Abstract
Integral membrane proteins (IMPs) contain localization signals necessary for targeting to their resident subcellular compartments. To define signals that mediate localization to the Golgi complex, we have analyzed a resident IMP of the Saccharomyces cerevisiae Golgi complex, guanosine diphosphatase (GDPase). GDPase, which is necessary for Golgi-specific glycosylation reactions, is a type II IMP with a short amino-terminal cytoplasmic domain, a single transmembrane domain (TMD), and a large catalytic lumenal domain. Regions specifying Golgi localization were identified by analyzing recombinant proteins either lacking GDPase domains or containing corresponding domains from type II vacuolar IMPs. Neither deletion nor substitution of the GDPase cytoplasmic domain perturbed Golgi localization. Exchanging the GDPase TMD with vacuolar protein TMDs only marginally affected Golgi localization. Replacement of the lumenal domain resulted in mislocalization of the chimeric protein from the Golgi to the vacuole, but a similar substitution leaving 34 amino acids of the GDPase lumenal domain intact was properly localized. These results identify a major Golgi localization determinant in the membrane-adjacent lumenal region (stem) of GDPase. Although necessary, the stem domain is not sufficient to mediate localization; in addition, a membrane-anchoring domain and either the cytoplasmic or full-length lumenal domain must be present to maintain Golgi residence. The importance of lumenal domain sequences in GDPase Golgi localization and the requirement for multiple hydrophilic protein domains support a model for Golgi localization invoking protein–protein interactions rather than interactions between the TMD and the lipid bilayer.
INTRODUCTION
The functional integrity of a cell depends on sorting of proteins to their appropriate subcellular compartments. Localization to a particular subcellular residence is achieved by a combination of sorting information within a protein, usually a specific sequence or structural motif (localization signal), and a cellular mechanism to recognize and engage the sorting signal. For resident membrane proteins of eukaryotic secretory pathway organelles, localization signals may direct proteins to their final location by specifying inclusion into vesicles destined for subsequent compartments (Pelham and Munro, 1993). Alternatively, the signals can maintain a protein in residence either by preventing incorporation into departing transport vesicles, or by signaling retrieval from distal compartments of the pathway (Pelham and Munro, 1993).
The Golgi complex, which comprises multiple subcompartments, is the central sorting station for membrane proteins in the secretory pathway (Mellman and Simons, 1992). In the cis-Golgi network (CGN),1 endoplasmic reticulum (ER) proteins are distinguished from proteins destined for transport to more distal compartments, and are incorporated into transport vesicles for return to the ER. Proteins in transit through the Golgi pass from the CGN to the medial Golgi compartment which serves as a major site for modifying glycoprotein oligosaccharides. In the following subcompartment, the trans Golgi network (TGN), proteins are sorted into distinct transport vesicles bound for the plasma membrane or for endosomes. Confronted by the dynamic flux of proteins and lipids through the Golgi complex, resident proteins also must be distinguished, and properly localized to their Golgi subcompartment. This process then represents a distinct sorting function of the Golgi complex.
Resident proteins of the Golgi complex in the yeast Saccharomyces cerevisiae are organized into distinct subcompartments that are functionally analogous to those defined in mammalian cells, although the yeast organelle is not arranged into the cisternal stacks characteristic of the mammalian Golgi complex (Cunningham and Wickner, 1989; Franzusoff and Schekman, 1989; Franzusoff et al., 1991; Bowser and Novick, 1991; Graham and Emr, 1991; Preuss et al., 1992; Bryant and Boyd, 1993; Wilsbach and Payne, 1993; Whitters et al., 1994; Gaynor et al., 1994). In the CGN-like early compartment, the α-1,6-mannosyltransferase Och1p initiates the addition of mannose residues to core oligosaccharides (Nakayama et al., 1992, 1997; Nakanishi-Shindo et al., 1993; Gaynor et al., 1994; Harris and Waters, 1996), and other α1,6-mannosyltransferases extend the α-1,6-mannose chains (Jungmann and Munro, 1998). In the medial Golgi subcompartment, α1,2-mannosyltransferases, including Kre2p/Mnt1p, and a chain-terminating α1,3-mannosyltransferase encoded by MNN1 elaborate the side chains (Herscovics and Orlean, 1993). Proteases required for maturation of the α-factor–mating pheromone precursor, such as Kex2p, are localized to a TGN-like compartment which is also the site of protein sorting to the lysosome-like vacuole (Graham and Emr, 1991).
Both mammalian and yeast glycosyltransferases have been the subject of studies to identify protein domains required for localization in Golgi subcompartments. These proteins have in common a type II topology with a relatively short amino-terminal cytoplasmic domain, a single-transmembrane domain (TMD), and a large lumenal domain. Mammalian transferases generally require the TMD and an adjacent lumenal region, referred to as the stem domain, for efficient Golgi localization (reviewed in Machamer, 1993; Pelham and Munro, 1993; Nilsson and Warren, 1994; Colley, 1997). In yeast, multiple domains contribute to localization of the two transferases examined in most detail, Kre2p/Mnt1p and Mnn1p. Localization of Kre2p to the medial Golgi compartment requires the cytoplasmic domain, whereas the TMD can be substituted and the stem region can be deleted without dramatically affecting Golgi localization. However, all three regions are necessary for proper localization of a chimeric protein containing reporter sequences (Lussier et al., 1995). In contrast, the sorting of Mnn1p to its resident Golgi compartment does not require the cytoplasmic domain; rather, the TMD and lumenal regions contain separate signals sufficient for localization (Graham et al., 1994; Graham and Krasnov, 1995).
To extend the characterization of Golgi localization signals in yeast, we have characterized guanosine diphosphatase (GDPase), a type II Golgi membrane glycoprotein of ∼60 kDa (Berninsone et al., 1995). GDPase cleaves the GDP product that originates from GDP-mannose after mannosylation of glycoproteins in the Golgi complex. Production of GMP by GDPase is critical for glycosylation, because import of GDP-mannose into the Golgi lumen is coupled to GMP export (Abeijon et al., 1993). As expected from its general role in mannosylation, GDPase activity colocalizes by subcellular fractionation with both the CGN enzyme Och1p and with the medial Golgi resident Mnn1p (Graham et al., 1994; Lupashin et al., 1996). In contrast, GDPase did not fractionate with Kex2p in a purified preparation of the yeast TGN (Whitters et al., 1994), nor was GDPase found in ER fractions (Abeijon et al., 1989). Thus, GDPase is specifically localized to the yeast Golgi complex but distributes in multiple compartments.
Here we characterize the Golgi localization determinants in GDPase through an analysis of mutant and chimeric proteins. Localization of GDPase to the Golgi requires the lumenal membrane-adjacent region (stem) anchored to the membrane through a TMD. The specific sequence of the TMD has only a minor role in targeting to the Golgi complex. However, the TMD and stem regions are not sufficient to mediate localization of reporter sequences; either the cytoplasmic domain or full-length lumenal domain of GDPase also must be present to maintain Golgi localization of a chimeric protein. Thus, the localization determinants of GDPase are distinct from, but share features with, those of proteins with more restricted distribution in the Golgi complex.
MATERIALS AND METHODS
Media and strains
YPD media contains 1% Bacto-yeast extract, 2% Bactopeptone (Difco Laboratories, Detroit, MI), and 2% dextrose. SD media contains 0.67% yeast nitrogen base without amino acids (Difco) and 2% dextrose. Supplemented SD is SD containing 40 μg/ml adenine, 30 μg/ml leucine and lysine, and 20 μg/ml histidine, uracil, and tryptophan (Sigma, St. Louis, MO). SD CAA is supplemented SD containing 5 mg/ml vitamin assay casamino acid (CAA) mix (Difco). SD CAA-ura is SD CAA lacking uracil. SDYE is SD with 0.2% yeast extract. Cell densities in liquid culture were measured in a 1-cm plastic cuvette in a Beckman Instruments (Palo Alto, CA) DU-62 spectrophotometer. One OD500 unit is equivalent to 2.3 × 107 cells/ml.
The genotypes of yeast strains used in this study are listed in Table 1.
Table 1.
List of Saccharomyces cerevisiae strains
| Strain | Genotype | Reference |
|---|---|---|
| G2-11 | MATα gda1::LEU2 ade2-101(oc) his3-Δ200 leu2-Δ1 lys2-801(am) trp1-1Δ1 ura3-52 | Abeijon et al. (1993) |
| GPY1452 | MATα gda1::LEU2 ade2-101(oc) his3-Δ200 leu2-Δ1 lys2-801(am) trp1-1Δ1 ura3-52 pep4Δ | This study |
| DKY6280 | MATa pho8::TRP1 ade2-101 his3-Δ200 leu2-3, 112 suc2Δ9 trp1-Δ901 ura3-52 | Klionsky and Emr (1989) |
| GPY1250 | MATa pho8::TRP1 ade2-101 his3-Δ200 leu2-3, 112 suc2Δ9 trp1-Δ901 ura3-52 pep4::LEU2 | This study |
| SEY5188 | MATa sec18-1 leu2-3, 112 suc2-Δ9 ura3-52 | Graham and Emr (1991) |
| SEY6210 | MATα his3-Δ200 leu2-3, 112 lys2-801 suc2-Δ9 trp1-Δ901 ura3-52 | Robinson et al. (1988) |
The PEP4 gene was disrupted in DKY6280 and G2-11 to generate strains GPY1250 and GPY1452, respectively, by single-step gene replacement (Rothstein, 1991) using plasmid pTS17 (PEP4::LEU2) for GPY1250 and pTS15 (PEP4::URA3) for GPY1452 (plasmids were gifts from Tom Stevens, University of Oregon, Eugene, OR). Loss of PEP4 function was confirmed in these strains by immunoblotting for mature carboxypeptidase Y (CPY). All plasmids transformed into these strains used uracil-based selection; therefore, to convert GPY1452 to ura3, cells were grown on 5-fluororotic acid to select for cells that had converted the URA3 marker present in the disrupted PEP4 gene to ura3.
Transformations into yeast were performed by the lithium acetate procedure (Ito et al., 1983).
Plasmid constructions
DNA manipulations were carried out essentially as described by Sambrook et al. (1989). A plasmid containing the GDA1 gene, which encodes GDPase (p13HB in Abeijon et al., 1993) on a pBluescript vector (Stratagene, La Jolla, CA) was a generous gift from C. Hirschberg (University of Massachusetts Medical Center, Boston, MA). To generate pRS GGG, a 2168-base pair (bp) HindIII–NheI (filled-in) fragment containing the 1557 bp GDA1 open reading frame was subcloned into the HindIII and SmaI sites in pRS316 (URA3) (Sikorski and Hieter, 1989). A BamHI–KpnI fragment from pRS GGG was subcloned into pRS314 (TRP1) to generate the GDPase-expressing plasmid used in the double-label immunofluorescence experiments. The numbering system for GDA1 DNA sequences is as follows: 1–132, promoter sequences; 133–159, cytoplasmic sequences; 160–204, TMD sequences; and 205-1689, lumenal sequences. Similarly for PHO8, which encodes alkaline phosphatase (ALP) (Kaneko et al., 1987), a 4-kilobase BamHI fragment containing the 1702-bp open reading frame of PHO8 was inserted into the BamHI site of pRS316. The numbering system for PHO8 DNA sequences is as follows: 1–99, cytoplasmic sequences; 100–177, TMD sequences; and 178-1702, lumenal sequences.
The PCR (Saiki et al., 1988) was used in constructing the plasmids for our analysis. All PCR fragments were sequenced to confirm that errors had not occurred during amplification. Sequencing was either performed by the dideoxy chain termination procedure (Sanger et al., 1977) using the Sequenase enzyme (United States Biochemical, Cleveland, OH), α-35S-dATP, and appropriate DNA primers or performed in the University of California Los Angeles (UCLA) DNA Sequencing Facility using the ABI PRISM dye terminator cycle sequencing ready reaction kit and AmpliTaq DNA polymerase from Perkin Elmer (Foster City, CA). All constructs were subcloned into the pRS316 vector and have GDA1 promoter sequences except pRS PHO8, which has native PHO8 promoter sequences. The procedure for construction of each chimeric protein is detailed below, and a partial peptide sequence for each construct is shown in Table 2.
Table 2.
Partial amino acid sequence of GDPase constructs
| Construct | Cytoplasmic tail sequence | Transmembrane domain sequence | Partial lumenal domain sequence |
|---|---|---|---|
| GGG | MAPIFRNYR | FAIGAFAVIMLILLI | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND |
| - -GG | MR | FAIGAFAVIMLILLI | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND |
| A13-33GG | MLVPGSDSSSRPKKRRISKRSKHMR | FAIGAFAVIMLILLI | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND |
| GAA | MAPIFRNYRK | LIVSTVVCIGLLLVLVQL AFPSSFAL | RSASHKKKN |
| GAG | MAPIFRNYRK | LIVSTVVCIGLLLVLVQL AFPSSFALVN | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND |
| GDG | MAPIFRNYRK | LVGIILVLLIWGTVLLLVN | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND |
| GGA | MAPIFRNYR | FAIGAFAVIMLILLI | KTSSEFRSASHKKKN |
| GGG34A | MAPIFRNYR | FAIGAFAVIMLILLI | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND EFRSASHKKKN |
| αF-G′ | MRFPSIFTAVLFAASSALAAPVD | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND | |
| - -GG34A | MR | FAIGAFAVIMLILLI | KTSSIGPPSIARTVTPNASIPKTPEDISILPVND EFRSASHKKKN |
Amino acid sequence from GDPase is shown in regular font. Amino acids derived from ALP are underlined. Italicized amino acids in the TMD sequence of GDG come from DPAP B. In the cytoplasmic tail sequence of αF-G′, α-factor sequences are italicized. Bold-face letters are amino acids encoded for by restriction sites that were introduced during construction of the protein.
- -GG.
GDA1 sequences 157–465 were PCR amplified using a 5′ primer with an NdeI restriction site and a 3′ primer containing a KpnI site. The resulting 5′ sequence, CAT ATG CGG, codes for the initiating methionine and an arginine residue adjacent to the transmembrane domain. An NdeI–BspE1 fragment from a plasmid containing the PCR product was inserted into pBKS αFssG205–465 (see below), thus generating pBKS - -GG205–465. A HindIII–BspE1 fragment from pBKS - -GG205–465 was ligated into pRS GGG to create the full-length protein.
A13–33GG.
PHO8 sequences 37–100 were PCR amplified using primers containing engineered NdeI sites at both the 5′ and 3′ ends. The NdeI fragment was inserted into the NdeI site of pBKS - -GG205–465, and correct orientation was determined by sequencing the plasmid. An EcoRV–BspEI fragment from the resulting pBKS A13–33GG205–465 was ligated into pRS GGG digested with KpnI (filled in) and BspEI to generate the complete chimeric protein.
GAA.
PHO8 sequences 103–953 were amplified by PCR using a 5′ primer containing a HindIII site. The PCR fragment digested with HindIII and ClaI was subcloned into pBKS to generate pBKS - -AA103–723. The HindIII fragment from GAG (see below) containing the promoter and cytoplasmic tail of GDA1 was inserted upstream of PHO8 in pBKS - -AA103–723 to yield pBKS GAA103–723. Correct orientation of the GDA1 promoter and cytoplasmic domain was confirmed by sequencing the plasmid. A full-length construct was generated by subcloning the KpnI–BglII fragment from pBKS GAA into pRS GGGA (pRS GGGA includes GDA1 sequences 1–1681 fused via an EcoRI site to PHO8 sequences 178-1701).
GAG.
To construct GAG, the TMD of PHO8 (bp 103–177) was amplified by PCR using primers containing a 5′-HindIII site and a 3′-HincII site, respectively. The resulting HindIII–HincII fragment was inserted upstream of GDA1 lumenal sequences 205–465 present in a previously constructed plasmid, pBKS AAG205–465 (GDA1 promoter sequences 1–133 fused via an NdeI site to PHO8 bp 1–177 containing the cytoplasmic and transmembrane domains of ALP, which were in turn ligated via a HincII site to GDA1 lumenal sequences 205–465). The resulting plasmid, pBKS - -AG205–465, contained only PHO8 TMD sequences fused to 260 bp of the GDA1 lumenal domain. To insert GDA1 promoter and cytoplasmic domain sequences upstream of the ALP TMD, a PCR fragment of GDA1 sequences 1–160 was subcloned into the HindIII site of pBKS - -AG205–465, thus yielding pBKS GAG205–465. To generate the full-length protein, an EcoRV–BspEI fragment from pBKS GAG205–465 was subcloned into the KpnI (blunt-ended) and BspEI sites of pRS GGG.
GDG.
Base pairs 88–135 of dipeptidyl aminopeptidase B (DPAP B; encoded by the DAP2 gene; Roberts et al., 1989) were PCR amplified using primers with engineered 5′-HindIII and 3′-HincII sites, and the resulting fragment was inserted into pBKS GAG205–465 using HindIII and HincII. This replaced the ALP TMD with that of DPAP B but also removed the GDA1 promoter and cytoplasmic domains (because of a HindIII site at bp 1), thus generating pBKS - -DG205–465. A HindIII fragment from pBKS GAG205–465 carrying the GDA1 promoter and cytoplasmic tail sequences was inserted 5′ to the DPAP B TMD to generate pBKS GDG205–465. The full-length protein was generated by ligating an EcoRV–BspEI fragment from pBKS GDG205–465 into the SalI (filled-in) and BspEI sites of pRS GGG.
GGA and GGG34A.
For GGA, sequences upstream of the GDA1 promoter to bp 216 were amplified from pRS GGG by PCR using a 3′ primer containing an engineered EcoRI site; the resulting fragment was subcloned into the KpnI and EcoRI sites of plasmid pRS GGGαF to create pRS GGαF. pRS GGGαF, constructed for another study, contains GDA1 sequences 1–1681 fused via an EcoRI site to α- factor sequences 142–498 (stop) subcloned into the KpnI and BamHI sites of pRS316. For GGG34A, GDA1 sequences 121–309 containing the cytoplasmic tail, transmembrane domain, and 34 amino acids of the lumenal domain were amplified by PCR, and the resulting fragment was inserted into the NarI and EcoRI sites of pRS GGGαF, thus creating pRS GGG34αF. The α-factor sequences in both GGαF and GGG34αF were replaced with PCR-amplified PHO8 lumenal sequences 177-1702 using EcoRI and BamHI.
αF-G′.
In plasmid pBKS αFH (gift from Gay Bush, UCLA), an NdeI site had been engineered at the extreme 5′ end of the α-factor gene; the ATG within the NdeI site encodes the initiating methionine. GDA1 promoter sequences 1–133 were PCR amplified using a 5′ primer containing a BamHI site and a 3′ primer with an NdeI site. The PCR fragment cleaved with BamHI and NdeI was inserted upstream of the α-factor gene in pBKS αFH to generate pBKS GpαFH. GDA1 lumenal sequences 205–465 were amplified by PCR, using a 5′ primer containing a HincII site and a 3′ primer with a KpnI site. The PCR product was subcloned into the HincII and KpnI sites of pBKS GpαFH, placing GDA1 sequences immediately downstream of the α-factor signal sequence. This plasmid, pBKS αFssG205–465, was cleaved with HindIII and BstEII, and the resulting fragment was inserted into the pRS GGG gene to yield pRS αF-G. This fusion protein was found to be highly glycosylated. To determine whether the extensive glycosylation was due to a potential glycosylation site created by the fusion of the α-factor signal sequence to GDA1 lumenal sequences, oligonucleotide mutagenesis was performed (McClary et al., 1989), mutating the asparagine (amino acid 23) within the glycosylation site to an aspartic acid (pRS αF-G′). This mutation did not change the glycosylation status of the protein.
- -GG34A.
To generate - -GG34A, the KpnI–BspEI fragment from pRS - -GG was subcloned into the pRS GGG34A plasmid.
Immunofluorescence
For single-label experiments, cells were prepared, and indirect immunofluorescence microscopy was performed as described (Roberts et al., 1991) except that 0.02 mg/ml oxalyticase (Enzymogenetics, Corvallis, OR) was used to convert cells to spheroplasts. Tween buffer (1% nonfat dry milk, 0.5 mg/ml BSA, 150 mM NaCl, 50 mM HEPES, pH 7.5, 0.1% Tween 20, and 1 mM NaN3) was used in place of PBS-BSA as the preincubation solution for fixed cells (30 min at 37°C) and as the wash solution. Cells were exposed to 10 μl primary antibody (1:100 dilution in delete extract; see below) for 1 h at 37°C, washed, and then incubated with FITC-conjugated anti-rabbit IgG secondary antibody (Sigma; 1:160 dilution in delete extract) under similar conditions. GPY1452 (gda1Δ pep4Δ) cells expressing pRS GGG from a plasmid were incubated with polyclonal antibodies to GDPase (a gift from P. Berninsone and C. Hirschberg, University of Massachusetts Medical Center, Boston, MA; Berninsone et al., 1995). GPY1250 (pho8Δ pep4Δ) cells expressing ALP, GGA, GGG34A or - -GG34A from a plasmid were stained with affinity-purified antibodies to ALP (a gift from J. Shaw, University of Utah, Salt Lake City, UT). For double-label experiments, SEY6210 wild-type cells expressing GDPase from the pRS314 plasmid and either the pRS316-based centromeric Och1-HA plasmid (a gift from G. Waters, Princeton University, Princeton, NJ; Harris and Waters, 1996) or the YEp352-based multicopy Mnn1-HA plasmid (a gift from M. Lussier, McGill University, Montreal, Quebec, Canada) were analyzed. Both of these plasmids have been previously used for Golgi localization studies (Gaynor et al., 1994; Lussier et al., 1995). Preincubation in Tween buffer was increased to 2 h at room temperature to reduce the signal from background staining. Cells were incubated sequentially with a 1:50 dilution of a monoclonal antibody to the hemagglutinin antigen (12CA5; a gift from G. Weinmaster, UCLA) and a 1:100 dilution of the polyclonal antibody to GDPase (at least 2 h each). This was followed by sequential incubations (at least 2 h each) with the FITC-conjugated anti-mouse (Sigma; 1:128 dilution), and Texas Red-conjugated anti-rabbit (Jackson ImmunoResearch, West Grove, PA; 1:200 dilution) secondary antibodies. In all experiments, both primary and secondary antibodies were preabsorbed with cell extracts lacking either GDPase or ALP (delete extract) to reduce nonspecific binding. Cells were visualized with a 100× Zeiss (Thornwood, NY) oil immersion lens on a Nikon (Garden City, NY) FXA fluorescence microscope. Images were collected using a Photometrics cooled charge-coupled device camera and Isee software from Inovision (Durham, NC) and were adjusted using standard settings in Adobe Photoshop (Adobe Systems, Mountain View, CA). To quantitate the colocalization of GDPase with Och1p and Mnn1p, Texas Red and FITC images were overlaid using Isee software. At least 25 cells clearly expressing both antigens were analyzed for overlap of the FITC signal (Och1p or Mnn1p) with the Texas Red signal (GDPase).
Metabolic Labeling and Immunoprecipitation
Metabolic labeling and immunoprecipitation were performed essentially as previously described (Seeger and Payne, 1992b), except that labeling was carried out for 10 min unless otherwise noted. After subjecting proteins to SDS-PAGE, gels were incubated for 15 min in a 25% methanol, 10% acetic acid solution, dried, and analyzed using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager. Antibodies to GDPase were a gift from P. Berninsone and C. Hirschberg (Berninsone et al., 1995). Antibodies to ALP have been described by Seeger and Payne (1992a). Antibodies specific for α1,6- and α1,3-mannose linkages were a gift from Randy Schekman (University of California, Berkeley, CA), and antibodies to CPY were a gift from Scott Emr (University of California San Diego School of Medicine, La Jolla, CA).
For several experiments, modifications were made to the above protocol.
In cases in which proteins were to be treated with endoglycosidase H (Endo H; Boehringer Mannheim, Mannheim, Germany), antibody-bound antigen collected with protein A-Sepharose beads (Pharmacia Biotech, Uppsala, Sweden) was eluted in 100 μl 10 mM Tris-HCl (pH 7.5) and 0.1% SDS instead of Laemmli sample buffer (Laemmli, 1970). Samples were heated at 100°C for 3 min, and beads were sedimented by centrifugation. Protein was precipitated with 500 μl of acetone at −20°C for at least 2 h. Precipitated protein was sedimented and resuspended in Endo H buffer (0.1 M sodium citrate, pH 6.0, 0.075% SDS, and 0.2% β-mercaptoethanol) plus 2 μl Endo H and incubated at 37°C overnight. Laemmli sample buffer was added, and proteins were analyzed as described above.
For sequential immunoprecipitations, immunoprecipitated proteins were dissociated from protein A-Sepharose beads at 100°C for 3 min in 50 μl of 2% SDS and 0.5% β-mercaptoethanol. One milliliter of PBS (4 mM KH2PO4, 16 mM Na2HPO4, and 115 mM NaCl, pH 7.3) containing 1% Triton X-100 was added, beads were sedimented, and the supernatant was transferred to a new tube containing the second antibody and fresh protein A-Sepharose.
Immunoprecipitation of αF-G′ from internal and external cellular fractions was performed as described (Seeger and Payne, 1992b), except that 1 mg/ml BSA and 10 μg/ml α2-macroglobulin were added to the labeling medium (pH 5.7) to protect secreted proteins from degradation.
RESULTS
Localization and Biosynthesis of GDPase
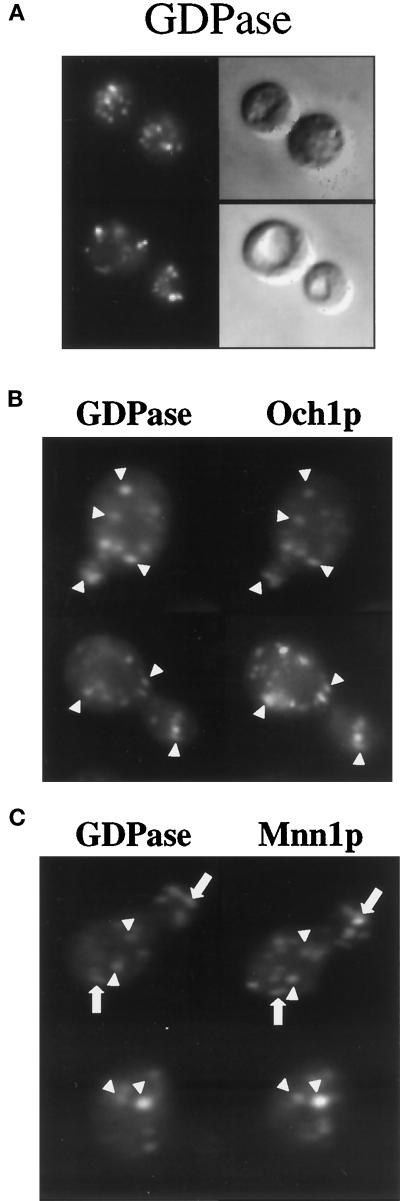
Both functional analyses and cell fractionation studies assaying enzyme activity indicate that GDPase is a resident of multiple compartments of the yeast Golgi complex (Abeijon et al., 1989; Bowser and Novick, 1991; Abeijon et al., 1993). To further characterize the subcellular localization of GDPase, we analyzed the distribution of the protein by indirect immunofluorescence. In addition, double-label immunofluorescence was performed to assess the degree of colocalization between GDPase and either the cis-Golgi protein Och1p or the medial and trans-Golgi protein Mnn1p (see MATERIALS AND METHODS). A punctate staining pattern characteristic of association with yeast Golgi membranes was observed in cells expressing GDPase (Figure 1A) but not in cells lacking GDPase because of a gene disruption (gda1Δ) (our unpublished observatons). Similar punctate patterns were found in cells stained for either Och1p or Mnn1p (Figure 1, B and C). In double-labeled cells, quantitative analysis of colocalization indicated that 62% of the Texas Red-labeled GDPase spots overlapped with FITC-labeled Och1p spots (Figure 1B; 234 spots, 36 cells). A similar evaluation of cells coexpressing GDPase and Mnn1p revealed that 31% of GDPase spots colocalized with Mnn1p structures (Figure 1C; 89 spots, 26 cells). Although not strongly overlapping, GDPase and Mnn1p fluorescent structures were often found adjacent to each other (Figure 1C), perhaps reflecting the small stacks of Golgi cisternae observed by electron microscopy (Preuss et al., 1992). These results reveal that GDPase is distributed in multiple Golgi compartments with a bias toward the cis compartments.
Figure 1.
Immunofluorescent localization of GDPase. (A) Representative GPY1452 (gda1Δ pep4Δ) cells expressing GDPase from the pRS GGG plasmid. Cells were stained with antibody to GDPase. Immunofluorescent images appear on the left; Nomarski optics images are on the right. (B and C) Double-immunofluorescent labeling of representative SEY6210 cells (wild type) expressing GDPase from the centromeric pRS GGG plasmid and either Och1p from the centromeric Och1-HA plasmid (B) or Mnn1p from the multicopy Mnn1-HA plasmid (C). Cells were sequentially stained with antibody to the hemagglutinin antigen and with antibody to GDPase. GDPase images are on the left; Och1p or Mnn1p images are on the right. Arrowheads indicate colocalization of fluorescent-labeled proteins. Arrows in the Mnn1p panels indicate adjacent GDPase and Mnn1p fluorescent spots.
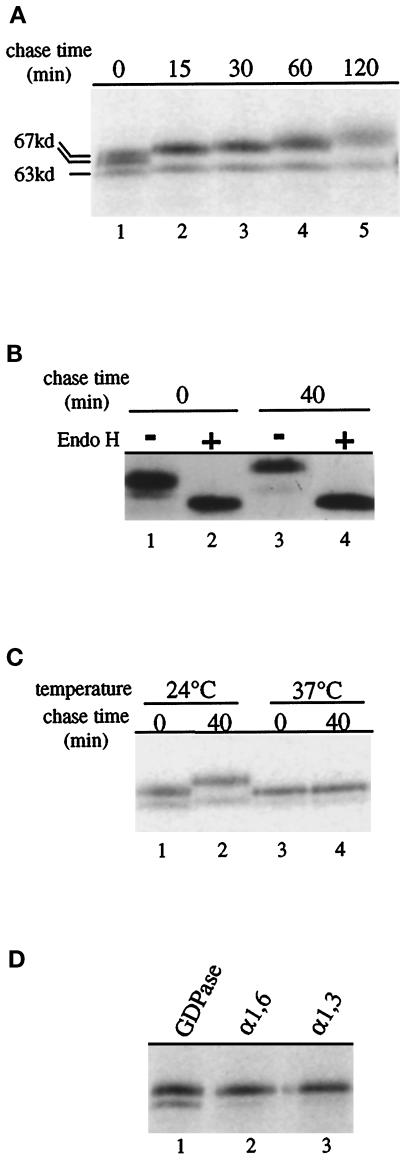
GDPase isolated from wild-type yeast membranes has been characterized as a heterogeneously glycosylated protein of 59–65 kDa (Berninsone et al., 1995). To investigate the kinetics of GDPase biosynthesis, we used a pulse–chase immunoprecipitation regimen. After a 10-min incubation of cells with 35S-labeled methionine and cysteine (pulse), excess unlabeled amino acids were added (chase), and samples were harvested at designated time intervals. GDPase was immunoprecipitated from cell lysates and analyzed by SDS-PAGE. After the 10-min labeling period, a predominant doublet at ∼67 kDa and a minor species of 63 kDa were observed (Figure 2A, lane 1). By the 15-min chase period, the doublet was replaced by a single species of slightly higher molecular mass, which exhibited a progressive increase in size over the remainder of the 120-min chase period. The minor 63-kDa form persisted throughout the chase period and also slightly increased in size. In a separate experiment, incubation of the immunoprecipitates with Endo H to remove N-linked oligosaccharides resulted in conversion of the multiple GDPase species to a single band with the same mobility at both 0 and 40 min (Figure 2B, lanes 2 and 4). These results indicate that the multiple species of GDPase at a given time point differ only in the extent of N-linked glycosylation. Because there are three potential sites of N-linked glycosylation in GDPase (Abeijon et al., 1993), it is likely that the two forms that persist during the chase period differ in the number of oligosaccharide side chains. In addition, because Endo H treatment eliminates the size difference between GDPase at the 0- and 40-min chase points (Figure 2B, compare lanes 1 and 3 with lanes 2 and 4), the progressive size increase of GDPase can be attributed to continued extension of the N-linked oligosaccharide side chains.
Figure 2.
Characterization of GDPase synthesis and glycosylation. (A) Pulse–chase analysis of GDPase. G2-11 cells expressing GDPase from a plasmid (pRS GGG) were incubated with 35S-labeled methionine for 10 min, and then a chase with unlabeled amino acids was initiated, and cells harvested at the indicated chase times. Protein was immunoprecipitated with antibody to GDPase and analyzed by SDS-PAGE. Hash marks indicate the major (67 kDa), and minor forms (63 kDa) of GDPase. (B) N-linked glycosylation of GDPase. Cells were treated as described above, except that after immunoprecipitation, protein was incubated with (+; lanes 2 and 4) or without (−; lanes 1 and 3) endoglycosidase H to remove N-linked glycosyl residues before analysis by SDS-PAGE. (C) Glycosylation of GDPase is blocked in a sec18-ts mutant. SEY5188 cells were incubated for 15 min at either the permissive (24°C; lanes 1 and 2) or restrictive (37°C; lanes 3 and 4) temperature before labeling. Native GDPase was immunoprecipitated and analyzed as described above. (D) Outer chain glycosylation of GDPase includes both α-1,6- and α-1,3-linked mannoses. Labeled, immunoprecipitated GDPase was reimmunoprecipitated with either GDPase antibody (GDPase; lane 1), antibodies specific for α-1,6-mannose residues (α-1, 6; lane 2), or antibodies that specifically recognize α-1,3-mannose extensions (α-1, 3; lane 3).
To determine whether the time-dependent size increase of GDPase requires localization to the Golgi complex, GDPase was examined in cells carrying the temperature-sensitive sec18 mutation, which inhibits protein transport from the ER to the Golgi complex at the nonpermissive temperature (Novick et al., 1981). When sec18 cells were incubated at the permissive temperature (24°C), the two differentially glycosylated GDPase forms increased in size during the chase period (Figure 2C, lanes 1 and 2). In contrast, no increase in size was observed when ER-to-Golgi traffic was blocked by incubating the sec18 cells at the restrictive temperature (37°C; Figure 2C, lanes 3 and 4). Thus, extension of the GDPase oligosaccharide side chains depends on delivery to the Golgi complex. Furthermore, the presence of two GDPase species in the 37°C samples (Figure 2C, lanes 3 and 4) suggests that the major and minor forms are due to differential core glycosylation of GDPase in the ER.
The slow incremental size increase exhibited by GDPase is also characteristic of the Golgi-localized mannosyltransferases Kre2p/Mnt1p and Mnn1p (Graham et al., 1994; Lussier et al., 1995). This size increase is thought to result from constant exposure to glycosylation reactions during residence in the Golgi complex. We analyzed the oligosaccharide modifications on GDPase to determine whether the protein acquires Golgi-specific α1,6- and α1,3-mannose linkages (Figure 2D). Lysates prepared from labeled cells after a 40-min chase period were first subjected to immunoprecipitation with GDPase antibodies and then reprecipitated with either GDPase antibodies (Figure 2D, lane 1) or antibodies specific for α1,6- or α1,3-linked mannose residues (Figure 2D, lanes 2 and 3). Each antibody efficiently recognized the major form of GDPase, indicating that the protein had been modified by Golgi-localized mannosyltransferases. Recognition of the minor form of GDPase is less efficient, perhaps because of the presence of fewer α1,6- and α1,3-linked mannose residues on this form of the protein. Combined with earlier studies, the results presented in Figures 1 and 2 characterize GDPase as a stable, Golgi-localized glycoprotein. The analyses also suggest that the progressive size increase displayed by GDPase can serve as a convenient indicator of residence in the Golgi complex.
Localization Assays
GDPase is predicted to be a type II membrane protein with an amino-terminal 9-amino acid (aa) cytoplasmic domain, a 15-aa TMD and a 495-aa lumenal domain (Abeijon et al., 1993). To investigate the role of each domain in localization of GDPase to the Golgi complex, we engineered deletion mutants and chimeric molecules containing topologically equivalent domains from vacuolar membrane proteins. Genes encoding these different versions of GDPase were introduced on centromere-containing (low-copy) plasmids into appropriate test strains. Subsequently, biochemical and functional assays were used to determine the subcellular distribution of the encoded proteins.
Localization to the Golgi complex was evaluated by monitoring the stability and glycosylation state of the deletion mutants and chimeric proteins over time using pulse chase analysis. Localization-defective forms of GDPase were predicted to be unstable based on ample documentation that Golgi membrane proteins with defects in localization determinants are transported to the vacuole where they are degraded (Wilcox et al., 1992; Cooper and Bussey, 1992; Roberts et al., 1992; Lussier et al., 1995). Vacuolar degradation was assessed by determining the stability of each protein in a congenic set of PEP4 and pep4Δ strains. In cells lacking the PEP4-encoded protease A, the zymogen forms of the principal vacuolar proteases are not matured to their active forms, and consequently the vacuoles are deficient in proteolytic activity (Jones, 1991). Thus, degradation in PEP4 cells but not pep4Δ cells signifies vacuolar delivery. Glycosylation was analyzed as an additional indicator of Golgi localization; localization-defective proteins should not undergo progressive size increases, because, even when stabilized in pep4Δ cells, delivery to the vacuole effectively sequesters the proteins away from the activity of the Golgi mannosyltransferases. Conversely, mutant or chimeric proteins that are faithfully localized to the Golgi complex should be stable and display progressive size increases similar to GDPase. Following the stability and glycosylation status of GDPase over time provides kinetic criteria for GDPase residence in the Golgi complex, thereby permitting a sensitive measure of proper localization.
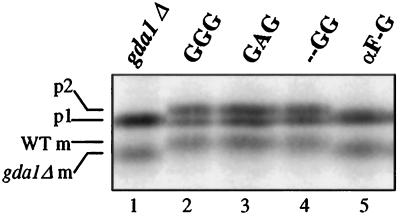
Functional complementation of the glycosylation defects of gda1Δ cells allowed a third, independent test for Golgi localization. Complementation was assayed by monitoring the glycosylation status of the soluble vacuolar protease CPY. Normally, the oligosaccharide side chains of the 67-kDa core-glycosylated form of CPY (p1 form) are extended in a limited manner by Golgi mannosyltransferases to yield a 69-kDa species (p2 form). Upon delivery to the vacuole, p2 CPY is proteolytically matured into the 61-kDa, active carboxypeptidase (Stevens et al., 1982). All three forms can be detected when CPY is immunoprecipitated from cells labeled for 5 min (Figure 3, lane 2). The glycosylation defect in gda1Δ cells that results from the inability to convert GDP into GMP is manifested in CPY biosynthesis as an inability to generate p2 CPY and the appearance of a correspondingly smaller, 59-kDa mature form (Figure 3, lane 1; Abeijon et al., 1993). Complementation was used as an indication of Golgi localization for all proteins containing the GDPase lumenal domain, which carries the active site of the enzyme.
Figure 3.
Complementation of the gda1Δ CPY processing defect by mutant and chimeric GDPase proteins. G2-11 cells containing either no plasmid (lane 1) or plasmids expressing either the wild-type or mutant proteins as indicated (lanes 2–5) were labeled with 35S-methionine for 5 min, subjected to a chase period of 5 min, and lysed, and CPY was immunoprecipitated. The immature forms of CPY (p1 and p2), and mature forms of CPY from wild-type (WT m) or gda1Δ mutant (gda1Δ m) cells are indicated on the left.
The GDPase Cytoplasmic Domain Is Not Required for Golgi Localization
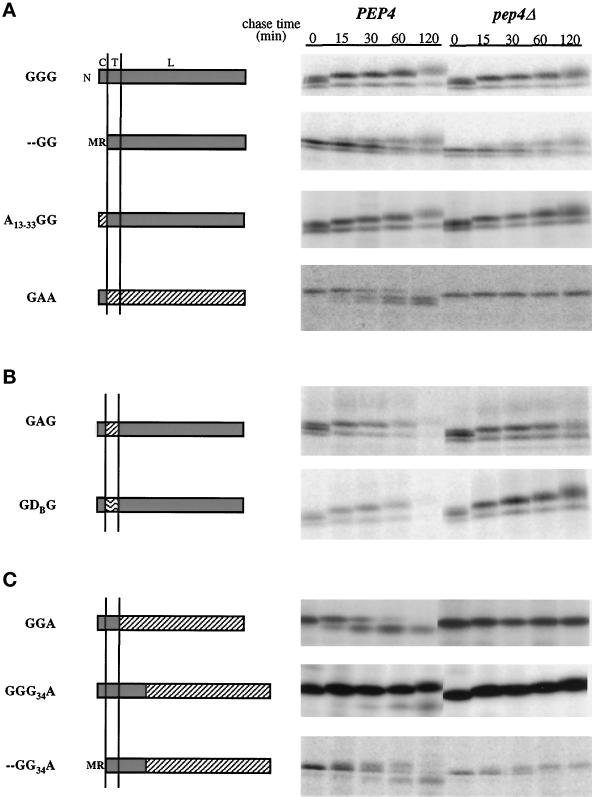
A deletion of the GDPase cytoplasmic tail was engineered to examine the role of this domain in Golgi localization. In the mutant, the initiating methionine was placed next to the arginine residue that defines the predicted N-terminal boundary of the TMD, thereby eliminating the intervening seven amino acids of the nine-amino acid cytoplasmic domain. We refer to this protein as - -GG; beginning with the N-terminal cytoplasmic domain, each letter (or - -) represents a topological domain. In chimeric proteins, letters indicating different domains refer to the protein of origin. This terminology will be applied to all subsequent proteins. Pulse–chase analysis of cells expressing - -GG revealed that the protein was stable and increased in size (Figure 4A). The - -GG protein also was able to complement the gda1Δ glycosylation defect (Figure 3, lane 4). However, when compared with the wild-type GDPase and with other constructs, a delay was noted in the rate at which the two forms present immediately after the labeling period were converted into larger species (in Figure 4A, compare - -GG with GGG). We interpret this delay as an indication that - -GG exits the ER less efficiently than native GDPase. A similar delay has been observed for another Golgi resident lacking a cytoplasmic domain, Kre2p (Lussier et al., 1995). To circumvent this problem, the A13–33GG chimera was constructed by replacing the GDPase cytoplasmic domain with amino acids 13–33 from the cytoplasmic domain of vacuolar ALP encoded by the PHO8 gene (Kaneko et al., 1987). Recent studies have demonstrated that the cytoplasmic domain of ALP contains vacuolar targeting information (Cowles et al., 1997; Piper et al., 1997; Vowels and Payne, 1998). Although the major sorting signal in ALP is located in the N-terminal 16 amino acids of the cytoplasmic tail (Cowles et al., 1997; Vowels and Payne, 1998), a weak vacuolar-targeting determinant may be present in the A13–33GG chimera (Piper et al., 1997). However, A13–33GG was efficiently localized to the Golgi complex without the ER to Golgi transport delay observed for - -GG (Figure 4A). These results indicate that vacuolar sorting information present in ALP residues 13–33 is not sufficient to override GDPase Golgi localization information and confirm that the cytoplasmic tail of GDPase is not necessary for Golgi localization in the context of the TMD and full-length lumenal domain.
Figure 4.
Pulse–chase analysis of wild-type and mutant GDPase proteins in PEP4 and pep4Δ cells. Proteins are named from amino (N) to carboxy terminus, and letters refer to the protein of origin, where G represents GDPase, A represents ALP, and D represents DPAP B. The domain structure of each protein is diagrammed with GDPase domains shaded, ALP domains diagonally hatched, and DPAP B domains wave hatched. Above the GGG diagram, C refers to the cytoplasmic domain, T to the transmembrane domain, and L to the lumenal domain. Vertical lines represent the membrane. Proteins GGG, - -GG, A13–33GG, GAG, and GDG were expressed and analyzed in strains G2-11 (gda1Δ PEP4) and GPY1452 (gda1Δ pep4Δ), and proteins were precipitated with antibodies to GDPase. Chimeric molecules GAA, GGA, GGG34A, and - -GG34A were expressed and analyzed in strains DKY6280 (pho8Δ PEP4) and GPY1250 (pho8Δ pep4Δ), and antibodies to ALP were used to precipitate the proteins of interest. Labelings and immunoprecipitations were as described for Figure 2A. (A) Wild-type GDPase and mutant and chimeric proteins used to analyze the role of the GDPase cytoplasmic domain in Golgi localization. (B) Chimeric proteins used to test the role of the GDPase transmembrane domain in Golgi localization. (C) Chimeric GDPase proteins used to analyze the role of the lumenal domain in GDPase localization.
To test the sufficiency of the GDPase cytoplasmic domain, the GAA protein was constructed by fusion of the GDPase cytoplasmic domain to the TMD and lumenal domains of ALP. ALP is normally synthesized as an inactive precursor that is activated upon delivery to the vacuole by PEP4-dependent cleavage at the carboxy terminus of the lumenal domain (Klionsky and Emr, 1989). The ALP transmembrane and lumenal domains do not contain vacuolar localization signals, and the lumenal domain retains enzymatic activity in chimeric molecules, making it an effective reporter for vacuolar delivery (Klionsky and Emr, 1990; Nothwehr et al., 1993; Chapman and Munro, 1994). The GAA protein was rapidly cleaved in a PEP4-dependent manner and showed only a marginal size increase (Figure 4A). The two degradation products probably represent cleavage of ALP to remove the carboxy-terminal “pro” region as well as scission at or near the ALP–GDPase fusion site. The analysis of GAA, along with the results from GGA (see below), indicates that the cytoplasmic domain of GDPase is not sufficient for Golgi localization.
The GDPase Transmembrane Domain Plays a Minor Role in Golgi Localization
Transmembrane domains differ in length and hydrophobicity, characteristics that have been proposed to affect the localization of proteins to different subcellular compartments (Bretscher and Munro, 1993; Masibay et al., 1993; Munro, 1995; Rayner and Pelham, 1997). The predicted 15-amino acid TMD of GDPase is relatively short, similar to other Golgi membrane proteins (e.g., Och1p [Nakayama et al., 1992] and Mnn1p [Yip et al., 1994]). The importance of the GDPase TMD in Golgi localization was investigated by replacement with the 26-amino acid TMD from ALP to generate the GAG protein. Pulse–chase analysis showed that GAG is relatively stable and undergoes a progressive size increase (Figure 4B), indicating Golgi localization. In agreement with this conclusion, GAG restores wild-type glycosylation of CPY when introduced into a gda1Δ strain (Figure 3, lane 3). However, GAG Golgi localization is not quite as efficient as native GDPase, because the majority of GAG is degraded by the 120-min chase point, whereas GDPase remains stable through this time point (Figure 4A, GGG PEP4). To address the possibility that the length of the TMD was the determining factor in this minor Golgi localization defect, the GDPase TMD was replaced by the 16-amino acid TMD from vacuolar DPAP B to create GDG. The results obtained with GDG were the same as with GAG (Figure 4B and Table 3), arguing that the length of the TMD is not responsible for the slight perturbation in Golgi localization. Therefore, although the GDPase TMD is required for optimal Golgi localization, the transmembrane domains of ALP and DPAP B provide effective substitutes.
Table 3.
Summary of phenotypes of GDPase chimeric proteins
| Construct | Stability | Glycosylation | Complements gda1Δ | Localization to Golgi |
|---|---|---|---|---|
| GGG | + | + | + | + |
| - -GG | + | + | + | + |
| A13-33GG | + | + | + | + |
| GAA | − | − | NA | − |
| GAG | ± | + | + | ± |
| GDG | ± | + | + | ± |
| GGA | − | − | NA | − |
| GGG34A | + | + | NA | + |
| αF-G′ | − | +++ | − | − |
| - -GG34A | ∓ | + | NA | ∓ |
In the stability column, + designates protein stability within the 2-h period of the experiment; decreases in stability are designated as ±, ∓, or − and correspond to increasing levels of PEP4-dependent processing of the protein. For glycosylation, + indicates Golgi-dependent glycosylation of the protein over time, whereas − indicates lack of glycosylation. αF-G′ is abnormally hyperglycosylated, as indicated by +++. In the Complements gda1Δ column, + indicates the ability of the chimeric protein to rescue the CPY glycosylation defect of a gda1Δ mutant, − indicates failure to rescue the mutant phenotype, and NA indicates not applicable. Localization to the Golgi, designated by +, is inferred on the basis of localization assay results; ±, ∓, and − indicate decreasing ability to localize to the Golgi complex.
The Lumenal Domain Is Required for GDPase Golgi Localization
Two GDPase–ALP chimeric proteins were constructed to test the role of the GDPase lumenal domain in Golgi localization. In the first, the cytoplasmic domain and TMD from GDPase were fused to the ALP lumenal domain; this protein is designated GGA (Figure 4C); In the second chimera, the ALP lumenal domain was fused to a site in the GDPase lumenal domain 34 amino acids from the TMD (GGG34A; Figure 4C). This short, membrane-adjacent lumenal domain will be referred to as the stem region by analogy with mammalian glycosyltransferases (Colley, 1997). When PEP4 cells expressing GGA were analyzed by pulse–chase immunoprecipitation, GGA was found to be rapidly converted into a lower molecular weight form (Figure 4C). In pep4Δ cells, the processed form was absent, and the accumulated GGA did not display a progressive size increase. These properties argue that GGA is not localized to the Golgi complex but instead is delivered to the vacuole. Compared with GGA, GGG34A was substantially more stable in PEP4 cells and progressively increased in size, indicating a predominantly Golgi localization. There was a minor degree of PEP4-dependent processing of GGG34A, reflecting slow leakage of this protein to the vacuole (Figure 4C). The different patterns of GGA and GGG34A implicate the stem region of the GDPase lumenal domain in Golgi localization.
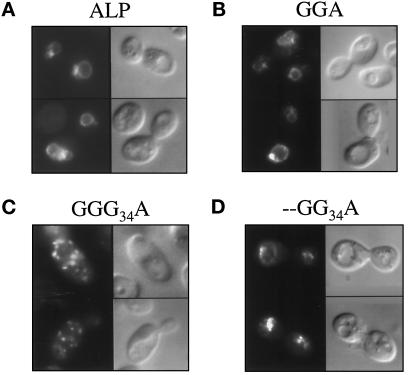
Because GGA and GGG34A lack the enzymatically functional domain of GDPase, gda1Δ complementation could not be performed. Instead, pho8Δ cells expressing either wild-type ALP or one of the two chimeric proteins were subject to immunofluorescence to directly observe the steady-state location of each protein. Wild-type ALP and GGA localize predominantly to the vacuolar membrane, as evidenced by prominent ring staining that coincides with the vacuoles observed by differential interference contrast microscopy (Figure 5, A and B). In contrast, the majority of GGG34A-expressing cells showed the punctate staining pattern associated with Golgi protein localization (Figure 5C); occasional cells also had vacuolar membrane staining (our unpublished observations). These immunofluorescence data confirm the interpretation of the results of the pulse–chase analysis and document the reliability of stability and glycosylation as indicators of protein localization.
Figure 5.
Immunofluorescent localization of wild-type ALP and selected GDPase–ALP chimeric proteins. GPY1250 (pho8Δ pep4Δ) cells expressing wild-type ALP (A), GGA (B), GGG34A (C), or - -GG34A (D) were stained with antibody to the lumenal domain of ALP. In each panel, fluorescent images are on the left; Nomarski images are on the right.
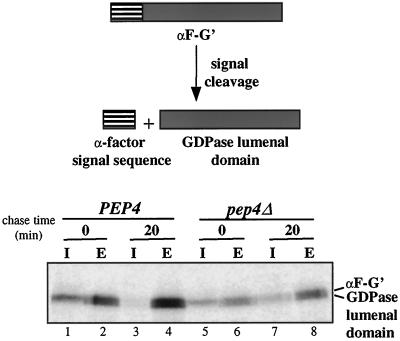
To determine whether the GDPase lumenal domain is sufficient to direct localization to the Golgi complex, the cytoplasmic and transmembrane domains of GDPase were replaced by the cleavable signal sequence from the secreted mating pheromone α-factor (αF-G′). Cleavage of the signal sequence from αF-G′ should result in a soluble protein containing only GDPase lumenal sequences. Cells expressing αF-G′ were labeled for 10 min and then subjected to 0- or 20-min chase periods. At each time point, the cells were separated into cellular and extracellular fractions, and GDPase antibodies were used to precipitate the GDPase lumenal domain from both fractions. Because initial experiments indicated that the αF-G′ protein is extensively glycosylated, each sample was treated with Endo H to reveal the protein backbone. After the 10-min labeling period, a 57-kDa form was present in internal fractions, and 57- and 55-kDa species were present in external fractions (Figure 6, lanes 1 and 2). The sizes of the two forms correspond to the predicted molecular weights of the protein before and after signal sequence cleavage. The presence of uncleaved αF-G′ suggests that signal cleavage is inefficient. Nevertheless, the protein is rapidly secreted, because the external fraction already contained significant levels of the protein at the beginning of the chase period (Figure 6, lane 2). After the 20-min chase period the majority of αF-G′ was secreted into the culture supernatant. A similar pattern is observed in pep4Δ cells, except that more protein was detected in the internal fraction at the 20-min time point than was observed in PEP4 cells (Figure 6, compare lane 7 with lane 3). This finding suggests that a small amount of αF-G′ is delivered to the vacuole. Additionally, αF-G′ is unable to functionally complement the gda1Δ mutant phenotype (Figure 3, lane 5). Thus, the lumenal domain by itself cannot mediate Golgi localization. Taken together, our results suggest that the stem region of the lumenal domain, when tethered to the membrane, plays a critical role in localization of GDPase to the Golgi complex.
Figure 6.
Metabolic labeling and immunoprecipitation of αF-G′ from internal and external cellular fractions. G2-11 and GPY1452 cells expressing αF-G′ were labeled as described in Figure 2A. Cells were converted to spheroplasts, and the released periplasmic fraction was combined with the culture supernatant to produce the external (E) fraction. Spheroplasts were lysed to generate the internal (I) fraction. Proteins were immunoprecipitated and treated with Endo H as described in Figure 2B before being subject to SDS-PAGE.
The - -GG34A chimera was constructed to determine whether the GDPase TMD and stem regions, which are domains necessary for optimal Golgi localization, are also sufficient. Based on pulse–chase immunoprecipitation analysis, - -GG34A displayed properties intermediate between - -GG and GGA. There was a clearly discernible increase in the size of - -GG34A over time (most apparent in pep4Δ cells; Figure 4C), suggestive of prolonged exposure to Golgi-localized glycosyltransferases, but the protein was also subject to PEP4-dependent cleavage (Figure 4C). Analysis of a construct containing ALP sequences 13–33 fused to - -GG34A gave similar results without the ER-to-Golgi transport delay (our unpublished observations). Another protein, - -AG34A, was also characterized to confirm that the lumenal stem region was insufficient for Golgi localization. - -AG34A showed a pattern of processing quite similar to that of - -GG34A (our unpublished results). Because - -GG34A lacks the enzymatically active region of GDPase, it was not possible to determine whether the protein resides in the Golgi complex long enough to complement gda1Δ defects. However, immunofluorescence analysis revealed that the majority of - -GG34A localizes to the vacuole (Figure 5D), as expected from the pattern seen by pulse–chase immunoprecipitation. In a significant number of cells, staining was also noted in regions abutting the vacuolar membrane and may represent protein present in prevacuolar compartments. Obvious Golgi complex staining was not observed. These findings show that although the TMD and stem domain are necessary for Golgi localization, they are not sufficient.
Characterization of the - -GG34A protein uncovered subtle contributions of the cytoplasmic tail and full-length lumenal domain to Golgi localization. When - -GG34A is compared with GGG34A (Figure 4C), it is apparent that the cytoplasmic domain is important for Golgi localization in molecules lacking the full-length lumenal domain. Similarly, comparison of - -GG (Figure 4A) with - -GG34A reveals that sequences in the lumenal domain beyond the stem region are required for Golgi localization if the cytoplasmic tail is absent. Thus, the cytoplasmic domain and the membrane-distal lumenal domain sequences exhibit functional redundancy in Golgi localization; in the presence of the TMD and stem region, either the cytoplasmic domain or the lumenal domain will suffice for optimal Golgi localization.
DISCUSSION
The molecular analysis presented here extends the characterization of S. cerevisiae GDPase and identifies localization information within the protein. The function of GDPase as an enzyme involved in Golgi-localized protein glycosylation reactions, as well as subcellular fractionation studies, suggested that GDPase is a resident of the Golgi complex (Abeijon et al., 1989, 1993; Bowser and Novick, 1991). In agreement with those findings, GDPase colocalizes with cis and medial Golgi proteins by immunofluorescence and is itself subject to Golgi-specific glycosylation. As summarized in Table 3, within GDPase, lumenal sequences adjacent to the membrane are most critical for proper localization of the protein. Every other domain of GDPase can be substituted or deleted without substantial mislocalization. However, the stem region of GDPase is not sufficient for Golgi residence. The lumenal domain must be anchored to the membrane via a transmembrane domain. Additional sequences in either the cytoplasmic domain or the lumenal domain also contribute to efficient Golgi localization.
Golgi localization determinants have been extensively characterized in two other yeast Golgi residents, Mnn1p and Kre2p/Mnt1p (Graham et al., 1994; Graham and Krasnov, 1995; Lussier et al., 1995). Comparison of the Mnn1p, Kre2p/Mnt1p, and GDPase localization signals reveals that each protein harbors a unique combination of localization determinants. These differences suggest that diverse localization mechanisms are involved, a diversity that potentially could play a role in governing the distinct distributions within this set of Golgi residents. Mnn1p contains two signals, one in the lumenal domain and one in the TMD. Although both Mnn1p and GDPase lumenal domains can mediate localization, only the Mnn1p signal can act autonomously. This is evident from the observation that a soluble form of Mnn1p is retained in the Golgi, whereas a similar soluble version of the GDPase lumenal domain is rapidly secreted (Graham and Krasnov, 1995; αF-G′; Table 3). It should be noted that the soluble GDPase lumenal domain is more extensively glycosylated than native GDPase (our unpublished observations), raising the possibility that aberrant extension of the oligosaccharide side chains in the αF-G′ construct might conceal the localization signal in the stem domain, which otherwise could act autonomously. However, this is unlikely, because both the - -GG34A and - -AG34A proteins, which contain the lumenal domain localization signal, are not glycosylated to the same extent as αF-G′ and yet do not localize to the Golgi complex. Instead, we favor the idea that the GDPase lumenal domain localization determinants are not sufficiently strong to mediate Golgi localization without the aid of mechanisms acting through other domains.
The TMD of Mnn1p, unlike that of GDPase, is sufficient to localize a heterologous lumenal domain to the Golgi complex (GGA, Table 3; Graham and Krasnov, 1995). Thus, Mnn1p but not GDPase carries a major Golgi localization signal in the TMD. Recently, Graham and Krasnov (1995) found that the TMD-mediated Golgi localization of an Mnn1p chimeric protein is clathrin dependent. In contrast, GDPase localization is unaffected upon inactivation of a temperature-sensitive form of clathrin heavy chain (Graham et al., 1994; our unpublished observations). These results suggest that the differential sensitivity of Mnn1p and GDPase to clathrin inactivation could be due, at least in part, to the difference in the role of the TMD in localizing these two proteins.
Kre2p/Mnt1p differs from GDPase (and Mnn1p) in its reliance on the cytoplasmic domain for Golgi localization. Deletion or substitution of the nine-amino acid cytoplasmic domain of Kre2p resulted in steady-state localization of the mutant protein to the vacuole (Lussier et al., 1995). In contrast, similar manipulations of the GDPase cytoplasmic domain had little effect on Golgi localization (- -GG, Table 3). An impact of the GDPase cytoplasmic domain on Golgi localization was not apparent until assessed in a construct lacking a segment of the lumenal domain (- -GG34A). By comparison, elimination of the cytoplasmic domain of Mnn1p, even in the absence of lumenal sequences, did not alter Golgi localization. Thus, of the three well-studied type II Golgi membrane proteins discussed here, the uniquely strong dependence of Kre2p localization on the cytoplasmic domain makes it the only likely candidate to directly interact with cytoplasmic factors as part of the localization process.
Despite the apparent differences in the localization signals of GDPase, Mnn1p, and Kre2p/Mnt1p, a role for the lumenal domain in Golgi localization is emerging as a common principal. In the cases of GDPase and Kre2p, analogous chimeric constructs revealed a role for the stem domain. Fusing the cytoplasmic domain, TMD, and stem sequences to the ALP lumenal domain resulted in efficient Golgi localization, whereas removal of the stem region from the chimeric proteins caused mislocalization to the vacuole (GGA and GGG34A, Table 3; Lussier et al., 1995). Additionally, lumenal sequences beyond the membrane-flanking region also make subtle contributions in these proteins, as these sequences become necessary for Golgi localization of GDPase lacking a cytoplasmic domain and Kre2p lacking the stem domain (- -GG34A, Table 3; Lussier et al., 1995). The Mnn1p lumenal domain is by itself Golgi localized, indicating an autonomous lumenal localization signal in this protein (Graham and Krasnov, 1995). In addition to being sufficient for Golgi localization, this signal is probably also necessary for optimal localization, because full-length Mnn1p is more strongly localized to the Golgi than chimeric forms of Mnn1p containing only the TMD (Graham et al., 1994; Graham and Krasnov, 1995). Together, these studies reveal a shared feature of yeast Golgi glycosylation enzymes, specifically the importance of the lumenal domain in specifying residence to the Golgi complex. This common property suggests that a key aspect of the localization process occurs within the lumen rather than the lipid bilayer or the cytoplasmic face of the Golgi cisternae. Although the lumenal domain in each protein is important in Golgi localization, there are no obvious sequence homologies between GDPase, Mnn1p, and Kre2p/Mnt1p in this region. It is not clear how the lumenal domains mediate Golgi localization, but we envision that protein–protein interactions within the lumen are likely to be important. Such interactions could, for example, localize glycosylation enzymes to the Golgi complex by 1) causing the formation of large oligomeric structures incapable of inclusion into transport vesicles, as suggested in the “kin recognition” hypothesis (Nilsson et al., 1993); 2) allowing the enzymes to bind to transmembrane proteins themselves anchored to cytoplasmic elements or insoluble lipid domains; or 3) allowing association with receptors that act to recycle cargo to the Golgi from more distal compartments. Identification of lumenal domain-interacting partners will be critical in evaluating these and other models.
Several mammalian Golgi-localized glycosyltransferases have been dissected to define Golgi localization signals (for review, see Colley, 1997). Interpretation of these studies has been complicated by the observation that requirements for Golgi localization can differ between cell types (Teasdale et al., 1994; Tang et al., 1995), but there is a general consensus that the TMD plays a key role. Differences in the lengths of Golgi (shorter) and plasma membrane protein (longer) TMDs led to the proposal that Golgi proteins, by virtue of their shorter TMDs, are localized to Golgi membranes by exclusion from the thicker lipid bilayer of the plasma membrane (Bretscher and Munro, 1993; Masibay et al., 1993). The major role of the TMD in localization of mammalian Golgi glycosyltransferases contrasts with the relatively inconsequential involvement of the TMD in localization of yeast GDPase and Kre2p. The TMD of GDPase or Kre2p can be replaced with heterologous TMDs of different sequence and length without substantial effects on Golgi localization. Also, neither the GDPase or Kre2p TMD is sufficient for localization. This difference between the major determinants of Golgi localization in mammalian and yeast proteins may implicate different localization mechanisms. For example, the importance of lumenal domains, not TMDs, in localization of GDPase and Kre2p suggests that lipid bilayer-based sorting is probably not a major factor in the Golgi localization process in yeast. A similar conclusion was reached in a study of t-SNARE localization in yeast (Rayner and Pelham, 1997).
More recent studies of the mammalian proteins have revealed that sequences flanking the TMD can also be required for Golgi localization and prompted a reevaluation of Golgi localization signals (Munro, 1995; Colley, 1997). In particular, several analyses demonstrated that lumenal stem sequences are necessary and/or sufficient Golgi localization signals in N-acetylglucosaminyltransferase I and α2–6-sialyltransferase (Colley et al., 1992; Dahdal and Colley, 1993; Munro, 1995; Tang et al., 1995; Nilsson et al., 1996). In the case of N-acetylglucosaminyltransferase I, the lumenal domain localization signal interacts with another medial Golgi glycosylation enzyme, mannosidase II, supporting oligomerization models (Nilsson et al., 1994, 1996). Additionally, studies of TMDs show that in some cases changing the composition or length of the TMDs in the mammalian proteins has only a partial effect, or no effect, on Golgi localization (Munro, 1991; Dahdal and Colley, 1993; Nilsson et al., 1996). These results, combined with the characterization of yeast Golgi proteins, support a concordant model of Golgi localization in both yeast and mammalian cells. This model derives from two common features: a prominent role for lumenal domain sequences and a frequent requirement for multiple domains for optimal localization. In this view, lumenal interactions play a key role in dictating Golgi residence, whereas one role of the other domains is to augment the formation of the lumenal domain interactions, perhaps by slowing transport through the Golgi complex or by allowing for the optimal presentation of the lumenal domain localization signals. Thus, optimal Golgi localization would require the cooperation of multiple, topologically distinct domains within each protein.
ACKNOWLEDGMENTS
We are especially grateful to Carlos Hirshberg and Patricia Berninsone, whose generous donation of plasmids, strains, and antibodies allowed us to initiate these studies. We thank Janet Shaw, Scott Emr, and Randy Schekman for contributing antibodies. We are grateful to Ken Oyadomari for technical assistance and to Diana Chu and James Howard for critically reading and providing helpful suggestions on the manuscript. This work was supported by grants to J.J.V. from the Jonsson Center Cancer Foundation at UCLA and from National Institutes of Health (National Research Service Award GM18242) and to G.S.P. from National Institutes of Health (GM39040).
Footnotes
Abbreviations used: ALP, alkaline phosphatase; aa, amino acid; bp, base pair; CAA, casamino acid; CGN, cis-Golgi network; CPY, carboxypeptidase Y; DPAP B, dipeptidyl aminopeptidase B; Endo H, endoglycosidase H; ER, endoplasmic reticulum; TGN, trans-Golgi network; TMD, transmembrane domain.
REFERENCES
- Abeijon C, Orlean P, Robbins PW, Hirschberg CB. Topography of glycosylation in yeast: characterization of GDPmannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeijon C, Yanagisawa K, Mandon EC, Hausler A, Moremen K, Hirschberg CB, Robbins PW. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J Cell Biol. 1993;122:307–323. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone P, Lin Z-Y, Kempner E, Hirschberg CB. Regulation of yeast Golgi glycosylation. J Biol Chem. 1995;270:14564–14567. doi: 10.1074/jbc.270.24.14564. [DOI] [PubMed] [Google Scholar]
- Bowser R, Novick P. Sec15 protein, an essential component of the exocytotic apparatus, is associated with the plasma membrane and with a soluble 19.5s particle. J Cell Biol. 1991;112:1117–1131. doi: 10.1083/jcb.112.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Bryant NJ, Boyd A. Immunoisolation of Kex2p-containing organelles from yeast demonstrates colocalisation of three processing proteinases to a single Golgi compartment. J Cell Sci. 1993;106:815–822. doi: 10.1242/jcs.106.3.815. [DOI] [PubMed] [Google Scholar]
- Chapman RE, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley KJ. Golgi localization of glycosyltransferases: more questions than answers. Glycobiology. 1997;7:1–13. doi: 10.1093/glycob/7.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley KJ, Lee EU, Paulson JC. The signal anchor and stem regions of the β-galactoside α-2,6,-sialyltransferase may each act to localize the enzyme to the Golgi apparatus. J Biol Chem. 1992;267:7784–7793. [PubMed] [Google Scholar]
- Cooper A, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KW, Wickner WT. Yeast KEX2 protease and mannosyltransferase I are localized to distinct compartments of the secretory pathway. Yeast. 1989;5:25–33. doi: 10.1002/yea.320050105. [DOI] [PubMed] [Google Scholar]
- Dahdal RY, Colley KJ. Specific sequences in the signal anchor of the β-galactoside α-2,6-sialyltransferase are not essential for Golgi locailzation. J Biol Chem. 1993;268:26310–26319. [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Krasnov VA. Sorting of yeast α1,3 mannosyltransferase is mediated by a lumenal domain interaction, and a transmembrane domain signal that can confer clathrin-dependent Golgi localization to a secreted protein. Mol Biol Cell. 1995;6:809–824. doi: 10.1091/mbc.6.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Seeger M, Payne GS, MacKay VL, Emr SD. Clathrin-dependent localization of α1,3 mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J Cell Biol. 1994;127:667–678. doi: 10.1083/jcb.127.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannoxyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Hayashi N, Toh-e A, Banno I, Oshima Y. Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. Gene. 1987;58:137–148. doi: 10.1016/0378-1119(87)90036-9. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. A new class of lysosomal/vacuolar protein sorting signals. J Biol Chem. 1990;265:5349–5352. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Hamamoto S, Schekman RW. Biochemical requirements for the targeting and fusion of ER-derived transport vesicles with purified yeast Golgi membranes. J Cell Biol. 1996;132:277–289. doi: 10.1083/jcb.132.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M, Sdicu A-M, Ketela T, Bussey H. Localization and targeting of the Saccharomyces cerevisiae Kre2p/Mnt1p medial-Golgi compartment. J Cell Biol. 1995;131:913–927. doi: 10.1083/jcb.131.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE. Targeting and retention of Golgi membrane proteins. Curr Opin Cell Biol. 1993;5:606–612. doi: 10.1016/0955-0674(93)90129-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masibay AS, Balaji PV, Boeggeman EE, Qasba PK. Mutational analysis of the Golgi retention signal of bovine β-1,4-galactosyltransferase. J Biol Chem. 1993;268:9908–9916. [PubMed] [Google Scholar]
- McClary JA, Witney F, Geisselsoder J. Efficient site-directed in vitro mutagenesis using phagemid vectors. BioTechniques. 1989;7:282–287. [PubMed] [Google Scholar]
- Mellman I, Simons K. The Golgi complex: in vitro veritas? Cell. 1992;68:829–840. doi: 10.1016/0092-8674(92)90027-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Sequences within and adjacent to the transmembrane segment of α-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi-Shindo Y, Nakayama K-I, Tanaka A, Toda Y, Jigami Y. Structure of the N-linked oligosaccharides that show the complete loss of α-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J Biol Chem. 1993;268:26338–26345. [PubMed] [Google Scholar]
- Nakayama K, Nakanishi-Shindo Y, Tanaka A, Haga-Toda Y, Jigami Y. Substrate specificity of alpha-1,6-mannosyltransferase that initiates N-linked mannose outer chain elongation in Saccharomyces cerevisiae. FEBS Lett. 1997;412:547–550. doi: 10.1016/s0014-5793(97)00634-0. [DOI] [PubMed] [Google Scholar]
- Nakayama K-I, Nagasu T, Shimma Y-I, Kuromitsu J-R, Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Rabouille C, Hui N, Watson R, Warren G. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J Cell Sci. 1996;109:1975–1989. doi: 10.1242/jcs.109.7.1975. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. Kin recognition: a model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Warren G. Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus. Curr Opin Cell Biol. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Munro S. Sorting of membrane proteins in the secretory pathway. Cell. 1993;75:603–605. doi: 10.1016/0092-8674(93)90479-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Pelham HRB. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Pohlig G, Rothman JH, Stevens TH. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989;108:1363–1373. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne G. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992a;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992b;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Tang BL, Low SH, Wong SH, Hong W. Cell type differences in Golgi retention signals for transmembrane proteins. Eur J Cell Biol. 1995;66:365–374. [PubMed] [Google Scholar]
- Teasdale RD, Matheson F, Gleeson PA. Post-translational modifications distinguish cell surface from Golgi-retained β1,4 galactosyltransferase molecules. Golgi localization involves active retention. Glycobiology. 1994;4:917–928. doi: 10.1093/glycob/4.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels, J.J., and Payne, G.S. (1998). A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Whitters EA, McGee TP, Bankaitis VA. Purification and characterization of a late Golgi compartment from Saccharomyces cerevisiae. J Biol Chem. 1994;269:28106–28117. [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Dynamic retention of TGN membrane proteins in Saccharomyces cerevisiae. Trends Cell Biol. 1993;3:426–431. doi: 10.1016/0962-8924(93)90031-u. [DOI] [PubMed] [Google Scholar]
- Yip CL, Welch SK, Klebl F, Gilbert T, Seidel P, Grant FJ, O’Hara PJ, MacKay VL. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc Natl Acad Sci USA. 1994;91:2723–2727. doi: 10.1073/pnas.91.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]