Abstract
The blocking of G1 progression by fission yeast pheromones requires inhibition of the cyclin-dependent kinase cdc2p associated with the B-cyclins cdc13p and cig2p. We show that cyclosome-mediated degradation of cdc13p and cig2p is necessary for down-regulation of B-cyclin–associated cdc2p kinase activity and for phermone-induced G1 arrest. The cyclin-dependent kinase inhibitor rum1p is also required to maintain this G1 arrest; it binds both cdc13p and cig2p and is specifically required for cdc13p proteolysis. We propose that rum1p acts as an adaptor targeting cdc13p for degradation by the cyclosome. In contrast, the cig2p–cdc2p kinase can be down-regulated, and the cyclin cig2p can be proteolyzed independently of rum1p. We suggest that pheromone signaling inhibits the cig2p–cdc2p kinase, bringing about a transient G1 arrest. As a consequence, rum1p levels increase, thus inhibiting and inducing proteolysis of the cdc13p–cdc2p kinase; this is necessary to maintain G1 arrest. We have also shown that pheromone-induced transcription occurs only in G1 and is independent of rum1p.
INTRODUCTION
Entry into S-phase and mitosis in the eukaryotic cell cycle is controlled by the activation of cyclin-dependent kinases (CDKs). In the yeasts, both processes are initiated by a single CDK core enzyme encoded by cdc2 in fission yeast and CDC28 in budding yeast. Cdc2p and Cdc28p associate with mitotic B-type cyclins to initiate mitosis, cdc13p in fission yeast (Booher and Beach, 1988; Hagan et al., 1988; Booher et al., 1989; Moreno et al., 1989), and Clb1–4p in budding yeast (Ghiara et al., 1991; Surana et al., 1991; Fitch et al., 1992; Richardson et al., 1992) and with S-phase B-cyclins to trigger S-phase, usually cig2p in fission yeast (Fisher and Nurse, 1996; Martin-Castellanos et al., 1996; Mondesert et al., 1996) and Clb5–6p in budding yeast (Epstein and Cross, 1992; Kühne and Linder, 1993; Schwob and Nasmyth, 1993; Schwob et al., 1994). There is considerable overlap between mitotic and S-phase B-cyclins (Schwob et al., 1994; Fisher and Nurse, 1996; Mondesert et al., 1996), and in fission yeast a single cyclin cdc13p can bring about both S-phase and mitosis (Fisher and Nurse, 1996; Mondesert et al., 1996). In budding yeast, activation of S-phase Clbp–Cdc28p protein kinase depends on the prior activation of Cdc28p associated with another class of G1 cyclins, Cln1–3p.
The mechanisms ensuring the timely inactivation and activation of cyclin B–CDK in G1 have been studied mainly in budding yeast. S-phase Clbp–Cdc28p protein kinase is up-regulated by three independent mechanisms, all of which involve Clnp-Cdc28p kinase activity. Clnp–Cdc28p protein kinase 1) activates transcription of CLB genes (Epstein and Cross, 1992; Schwob and Nasmyth, 1993) and 2) inactivates Clbp proteolysis (Amon et al., 1994). The latter involves ubiquitin-mediated degradation of B-type cyclins, which requires the cyclosome (Sudakin et al., 1995) or anaphase-promoting complex consisting of eight subunits, including Apc1p/bimEp/cut4p (Peters et al., 1996; Yamashita et al., 1996; Zachariae et al., 1996), Cdc16p, Cdc23p, and Cdc27p (Irniger et al., 1995; King et al., 1995; Tugendreich et al., 1995). Cyclosome-mediated proteolysis is activated at the metaphase–anaphase transition, and its activity is maintained during early G1 where it contributes to the prevention of a premature rise of Clbp–Cdc28p kinase activity (Irniger et al., 1995). 3) Clnp–Cdc28p protein kinase phosphorylates the cyclin-dependent kinase inhibitor (CKI) Sic1p, targeting it for ubiquitin-mediated degradation via the ubiquitin-conjugating enzyme Cdc34p (Schwob et al., 1994; Schneider et al., 1996). Sic1p is present in early G1 (Donovan et al., 1994; Schwob et al., 1994) and specifically inhibits Clbp–Cdc28p protein kinase activity (Mendenhall, 1993; Schwob et al., 1994). Thus in budding yeast, down-regulation of Clbp-associated kinase is brought about by transcriptional, proteolytic, and CKI mechanisms that are relieved in late G1 by Clnp–Cdc28p protein kinase activity. A second CKI in budding yeast, Far1p, directly inhibits the Clnp–Cdc28p protein kinase activity in response to pheromone and causes G1 arrest (Chang and Herskowitz, 1990). Far1p is activated by the pheromone-dependent MAP kinase Fus3p, allowing Far1p to bind and inhibit the Clnp–Cdc28p protein kinase (Peter et al., 1993; Peter and Herskowitz, 1994).
In fission yeast, the CKI encoded by the rum1 gene plays a crucial role in regulating the cyclin B–CDK activity in G1 (Moreno and Nurse, 1994). rum1p is a potent in vitro inhibitor of cdc2p associated with the mitotic B-type cyclin cdc13p (Correa-Bordes and Nurse, 1995; Jallepalli and Kelly, 1996) and also partly inhibits the in vitro kinase activity associated with the G1 B-cyclin cig2p (Correa-Bordes and Nurse, 1995; Martin-Castellanos et al., 1996). A rum1Δ mutant initiates mitosis from G1 when S-phase is blocked (Moreno and Nurse, 1994). In these cells, the mitotic cdc13p–cdc2p kinase is activated prematurely (Correa-Bordes and Nurse, 1995), suggesting that one function of rum1p is to provide a safeguard that prevents mitosis from taking place in G1 cells. rum1p is also required to extend G1 during nitrogen starvation or in a wee1 mutant background (Moreno and Nurse, 1994).
To better understand the mechanisms that control the activation of the G1 cyclin B–cdc2p kinases in fission yeast, we have investigated the cell cycle controls that bring about pheromone-induced G1 arrest (Davey and Nielsen, 1994; Imai and Yamamoto, 1994). We have shown previously that the fission yeast-mating pheromone P-factor blocks entry into S-phase by inhibiting both the cig2p- and cdc13p-associated cdc2p kinase activity in G1 (Stern and Nurse, 1997). Here we show that rum1+ is required for this pheromone-induced G1 arrest. Our data establish that the cdc13p–cdc2p kinase is the main target for rum1p, whereas down-regulation of the cig2p-associated kinase activity can occur by another mechanism. Mutants in the cyclin B degradation machinery accumulate both cig2p and cdc13p and fail to arrest in G1 in response to pheromones. Turnover of cdc13p requires rum1p, whereas cig2p turnover can occur in the absence of rum1p, suggesting that rum1p may act as an adaptor specifically targeting cdc13p for cyclosome-dependent degradation.
MATERIALS AND METHODS
Fission Yeast Strains and Methods
The following strains were constructed: h−cdc22-M45cyr1Δ::LEU2 sxa2Δ::ura4+leu1–32ura4-D18; h−rum1-HAcyr1Δ::LEU2sxa2Δ::ura4+ leu1–32ura4-D18ade6–704his3-D1; h−cyr1Δ::LEU2sxa2Δ::ura4+:: REP6Xrum1leu1–32ura4-D18ade6–704; h−rum1Δ::his3+cyr1Δ:: LEU2sxa2Δ::ura4+leu1–32ura4-D18his3-D1ade6–704; h−cdc10– 129rum1Δ::his3+cyr1Δ::LEU2sxa2Δ::ura4+leu1–32ura4-D18his3D1ade6–704; h−rum1Δ::his3+cig2Δ::ura4+cyr1Δ::LEU2sxa2Δ:: ura4+leu1–32ura4-D18his3-D1ade6–704; h−nuc2–663 cyr1Δ::ura4+ sxa2Δ::ura4+leu1–32ura4-D18ade6–704.
Strains were constructed using random spore analysis. Candidate colonies with the appropriate selectable markers and mutations were tested for formation of conjugation tubes on agar plates containing 3 μg/ml P-factor. Only h−cyr1Δsxa2Δ mutant cells will respond to P-factor and grow conjugation tubes. rum1Δ:his3+ and nuc2ts mutants were crossed into the h−cyr1Δsxa2Δ background in the presence of a nuc2 plasmid or rum1 plasmid, respectively. The plasmids were lost after selection of the h−rum1Δcyr1Δsxa2Δ and h−nuc2–663cyr1Δsxa2Δ triple mutants using marker selection or temperature-sensitive phenotype and response to P-factor on agar plates or both. For the construction of the cig2Δrum1Δcyr1Δsxa2Δ, quadruple mutant colonies were tested by PCR for the absence of cig2+. A pheromone-responsive strain carrying an N-terminal hemagglutinin peptide (HA)-tagged rum1 at the rum1 locus (Correa-Bordes and Nurse, 1995) was selected on the basis of an increased size of a rum1-HA PCR product compared with the rum1 wild-type allele.
Media and growth conditions were as described by Moreno et al. (1991). Physiological experiments with P-factor, flow cytometric analysis (FACS), cell number, and cell size measurements were as described by Stern and Nurse (1997).
Construction of a rum1Δ::his3+ Mutant Strain
A 1.9-kb SpeI fragment containing the whole rum1+ open reading frame was removed from a 6.1-kb genomic rum1+ clone in pTZ18R and replaced by blunt-end ligation with a 1.8-kb EcoRV–DraI fragment of the his3+ gene. The linearized 6.1-kb rum1Δ::his3+ deletion construct was transformed into a his3-D1 strain, and a stable his prototroph colony was isolated. Southern blotting established that the integration had taken place at the rum1+ locus. The rum1Δ::his3+ mutant was sterile and synthetically lethal with a cdc10.129 allele like the previously described rum1::ura4+ strain (Moreno and Nurse, 1994). Both phenotypes were rescued in the presence of a rum1+-containing plasmid. A single copy of the EcoRV–DraI his3+ fragment does not fully rescue the his3-D1 deletion in liquid culture. We therefore supplemented the medium with histidine for physiological experiments.
RNA Preparation and Northern Blot
RNA was prepared by glass bead lysis in the presence of phenol and SDS and was subsequently separated using a formaldehyde gel. Ten micrograms as measured by OD260 were loaded in each track. Probes for blotting were prepared by random oligo priming with [32P]dATP using a Prime-It kit (Stratagene, La Jolla, CA).
Cloning of mat1-Mm and fus1
A 210-bp fragment of the mat1-Mm gene was amplified from genomic DNA by PCR using the following primers: CATATGCATTTGTATAGCAT and AATAATGTCAGCAGAAGACC. The resulting PCR fragment was cloned into the REP5 vector using the NdeI and BamHI sites in the primers. A 1.1-kb fragment of the fus1 gene was amplified in a similar manner using the following primers: CGGGATCCGGGGTACTCAAGTGTTACGTCTGG and CGGGATCCAGCTGCTTTAGCCGTTTAGAAGG. The resulting PCR fragment was cloned into pKS+ using the BamHI sites in the primers.
Protein Kinase Assays and Immunoprecipitations
Kinase assays were performed as described by Stern and Nurse (1997). Cig2p-associated cdc2p kinase activity was immunoprecipitated from 3.8 mg (Figure 3A) and 2.5 mg (Figure 5B) of soluble extract with 10 μl of anti-cig2p polyclonal serum MOC8 (Stern and Nurse, 1997). Cdc13p-associated kinase activity was immunoprecipitated with 10 μl of anti-cdc13p serum SP4 (Moreno et al., 1989) from 1 mg (Figure 3A) and 2.5 mg (Figure 5B) of soluble extract.
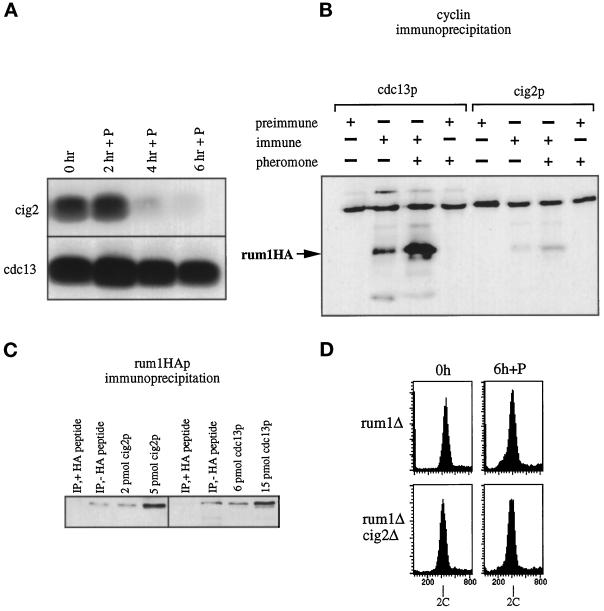
Figure 3.
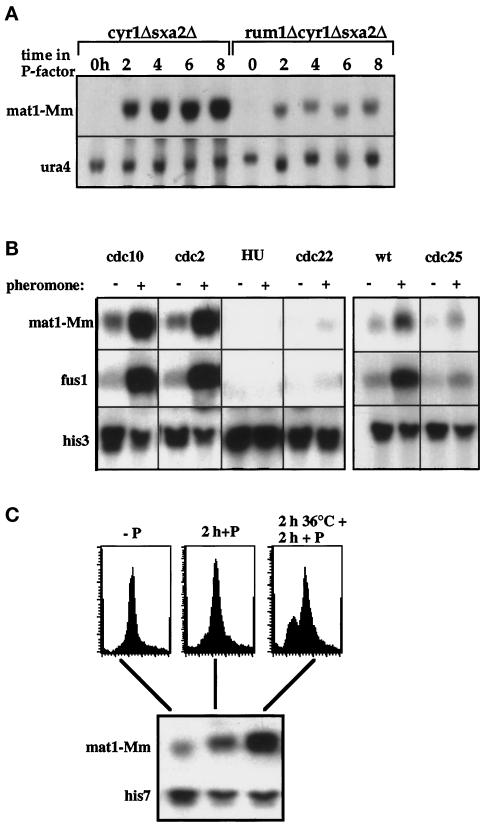
rum1p binds to and inhibits the cdc13p–cdc2p kinase in P-factor. (A) cig2p- and cdc13p-associated H1 kinase activities in a rum1Δcyr1Δsxa2Δ after exposure to P-factor at 25°C. (B) Immunoprecipitations from a rum1-HAcyr1Δsxa2Δ strain with anti-cdc13p and anti-cig2p antibodies before and 3 h after P-factor addition, Western blotted, and probed with anti-HA-antibody. (C) Immunoprecipitations from a rum1-HAcyr1Δsxa2Δ strain with anti-HA antibodies 3 h after P-factor addition, Western blotted, and probed with antibodies raised against cig2p (left panel) and cdc13p (right panel). Extracts (2 and 5 μl) containing the indicated amounts of cig2p and cdc13p were loaded to quantify the coimmunoprecipitated cyclins. (D) FACS analysis of h−rum1Δcyr1Δsxa2Δ, h−cig2Δrum1Δcyr1Δsxa2Δ mutants incubated in P-factor for 6 h at 25°C.
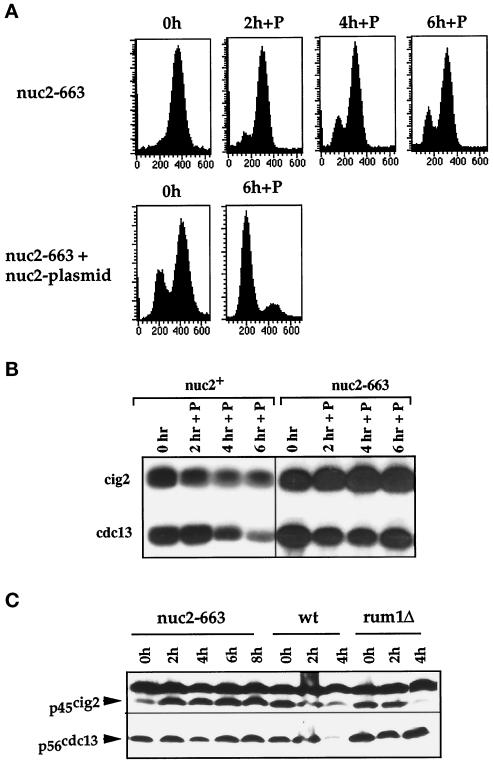
Figure 5.
A mutant defective in the cyclosome/APC fails to arrest in G1 and does not down-regulate B-cyclin protein levels and associated kinase activities. (A) FACS analysis of nuc2–663cyr1Δsxa2Δ and nuc2–663cyr1Δsxa2Δ containing a nuc2+ plasmid exposed to P-factor at 25°C. 1C peak seen at 0 h in nuc2–663 with nuc2 plasmid is due to plasmid loss. (B) Cig2p- and cdc13p-associated H1 kinase activities in a nuc2+ and a nuc2–663 mutant after P-factor addition at 25°C. (C) Cig2p and cdc13p levels in nuc2–663cyr1Δsxa2Δ (nuc2–663), cyr1Δsxa2Δ (wt) and rum1Δcyr1Δsxa2Δ (rum1Δ) cells after exposure to P-factor. A nonspecific, 50-kDa protein cross-reacted with cig2p antibodies at a 1:1000 dilution and served as a loading control.
For cyclin immunoprecipitations (Figure 3B), 6 mg of soluble extract were incubated for 15 min at 4°C with 20 μl of polyclonal rabbit anti-cig2p serum (MOC8), with polyclonal rabbit anti-cdc13p serum (SP4), or with the respective preimmune serum. Rum1-HAp was immunoprecipitated for 30 min at 4°C from 9.5 mg of soluble extract (Figure 3C) with 30 μl of 12CA5 mAbs coupled to AffiGel beads (Bio-Rad, Richmond, CA; 4.3 mg/ml).
The immunoprecipitations were washed three times with 1 ml of HB buffer, boiled in 1× SDS sample buffer, separated on 10% SDS-PAGE, and Western blotted. The filters were probed for rum1HAp (Figure 3B) with the 12CA5 mAb (1:500 dilution), for cig2p (Figure 3C) with affinity-purified rabbit anti-cig2p antibody MOC6 (1:2000), and for cdc13p with affinity-purified rabbit anti-cdc13p SP4 (1:1000) (Figure 3C).
Western Blot
For Western blotting, 1–4 × 108 cells were harvested by centrifugation, washed once with ice-cold STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3, pH 8.0), resuspended in 50–100 μl of HB buffer, and boiled for 4 min. Glass beads were added to the meniscus, and cells were broken by vortexing on an IWAKI TWM 4836 microtube mixer (Iwaki Glass, Ikuta, Japan) for 2–5 min. Extract (50 μg) was separated on 10% SDS-PAGE (Laemmli, 1970) and transferred to ECL nitrocellulose or an Immobilon-P membrane (Millipore, Bedford, MA), and the protein of interest was detected using ECL (Amersham, Arlington Heights, IL). Dilutions of the antibodies were 1:1000 (Figure 5C) and 1:2000 (Figure 3C) for the anti-cig2p affinity-purified polyclonal antibody, 1:1000 for rabbit anti-cdc13p antibodies (SP4), 1:1000 for rabbit anti-rum1p antibodies (Correa-Bordes and Nurse, 1995), and 1:50,000 for the anti-α-tubulin monoclonal antibody (T5168; Sigma, St. Louis, MO).
In Vivo 35S-Methionine Labeling
Cells were grown in minimal medium with glutamate (1 g/l) as a nitrogen source, to an OD595 of 0.5 (6 × 106 cells/ml) in the presence or absence of P-factor. Cells (10 ml) were incubated with 600 μCi of 35S-methionine (Amersham Promix) for 10 min. After labeling, cells were harvested and washed with 10 ml of cold STOP buffer, resuspended in 50 μl of HB buffer, and broken with 1 ml of glass bead for 1 min. After cell breakage, the crude extract was recovered with 1 ml of cold HB buffer. Cell debris was removed by a 5-min spin in a microcentrifuge, and rum1p was isolated by immunoprecipitation with 10 μl of rum1p antiserum.
RESULTS
CDK Inhibitor rum1p Is Required for Pheromone-induced G1 Arrest
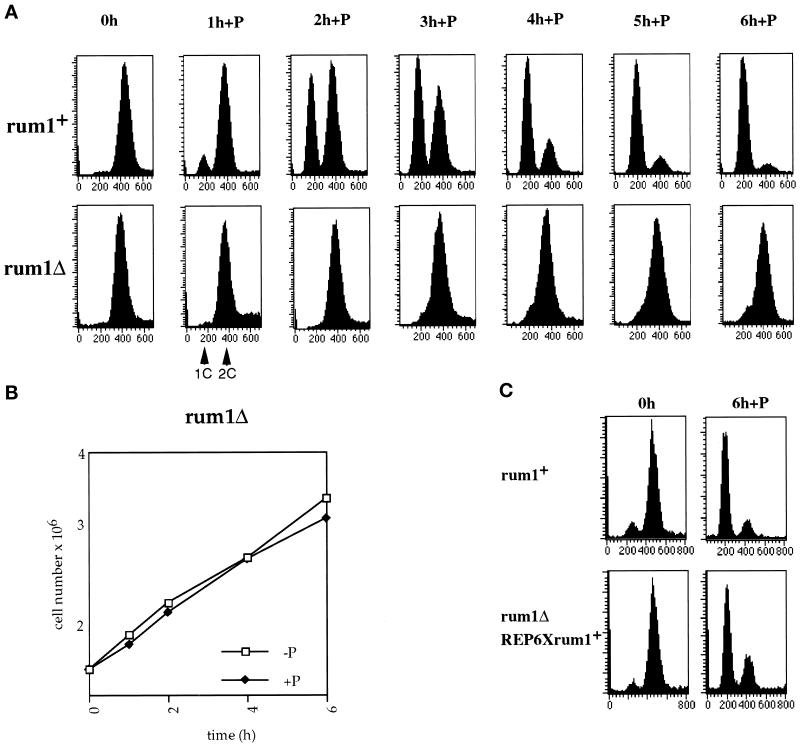
The mating pheromone P-factor brings about G1 arrest by inhibiting the cdc2p protein kinase activity (Stern and Nurse, 1997). Given that the CKI rum1p is present during G1 and is required for pheromone-induced conjugation (Moreno and Nurse, 1994), it is possible that rum1p may have an analogous role to budding yeast Far1p in bringing about pheromone-induced G1 arrest (Imai and Yamamoto, 1994). To test this possibility, we crossed a rum1Δ into a cyr1Δsxa2Δ background. This genetic background is required to observe P-factor–induced G1 arrest in exponentially growing cells. Elimination of the adenylate cyclase cyr1+ gene leads to constitutive expression of nutritionally controlled genes, including components of the pheromone signal transduction cascade (Maeda et al., 1990; Kawamukai et al., 1991; Sugimoto, 1991); sxa2+ encodes a P-factor–degrading protease (Imai and Yamamoto, 1992; Ladds et al., 1996). Control h−rum1+cyr1Δsxa2Δ cells were completely arrested in G1 6 h after addition of P-factor (Figure 1A). In contrast, addition of P-factor to h−rum1Δcyr1Δsxa2Δ cells failed to arrest them in G1 (Figure 1A), and the cells continued to divide like untreated cells (Figure 1B). Similar results were obtained with h+rum1Δcyr1Δ, which did not arrest in G1 in response to the mating pheromone M-factor (our unpublished results). We conclude that rum1 is required for pheromone-induced G1 arrest.
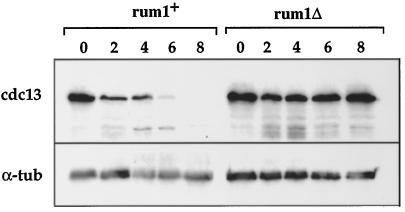
Figure 1.
The rum1Δ mutant does not arrest in G1 in response to pheromone. (A) FACS analysis of cyr1Δsxa2Δ and rum1Δcyr1Δsxa2Δ strains exposed to P-factor at 25°C. (B) Growth curve of rum1Δcyr1Δsxa2Δ in the absence and presence of P-factor. (C) FACS analysis of a cyr1Δsxa2Δ and a rum1Δcyr1Δsxa2Δ::REP6Xrum1+integrant exposed to P-factor for 6 h in medium containing thiamine.
To investigate whether ectopic expression of rum1 rescues the G1 arrest defect, we integrated a REP6Xrum1 plasmid with the rum1 cDNA under the control of the thiamine-repressible nmt promoter (Maundrell, 1993) into a rum1Δ strain. In this strain, pheromone-induced G1 arrest was restored even when the promoter was switched off in the presence of thiamine (Figure 1C), indicating that low-level ectopic expression of rum1 is sufficient to rescue the G1 arrest defect.
rum1p is barely detectable in exponentially growing cells. If rum1p has a physiological role in bringing about G1 arrest in response to pheromone, then rum1p levels need to increase after pheromone addition to cells. As expected, rum1p levels increased rapidly and became maximal within 2–3 h (Figure 2A, left panel) after pheromone addition to cells. Previous work has shown that rum1p levels increase when cells are arrested in G1 (Correa-Bordes and Nurse, 1995). Therefore, the increase in rum1p levels after pheromone addition could be an effect of the G1 arrest induced by pheromone. To investigate this further, P-factor was added to cells arrested in G2 using a cdc25ts (cdc25–22) mutant. These G2-arrested cells are capable of responding to pheromone (Stern and Nurse, 1997) but did not accumulate rum1p (Figure 2A, middle panel), suggesting that cells need to be in G1 for rum1p to be induced. Furthermore, the addition of P-factor to nitrogen-starved cells already blocked in G1 did not lead to a further increase of rum1p levels (Figure 2B), indicating that pheromone addition to these G1 cells had no further direct effect on rum1p induction. This suggests that pheromone may not directly induce rum1p accumulation but, rather, that rum1p is induced after pheromone addition as an indirect consequence of cells arresting in G1. We conclude that the primary role for rum1p may be in maintaining G1 arrest after pheromone addition rather than in bringing about the initial G1 arrest.
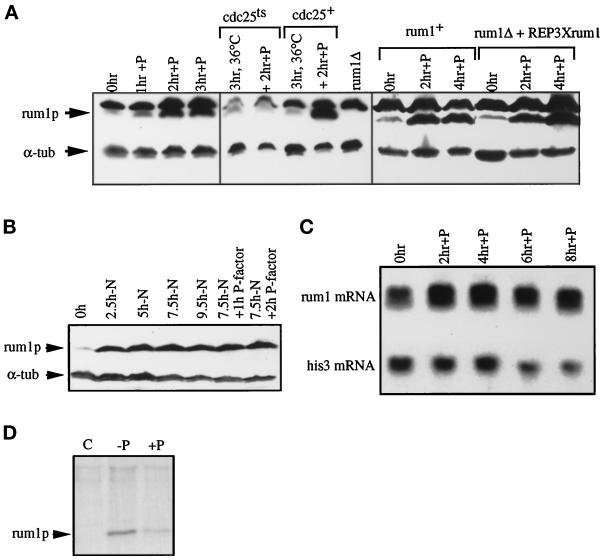
Figure 2.
rum1p is induced in G1 in pheromone. (A) Western blot of extracts from P-factor–treated cells probed with anti-rum1p and control anti-α-tubulin antibodies. A lane with extracts from isogenic rum1Δ cells demonstrates that the slower migrating band cross-reacting with anti-rum1p is nonspecific. A cyr1Δsxa2Δ strain exposed to P-factor for 3 h at 25°C (left panel), cdc25–22cyr1Δsxa2Δ and cyr1Δsxa2Δ strains incubated at 36°C for 3 h and exposed to P-factor for 2 h at 36°C (middle panel), and a rum1Δcyr1Δsxa2Δ::REP6Xrum1+ integrant and a rum1+ control exposed to P-factor at 25°C for 4 h (right panel). (B) Cyr1Δsxa2Δ cells were nitrogen starved, and after 7.5 h, P-factor was added to half the culture. Extracts were Western blotted and probed with anti-rum1p and anti-α-tubulin antibodies. This anti-rum1p antibody shows no nonspecific cross-reacting band. (C) Northern blot of RNA samples from a cyr1Δsxa2Δ strain exposed to P-factor for 8 h at 29°C, probed for rum1+ and his3+ mRNA. (D) Protein extracts from 35S-methionine pulse-labeled cyr1Δsxa2Δ cells before (C, −P) or after 3 h exposure to P-factor (+P), immunoprecipitated with preimmune serum (C), or anti-rum1p antibody (−P and +P). Because there was considerably more 35S-methionine–labeled protein in the −P extract than in the +P extract, the rum1p-specific band was quantified and normalized with respect to an unspecific band. With this quantification, 120 relative units of 35S were incorporated in the absence of pheromone, compared with 97 relative units in the presence of pheromone.
The increase of rum1p levels in cells blocked in G1 by P-factor still occurred in cells expressing a constant low level of rum1 from the nmt promotor (Figure 2A, right panel). This suggests that rum1p up-regulation involves primarily posttranscriptional mechanisms. Increased rum1 transcription might contribute, however, to the increased level of rum1p in pheromone because rum1 transcript levels increased ∼1.6-fold after P-factor addition (Figure 2C). The posttranscriptional mechanism probably involves changes in rum1p turnover, as pulse labeling of cells with 35S-methionine for 10 min showed that the levels of rum1p translation were not increased in pheromone-treated cells (Figure 2D). This conclusion is supported by the recent observation that rum1p accumulates in proteasome mutants that are defective in ubiquitin-mediated proteolysis (Benito et al., 1998).
Cdc13p-associated cdc2p Kinase Is Deregulated in a rum1 Mutant
Next we investigated further the effects of rum1p on the cdc2p kinase during G1 cell cycle arrest in pheromone. In vitro, rum1p inhibits the cdc13p-associated kinase and to a lesser extent the cig2p-associated kinase (Correa-Bordes and Nurse, 1995). Both cyclins can promote S-phase (Fisher and Nurse, 1996; Mondesert et al., 1996), and so the cdc2p kinase activity associated with both cyclins needs to be inhibited to bring about and maintain G1 arrest. Thus, the failure of rum1Δ cells to undergo pheromone-induced G1 arrest could be due to a lack of inhibition of either the cig2p- or cdc13p-associated cdc2p protein kinase activity. We monitored both kinase activities in a rum1Δ mutant after addition of P-factor. The cig2p-associated activity still responded to P-factor falling to a low level within 4 h, whereas the cdc13p-associated activity remained high (Figure 3A). This indicates that the rum1p inhibitor is required to inhibit cdc13p-associated kinase activity but not to inhibit cig2p-associated kinase activity.
We confirmed that the inappropriate entry into S-phase in pheromone-treated rum1Δ cells does not require cig2 by using a cig2Δrum1Δcyr1Δsxa2Δ quadruple mutant. We found that a cig2Δ background did not restore the ability of a rum1Δ to G1 arrest in response to P-factor (Figure 3D), indicating that cig2 is not required in a rum1Δ mutant to overcome the G1 block. Because cdc13p is the major B-type cyclin compensating for the loss of cig2, these results indicate that the premature onset of S-phase observed in pheromone-treated rum1Δ cells results from deregulation of the cdc13p-associated kinase activity rather than the cig2p-associated kinase activity.
Given these results, we used a rum1-HAcyr1Δsxa2Δ strain to test whether rum1p physically associated with cdc13p after pheromone addition. Immunoprecipitations of cdc13p and cig2p before and after addition of pheromone were analyzed by Western blotting with anti-HA antibodies. rum1p was found associated with cdc13p, and this association was increased after 3 h in P-factor (Figure 3B). rum1p was also found associated with cig2p, although the amount detected was lower (Figure 3B). In a reciprocal experiment, rum1HAp immunoprecipitation was found to coprecipitate 2 pmol of cig2p and 6 pmol of cdc13p (Figure 3C). Similar results were obtained by immunoprecipitations of rum1p from a cig2-HAcyr1Δsxa2Δ strain (our unpublished results). The different amounts of cig2p and cdc13p in rum1p precipitations are similar to the different cyclin B concentrations in the cell, because the level of cdc13p is about three times that of cig2p (Figure 3C and our unpublished results). The increased amount of cdc13p associated with rum1p is consistent with rum1p having an effect on the cdc13p-associated cdc2p protein kinase.
rum1p Is Required for Cyclosome-dependent Proteolysis of cdc13p during Pheromone-induced G1 Arrest
We also found that rum1p is required for cdc13p proteolysis during the response to pheromone. We monitored cdc13p levels after P-factor addition in rum1+ cells and found that they became reduced within 2 h of P-factor addition and were very low by 6 h after addition (Figure 4). This drop in level contributed to the observed inhibition of cdc13p-associated kinase activity. In rum1Δ cells, cdc13p levels remained completely constant (Figure 4). Cdc13 transcript levels were unchanged after addition of P-factor to rum1+ and rum1Δ cells, indicating that transcriptional control does not contribute to regulation of cdc13p in pheromone (our unpublished observations). These results establish that rum1p is required for the reduction in cdc13p levels observed after pheromone addition.
Figure 4.
rum1p is required for cdc13p degradation in pheromone. Cdc13p and α-tubulin protein levels in rum1+cyr1Δsxa2Δ and rum1Δcyr1Δsxa2Δ strains after addition of P-factor.
Given this result, we investigated whether cyclosome-mediated cyclin B degradation was required for pheromone-induced G1 arrest. The nuc2+ gene encodes a component of the cyclosome and is homologous to the budding yeast CDC27 gene (Hirano et al., 1990; Goebl and Yanagida, 1991). When pheromone was added to the temperature-sensitive nuc2–663 mutant at the permissive temperature of 25°C, only 10% of cells arrested in G1, demonstrating that complete nuc2+ activity is required to bring about G1 arrest (Figure 5A). This effect was completely reversed by nuc2+ expression from a plasmid (Figure 5A). Cdc13p level and associated kinase activities remained high (Figure 5, B and C, bottom panels), indicating that the nuc2 gene product and thus the cyclosome are required for pheromone-induced proteolysis of cdc13p. We have shown previously that nondegradable cdc13p prevents pheromone-induced G1 arrest (Stern and Nurse, 1997). Together these data suggest that in pheromone-treated cells cdc13p undergoes proteolysis by a mechanism that requires both rum1p and the cyclosome. This proteolysis keeps cdc13p-associated kinase activity low, allowing G1 arrest to be maintained in the presence of pheromone.
The Cyclosome Mediates cig2p Degradation in the Pheromone-induced G1 Arrest
The B-cyclins cig2p and cdc13p are redundant for promoting entry into S-phase, and so the cdc2p kinase activity associated with both cyclins needs to be inhibited to bring about G1 arrest in pheromone. Therefore we investigated the mechanism by which pheromone inhibits cig2p-associated kinase activity by monitoring cig2p levels. Cig2p levels and associated kinase activity decreased in pheromone-treated nuc2+ cells and reached low levels 4 h after P-factor addition (Figure 5, B and C, wt). In the nuc2–663 mutant, cig2p levels and cig2p-associated kinase activities remained high (Figure 5, B and C, top panels). This contrasts to the situation in a rum1Δ, where the cig2p-associated kinase activity was down-regulated in response to pheromone (Figure 3A). Cig2p levels were also down-regulated in a rum1Δ after addition of P-factor (Figure 5C). This was not due to reduced cig2 transcription, which was maintained at a constant level after addition of P-factor to rum1Δ cells (our unpublished observations). Thus, rum1Δ and nuc2–663 mutants are similar in that they fail to arrest in G1 in response to pheromone but differ in cig2p turnover, which can occur in a rum1Δ but not a nuc2–663 mutant. We conclude that cig2p-associated kinase activity is down-regulated in pheromone by cyclosome-induced cig2p proteolysis, but unlike the situation with cdc13p, this proteolysis does not require rum1p.
Effects of rum1 on Pheromone-induced Transcription
The experiments described above identify a role for rum1p in maintaining G1 arrest after pheromone addition. We next investigated whether lack of rum1 also affects the pheromone-induced transcription using the mating type gene mat1-Mm, which is specifically induced by P-factor (Willer et al., 1995). Figure 6A shows that mat1-Mm transcript was induced in a cyr1Δsxa2Δ after addition of P-factor. In a rum1Δcyr1Δsxa2Δ triple mutant, mat1-Mm transcripts were still induced by P-factor, but to a much lower level (Figure 6A).
Figure 6.
Expression of pheromone-dependent genes is restricted to G1. (A) Northern blot of RNA samples from cyr1Δsxa2Δ and rum1Δcyr1Δsxa2Δ strains after addition of P-factor probed for mat1-Mm and for ura4 as a loading control. (B) Cdc10–129cyr1Δsxa2Δ and cdc22-M45cyr1Δsxa2Δ strains arrested in G1 or S, respectively, after 6 h at 36°C; cdc2-M26cyr1Δsxa2Δ strain after nitrogen starvation for 20 h followed by 5 h at 36.5°C in nitrogen; cyr1Δsxa2Δ cells after 6 h in hydroxyurea. All strains were released from the block and incubated in P-factor for 90 min. Cyr1Δsxa2Δ and cdc25–22cyr1Δsxa2Δ strains were incubated at 36°C for 4 h; P-factor was added for 90 min at 36°C. Northern blots were probed for the P-factor–inducible genes mat1-Mm and fus1 and for his3 as a loading control. (C) A rum1Δcdc10–129cyr1Δsxa2Δ mutant was exposed to P-factor with or without a 2-h preincubation at 36°C. DNA content analysis (top panel) and Northern blot probed with mat1-Mm- and his7-specific DNA (bottom panel).
This reduction could have been either because rum1 was directly required for activation of pheromone-dependent transcription or because full induction of pheromone-dependent genes required G1 arrest, which was defective in the rum1 mutant. To distinguish between these two explanations, we assessed the expression of P-factor–induced transcription at various stages in the cell cycle. A cyr1Δsxa2Δ strain was arrested in G1 using the temperature-sensitive cdc10–129 and cdc2-M26 mutants or in early S-phase using hydroxyurea and the temperature-sensitive cdc22-M46 mutant. The cultures were shifted to 25°C after cell cycle arrest and incubated in P-factor for 90 min. Samples for RNA preparation were taken at the beginning and the end of the P-factor treatment, and the transcript levels of two P-factor–dependent genes, mat1-Mm (Willer et al., 1995) and fus1 (Petersen et al., 1995), were assessed. Both genes were induced by P-factor in G1-arrested cdc10–129 and cdc2-M26 mutant cells, but little induction was observed in cells released from the S-phase blocks (Figure 6B). To test pheromone-dependent transcription in G2 cells, a cdc25–22cyr1Δsxa2Δ strain was arrested in G2 for 4 h at 36°C and kept at the restrictive temperature during the subsequent 90 min exposure to P-factor. Although fus1 and mat1-Mm transcripts were induced in a cdc25+ control strain, induction was severely reduced in cdc25–22 mutant cells. These results indicate that P-factor can induce expression of pheromone-dependent genes in the G1 phase of the cell cycle, but expression is much reduced at later stages of the cell cycle. Therefore the low level of mat1-Mm expression in rum1Δ cells is likely to be caused by the failure of this strain to arrest in G1.
To test this conclusion, a cdc10–129rum1Δcyr1Δsxa2Δ quadruple mutant was exposed to P-factor after a 2 h incubation at 36°C. P-factor induced a higher level of mat1-Mm transcript in cells that were prearrested in G1 than in cells without the 36°C preincubation (Figure 6C). This result indicates that the level of mat1-Mm transcript in a rum1Δ is reduced because of the shortened G1 and can be elevated by prearresting rum1Δ cells in G1. We conclude that pheromone can induce transcription only in G1-arrested cells and that the effects of rum1 on pheromone-induced transcription are because rum1p is required to maintain cells in G1 for that induction to take place.
DISCUSSION
In this article we have investigated the effects of the CKI rum1p and cyclosome-dependent cyclin B degradation on pheromone-induced inhibition of the CDK cdc2p. Our major observations are as follows: 1) cyclosome-mediated degradation of cig2p and cdc13p is essential for down-regulation of cyclin B–cdc2p kinase activity during pheromone-induced G1 arrest; 2) rum1p is required to maintain this G1 arrest and specifically inhibits the cdc13p-cdc2p kinase; 3) rum1p mediates cdc13p turnover, whereas cig2p turnover can occur in a rum1-independent manner, indicating that rum1p is specifically required for cdc13p degradation by the cyclosome; and 4) pheromone-induced transcription requires cells to be in G1 and is independent of rum1p.
Proteolysis of both cig2p and cdc13p B-cyclins in pheromone was shown to require the cyclosome by the lack of proteolysis in cells defective for the nuc2p cyclosome subunit (Figure 5). Thus cyclosome-mediated degradation of these B-cyclins is an important mechanism for pheromone-induced G1 arrest. The maintenance of cyclosome activity during pheromone-induced G1 arrest may involve cAMP. The cyclosome is stabilized by low cAMP levels, and mutants in cut4, the fission yeast Apc1/BimE cyclosome subunit, are sensitive to high levels of cAMP (Yamashita et al., 1996). Pheromone response requires low levels of cAMP, and this could act in part by maintaining the cyclosome activity required to bring about cig2p and cdc13p proteolysis.
The CKI rum1p is also required for cdc13p cyclin B proteolysis and for down-regulation of cdc13p–cdc2p CDK activity. Levels of cdc13p and cdc13p–cdc2p CDK activity remain high in pheromone-treated rum1Δ cells, and rum1p physically interacts with cdc13p (Figure 3). This effect on proteolysis is specific because cig2p cyclin degradation does not require rum1p, even though rum1p can associate with cig2p (Figure 3). The fact that cig2p proteolysis still occurs in rum1Δ cells in a cyclosome-dependent manner indicates that the failure to turn over cdc13p is not simply due to the rum1Δ cells proceeding to a later stage in the cell cycle when the cyclosome is inactive. These data corroborate recent results that suggest that rum1p is required for cdc13p degradation in G1 cells arrested at the cdc10 block point (Correa-Bordes et al., 1997). We propose that rum1p in pheromone-treated cells acts as an adaptor protein specifically targeting cdc13p for degradation by the cyclosome during G1 and thus maintaining G1 arrest. rum1p is not required for the cdc13p proteolysis occurring at mitotic exit but may be necessary for inhibiting and degrading the cdc13p kinase during G1. In contrast, rum1p is not required for cig2p proteolysis, suggesting either that no adapter protein is necessary or that one still has to be identified. Similar to rum1Δ, mutants in the srw1+ gene specifically stabilize cdc13p but not cig2p (Yamaguchi et al., 1998). rum1+ and srw1+ might act together to target cdc13p for degradation.
The initial G1 arrest brought about by pheromone is likely to involve inhibition of the cig2p–cdc2p protein kinase by a mechanism that is independent of rum1p, although the molecular mechanism underlying pheromone signaling and the inhibition and proteolysis of the cig2p–cdc2p protein kinase remain to be elucidated. We imagine that these mechanisms bring about a transient G1 arrest but that this cannot be maintained without further inhibition of the cdc13p–cdc2p protein kinase, because the latter can substitute for cig2p–cdc2p in bringing about S-phase (Fisher and Nurse 1996; Stern and Nurse, 1996). The transient G1 arrest leads to a rise in rum1p levels that in turn prevents cdc13p–cdc2p protein kinase activity from increasing.
rum1p may also be able to inhibit cig2p–cdc2p activity at least temporarily, as suggested previously (Martin-Castellanos et al., 1996), given that cig2p and rum1p physically interact; however, cig2p is unlikely to be an important long-term target of rum1p given that a cig2Δ does not rescue the G1 arrest defect of a rum1Δ. The fact that a cig2Δ can rescue the sterility of a rum1Δ may be because conjugation and sporulation require both a pheromone and a starvation signal, and starvation-induced G1 arrest is partially restored in a cig2Δ (Martin-Castellanos et al., 1996).
The rum1Δ phenotype in pheromone is superficially reminiscent of the pheromone response of far1 mutants in budding yeast. Although both rum1 and FAR1 encode CKIs that are essential for pheromone-induced G1 arrest, there are important differences between their activities. Far1p inhibits the Cdc28p activity associated with the G1 Clnp cyclins (Peter and Herskowitz, 1994), whereas rum1p specifically inhibits cdc2p associated with the mitotic B-cyclin cdc13p (Correa-Bordes and Nurse, 1995). Far1p is required exclusively for the pheromone response and is only active as a CDK inhibitor after phosphorylation by the pheromone-dependent MAP kinase Fus3p (Peter et al., 1993). In contrast, the rum1 function is not confined to pheromone response, being required in other situations with a prolonged G1 phase, such as the extended G1 in a wee1–50 mutant or after nitrogen starvation (Moreno and Nurse, 1994), and in cells arrested in G1 by a cdc10.129 block (Correa-Bordes and Nurse, 1995). Also there is no evidence that rum1p needs an MAP kinase-dependent phosphorylation event for activation. Bacterially produced rum1p is fully active as an inhibitor of cdc13p–cdc2p kinase (Correa-Bordes and Nurse, 1995), and a truncated rum1 lacking all putative MAP kinase phosphorylation sites is able to rescue the sterility of a rum1Δ (Stern and Nurse, unpublished observations). rum1p has more in common with the second budding yeast CKI, Sic1p. Both are induced in G1 and inhibit cyclin B-associated CDK to prevent premature onset of S-phase. However, despite these similarities, there is only very limited sequence homology between Sic1p and rum1p. The phenotypic consequences of loss of rum1 and SIC1 are also different, because SIC1 is not required for sexual differentiation.
In this study we also found that pheromone induces transcription of the pheromone-dependent genes mat1-Mm and fus1 only in G1 cells (Figure 6). The cell cycle regulation of pheromone-dependent transcription might help restrict conjugation to the G1 phase of the cell cycle. Yeast cells cannot conjugate when arrested in G2, and mutants such as rum1Δ and nuc2–663 that fail to arrest in G1 under mating conditions are sterile (Moreno and Nurse, 1994; Kumada et al., 1995). The failure to express pheromone-dependent genes later in the cell cycle could be due to reduced pheromone signaling or because a component of the transcriptional apparatus can only be activated in G1. A possible candidate is the transcription factor ste11p (Sugimoto et al., 1991), which is required for both nitrogen starvation and pheromone-induced transcription (Aono et al., 1994; Petersen et al., 1995). Pheromone-dependent transcription is also cell cycle regulated in budding yeast (Oehlen and Cross, 1994). The expression profile of pheromone-dependent transcripts is controlled by the activity of the G1 CDK activity, Clnp–Cdc28p (Oehlen and Cross, 1994). Transcript levels are high in early G1 and in S and G2 when Clnp–Cdc28p protein kinase activity is low, and they dip in late G1 when Clnp–Cdc28p protein kinase activity is high. Fission yeast may use a similar mechanism with cyclin B–cdc2p kinase activity, which is present from late G1 until the end of mitosis, to restrict pheromone-induced transcription to G1.
Fission yeast appears to use quick and reversible CKI action with irreversible cyclin turnover to inhibit B-cyclin kinases and maintain pheromone-induced G1 arrest. A combination of CKI-mediated inhibition and proteolysis also controls the Clbp-associated kinase in budding yeast. Overexpression of nondegradable mitotic Clb2p can overcome pheromone-induced G1 arrest (Amon et al., 1994), and mutants in the cyclin B- and CDK-specific CKI Sic1p undergo premature S-phase after expression of nondegradable Clb5p in early G1 cells (Schwob et al., 1994). A recent study shows that cyclosome mutants in budding yeast are defective in pheromone-induced G1 arrest similar to fission yeast nuc2 mutants (Irniger and Nasmyth, 1997). Precocious S-phase in cyclosome mutants can be rescued by ectopic expression of Sic1p (Irniger and Nasmyth, 1997), indicating that CKI and cyclin proteolysis cooperate in G1 regulation as in fission yeast. A major difference with fission yeast is that budding yeast secures a low cyclin B–associated kinase in early G1 by both transcriptional and posttranscriptional mechanisms. In fission yeast cdc13+ and cig2+ transcription are not down-regulated in G1 (Correa-Bordes and Nurse, 1995; Stern and Nurse, 1997), leaving the posttranscriptional mechanisms of cyclin B degradation and the CKI rum1p as the sole control of cyclin B-associated kinase in G1. Posttranscriptional control using CKIs and regulation of cyclin B turnover may be the more generally used mechanism to control cyclin B–CDKs in G1, with a transcriptional control providing a more robust control system.
It will be important to determine whether both CKIs and cyclosome-mediated proteolysis are involved in down-regulating cyclin B–CDKs or the related cyclin A–CDK in G1 in higher eukaryotes. Cyclin A-associated CDKs have been implicated in the control of both S-phase (Girard et al., 1991; Pagano et al., 1992) and mitosis (Lehner and O’Farrell, 1989; Minshull et al., 1989) in the Metazoa, and it may be crucial to tightly control its activity during G1. Loss of the Drosophila gene fizzy-related, which is involved in degradation of A and B cyclins, results in cells failing to exit the cell cycle in G1, suggesting that down-regulation of mitotic cyclins in G1 might be equally important in higher eukaryotes as in yeast (Sigrist and Lehner, 1997). The Drosophila roughex (rux) gene also controls cyclin A kinase activity in G1 (Gönczy et al., 1994; Thomas et al., 1994; Dong et al., 1997). Like the rum1Δ mutant, rux mutant cells fail to arrest in G1, and they enter S-phase prematurely, with elevated cyclin A-associated kinase activity (Thomas et al., 1994; Sprenger et al., 1997; Thomas et al., 1997). rux may have a task similar to that of rum1 in fission yeast or SIC1 in budding yeast by preventing cyclin A from activating S-phase in early G1.
ACKNOWLEDGMENTS
We thank Jacky Hayles, Benjamin Baum, Patrick Zarzov, and Takashi Toda for critical comments and suggestions, J. Correa-Bordes for his assistance concerning kinase assays and rum1p biochemistry, Takashi Toda for providing a nuc2–663 strain, and all members of the ICRF Cell Cycle Laboratory for their help and support. This work was funded by ICRF. B.S. was also supported by a Boehringer Ingelheim Fonds fellowship.
REFERENCES
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Aono T, Yanai H, Miki F, Davey J, Shimoda C. Mating pheromone induced expression of the mat1-Pm gene of Schizosaccharomyces pombe: identification of signalling components and characterisation of upstream controlling elements. Yeast. 1994;10:757–770. doi: 10.1002/yea.320100607. [DOI] [PubMed] [Google Scholar]
- Benito J, Martin-Castellanos C, Moreno S. Regulation of the G1 phase of the cell cycle by periodic stabilisation and degradation of the p25rum1 CDK inhibitor. EMBO J. 1998;17:482–497. doi: 10.1093/emboj/17.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R, Beach D. Involvement of cdc13+ in mitotic control in Schizosaccharomyces pombe: possible interaction of the gene product with microtubules. EMBO J. 1988;7:2321–2327. doi: 10.1002/j.1460-2075.1988.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Alfa CE, Hyams JS, Beach DH. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Correa-Bordes J, Gulli MP, Nurse P. p25rum1 promotes proteolysis of the mitotic B-cyclin p56cdc13 during G1 of the fission yeast cell cycle. EMBO J. 1997;16:4657–4664. doi: 10.1093/emboj/16.15.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- Davey J, Nielsen O. Mutations in cyr1 and pat1 reveal pheromone-induced G1 arrest in the fission yeast Schizosaccharomyces pombe. Curr Genet. 1994;26:105–112. doi: 10.1007/BF00313796. [DOI] [PubMed] [Google Scholar]
- Dong X, Zavitz KH, Thomas BJ, Lin M, Campbell S, Zipursky SL. Control of G1 in the developing Drosophila eye: rca1 regulates cyclin A. Genes Dev. 1997;11:94–105. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- Donovan JD, Toyn JH, Johnson AL, Johnston LH. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Cross F. A novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. A single fission yeast mitotic cyclin B-p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher AB. Characterization of four B-type cyclin genes of the budding yeast S. cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiara JB, Richardson HE, Sugimoto K, Henze M, Lew DJ, Wittenberg C, Reed SI. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991;65:163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- Girard F, Strausfeld U, Fernandez A, Lamb NJC. Cyclin-A is required for the onset of DNA-replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Thomas BJ, DiNardo S. roughex is a dose-dependent regulator of the second meiotic division during Drosophila spermatogenesis. Cell. 1994;77:1015–1025. doi: 10.1016/0092-8674(94)90441-3. [DOI] [PubMed] [Google Scholar]
- Hagan I, Hayles J, Nurse P. Cloning and sequencing of the cyclin-related cdc13+ gene and a cytological study of its role in fission yeast mitosis. J Cell Sci. 1988;91:587–595. doi: 10.1242/jcs.91.4.587. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- Imai Y, Yamamoto M. Schizosaccharomyces pombe sxa1+ and sxa2+ encode putative proteases involved in the mating response. Mol Cell Biol. 1992;12:1827–1834. doi: 10.1128/mcb.12.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Kelly TJ. Rum1 and Cdc18 link inhibition of cyclin-dependent kinase to the initiation of DNA replication in Schizosaccharomyces pombe. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- Kawamukai M, Ferguson K, Wigler M, Young D. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell Regul. 1991;2:155–164. doi: 10.1091/mbc.2.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters J, Tugendreich S, Rolfe M, Hieter P, Krischner M. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kühne C, Linder P. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 1993;12:3437–3447. doi: 10.1002/j.1460-2075.1993.tb06018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada K, Su S, Yanagida M, Toda T. Fission yeast TPR-family protein nuc2 is required for G1-arrest upon nitrogen starvation and is an inhibitor of septum formation. J Cell Sci. 1995;108:895–905. doi: 10.1242/jcs.108.3.895. [DOI] [PubMed] [Google Scholar]
- Ladds G, Rasmussen EM, Young T, Nielsen O, Davey J. The sxa2-dependent inactivation of the P-factor mating pheromone in the fission yeast Schizosaccharomyces pombe. Mol Microbiol. 1996;20:35–42. doi: 10.1111/j.1365-2958.1996.tb02486.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehner CF, O’Farrell PH. Expression and function of Drosophila cyclin A during embryonic cell cycle progression. Cell. 1989;56:957–968. doi: 10.1016/0092-8674(89)90629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Maeda T, Mochizuki N, Yamamoto M. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1990;87:7814–7818. doi: 10.1073/pnas.87.20.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Labib K, Moreno S. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 CDK inhibitor. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD. An inhibitor of p34(cdc28) protein-kinase activity from Saccharomyces-cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- Minshull J, Blow JJ, Hunt T. Translation of cyclin mRNA is necessary for extracts of activated Xenopus eggs to enter mitosis. Cell. 1989;56:947–956. doi: 10.1016/0092-8674(89)90628-4. [DOI] [PubMed] [Google Scholar]
- Mondesert O, McGowan C, Russell P. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–972. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Peters J-M, King RW, Höög C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- Petersen J, Weilguny D, Egel R, Nielsen O. Characterisation of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI. Cyclin-B homologs in Saccharomyces cerevisiae function in S-phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Yang Q, Futcher AB. Linkage of replication to Start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40Sic1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Sigrist S, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Sprenger F, Yakubovich N, O’Farrell PH. S-phase function of Drosophila cyclin A and its downregulation in G1 phase. Curr Biol. 1997;7:488–499. doi: 10.1016/s0960-9822(06)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B, Nurse P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996;12:345–350. [PubMed] [Google Scholar]
- Stern B, Nurse PN. Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclin B-p34cdc2 kinase. EMBO J. 1997;16:534–544. doi: 10.1093/emboj/16.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Horshko J, Iuca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A, Iino Y, Wantanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11 encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Gunning DA, Cho J, Zipursky SL. Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Zavutz KH, Dong X, Lane ME, Weigmann K, Finley RL, Brent R, Lehner CF, Zipursky SL. Roughex down-regulates G2 cyclins in G1. Genes Dev. 1997;11:1289–1298. doi: 10.1101/gad.11.10.1289. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs Colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol Cell Biol. 1995;15:4964–4970. doi: 10.1128/mcb.15.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Murakami H, Okayama H. A WD repeat protein controls the cell cycle and differentiation by negatively regulating cdc2/B-type cyclin complexes. Mol Biol Cell. 1998;8:2475–2486. doi: 10.1091/mbc.8.12.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature. 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]