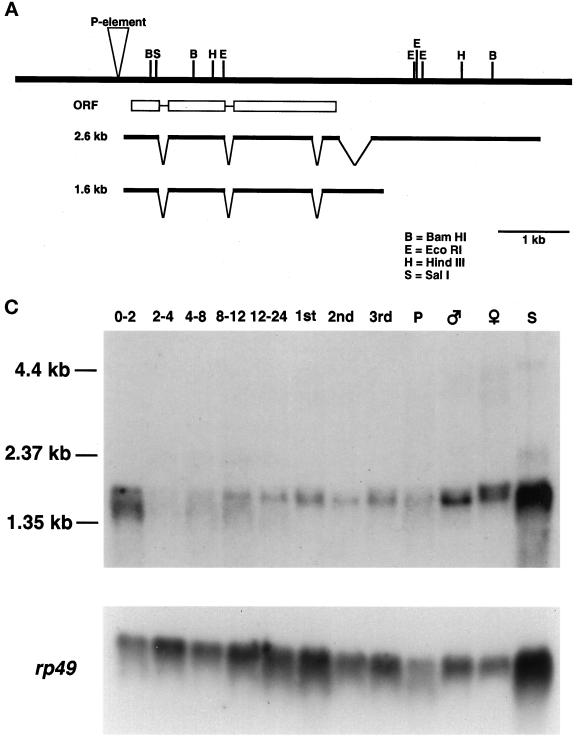
Figure 2.
WRS-85D encodes a protein homologous to mammalian tryptophanyl-tRNA synthetases. (A) Genomic map of WRS-85D showing insertion site of l(3)03559/l(3)04410, cDNA mapping, and intron–exon boundaries. (B) Alignment between WRS-85D and mammalian TrpRS. Residues that are identical are black; residues that are similar are gray. The HVGH and KMSAS signature motifs are indicated with black lines above the sequence. Sequences for two WRS-85D splice forms are available at GenBank (accession numbers BankIt250528 AF125156 and BankIt250542 AF125157). (C) Developmental Northern blot showing expression profile throughout development. 0–2, 2–4, 4–8, 8–12, and 12–24, hours of embryogenesis; 1st, 2nd, and 3rd, three larval stages; P, pupae; S, Drosophila Schneider cells. The same Northern blot is shown hybridized to rp49 probe as a control for loading.
To characterize the expression pattern of WRS-85D, we carried out both developmental Northern analysis and in situ hybridizations to whole-mount embryos. We detected several WRS-85D transcripts throughout development by Northern blot analysis (Figure 2C). The most abundant transcripts are estimated to be 1.65 and 1.8 kb, based on the migration of RNA standards. Because WRS-85D transcripts were detected in 0- to 2-h embryos, the mRNA is most likely to be provided maternally, because zygotic transcription does not begin until ∼2.5 h after egg laying. We also detected WRS-85D transcripts in Drosophila Schneider tissue culture cells.
To examine the spatial expression pattern of WRS-85D, we performed in situ hybridizations on whole-mount embryos. As shown in Figure 3, A–D, we detected abundant transcript levels in the secretory portion of the salivary gland primordia. This high level of expression in salivary gland secretory cells was visible throughout embryogenesis and was consistent with the expression of β-gal from the two P-element insertions in the WRS-85D gene. We were unable to detect WRS-85D transcripts in the PNS probably because of differences in the sensitivity of β-gal versus RNA detection or because of the response of the enhancer trap line to enhancers from other nearby gene(s). At present, we cannot distinguish between these two possibilities. However, consistent with the β-gal expression from the two enhancer trap lines, salivary gland expression of WRS-85D is SCR dependent (our unpublished results).
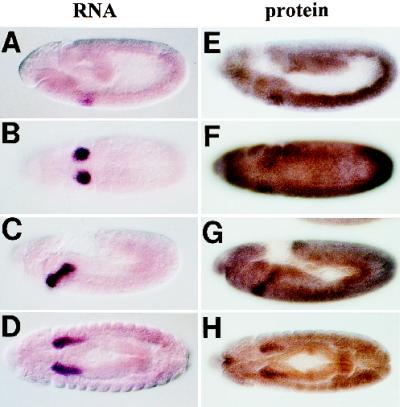
Figure 3.
WRS-85D RNA and protein show similar patterns of accumulation. (A–D) Embryos that have been hybridized with a WRS-85D antisense digoxygenin-labeled RNA probe. (E–H) show embryos that have been immunostained with polyclonal antisera raised against the WRS-85D protein. (A, C, E, and G) Lateral views. (B, D, F, and H) Ventral views. Note the similarity in the patterns observed with the WRS-85D transcript and protein.
We generated antisera to the WRS-85D protein and used it to immunostain whole-mount embryos (Figure 3, E–H). We detected elevated expression of the WRS-85D protein in the salivary gland and its primordia. However, we also detected a global expression pattern of relatively high levels of the protein. We propose that the protein detected in all cells may be partially due to translation of maternally provided WRS-85D transcripts that would be distributed at equivalent levels to all cells of the embryo. Low levels of zygotically transcribed WRS-85D, visible when the detection step of our in situ hybridization reactions was allowed to continue for several hours, might also contribute to overall protein levels. So, although WRS-85D expression is not limited to the salivary gland secretory cells, it is much higher in these cells than in other embryonic tissues.
To determine whether WRS-85D is the only TrpRS gene in Drosophila, we first performed a genomic Southern analysis using four different restriction enzymes and a probe encoding the N-terminal 200 residues, which include the conserved HVGH ATP-binding motif. Even under reduced stringency hybridization conditions, the probe hybridized only to DNA bands that corresponded in size to the WRS-85D locus (our unpublished results). Additionally, our search of the Berkeley Drosophila Genome Project Expressed Sequence Tags database (http://www.fruitfly.org/EST) identified two new WRS cDNAs (LD24552 and GH06221). The ORF sequences available from these two clones were identical matches to the WRS-85D sequence (our unpublished results). Finally, all the cDNAs isolated in our screen hybridized to a single, resolvable locus, WRS-85D at 85D7,8, on polytene chromosomes (our unpublished results). Based on these data, we conclude that WRS-85D is the only tryptophanyl-tRNA synthetase gene in D. melanogaster.
WRS-85D Has tRNATrp Charging (Aminoacylation) Activity
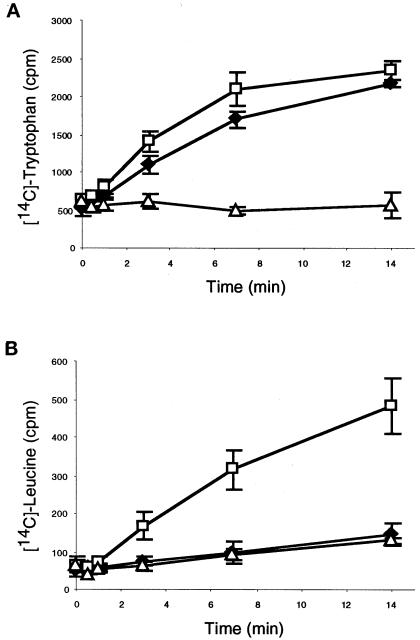
To demonstrate that WRS-85D encodes a functional tryptophanyl-tRNA synthetase, we expressed and purified WRS-85D from bacteria as an N-terminal His6-tagged fusion protein. The fusion protein, purified by Ni-NTA affinity chromatography, migrated as a polypeptide with an apparent molecular mass of 55 kDa on SDS-PAGE gels, close to its predicted mass of 51 kDa (our unpublished results). If WRS-85D possesses tRNATrp charging activity, then we would expect [14C]l-tryptophan to be covalently linked to tRNATrp in an enzymatically dependent manner. When we incubate [14C]l-tryptophan with recombinant WRS-85D, the covalent linkage of [14C]l-tryptophan to tRNATrp can be measured as acid-precipitatable counts on GF/C filters.
We observed increasing amounts of TCA-precipitatable counts over the time course of the assay, indicating that yeast tRNA was tryptophanylated by WRS-85D (Figure 4A). This activity was substrate dependent, because only a background level of counts was obtained when using [14C]l-leucine instead of [14C]l-tryptophan (Figure 4B). As expected, the reaction was ATP dependent; omission of ATP resulted in a complete loss of activity (our unpublished results). The tRNATrp charging activity could be assigned specifically to WRS-85D, because mock-purified protein from bacteria containing only the expression vector pTrcHisB gave a low level of counts that did not increase over time. Therefore, beyond the significant sequence homology to other TrpRS, WRS-85D tryptophanylates tRNA and is a tryptophanyl-tRNA synthetase.
Figure 4.
WRS-85D has aminoacylation activity. Bacterially purified WRS-85D was incubated with total tRNA and [14C]l-tryptophan (A) or [14C]l-leucine (B). Protein purified from cells containing the expression vector pTrcHisB alone was used as a negative control. Reticulocyte lysate was used as a positive control. ♦, WRS-85D; □, reticulocyte lysate; ▵, pTrcHisB. Data are the mean ± SD of three independent experiments for each sample.
WRS-85D Is an Essential Gene
Sequencing the plasmid rescue DNA from l(3)03559 and l(3)04410 revealed that the P-element had inserted, in both cases, 103 bp upstream of the WRS-85D ORF. Both lines are homozygous lethal. To test whether the lethality is due to the insertions and to generate additional alleles of the gene, we initiated an excisional mutagenesis screen selecting for the loss of the rosy+ (ry+) eye color marker contained within the P-element (Hamilton and Zinn, 1994). From 115 independent lines, we obtained 43 lethal lines, 22 semilethal lines, and 29 homozygous viable lines. The recovery of 29 viable lines associated with loss of the P-element suggests that the lethality in both l(3)03559 and l(3)04410 is due to the insertion of the P-element disrupting WRS-85D function and not due to a mutation at another site.
We used complementation tests among the lethal excision lines derived from l(3)03559 and l(3)04410 to identify a set of potential WRS-85D alleles. Based on immunostaining of homozygous WRS-85D mutant embryos with anti-WRS-85D antisera, neither the original insertion alleles l(3)03559 and l(3)04410 nor the lethal alleles that failed to complement the original insertions make detectable levels of WRS-85D in the salivary glands (Table 1). However, WRS-85D levels in the salivary glands in embryos from the viable excisant lines were equivalent to the levels observed in wild-type embryos. These results establish that loss-of-function mutations in WRS-85D are lethal.
Table 1.
WRS-85D expression in excision alleles
| Allele | Original name | Viability | WRS-85D protein in salivary gland |
|---|---|---|---|
| WRS-85D1 | l(3)3559 | Lethal | − |
| WRS-85D2 | l(3)4410 | Lethal | − |
| WRS-85D3 | 3559exc35 | Lethal | − |
| WRS-85D4 | 3559exc50 | Lethal | − |
| WRS-85D5 | 3559exc74 | Lethal | − |
| WRS-85D6 | 4410exc14 | Lethal | − |
| WRS-85D7 | 4410exc18 | Lethal | − |
| WRS-85D8 | 4410exc28B | Lethal | − |
| WRS-85D9 | 4410exc41 | Lethal | − |
| Wild-type | 3559exc25 | Viable | + |
| Wild-type | 4410exc30 | Viable | + |
Immunostaining of embryos homozygous mutant for the lethal WRS-85D alleles with antibodies to nuclear (dCREB-A) or lumenal (CRUMBS) salivary gland markers revealed no overt defects in the salivary gland (our unpublished results). Because these homozygous mutants do not survive to adulthood, we determined the lethal phase by using the Tubby-containing balancer chromosome TM6B to distinguish the homozygous WRS-85D mutant larvae (non-Tb) from their heterozygous (Tb/+) or homozygous (Tb/Tb) balancer siblings. Tb heterozygotes and homozygotes are short and squat relative to non-Tb larvae and pupae. Animals homozygous for two of the four tested WRS-85D alleles died during the larval stages, whereas animals homozygous for the other two alleles died during the larval–pupal transition (our unpublished results). We also examined the cuticles of homozygous mutant larvae and found no overt defects (our unpublished results).
Because WRS-85D encodes a housekeeping gene, we were surprised that the homozygous mutant animals survived embryogenesis and the early larval stages. However, results from the Northern blot analysis and WRS-85D immunostaining indicate that WRS-85D is maternally contributed. To determine whether this maternal contribution is allowing the WRS-85D mutant animals to survive beyond embryogenesis, we used the FLP-FRT system to generate germ line clones (Chou and Perrimon, 1996) that removed the maternal contribution of WRS-85D transcripts. Although we collected ∼3200 females from larvae heterozygous for two protein-null alleles that had been subjected to germ line clone induction, we failed to obtain any eggs from these animals. Thus, WRS-85D must be essential in oogenesis and is likely to be essential in all cells. This result suggests that the maternal contribution of WRS-85D allows animals missing zygotic WRS-85D function to survive to late larval–pupal stages.
Other Aminoacyl-tRNA Synthetases in Drosophila Have Elevated Expression Levels in the Salivary Gland
A possible explanation for the elevated levels of WRS-85D expression in the salivary gland secretory cells is that these cells may synthesize very high levels of protein compared with most cells in the embryo. To accommodate increased levels of protein production in the salivary gland, genes encoding enzymes such as aminoacyl-tRNA synthetases that are required for protein synthesis might be expressed to elevated levels. We thus decided to examine the expression pattern of all 20 aminoacyl-tRNA synthetases. To obtain probes for these genes, we first searched the Berkeley Drosophila Genome Project Expressed Sequence Tags database for cDNAs with homology to aminoacyl-tRNA synthetase genes. We found multiple expressed sequence tags (ESTs) corresponding to each aminoacyl-tRNA synthetase. We then used these ESTs to search GenBank and selected the EST with the highest homology to each aminoacyl-tRNA synthetase. Once we obtained cDNAs corresponding to each of the 20 aminoacyl-tRNA synthetases (Genome Systems, St. Louis, MO; Research Genetics, Huntsville, AL), we sequenced the 5′ ends of each clone and verified that we had indeed obtained the correct cDNA.
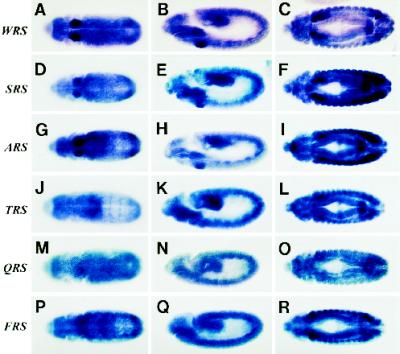
To determine the expression patterns of each Drosophila aminoacyl-tRNA synthetase, we prepared antisense RNA probes for each gene and performed in situ hybridizations to whole-mount embryos. The experiment was carried out in parallel for all 20 genes to eliminate any potential experimental variation in transcript detection. The results are shown in Figure 5 and Table 2. Transcripts were detected in embryos with all 20 probes. Besides WRS-85D, two other aminoacyl tRNA synthetases, seryl-tRNA synthetase and alanyl-tRNA synthetase had elevated levels of expression in the salivary gland primordia that persisted throughout embryogenesis (Figure 5, D–I). Threonyl-tRNA synthetase showed a slight elevation in expression in the salivary gland primordia (Figure 5K), which increased at later stages (Figure 5L). Several other aminoacyl-tRNA synthetase genes showed transiently increased levels of expression in the salivary gland primordia that persisted from approximately embryonic stage 11–12 (stages according to Campos-Ortega and Hartenstein, 1997). Some aminoacyl-tRNA synthetase genes showed no increase in expression in the salivary gland or salivary gland primordia (Figure 5, M–O), and at least two others had high-level expression in other cells, specifically in the embryonic muscle precursors (Figure 5, P–R).
Figure 5.
A limited number of aminoacyl-tRNA synthetases show elevated transcript levels in the early salivary gland. Embryos have been hybridized with digoxygenin-labeled antisense RNA probes to different Drosophila aminoacyl-tRNA synthetase genes. (A–C) WRS-85D; (D–F) seryl-tRNA synthetase; (G–I) alanyl-tRNA synthetase; (J–L) threonyl-tRNA synthetase; (M-O) glutaminyl-tRNA synthetase; (P–R) phenylalanyl-tRNA synthetase. Embryos shown in the left column are stage 11 ventral views. Embryos in the middle column are stage 11 lateral views. Embryos in the right column are stage 13 ventral views.
Table 2.
Expression of aminoacyl-tRNA synthetases during embryogenesis
| Expression pattern during embryogenesis | Aminoacyl-tRNA synthetases |
|---|---|
| Highly and persistently elevated in the salivary gland | WRS, ARS, SRS |
| Moderately and persistently elevated in the salivary gland | TRS |
| Transiently elevated in the salivary gland | RRS, GRS, IRS, YRS, VRS, NRS, DRS, KRS |
| Elevated levels in other tissues | HRS, FRS |
| Equivalent levels in all embryonic tissues | CRS, QRS, ERS, LRS, MRS, PRS |
DISCUSSION
We have identified a tryptophanyl-tRNA synthetase gene (WRS-85D) whose expression is dependent on activity of the homeotic gene Scr using an enhancer trap screen for genes regulated by Scr. Scr is required for salivary gland formation in D. melanogaster. The identity of WRS-85D was confirmed by strong homology with mammalian TrpRS, by conservation of HVGH and KMSAS motifs, and by the ability of recombinant WRS-85D protein to aminoacylate tRNATrp. We have shown that WRS-85D is essential for embryos to survive to adulthood. In addition, we found that a defining feature of WRS-85D is its ele-vated levels in the embryonic salivary gland. Finally, we isolated the remaining 19 aminoacyl-tRNA synthetases and found, by in situ hybridization, accumulation of three other aminoacyl-tRNA synthetases, seryl-, alanyl-, and threonyl-tRNA synthetases, in the salivary gland. However, none approached the level of WRS-85D.
Aminoacyl-tRNA synthetases catalyze the ligation of an amino acid to its cognate tRNA. This reaction requires ATP and occurs in a two-step process with the formation of an enzyme-bound aminoacyladenylate (E:aa∼AMP) intermediate, followed by esterification of the activated amino acid to the tRNA and release of AMP (Meinnel et al., 1995). Aminoacyl-tRNA synthetases are divided into two classes. Class I enzymes contain HVGH and KMSAS signature motifs, which help stabilize the transition state during formation of the aminoacyladenylate (Arnez and Moras, 1997). These housekeeping enzymes are presumably expressed in all cells, which raises the question of why four synthetases are expressed at elevated levels in the developing salivary gland. A simple explanation is that the salivary gland, a secretory organ, requires elevated levels of aminoacyl-tRNA synthetases to accommodate high levels of protein synthesis. A prominent example is the set of glue proteins, secreted by the third instar larval salivary gland, which allow the pupae to adhere to solid substrates in preparation for metamorphosis. Because based on β-gal expression from the P-element insertions, WRS-85D expression is maintained at high levels in the salivary gland throughout larval life (our unpublished results), WRS-85D may be important for the high level of synthesis of the glue proteins. We analyzed the amino acid use among the seven D. melanogaster glue proteins for which sequence was available (NG-1, NG-2, SGS-3, SGS-4, SGS-5, SGS-7, and SGS-8; GenBank accession numbers 134473, 730133, 134467, 1711388, 134470, 72268, and 134472, respectively) and found that the amino acids alanine, serine, and threonine are relatively abundant in these proteins, with mean frequencies of 6.4, 8.1, and 16.2%, respectively. Thus, the elevated expression levels of alanyl-, seryl-, and threonyl-tRNA synthetases observed in the salivary gland may correspond to the production of proteins enriched in the cognate amino acids. However, tryptophan had the lowest amino acid use frequency among all 20 amino acids, with a mean frequency of 0.7%. Similarly, among proteins known to be expressed in the embryonic salivary gland, including αPS3 integrin, CREB, CRUMBS, DHR78, fork head, huckebein, semaphorin II, and sulfurylase (GenBank accession numbers 2914733, 345483, 103119, 1036839, 120228, 743794, 436557, and 2073406, respectively), the mean trp composition is 0.7%. Although this analysis does not rule out the possibility of an undiscovered trp-rich salivary gland protein in Drosophila, it suggests that there may be a different requirement for the elevated levels of tryptophanyl-tRNA synthetase in the salivary gland.
Tryptophanyl-tRNA synthetases possess unusual properties in other eukaryotic systems. One of the most striking is the up-regulation of TrpRS by interferon gamma (IFN-γ) in a number of cell types, including several human cell culture lines (Kisselev et al., 1993; Reano et al., 1993). IFN-γ is a cytokine that mediates both antiproliferative and antiviral effects (Burke et al., 1995). Human WRS contains IFN-stimulating response elements and IFN-γ activation sites, which are cis-acting elements upstream of the start of transcription that are bound by transcription factors activated by IFN stimulation (Frolova et al., 1993; Strehlow et al., 1993; Eilers et al., 1994; Tolstrup et al., 1995). In WRS-85D, we found the sequences TTTCTGTGAA, a very close match to the IFN-γ activation site consensus TTNCNNNA, and CCAATCG in inverse orientation, a perfect match to the Y-box (CTGATTGG), which is necessary for IFN-γ induction of major histocompatibility complex class II genes (Tolstrup et al., 1995). Mammalian TrpRS transcripts are also known to be alternatively spliced and polyadenylated, leading to differences in transcript size of 800 bp (Pajot et al., 1994; Tolstrup et al., 1995; Shen et al., 1996; Turpaev et al., 1996). Likewise, we also isolated an alternatively spliced form of WRS-85D in which the fourth intron is retained; this mRNA is 1 kb shorter and contains an alternative polyadenylation signal (Figure 2A).
Drosophila secrete both antibacterial and antifungal peptides as part of their immune response to infection. A sequence with similarity to the mammalian IFN response element has been shown to positively regulate the promoter of the Drosophila gene for the antibacterial peptide diptericin (Georgel et al., 1995). Additionally, when larvae containing a transgene of the antifungal peptide drosomycin promoter fused to green fluorescnet protein were exposed to a concentrated fungal solution, green fluorescent protein expression was induced in the salivary gland and other tissues (Ferrandon et al., 1998). However, the question of whether a link exists between the induction of the immune response in Drosophila and the expression of the Drosophila TrpRS WRS-85D remains to be answered.
The functional relationship between up-regulation of a housekeeping gene and the pleiotropic effects of IFN-γ is unknown. A possible link is that IFN-γ also up-regulates expression of the enzyme indoleamine 2,3-dioxygenase (IDO), which catabolizes tryptophan (Pfefferkorn et al., 1986). The growth-inhibitory effects of IFN-γ on intracellular parasites have been attributed to the induction of IDO and subsequent depletion of tryptophan (Byrne et al., 1986; Pfefferkorn et al., 1986). In cell lines, the antiproliferative effect of IFN-γ was shown to be most potent in lines in which IDO was induced; the addition of tryptophan to the medium reversed these antiproliferative effects (Burke et al., 1995). It has been proposed that the up-regulation of WRS by IFN-γ may allow host cells to continue protein synthesis in an environment with depleted levels of tryptophan (Flohr et al., 1992). Alternatively, WRS may help sequester TRP into a form that cannot be used for parasite proliferation. The effects of IDO have recently been implicated in prevention of fetal rejection in mice by showing that an IDO inhibitor increases fetal rejection (Munn et al., 1998). It will be interesting to determine whether WRS levels are altered at the maternal–fetal interface and whether addition of exogenous tryptophan also causes an increase in fetal rejection.
All aminoacyl-tRNA synthetases can catalyze the formation of dinucleotide oligophosphates by the back-reaction of ATP or ADP with the E:aa∼AMP intermediate to produce AppppA (Ap4A) or ApppA (Ap3A) (Goerlich et al., 1982). These molecules are called “alarmones,” in reference to the molecules in prokaryotes that accumulate in response to metabolic stress, such as amino acid starvation. In prokaryotes, alarmones initiate cellular changes such as the stringent response, which results in the shutdown of rRNA and tRNA synthesis (Gallant, 1979). Among its effects in eukaryotic systems, ApnA is associated with nuclear functions such as stimulation of DNA synthesis, mitogenic activity, and activation of transcription (Kisselev et al., 1998). Treatment of cell lines with IFN-γ increases the levels of intracellular Ap3A through induction of WRS expression (Merkulova et al., 1994; Vartanian et al., 1996). However, unlike the majority of aminoacyl-tRNA synthetases, mammalian WRS only synthesizes Ap3A.
Furthermore, mutations in a putative tumor suppressor gene, FHIT, which is an Ap3A hydrolase, are found in esophageal, stomach, and colon carcinomas (Barnes et al., 1996; Ohta et al., 1996). Although the cellular pathways of FHIT are not understood, FHIT-substrate–bound complex is likely to be the signaling form of the enzyme (Pace et al., 1998). Whether there is a functional relationship between the antiproliferative effects of IFN-γ and the induction of WRS, an enzyme that can synthesize Ap3A, and the function of FHIT, a protein that potentially requires Ap3A for its activity as a tumor suppressor, remains to be seen. The recent identification of the Drosophila FHIT homologue will allow us to study the interaction between WRS-85D and FHIT (Pekarsky et al., 1998).
The elevated levels of WRS-85D in the salivary gland do not indicate a requirement for synthesis of proteins enriched in tryptophan. However, WRS-85D may be involved in noncanonical functions, such as immune response and control of cell growth. Because Drosophila is highly amenable to genetic analysis, and gene function can be studied within the context of an organ, the salivary gland is a useful model system to determine the roles of tryptophanyl-tRNA synthetase.
ACKNOWLEDGMENTS
We thank Bret Miller for contributions to the early stages of this project. We thank Pierre Coulombe, Cristina Machado, and Wendy Yee for critical comments on the manuscript. We thank M. Fuller, C. Goodman, G. Rubin, M. Scott, and A. Spradling for enhancer trap stocks and the Bloomington Stock Center for FLP-DFS stocks. We thank C. Machado for the Drosophila developmental Northern blot. This work was supported by National Institutes of Health grant RO1-GM51311 and by an institutional research grant from the Johns Hopkins University School of Medicine.
Abbreviations used:
- β-gal
β-galactosidase
- EST
expressed sequence tag
- FLP
flippase
- IDO
indoleamine 2,3-dioxygenase
- IFN
interferon
- PNS
peripheral nervous system
- PS
parasegment
- SCR
sex combs reduced
- TCA
trichloroacetic acid
- Trp
tryptophan
- TrpRS
tryptophanyl-tRNA synthetase
- WRS
tryptophanyl-tRNA synthetase
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DJ. Regulation and formation of the Drosophila salivary glands. Ann NY Acad Sci. 1998;842:55–69. doi: 10.1111/j.1749-6632.1998.tb09632.x. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Horner MA, Petitt MG, Smolik SM, Scott MP. Setting limits on homeotic gene function: restraint of Sex combs reduced activity by teashirt and other homeotic genes. EMBO J. 1994;13:1132–1144. doi: 10.1002/j.1460-2075.1994.tb06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DJ, Scott MP. Downstream of the homeotic genes. New Biol. 1992;4:5–15. [PubMed] [Google Scholar]
- Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- Bange FC, Flohr T, Buwitt U, Bottger EC. An interferon-induced protein with release factor activity is a tryptophanyl-tRNA synthetase. FEBS Lett. 1992;300:162–166. doi: 10.1016/0014-5793(92)80187-l. [DOI] [PubMed] [Google Scholar]
- Barnes LD, Garrison PN, Siprashvili Z, Guranowski A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K. Fhit, a putative tumor suppressor in humans, is a dinucleoside 5′:5‴-P1,P3-triphosphate hydrolase. Biochemistry. 1996;35:11529–11535. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, O’Kane C, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes & Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Bier E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes & Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Biggin MD, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- Burke F, Knowles RG, East N, Balkwill FR. The role of indoleamine 2,3-dioxygenase in the antitumor activity of human interferon-gamma in vivo. Int J Cancer. 1995;60:115–122. doi: 10.1002/ijc.2910600117. [DOI] [PubMed] [Google Scholar]
- Byrne GI, Lehmann LK, Landry GJ. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect Immun. 1986;53:347–351. doi: 10.1128/iai.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. 2nd ed. Berlin: Springer–Verlag; 1997. [Google Scholar]
- Chou T, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan IM. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- Eilers A, Baccarini M, Horn F, Hipskind RA, Schindler C, Decker T. A factor induced by differentiation signals in cells of the macrophage lineage binds to the gamma interferon activation site. Mol Cell Biol. 1994;14:1364–1373. doi: 10.1128/mcb.14.2.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohr T, Bange FC, von Euch A, Kiekenbeck M, Bottger EC. Depletion of tryptophan is not involved in expression of tryptophanyl-tRNA synthetase mediated by interferon. Infect Immun. 1992;60:4418–4421. doi: 10.1128/iai.60.10.4418-4421.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Grigorieva AY, Sudomoina MA, Kisselev LL. The human gene encoding tryptophanyl-tRNA synthetase: interferon-response elements and exon-intron organization. Gene. 1993;128:237–245. doi: 10.1016/0378-1119(93)90568-n. [DOI] [PubMed] [Google Scholar]
- Gallant JA. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- Georgel P, Kappler C, Langley E, Gross I, Nicolas E, Reichhart JM, Hoffmann JA. Drosophila immunity: a sequence homologous to mammalian interferon consensus response element enhances the activity of the diptericin promoter. Nucleic Acids Res. 1995;23:1140–1145. doi: 10.1093/nar/23.7.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerlich O, Foeckler R, Holler E. Mechanism of synthesis of adenosine(5′)tetraphospho(5′)adenosine (AppppA) by aminoacyl-tRNA synthetases. Eur J Biochem. 1982;126:135–142. doi: 10.1111/j.1432-1033.1982.tb06757.x. [DOI] [PubMed] [Google Scholar]
- Graba Y, Aragnol D, Pradel J. Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays. 1997;19:379–388. doi: 10.1002/bies.950190505. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Bellen HJ, Wilson C, Gehring WJ. P-element-mediated enhancer detection applied to the study of oogenesis in Drosophila. Development. 1989;107:189–200. doi: 10.1242/dev.107.2.189. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Zinn K. From Clone to Mutant Gene. 1st ed. Vol. 44. L.S.B. Goldstein and E.A. Fyrberg, New York: Academic Press; 1994. [DOI] [PubMed] [Google Scholar]
- Henderson KD, Andrew DJ. Identification of a novel Drosophila SMAD on the X-chromosome. Biochem Biophys Res Commun. 1998;252:195–201. doi: 10.1006/bbrc.1998.9562. [DOI] [PubMed] [Google Scholar]
- Henderson KD, Isaac DD, Andrew DJ. Cell fate specification in the Drosophila salivary gland: the integration of homeotic gene function with the DPP signaling cascade. Dev Biol. 1998;205:10–21. doi: 10.1006/dbio.1998.9113. [DOI] [PubMed] [Google Scholar]
- Higgins DG. ClustalV: Multiple Alignment of DNA and Protein Sequence. Totowa, NJ: Human Press; 1993. [DOI] [PubMed] [Google Scholar]
- Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes & Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organization of the Antennapedia gene complex of Drosophila melanogaster. Adv Genet. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- Kisselev L, Frolova L, Haenni AL. Interferon inducibility of mammalian tryptophanyl-tRNA synthetase: new perspectives. Trends Biochem Sci. 1993;18:263–267. doi: 10.1016/0968-0004(93)90178-p. [DOI] [PubMed] [Google Scholar]
- Kisselev LL, Justesen J, Wolfson AD, Frolova LY. Diadenosine oligophosphates (Ap(n)A), a novel class of signaling molecules? FEBS Lett. 1998;427:157–163. doi: 10.1016/s0014-5793(98)00420-7. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–200. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Krumlauf R, Holland P, McVey J, Hogan B. Developmental and spatial patterns of expression of the mouse homeobox gene Hox 2.1. Development. 1987;99:603–617. doi: 10.1242/dev.99.4.603. [DOI] [PubMed] [Google Scholar]
- Laird C, McConaghy B, McCarthy B. Nucleic acid reassociation in formamide. Biochemistry. 1969;8:3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. San Diego: Academic Press; 1994. , 755. [DOI] [PubMed] [Google Scholar]
- Levine M, Hoey T. Homeobox proteins as sequence-specific transcription factors. Cell. 1988;55:537–540. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd ed. Vol. 1. 1989. –3, ed. C. Nolan, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Mechulam Y, Blanquet S. Aminoacyl-tRNA synthetases: occurrence, structure, and function. In: Soll D, Rajbhandary U, editors. tRNA: Structure, Biosynthesis, and Function. Washington, DC: American Society for Microbiology; 1995. pp. 251–293. [Google Scholar]
- Merkulova T, Kovaleva G, Kisselev L. P1,P3-bis(5′-adenosyl)triphosphate (Ap3A) as a substrate and a product of mammalian tryptophanyl-tRNA synthetase. FEBS Lett. 1994;350:287–290. doi: 10.1016/0014-5793(94)00764-0. [DOI] [PubMed] [Google Scholar]
- Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- O’Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Pace HC, et al. Genetic, biochemical, and crystallographic characterization of Fhit-substrate complexes as the active signaling form of Fhit. Proc Natl Acad Sci USA. 1998;95:5484–5489. doi: 10.1073/pnas.95.10.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajot B, Sarger C, Bonnet J, Garret M. An alternative splicing modifies the C-terminal end of tryptophanyl-tRNA synthetase in murine embryonic stem cells. J Mol Biol. 1994;242:599–603. doi: 10.1006/jmbi.1994.1608. [DOI] [PubMed] [Google Scholar]
- Panzer S, Weigel D, Beckendorf SK. Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development. 1992;114:49–57. doi: 10.1242/dev.114.1.49. [DOI] [PubMed] [Google Scholar]
- Pardue M-L. Looking at polytene chromosomes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. San Diego: Academic Press; 1994. , 755. [Google Scholar]
- Pekarsky Y, et al. Nitrilase and Fhit homologs are encoded as fusion proteins in Drosophila melanogaster and Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:8744–8749. doi: 10.1073/pnas.95.15.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER, Rebhun S, Eckel M. Characterization of an indoleamine 2,3-dioxygenase induced by gamma-interferon in cultured human fibroblasts. J Interferon Res. 1986;6:267–279. doi: 10.1089/jir.1986.6.267. [DOI] [PubMed] [Google Scholar]
- Reano A, Richard MH, Denoroy L, Viac J, Benedetto JP, Schmitt D. Gamma interferon potently induces tryptophanyl-tRNA synthetase expression in human keratinocytes. Journal of Investigative Dermatology. 1993;100:775–779. doi: 10.1111/1523-1747.ep12476463. [DOI] [PubMed] [Google Scholar]
- Rio DC, Laski FA, Rubin GM. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell. 1986;44:21–32. doi: 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Scott MP, Tamkun JW, Hartzell GW., III The structure and function of the homeodomain. Biochim Biophys Acta Rev Cancer. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Shen T, Anderson SL, Rubin BY. Use of alternative polyadenylation sites in the synthesis of mRNAs encoding the interferon-induced tryptophanyl tRNA synthetase. Gene. 1996;179:225–229. doi: 10.1016/s0378-1119(96)00361-7. [DOI] [PubMed] [Google Scholar]
- Strehlow I, Seegert D, Frick C, Bange FC, Schindler C, Bottger EC, Decker T. The gene encoding IFP 53/tryptophanyl-tRNA synthetase is regulated by the gamma-interferon activation factor. J Biol Chem. 1993;268:16590–16595. [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Kahn RA, Kissinger M, Bruzuela BJ, Rulka C, Scott MP, Kennison JA. The arf-like gene encodes an essential GTP-binding protein in Drosophila. Proc Natl Acad Sci USA. 1991;88:3120–3124. doi: 10.1073/pnas.88.8.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup AB, Bejder A, Fleckner J, Justesen J. Transcriptional regulation of the interferon-gamma-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J Biol Chem. 1995;270:397–403. doi: 10.1074/jbc.270.1.397. [DOI] [PubMed] [Google Scholar]
- Turpaev KT, Zakhariev VM, Sokolova IV, Narovlyansky AN, Amchenkova AM, Justesen J, Frolova LY. Alternative processing of the tryptophanyl-tRNA synthetase mRNA from interferon-treated human cells. Eur J Biochem. 1996;240:732–737. doi: 10.1111/j.1432-1033.1996.0732h.x. [DOI] [PubMed] [Google Scholar]
- Vartanian A, Narovlyansky A, Amchenkova A, Turpaev K, Kisselev L. Interferons induce accumulation of diadenosine triphosphate (Ap3A) in human cultured cells. FEBS Lett. 1996;381:32–34. doi: 10.1016/0014-5793(96)00073-7. [DOI] [PubMed] [Google Scholar]
- Wilson C, Pearson RK, Bellen HJ, O’Kane CJ, Grossniklaus U, Gehring WJ. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes & Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- Zeng W, Andrew DJ, Mathies LD, Horner MA, Scott MP. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functionall specificity. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- Zinn K, McAllister L, Goodman CS. Sequence analysis and neuronal expression of Fasciclin I in grasshopper and Drosophila. Cell. 1988;53:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]