Abstract
Addition of ammonium ions to yeast cells growing on proline as the sole nitrogen source induces rapid inactivation and degradation of the general amino acid permease Gap1 through a process requiring the Npi1/Rsp5 ubiquitin (Ub) ligase. In this study, we show that NH4+ induces endocytosis of Gap1, which is then delivered into the vacuole where it is degraded. This down-regulation is accompanied by increased conversion of Gap1 to ubiquitinated forms. Ubiquitination and subsequent degradation of Gap1 are impaired in the npi1 strain. In this mutant, the amount of Npi1/Rsp5 Ub ligase is reduced >10-fold compared with wild-type cells. The C-terminal tail of Gap1 contains sequences, including a di-leucine motif, which are required for NH4+-induced internalization and degradation of the permease. We show here that mutant Gap1 permeases affected in these sequences still bind Ub. Furthermore, we provide evidence that only a small fraction of Gap1 is modified by Ub after addition of NH4+ to mutants defective in endocytosis.
INTRODUCTION
In eukaryotic cells, the degradation of many proteins requires their initial modification by conjugation to a 76-amino acid peptide called ubiquitin (Ub). In general, the covalent bond between Ub and lysine residues marks the substrate proteins for degradation by the 26S proteasome (reviewed by Hochstrasser, 1996). In the yeast Saccharomyces cerevisiae, involvement of the Ub pathway has been demonstrated in the degradation of several soluble proteins such as the Matα2 repressor (Chen et al., 1993), the Gcn4 transcriptional activator (Kornitzer et al., 1994), the fructose 1,6-bisphosphatase (Schork et al., 1995), and several cyclins (Deshaies et al., 1995; Seufert et al., 1995; Yaglom et al., 1995). There is now growing evidence, both in higher and lower eukaryotes, that Ub may also be used as a signal for endocytosis of some cell surface proteins and their subsequent degradation in the lysosome and vacuole (Hochstrasser, 1996). For instance, the yeast α-peptide transporter Ste6 is stabilized and accumulates in a ubiquitinated form in end4 cells defective in the internalization step of endocytosis (Kölling and Hollenberg, 1994). Evidence supporting a role of Ub in turnover of cell surface proteins was further provided by Hicke and Riezman (1996) in the case of ligand-induced endocytosis of the Ste2 pheromone receptor. In a C-terminally truncated form of Ste2 still competent for ligand-induced endocytosis, substitution of a single lysine residue for arginine within a DAKSS sequence was shown to impair both ligand-induced ubiquitination and endocytosis of the receptor (Hicke and Riezman, 1996). Other recent experiments in yeast suggest that Ub is used as a signal for endocytosis of the uracil permease Fur4 (Galan et al., 1996), the galactose permease Gal2 (Horak and Wolff, 1997), the pheromone receptor Ste3 (Roth and Davis, 1996), and the multidrug resistance protein Pdr5 (Egner and Kuchler, 1996). The mechanism by which Ub promotes endocytosis is still unknown. Whether binding of Ub to a cell surface protein constitutes a sufficient signal for endocytosis also remains undetermined. It has been suggested that Ub might be recognized by a component of the endocytosis machinery or might promote movement of ubiquitinated proteins into membrane regions that actively endocytose (Hicke and Riezman, 1996).
This report focuses on the regulation of turnover of the general amino acid permease (Gap1) in S. cerevisiae. The synthesis, the activity, and more recently the sorting of this permease in the late secretory pathway have been shown to be regulated according to the nitrogen source used by the cells (Wiame et al., 1985; Grenson, 1992; Roberg et al., 1997). Upon addition of NH4+ to cells grown on a less-favored nitrogen source such as proline, synthesis of the Gap1 permease is strongly reduced, and presynthesized permease is completely inactivated and degraded (Grenson, 1983a; Hein et al., 1995). The products of the NPI1 and NPI2 genes are required for both inactivation and degradation of Gap1. We have previously shown that NPI1 is an essential gene encoding the Ub–protein ligase Rsp5 (Huibregtse et al., 1995); this suggests that the Ub pathway is involved in the turnover regulation of Gap1 (Hein et al., 1995). In keeping with a role of Npi1/Rsp5 in permease turnover, this enzyme is required for basal and stress-induced ubiquitination and degradation of the uracil permease Fur4 (Galan et al., 1996). Several mutations affecting the C-terminal hydrophilic tail render the Gap1 permease resistant to NH4+-triggered inactivation and degradation (Hein and André, 1997); one is a di-leucine→di-alanine substitution (Gap1LL→AA). In higher eukaryotic cells, the di-leucine motif has been shown to act as a signal for internalization of several cell surface proteins (Shin et al., 1991; Letourneur and Klausner, 1992; Aiken et al., 1994; Haft et al., 1994; Dittrich et al., 1996). A role of di-leucine in targeting membrane proteins to endosome and lysosome has also been reported (Sandoval and Bakke, 1994). The di-leucine of Gap1 is located in a region predicted to adopt a stable α-helical conformation. This putative helix also contains a glutamate residue which, when replaced by a lysine, leads to resistance of Gap1 to NH4+-induced inactivation and degradation. Finally, a Gap1 permease lacking the last 11 amino acids directly following the putative α-helix also remains active and stable after addition of NH4+ to the medium (Hein and André, 1997).
The present work shows that a small fraction of Gap1 is ubiquitinated in cells grown on proline medium. Addition of NH4+ increases the conversion of Gap1 to Ub-conjugated forms. This conversion is followed by rapid internalization of the permease and subsequent degradation in the vacuole. In npi1 mutant cells, in which the level of Npi1/Rsp5 Ub ligase is much reduced, Gap1 ubiquitination is impaired, and the permease remains stable on the plasma membrane. Finally, mutations affecting the C-terminal tail of Gap1 impair NH4+-induced endocytosis and degradation of Gap1, but the mutant permeases are still ubiquitinated.
MATERIALS AND METHODS
Strains, Growth Conditions, and Plasmids
S. cerevisiae strains isogenic with the wild-type Σ1278b (Béchet et al., 1970) are 24346c (MATa, ura3), 27061b (MATa, ura3, trp1), 27038a (MATa, ura3, npi1) (Grenson, 1983a), JOD0097 (MATa, ura3, gap1::kanMX2), and RTY1 (MATa, ura3, trp1, pep4::kanMX4). Strains nonisogenic with Σ1278b are NY279 (MATa, ura3-52, act1-1) and its isogenic parental strain NY13 (MATa, ura3-52) (Shortle et al., 1984; Goud et al., 1988). Cells were grown in minimal buffered medium (pH 6.1) with 3% glucose as the carbon source (Jacobs et al., 1980). Nitrogen sources were added as indicated at the following final concentrations: (NH4)2SO4, 10 mM; proline, 0.1%. The YEpJYS-2 plasmid contains the NPI1 gene of the YCpJYS-1 plasmid (Hein et al., 1995), inserted into the 2μ-based multicopy vector pFL44 (Bonneaud et al., 1991). The YCpGAP1 plasmid contains the GAP1 gene (Jauniaux and Grenson, 1990) in the centromere-based vector pFL38 (Bonneaud et al., 1991). YCpGap1pgr, YCpGAP1LL→AA, and YCpGAP1Δ2 are modified versions of plasmid YCpGAP1, encoding altered Gap1 permeases (respective alterations: E582→K582, L575 L576→A575 A576, and truncation of the last 11 amino acids; Hein and André, 1997). The 2μ-based multicopy plasmid YEp96 contains a synthetic yeast Ub gene under the control of the copper-inducible CUP1 promoter; YEp105 is identical to YEp96 except that it encodes a c-Myc-tagged version of Ub (Hochstrasser et al., 1991). Yeast cells treated with lithium acetate (Ito et al., 1983) were transformed according to the method of Sherman et al. (1986). The Escherichia coli strain used was JM109. All procedures for manipulating DNA used standard methods (Ausubel et al., 1995; Sambrook et al., 1997).
Permease Assays
Gap1 permease activity was determined by measuring incorporation of 14C-labeled citrulline as described by Grenson (1966). All permease activities were measured in cells that had reached the state of balanced growth (Wiame et al., 1985). The permease was inactivated by adding prewarmed (NH4)2SO4 to the culture at a final concentration of 10 mM.
Yeast Cell Extracts and Immunoblotting
Crude cell extracts were prepared as previously described (Hein et al., 1995). For membrane-enriched preparations, ∼108 yeast cells were filtered (Millipore, Bedford, MA; 0.45 μm), washed with cold water plus NaN3 (10 mM), and resuspended in 0.2 ml lysis buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, 5 mM EDTA) containing the following proteinase inhibitors: 100 μg/ml PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 10 mM NaN3, 50 mM N-ethyl maleimide. An equal volume of glass beads (0.45 μm) was added, and the cells were lysed at 4°C in a 2.0-ml Eppendorf (Madison, WI) tube by vortex mixing for 2 min. The extracts were diluted with 2 volumes of lysis buffer, transferred to new tubes, and centrifuged for 3 min at 3000 rpm. The membrane-enriched fraction was obtained from the supernatant as described elsewhere (Galan et al., 1996). For Western blot analysis, 10 μl of solubilized proteins were loaded on a 12% SDS-polyacrylamide gel in a Tricine system (Schägger and von Jagow, 1987). For the experiment showing the shift of the ubiquitinated form of Gap1 upon expression of Ub-myc, the extracts were loaded on a 12% gel in Laemmli’s system (Laemmli, 1970). After transfer to nitrocellulose the proteins were probed with rabbit antiserum raised against the N-terminal region of GAP1 (1:20,000) or against the mouse protein Nedd4 (1:1000) (Kumar et al., 1997). Anti-Gap1 antibodies where shown elsewhere to be specific of Gap1 protein (De craene and André, unpublished data). Primary antibodies were detected with horseradish peroxidase–conjugated anti-rabbit-IgG secondary antibody followed by enhanced chemoluminescence (Amersham, Arlington Heights, IL). All Western blots were quantitated with a densitometer to measure protein amounts.
RESULTS
Ammonium-induced Endocytosis and Vacuolar Degradation of the Gap1 Permease
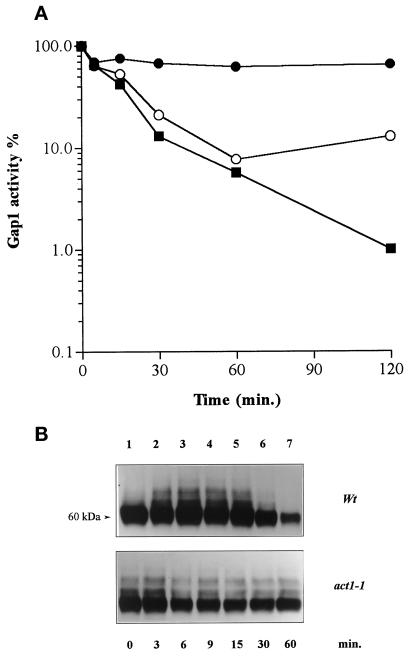
Addition of NH4+ to wild-type cells growing on proline as the sole nitrogen source induces loss of Gap1 permease activity and degradation of the permease (Grenson, 1983a; Hein et al., 1995). As previously reported for other yeast cell surface proteins (Schandel and Jenness, 1994; Volland et al., 1994; Egner et al., 1995; Riballo et al., 1995; Hicke and Riezman, 1996; Roth and Davis, 1996; Horak and Wolf, 1997), this degradation could take place in the vacuole after internalization of the permease. NH4+-induced loss of measurable Gap1 activity could thus reflect progressive removal of Gap1 from the plasma membrane. Alternatively, Gap1 could be inactivated before its internalization. To further elucidate the mechanisms underlying nitrogen-regulated turnover of the Gap1 permease, we assayed the activity of Gap1 in a strain carrying a thermosensitive mutation, act1-1, in the actin gene (Shortle et al., 1984). This mutant is defective in the internalization step of receptor-mediated endocytosis of the α-factor (Kübler and Riezman, 1993). It is also defective in endocytosis of the uracil permease (Fur4), observed after inhibition of protein synthesis by cycloheximide (Galan et al., 1996). The act1-1 mutation has a strong effect at 37°C, a temperature at which Gap1 is partially inactivated, so we performed the experiment at 29°C, at which temperature act1-1 cells still exhibit defective endocytosis, although the effect is less pronounced than at 37°C (Kübler and Riezman, 1993; Galan et al., 1996). The results show that NH4+-induced inactivation of Gap1 is severely impaired in the act1-1 mutant compared with the wild type (Figure 1A). An internalization step thus seems required for complete NH4+-triggered loss of Gap1 activity. In other words, the so-called nitrogen catabolite inactivation of Gap1 (Grenson, 1983a) is likely the result of progressive internalization of the permease by endocytosis.
Figure 1.
Ammonium-induced down-regulation of Gap1 is impaired in act1-1 mutant cells. (A) Cells were grown on proline medium, and Gap1 activity was assayed by measuring incorporation of [14C]citrulline (0.1 mM) before (t = 0) and at various times after addition of (NH4+)2SO4 (10 mM) in strains NY13 (wild type; ○), NY279 (act1-1; •), and 24346c (wild type derived from Σ1278b; ▪). The Gap1 activities were calculated in nanomoles per minute per milliliter to avoid dilution effect due to NH4+-triggered arrest of Gap1 synthesis. In proline-grown cells, initial Gap1 activity is lower in act1-1 cells than in isogenic wild-type cells (1.3 vs. 4.2 nmol · min−1 · ml−1). (B) Immunoblot of Gap1 from membrane-enriched cell fractions prepared before (t = 0) and at several times after addition of (NH4)2SO4. Note that the disapearance of Gap1 signal in the NY13 wild-type strain is slower compared with the wild type derived from Σ1278b.
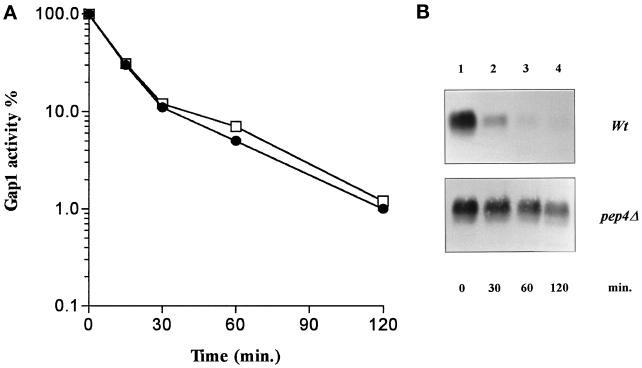
To determine whether degradation of Gap1 after internalization takes place within the vacuole, we examined Gap1 in yeast cells with a defective vacuolar proteinase A (pep4 mutant). This proteinase is required for maturation of several vacuolar proteases (Ammerer et al., 1986; Woolford et al., 1986). In the pep4Δ strain, Gap1 is inactivated by NH4+ as efficiently as in the isogenic wild type (Figure 2A). The amount of Gap1 protein in crude cellular extracts was analyzed using antibodies raised against the N-terminal region of the permease. The unique signal immunodected in wild-type cells corresponds to the Gap1 protein, as no signal is visible in a gap1Δ strain (our unpublished observations). Addition of NH4+ to cells growing on proline as the sole nitrogen source led to rapid degradation of the Gap1 protein. In the pep4Δ mutant, however, Gap1 was strongly protected against NH4+-induced degradation (Figure 2B). These results indicate that after internalization, Gap1 is targeted for vacuolar proteolytic breakdown. The phenomenon including internalization and subsequent degradation of preaccumulated Gap1 will be henceforth referred to as “down-regulation” of the permease.
Figure 2.
Ammonium-induced degradation of Gap1 is dependent on vacuolar proteases. (A) Cells were grown on proline medium, and Gap1 activity was measured by incorporation of [14C]citrulline (0.1 mM) before (t = 0) and at several times after addition of (NH4+)2SO4 (10 mM) in strains RTY1 (pep4Δ; •) and 27061b (wild type; □). The Gap1 activities were calculated in nanomoles per minute per milliliter. (B) Immunoblot of Gap1 in crude extracts prepared before (t = 0) and at several times after addition of (NH4)2SO4.
A High Amount of Npi1/Rsp5 Ub Ligase Is Required for Ammonium-induced Endocytosis and Degradation of Gap1 Permease
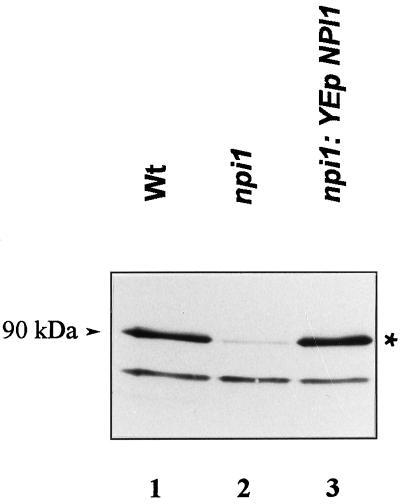
In npi1 mutant cells, the Gap1 permease is known to remain active (i.e., plasma membrane located) and stable after addition of NH4+ (Grenson, 1983a; Hein et al., 1995). In the mutant used here, a Ty1 transposon is inserted 500 bp upstream from the translation initiation codon of the NPI1/RSP5 gene. This Ty1 insertion results in a reduced NPI1 transcript level, raising the possibility that a fairly large amount of Npi1 is required for NH4+-induced down-regulation of Gap1 (Hein et al., 1995). This has now been confirmed using polyclonal antibodies raised against Nedd4, the mouse homologue of Npi1 (Kumar et al., 1992, 1997). In Western blot experiments performed with crude extract of wild-type cells, these antibodies detected two polypeptides, one at ∼80 kDa and one at ∼90 kDa (Figure 3, lane 1). The apparent mass of the upper band is consistent with the predicted molecular mass of Npi1 (91.8 kDa). The amount of this upper band is reduced >10-fold in the npi1 strain (Figure 3, lane 2), but it reaches a normal level in npi1 cells transformed with a high-copy number plasmid bearing a complete NPI1 gene demonstrating that the 90-kDa signal does correspond with the Npi1 Ub ligase (Figure 3, lane 3). npi1 cells transformed with the NPI1-bearing plasmid also show restored sensitivity of Gap1 to NH4+ regulation (Hein et al., 1995). Thus, the reduced amount of Npi1 present in npi1 cells, although sufficient to ensure cell viability, is limiting for NH4+-induced down-regulation of the permease. Also consistent with this conclusion is the observation that when Gap1 was assayed in npi1 cells transformed with either a low- or a high-copy number plasmid bearing the promoter-truncated npi1 gene, only multiple copies of the npi1 gene were able to restore NH4+-induced loss of Gap1 activity (our unpublished results).
Figure 3.
The amount of Npi1 Ub ligase is reduced in npi1 cells. Crude extracts were prepared from cells grown on proline medium, resolved by electrophoresis, and blotted onto a nitrocellulose membrane. The blot was probed with polyclonal Nedd4 antibodies (Kumar et al. 1997). The strains used were 24346c (wild type; lane 1), 27038a (npi1; lane 2), and 27038a (npi1) transformed with plasmid YEpJYS-2 carrying the NPI1 gene (lane 3). The signal corresponding to Ub ligase Npi1/Rsp5 is marked with an asterisk.
Ammonium Induces Npi1-dependent Ubiquitination of Gap1 Permease
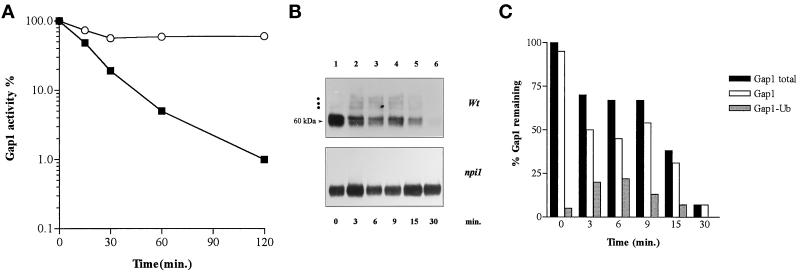
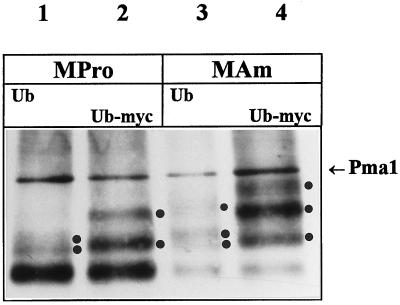
The fact that efficient NH4+-induced down-regulation of Gap1 requires the presence of Npi1 Ub ligase in relatively high amounts indicates that Ub must be involved in this regulatory process. As previously suggested for several other yeast cell surface proteins (Egner and Kuchler, 1996; Galan et al., 1996; Hicke and Riezman, 1996; Roth and Davis, 1996), conjugation of Ub to Gap1 might constitute a signal required for endocytosis of the permease. Ubiquitination of Gap1 was tested by Western blotting analysis of membrane-enriched extracts. These preparations contained >90% of the plasma membrane H+-ATPase Pma1 immunodetectable in crude extracts (our unpublished observations) and possibly membrane of other compartments. The Gap1 signal detected in the proline-grown wild-type strain consists of two intense bands at ∼60 kDa, which we shall call the major Gap1 signal, and at least one minor band at ∼70 kDa, which is visible if exposure is long enough (Figure 4B, lane 1). A second minor band just above the first and additional bands of still higher molecular weight were also detected upon still longer exposure (our unpublished observations). The difference of ∼10 kDa between the major Gap1 signal and the first minor band is about as one would expect if a Ub molecule (∼9 kDa) is linked to the permease. In accordance with this prediction, there appear no minor bands in the lanes corresponding to npi1 mutant cells. After addition of NH4+ to wild-type cells, in parallel with a decrease in the intensity of the major Gap1 signal, a ladder consisting of the first two minor bands plus a third one becomes clearly visible (Figure 4B). Quantitation of the immunoblot signals indicates that the upper minor bands represent up to 25% of the total Gap1 signal (Figure 4C). The three minor bands most probably correspond with Ub-conjugated forms of the permease, as they are barely detectable in extracts of npi1 cells. To test this assumption, we prepared membrane-enriched fractions from the wild-type strain overexpressing either normal Ub or an epitope-tagged form of Ub (Ub-myc). Because Ub-myc is larger than Ub, proteins binding Ub-myc instead of Ub are retarded in a gel mobility assay (Hochstrasser et al., 1991; Galan et al., 1996; Roth and Davis, 1996). We carried out a Western blot experiment with modifying electrophoresis conditions to improve signal detection. The results are shown in Figure 5. The Gap1 signal obtained with proline-grown cells overexpressing normal Ub consists of a major band, resolved as a doublet upon longer migration, and two additional minor bands of higher molecular weight (Figure 5, lane 1). Extracts of cells harvested 5 min after NH4+ addition display a less-intense major Gap1 signal and a third additional minor band of higher molecular weight (Figure 5, lane 3). These results are approximately similar to those obtained without overexpression of Ub (Figure 4). In contrast, overexpression of Ub-myc does alter the migration pattern of the minor bands, whether the cells are grown on proline or ammonium ions; these minor bands are more intense, and some are shifted to a higher molecular weight (Figure 5, lanes 2 and 4). Specifically, in both proline- and NH4+-grown cells, the second minor band is clearly retarded, indicating that this signal corresponds to a ubiquitinated form of the permease (Figure 5, lane 2 vs. lane 1 and lane 4 vs. lane 3). The third minor band appearing after NH4+ addition also shifts to higher molecular weight upon expression of Ub-myc. Although the first minor band, directly above the major Gap1 signal, is retarded only slightly if at all upon expression of Ub-myc, it does most likely correspond to a ubiquitinated form of Gap1, because it is not detected in npi1 cells, and the corresponding molecular weight is as expected for a monoubiquitinated form of Gap1. Taken together, these results show that minor bands just above the major Gap1 signal must indeed correspond with Ub-conjugated forms of the Gap1 permease. The fact that the intensity of the Ub-conjugated forms increases upon expression of Ub-myc suggests that the presence of the myc epitope on Ub leads to stabilization of at least some ubiquitinated forms of the Gap1 permease. A similar stabilization effect has been observed for Ub-conjugated forms of the uracil permease (Haguenauer-Tsapis and Galan, personal communication).
Figure 4.
Ubiquitination of Gap1 is impaired in npi1 cells. (A) Cells were grown on proline medium, and Gap1 activity was measured by incorporation of [14C]citrulline (0.1 mM) before (t = 0) and at several times after addition of (NH4)2SO4 (10 mM) in strains 24346c (wild type; ▪) and 27038a (npi1; ○). The Gap1 activities were expressed in nanomoles per minute per milliliter. (B) Immunoblot of Gap1 from membrane-enriched cell fractions prepared before (t = 0) and at several times after addition of (NH4)2SO4. The major Gap1 signal is composed of two bands at ∼60 kDa, and the positions of ubiquitinated forms are indicated with dots. (C) Quantitation of the Gap1 signal in immunoblot shown in the upper part of B (wild-type cells). The immunoblot was scanned with a densitometer to measure the amount of total and ubiquitinated Gap1 present at each time point. Each value is expressed as percentage of the Gap1 found in all forms at t = 0 min.
Figure 5.
Effect of Ub and Ub-myc expression on the ubiquitination profile of Gap1. Cells of strain 27061b (wild type) bearing plasmid YEp96 (Ub; lanes 1 and 3) or YEp105 (Ub-myc; lanes 2 and 4) were grown in the presence of CuSO4 to induce synthesis of Ub and Ub-myc from the CUP1 promoter. Two hours after induction, membrane-enriched cell fractions were prepared before (lanes 1 and 2) and 5 min after addition of ammonium (lanes 3 and 4) and subjected to Western analysis with anti-Gap1 antibodies. The positions of the ubiquitinated forms of Gap1 are indicated with dots, and the band marked with an arrow corresponds to the H+-ATPase Pma1 used as an internal control.
Ubiquitination of Gap1 Mutants Resistant to NH4+-induced Internalization and Degradation
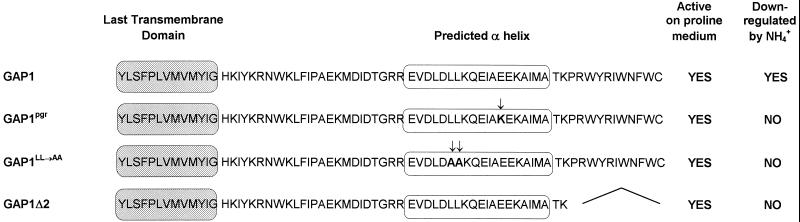
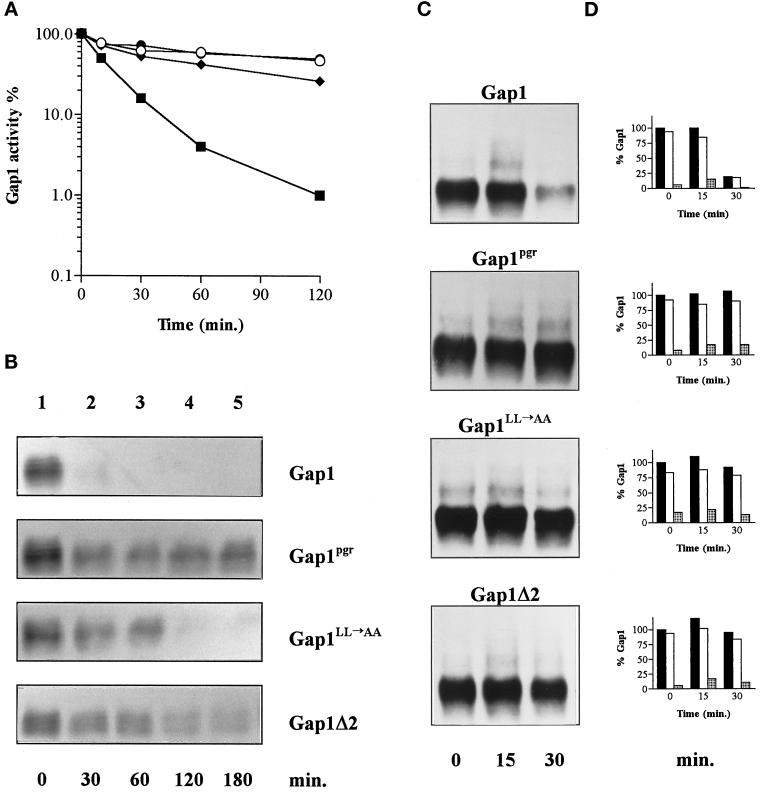
Mutations affecting the C terminus of Gap1 have been shown to protect the permease against NH4+-induced inactivation (Hein and André, 1997). Two such mutations, Gap1LL→AA and Gap1pgr, are located in a region predicted to adopt a stable α-helix conformation, whereas another (Gap1Δ2) results in a truncated permease lacking the last 11 C-terminal amino acids directly following this putative helix (Figure 6). The resistance of these mutant permeases to NH4+-induced down-regulation (Figure 7, A and B) might be due to a defect in ubiquitination, but alternatively, the permeases might be ubiquitinated but not down-regulated. To test whether these C-terminal mutations affect ubiquitination of Gap1, we prepared membrane-enriched extracts of gap1Δ cells expressing either wild-type Gap1 or the mutant Gap1pgr, Gap1LL→AA, or Gap1Δ2. We then compared these extracts by Western analysis (Figure 7C). In cells expressing wild-type Gap1, addition of NH4+ led to an intensification of the minor bands corresponding to ubiquitinated forms of the permease rapidly followed by a decrease of the major Gap1 signal. In contrast, in cells expressing the mutant Gap1pgr permease, the major Gap1 signal remained stable after addition of NH4+. Yet these cells did display bands corresponding to ubiquitinated forms of the permease; these bands are already visible in proline-grown cells, and addition of NH4+ increased their intensity. Thus, NH4+ enhances ubiquitination of Gap1pgr, but this modification does not lead to permease down-regulation. Perhaps the Gap1pgr permease is less efficiently ubiquitinated than the wild-type permease and is thus protected against degradation. This, however, seems unlikely, because quantitation of variously exposed blots revealed that the relative level of ubiquitinated permease detected 15 min after NH4+ addition is approximately similar for the wild-type and Gap1pgr permeases (Figure 7D). The Gap1LL→AA and Gap1Δ2 mutant permeases are also significantly protected against NH4+ down-regulation; after NH4+ addition, both are destabilized at a much lower rate than wild-type Gap1, and yet minor bands corresponding to ubiquitinated forms of the Gap1LL→AA and Gap1Δ2 permeases appear clearly, indicating that the reduced sensitivity of the two proteins to NH4+-induced down-regulation is not due to complete failure to be ubiquitinated.
Figure 6.
Sequence of the C-terminal region (residue 533–601) of the Gap1 permease and mutant derivatives. Arrows indicate the positions of the amino acid substitutions (shown in bold). The deleted region is indicated by joined lines.
Figure 7.
Ubiquitination of C-terminal Gap1 mutants defective in down-regulation. (A) Cells were grown on proline medium, and Gap1 activity was measured by incorporation of [14C]citrulline before (t = 0) and at several times after addition of (NH4)2SO4 (10 mM) in strain JOD0097 (gap1Δ) expressing the wild-type Gap1 permease (▪) or one of the three mutant permeases, Gap1pgr (○), Gap1LL→AA (♦), or Gap1Δ2 (•). The Gap1 activities were expressed in nanomoles per minute per milliliter. (B) Immunoblot of Gap1 from crude extracts prepared before (t = 0) and several times after (NH4)2SO4 addition. (C) Immunoblot of Gap1 from membrane-enriched fractions of cells prepared before (t = 0) and 15 and 30 min after addition of (NH4)2SO4. (D) Quantitation of blots shown in C. Measure of the amount of Gap1 present at each time point. Each value is expressed as a percentage of the total Gap1 found in all forms at (t = 0). Closed bars, all forms of Gap1; open bars, unconjugated forms of Gap1; stippled bars, ubiquitinated forms of Gap1.
The fact that Gap1pgr remains largely active after NH4+ addition indicates that it remains plasma membrane located. It is noteworthy, however, that addition of NH4+ does not lead to complete conversion of the Gap1pgr permease to ubiquitinated forms (Figure 7C and our unpublished results). This contrasts with the behavior of α-factor receptor Ste2 observed in end4 cells defective in the internalization step of endocytosis. In this latter case, ligand binding leads to nearly complete conversion of Ste2 to ubiquitinated forms (Hicke and Riezman, 1996). To further investigate this question, we examined the ubiquitination of wild-type Gap1 in act1-1 cells defective in endocytosis (Figure 1B). When the act1 cells were grown on proline medium, the minor bands corresponding to ubiquitinated forms of the permease were more intense than the corresponding bands of the proline-grown isogenic wild type. After addition of NH4+, the intensity of these minor bands did not increase significantly. In contrast, addition of NH4+ to wild-type cells led to strong intensification of the minor bands detectable in extracts from proline-grown cells. Thus, in contrast to the situation reported for the Ste2 receptor, the Gap1 permease does not seem to be completely converted to Ub-conjugated forms in a mutant defective in endocytosis.
DISCUSSION
In this article, we show that ammonium-triggered degradation of the Gap1 permease depends on vacuolar proteases and is preceded by internalization of the protein by endocytosis. In act1-1 cells defective in the internalization step of endocytosis, the Gap1 permease remains largely active (i.e., plasma membrane located) and stable after addition of NH4+, indicating that nitrogen catabolite inactivation of Gap1 (Grenson, 1983a) most likely results from removal of the permease from the plasma membrane. What we have called Gap1 down-regulation, i.e., the internalization and subsequent degradation of Gap1, thus seems to occur via the same pathway as for other cell surface proteins such as the uracil (Fur4), maltose (Mal61), and galactose (Gal2) permeases, the pheromone receptors Ste2 and Ste3, and the multidrug resistance protein Pdr5 (Singer and Riezman, 1990; Davis et al., 1993; Volland et al., 1994; Egner and Kuchler, 1995; Riballo et al., 1995; Horak and Wolf, 1997). In the Gap1 system, however, down-regulation is induced by adding a preferred nitrogen source to the medium, suggesting that regulatory factors responding to nitrogen must be specifically involved.
Our results show that ubiquitination of the Gap1 permease is required for its down-regulation. Ubiquitinated forms of Gap1 were detected on Western blots as several minor bands migrating to positions just above the major Gap1 signal. At least three minor bands were detected in most experiments; they likely correspond to mono-, di-, and tri-ubiquitinated forms of the permease. In proline-grown cells, these bands represent only a small fraction (∼5%) of the immunodetected Gap1 signal, but they markedly rise in proportion a few minutes after addition of NH4+. Both basal and NH4+-stimulated ubiquitination are severely impaired in npi1 mutant cells, in which the level of immunodetected Npi1/Rsp5, a Ub ligase essential to cell viability (Hein et al., 1995; Huibregtse et al., 1995), is reduced at least 10-fold compared with the wild type. This reduced level of Npi1/Rsp5 is due to a Ty element inserted 500 bp upstream from the initiation codon of the NPI1 gene (Hein et al., 1995). The reduced level of Npi1/Rsp5 in npi1 mutant cells is thus sufficient to ensure cell viability but limiting for ubiquitination of Gap1. It is likely that Npi1/Rsp5 is directly involved in ubiquitination of Gap1. In keeping with a relatively high level of Npi1/Rsp5 protein being needed for NH4+-induced permease down-regulation, Gap1 is a relatively abundant protein, and Npi1/Rsp5 is involved in NH4+-induced inactivation of other, probably numerous, nitrogen-sensitive permeases, including the proline (Put4) and allantoate/ureidosuccinate (Dal5) permeases (Grenson, 1983a). Furthermore, the Npi1/Rsp5 Ub ligase is also involved in turnover control of nitrogen-insensitive permeases such as the uracil (Fur4) (Hein et al., 1995; Galan et al., 1996) and maltose (Mal61) permeases (Lucero and Lagunas, 1997).
The mechanism by which Ub binding induces down-regulation of Gap1 remains unknown. As suggested for several other cell surface proteins, Ub molecules attached to Gap1 might provide a signal triggering endocytosis of the permease. This model finds support in the fact that ubiquitinated forms of Gap1 accumulate to some extent in act1-1 cells defective in endocytosis, and that Gap1 remains plasma membrane located after addition of NH4+ to npi1 cells defective in ubiquitination. It is also possible that ubiquitination would also be involved in another step of down-regulation, such as vacuolar sorting of internalized permease. For instance, deubiquitination of internalized permeases might lead to their recycling back to the plasma membrane, whereas maintenance of their ubiquitinated state might target them to vacuolar breakdown. Although recycling of protein after internalization is generally considered as the default pathway in higher eukaryotic cells (Mayor et al., 1993), no such pathway has been documented in yeast. Yet in the sec18 mutant strain recently shown to be impaired in forward progression of molecules from the plasma membrane to the vacuole (Hicke et al., 1997), the maltose permease remains active under conditions that normally induce its internalization and vacuolar degradation. This raises the possibility that at least some cell surface proteins might undergo recycling after endocytosis (Riballo et al., 1995). Clearly, elucidating the exact role of Ub in down-regulation of Gap1 requires further investigation.
The Gap1LL→AA and Gap1Δ2 permeases carrying mutations in the C-terminal tail of the permease (Figure 6) are significantly protected against NH4+-induced degradation, but this protection is not due to complete failure of the proteins to be ubiquitinated. These mutant Gap1 proteins might be less efficiently ubiquitinated and thus partially protected against down-regulation. Another possibility is that these C-terminal mutations alter down-regulation at a step downstream from ubiquitination. Another mutant, Gap1pgr, carries a glutamate-to-lysine substitution within the C-terminal tail (Figure 6). This mutant permease is strongly protected against NH4+-triggered degradation, but it is apparently nevertheless converted to ubiquitinated forms in a manner similar to that observed for the wild-type permease. This would mean that, in addition to Ub binding, sequences located within the C-terminal tail of the permease are required for normal down-regulation. The exact role of the glutamate residue substituted in the Gap1pgr mutant is unknown. It is located within an EEKAI sequence reminiscent of the DAKSS sequence. The latter is present in the cytosolic tail of the Ste2 receptor and is essential to ubiquitination and endocytosis of a truncated form of the receptor (Hicke and Riezman, 1996). Although the EEKAI sequence of Gap1 clearly differs from the DAKSS sequence of Ste2, mutagenesis experiments on the DAKSS motif have shown that EAKSS and DAKAS promote efficient internalization, whereas AAKSS does not (Rohrer et al., 1993). Replacing the lysine residue (DARSS) impairs both ubiquitination and endocytosis of the truncated form of Ste2 (Hicke and Riezman, 1996). That the E→R substitution in the EEKAI sequence of Gap1 markedly impairs down-regulation without apparently affecting ubiquitination suggests that this sequence plays a role in a subsequent step of the down-regulation pathway.
Our data also show that ubiquitinated forms of Gap1 are more abundant in the endocytosis-defective act1-1 strain, but that addition of NH4+ to these cells does not cause a rise in the level of ubiquitinated permease, the latter forms representing a relatively constant fraction of the immunodetected Gap1 protein. The situation of the Gap1pgr mutant permease is quite similar; added NH4+ triggers conversion of the permease to ubiquitinated forms, but only a fraction of the permease undergoes this modification. These results were unexpected in the light of the finding that Ste2 receptor is nearly completely converted to Ub-conjugated forms upon addition of α-factor to mutant cells defective in endocytosis (Hicke and Riezman, 1996). Perhaps the effects of the act1-1 and gap1pgr mutations are only partial, and residual down-regulation is sufficient to prevent accumulation of ubiquitinated forms of Gap1. However, should this interpretation be true, addition of NH4+ would lead to progressive loss of the preexisting Gap1 activity, an effect that has not been observed. Alternatively, a specific defect in endocytosis due to gap1pgr and act1-1 mutations could prevent further ubiquitination beyond a certain amount of permease. For instance, some limiting components of the ubiquitination machinery (such as the Npi1/Rsp5 Ub ligase itself) might be sequestrated in endocytosis-defective cells. Another explanation for the failure to accumulate Ub conjugated forms in endocytosis mutants is that deubiquitinating enzyme may impose a steady-state level of modification.
One particularity of the Gap1 system, compared with other cell surface proteins that undergo ubiquitination, is the ubiquitination-enhancing effect of added NH4+. This suggests that nitrogen-sensitive regulatory factors are involved in this process. One such factor is likely Npr1. Previous work has shown that Gap1 is inactive in an npr1 mutant grown on proline medium, and that this loss of activity requires Npi1/Rsp5 and the integrity of the C-terminal region of the permease (Grenson, 1983b; Hein and André, 1997). Npr1 thus seems to protect Gap1 against down-regulation in cells grown on a poor nitrogen source. This observation, together with the data of this study, suggests the following model for the regulation of Gap1 turnover according to the nitrogen source. In cells growing on proline medium, Gap1 is abundant and highly active. Only a small fraction of the permease is ubiquitinated. The presence of ubiquitinated Gap1 forms might reflect basal turnover of the permease. Under these growth conditions, the role of Npr1 might be to limit the conversion of the Gap1 proteins into ubiquitinated forms. Addition of NH4+ might either enhance the efficiency of the ubiquitination reaction itself or counter the putative protective action of Npr1 against ubiquitination. The result would be rapid ubiquitination of all preaccumulated Gap1 molecules followed by their down-regulation. Molecular analysis has shown that the NPR1 encodes a kinase homologue (Vandenbol et al., 1990), suggesting that phosphorylation could be involved in protecting Gap1 against down-regulation.
ACKNOWLEDGMENTS
We are grateful to Rosine Haguenauer-Tsapis for discussions during progress of this work and for critical comments on the manuscript. We thank Anne-Marie Marini and Stephan Vissers for critical reading of the manuscript. We also thank Howard Riezman, Rosine Haguenauer-Tsapis, and S. Kumar for providing strains, plasmid and antisera. This work was supported by Fund for Medical Scientific Research (Belgium) grant 3.4602.94. J.Y.S. is a recipient of a predoctoral fellowship from the Fond pour la formation à la Recherche dans l’Industrie et dans l’Agriculture.
REFERENCES
- Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995. [Google Scholar]
- Béchet J, Grenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bonneaud N, Ozier Kalogeropoulos O, Li GY, Labouesse M, Minvielle Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Davis NG, Horecka JL, Sprague GF., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich E, Haft CR, Muys L, Heinrich PC, Graeve L. A di-leucine motif and an upstream serine in the interleukin-6 (IL-6) signal transducer gp130 mediate ligand-induced endocytosis and down-regulation of the IL-6 receptor. J Biol Chem. 1996;271:5487–5494. doi: 10.1074/jbc.271.10.5487. [DOI] [PubMed] [Google Scholar]
- Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- Egner R, Mahe Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Moreau V, André B, Volland C, Haguenauer Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Grenson M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. II. Evidence for a specific lysine-transporting system. Biochim Biophys Acta. 1966;127:339–346. doi: 10.1016/0304-4165(66)90388-6. [DOI] [PubMed] [Google Scholar]
- Grenson M. Inactivation-reactivation process and repression of permease formation regulate several ammonia-sensitive permeases in the yeast Saccharomyces cerevisiae. Eur J Biochem. 1983a;133:135–139. doi: 10.1111/j.1432-1033.1983.tb07438.x. [DOI] [PubMed] [Google Scholar]
- Grenson M. Study of the positive control of the general amino-acid permease and other ammonia-sensitive uptake systems by the product of the NPR1 gene in the yeast Saccharomyces cerevisiae. Eur J Biochem. 1983b;133:141–144. doi: 10.1111/j.1432-1033.1983.tb07439.x. [DOI] [PubMed] [Google Scholar]
- Grenson M. Amino acid transporters in yeast: structure, function and regulation. In: De Pont JJLLM, editor. Molecular Aspects of Transport Proteins. Amsterdam: Elsevier Science; 1992. pp. 219–245. [Google Scholar]
- Haft CR, Klausner RD, Taylor SI. Involvement of dileucine motifs in the internalization and degradation of the insulin receptor. J Biol Chem. 1994;269:26286–26294. [PubMed] [Google Scholar]
- Hein C, André B. A C-terminal di-leucine motif and nearby sequences are required for NH4(+)-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol Microbiol. 1997;24:607–616. doi: 10.1046/j.1365-2958.1997.3771735.x. [DOI] [PubMed] [Google Scholar]
- Hein C, Springael JY, Volland C, Haguenauer Tsapis R, André B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hicke L, Zanolari B, Pypaert M, Rohrer J, Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol Biol Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Ellison MJ, Chau V, Varshavsky A. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J, Wolf DH. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. (erratum 92, 5249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs P, Jauniaux JC, Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J Mol Biol. 1980;139:691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
- Jauniaux JC, Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem. 1990;190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- Kölling R, Hollenberg CP. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Harvey KF, Kinoshita M, Copeland NG, Noda M, Jenkins NA. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics. 1997;40:435–443. doi: 10.1006/geno.1996.4582. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:608–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Klausner RD. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- Lucero P, Lagunas R. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Microbiol Lett. 1997;147:273–277. doi: 10.1111/j.1574-6968.1997.tb10253.x. [DOI] [PubMed] [Google Scholar]
- Mayor S, Presley JF, Maxfield FR. Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J Cell Biol. 1993;121:1257–1269. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Herweijer M, Wolf DH, Lagunas R. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol. 1995;177:5622–5627. doi: 10.1128/jb.177.19.5622-5627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg KJ, Rowley N, Kaiser CA. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer J, Benedetti H, Zanolari B, Riezman H. Identification of a novel sequence mediating regulated endocytosis of the G protein-coupled alpha-pheromone receptor in yeast. Mol Biol Cell. 1993;4:511–521. doi: 10.1091/mbc.4.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AF, Davis NG. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. In: Molecular Cloning. A Laboratory Manual. 2nd ed. Nolan C, editor. New York: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Sandoval IV, Bakke O. Targeting of membrane proteins to endosomes and lysysomes. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory Course Manual for Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Shin J, Dunbrack RL, Jr, Lee S, Strominger JL. Phosphorylation-dependent down-modulation of CD4 requires a specific structure within the cytoplasmic domain of CD4. J Biol Chem. 1991;266:10658–10665. [PubMed] [Google Scholar]
- Shortle D, Novick P, Botstein D. Construction and genetic characterization of temperature-sensitive mutant alleles of the yeast actin gene. Proc Natl Acad Sci USA. 1984;81:4889–4893. doi: 10.1073/pnas.81.15.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Riezman H. Detection of an intermediate compartment involved in transport of alpha-factor from the plasma membrane to the vacuole in yeast. J Cell Biol. 1990;110:1911–1922. doi: 10.1083/jcb.110.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbol M, Jauniaux JC, Grenson M. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encodes a protein kinase homologue. Mol Gen Genet. 1990;222:393–399. doi: 10.1007/BF00633845. [DOI] [PubMed] [Google Scholar]
- Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- Wiame JM, Grenson M, Arst HNJ. Nitrogen catabolite repression in yeasts and filamentous fungi. Adv Microbiol Physiol. 1985;26:1–87. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- Woolford CA, Daniels LB, Park FJ, Jones EW, Van Arsdell JN, Innis MA. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol Cell Biol. 1986;6:2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom J, Linskens MH, Sadis S, Rubin DM, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]