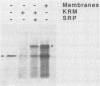
Abstract
Class II membrane glycoproteins share a common topology of the NH2 terminus inside and the COOH terminus outside the cell. Their transport to the cell surface is initiated by the function of a single hydrophobic domain near the NH2 terminus. This functional domain serves both as an uncleaved signal sequence and as a transmembrane anchor. We examined the signal and anchor functions of influenza virus neuraminidase, a prototype class II membrane glycoprotein, by deletion analysis of its long, uncharged amino-terminal region. The results presented here show that the entire stretch of 29 uncharged amino acids (7 to 35) is not required for either a signal sequence or an anchor sequence function. On the basis of translocation and membrane stability data for different mutants, we suggest that the first 20 amino acid residues (7 to 27) are likely to provide the hydrophobic core for these functions and that within this putative subdomain some sequences are more efficient than the other sequences in providing a translocation function. Finally, it appears that neuraminidase and its mutant proteins are translocated with the proper orientation, regardless of the characteristics of the flanking sequences.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams G. A., Rose J. K. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985 Jul;41(3):1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- Blackburn P., Wilson G., Moore S. Ribonuclease inhibitor from human placenta. Purification and properties. J Biol Chem. 1977 Aug 25;252(16):5904–5910. [PubMed] [Google Scholar]
- Bos T. J., Davis A. R., Nayak D. P. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2327–2331. doi: 10.1073/pnas.81.8.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. S., Gierasch L. M. Molecular mechanisms of protein secretion: the role of the signal sequence. Adv Protein Chem. 1986;38:109–180. doi: 10.1016/s0065-3233(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Colman P. M., Ward C. W. Structure and diversity of influenza virus neuraminidase. Curr Top Microbiol Immunol. 1985;114:177–255. doi: 10.1007/978-3-642-70227-3_5. [DOI] [PubMed] [Google Scholar]
- Cutler D. F., Melancon P., Garoff H. Mutants of the membrane-binding region of Semliki Forest virus E2 protein. II. Topology and membrane binding. J Cell Biol. 1986 Mar;102(3):902–910. doi: 10.1083/jcb.102.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R. E., Wickner W. Characterization of the internal signal-anchor domain of Escherichia coli leader peptidase. J Biol Chem. 1988 Jan 5;263(1):404–408. [PubMed] [Google Scholar]
- Davis A. R., Bos T. J., Nayak D. P. Active influenza virus neuraminidase is expressed in monkey cells from cDNA cloned in simian virus 40 vectors. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3976–3980. doi: 10.1073/pnas.80.13.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S. D., Silhavy T. J. Importance of secondary structure in the signal sequence for protein secretion. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4599–4603. doi: 10.1073/pnas.80.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982 Apr;93(1):97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiti A. L., Nayak D. P. Complete nucleotide sequence of the neuraminidase gene of human influenza virus A/WSN/33. J Virol. 1982 Feb;41(2):730–734. doi: 10.1128/jvi.41.2.730-734.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi I., Gentz R. A functional interaction between the signal peptide and the translation apparatus is detected by the use of a single point mutation which blocks translocation across mammalian endoplasmic reticulum. J Biol Chem. 1987 Jul 25;262(21):10189–10194. [PubMed] [Google Scholar]
- Kaiser C. A., Preuss D., Grisafi P., Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987 Jan 16;235(4786):312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipp J., Dobberstein B. Signal recognition particle-dependent membrane insertion of mouse invariant chain: a membrane-spanning protein with a cytoplasmically exposed amino terminus. J Cell Biol. 1986 Jun;102(6):2169–2175. doi: 10.1083/jcb.102.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp J., Dobberstein B. The membrane-spanning segment of invariant chain (I gamma) contains a potentially cleavable signal sequence. Cell. 1986 Sep 26;46(7):1103–1112. doi: 10.1016/0092-8674(86)90710-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L., Lin B. C., Sveda M. M., Lai C. J. Glycosylation and surface expression of the influenza virus neuraminidase requires the N-terminal hydrophobic region. Mol Cell Biol. 1984 Jan;4(1):8–16. doi: 10.1128/mcb.4.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mize N. K., Andrews D. W., Lingappa V. R. A stop transfer sequence recognizes receptors for nascent chain translocation across the endoplasmic reticulum membrane. Cell. 1986 Dec 5;47(5):711–719. doi: 10.1016/0092-8674(86)90514-3. [DOI] [PubMed] [Google Scholar]
- Morimoto T., Arpin M., Gaetani S. Use of proteases for the study of membrane insertion. Methods Enzymol. 1983;96:121–150. doi: 10.1016/s0076-6879(83)96013-5. [DOI] [PubMed] [Google Scholar]
- Perara E., Lingappa V. R. A former amino terminal signal sequence engineered to an internal location directs translocation of both flanking protein domains. J Cell Biol. 1985 Dec;101(6):2292–2301. doi: 10.1083/jcb.101.6.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian N., Nayak D. P. Mutational analysis of the signal-anchor domain of influenza virus neuraminidase. Proc Natl Acad Sci U S A. 1987 Jan;84(1):1–5. doi: 10.1073/pnas.84.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess M., Handschin C. Deletion analysis of the internal signal-anchor domain of the human asialoglycoprotein receptor H1. EMBO J. 1987 Sep;6(9):2683–2691. doi: 10.1002/j.1460-2075.1987.tb02560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess M., Lodish H. F. An internal signal sequence: the asialoglycoprotein receptor membrane anchor. Cell. 1986 Jan 17;44(1):177–185. doi: 10.1016/0092-8674(86)90496-4. [DOI] [PubMed] [Google Scholar]
- Tarentino A. L., Maley F. Purification and properties of an endo-beta-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974 Feb 10;249(3):811–817. [PubMed] [Google Scholar]
- Walter P., Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle: a ribonucleoprotein required for cotranslational translocation of proteins, isolation and properties. Methods Enzymol. 1983;96:682–691. doi: 10.1016/s0076-6879(83)96057-3. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981 Nov;91(2 Pt 1):557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. T., Lodish H. F. Multiple mechanisms of protein insertion into and across membranes. Science. 1985 Oct 25;230(4724):400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Zerial M., Huylebroeck D., Garoff H. Foreign transmembrane peptides replacing the internal signal sequence of transferrin receptor allow its translocation and membrane binding. Cell. 1987 Jan 16;48(1):147–155. doi: 10.1016/0092-8674(87)90365-5. [DOI] [PubMed] [Google Scholar]
- Zerial M., Melancon P., Schneider C., Garoff H. The transmembrane segment of the human transferrin receptor functions as a signal peptide. EMBO J. 1986 Jul;5(7):1543–1550. doi: 10.1002/j.1460-2075.1986.tb04395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Towards a comparative anatomy of N-terminal topogenic protein sequences. J Mol Biol. 1986 May 5;189(1):239–242. doi: 10.1016/0022-2836(86)90394-3. [DOI] [PubMed] [Google Scholar]