Abstract
We present a two-part system for conditional FLP-out of FRT-flanked sequences in Caenorhabditis elegans to control gene activity in a spatially and/or temporally regulated manner. Using reporters, we assess the system for efficacy and demonstrate its use as a cell lineage marking tool. In addition, we construct and test a dominant-negative form of hlh-12, a gene that encodes a basic helix-loop-helix (bHLH) transcription factor required for proper distal tip cell (DTC) migration. We show that this allele can be conditionally expressed from a heat-inducible FLP recombinase and can interfere with DTC migration. Using the same DTC assay, we conditionally express an hlh-12 RNAi-hairpin and induce the DTC migration defect. Finally, we introduce a set of traditional and Gateway-compatible vectors to facilitate construction of plasmids for this technology using any promoter, reporter, and gene/hairpin of interest.
TWO-COMPONENT gene expression systems are indispensable tools to probe molecular mechanisms underlying development. Because control can be exerted by each component independently, exquisite temporal and spatial control of gene activation or repression can be achieved. Using different promoter combinations to drive each component of these systems, additional control can be obtained beyond that afforded by heat-inducible or tissue-specific promoters alone. Site-specific recombination systems such as the FLP/FRT system have been used to control gene expression by “FLP-out”: a recombinase-catalyzed intramolecular excision of spacer DNA that lies between tandemly oriented FRT sites. The spacer includes a transcriptional stop so that prior to activation of the FLP recombinase (and subsequent FLP-out) the gene downstream of the spacer is not transcribed (Golic and Lindquist 1989; Struhl and Basler 1993; Figure 1A). After the FRT-containing cassette is excised by the FLP recombinase, the downstream gene is brought into proximity to the promoter and is expressed (reporter 2 in Figure 1A). This system and related systems have proven quite powerful and flexible in model organisms including Drosophila and mouse (see Branda and Dymecki 2004, for review; McGuire et al. 2004). However, prior to our study presented here, and a recently published study (Davis et al. 2008), these systems had not been developed for use in Caenorhabditis elegans.
Figure 1.—
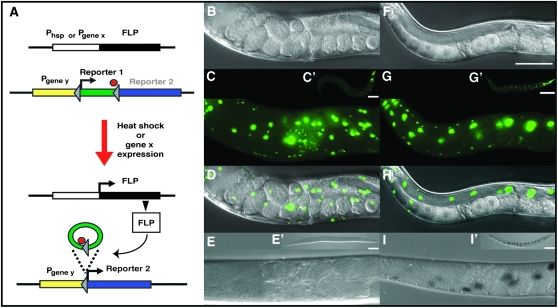
Control of gene expression by FLP-out reaction. (A) Schematic of FLP-out reaction induced by heat shock or gene x-specific expression of FLP recombinase. Target FRT sites are indicated by arrowheads; red stop sign indicates a transcriptional stop. (B–E) Control worms carrying naEx40[Phsp-16.41∷FLP, Pceh-22∷GFP] and naEx64[Ppro-1<GFP<lacZ] that were not subjected to heat shock (see Tables 1 and 2 and materials and methods for details on constructs and strains). (F–I) Heat-induced FLP-out visualized by acquisition of lacZ expression in worms with the same genotype as B–E. All images were captured at 400× magnification except where indicted. B and F, DIC; C and G, green channel. C′ and G′ are the same worms as in C and G, respectively, but at 100× magnification. D and H, merge of DIC and GFP sections above. E and I, DIC images of the same individual worms in the panels above after X-gal staining (that is, worm in E is the same individual as worm in B, C, and D and worm in I is the same individual as in F, G, and H). E′ and I′ are the same worms as in E and I, respectively, but at 100× magnification. Bar, 50 μm and applies to all panels except C′, G′, E′, and I′ in which the bar is 100 μm.
An ideal FLP-out system provides the means to generate both loss- and gain-of-function effects in a spatially and temporally controlled manner. In addition, wild-type gene expression can be turned on in particular cells at particular times in an otherwise mutant background. In organisms where transgenes can be reliably inserted in single copy, FLP-out can also be used to eliminate wild-type gene expression by excision of an FRT-flanked wild-type cassette in a mutant background. In C. elegans, the most common methods for generating transgenic C. elegans introduce multiple copies of transgenes on extrachromosomal arrays (Stinchcomb et al. 1985; Mello et al. 1991; Kelly et al. 1997). Even methods that generate genomic insertions such as microparticle bombardment do not reliably result in low-copy or single-copy insertions (Praitis et al. 2001). Therefore, unless excision is extremely efficient, only dominantly acting gene expression changes are amenable to this technology (e.g., induction of reporters, of wild-type or dominant forms, or ectopic gene activation). This limitation would appear to preclude the ability of a FLP-out expression system to provide an inducible “loss” of gene activity (e.g., by FLP-out of a wild-type gene in a loss-of-function genetic background). Fortunately, RNAi is a dominantly acting means to reduce gene activity (Fire et al. 1998), providing the theoretical possibility of reducing gene activity by FLP-out (see also discussion).
Here we show that the FLP/FRT system can provide both temporal and spatial control of gene expression in C. elegans by combining expression of the FLP recombinase from either a heat-shock promoter or a tissue-specific promoter and expression of the target FLP-out cassette from either a ubiquitous or a tissue-specific promoter. We demonstrate efficacy of the FLP recombinase in different tissue types and quantitate its effect in one cell type for a given set of transgenes. Using reporters, we show its potential use as a lineage tracer. In addition, we use the hlh-12 gene (also called mig-24, Tamai and Nishiwaki 2007) to assay for FLP-mediated induction of the distal tip cell (DTC) migration defect (Mig phenotype) in response to an hlh-12 dominant-negative allele and to an hlh-12 RNAi-inducing hairpin. Finally, we introduce a series of traditional cloning constructs and Gateway-compatible constructs to facilitate the use of this technique.
MATERIALS AND METHODS
Vector construction
Construction of plasmids listed in Table 1 that were used directly in this study is described here. Table 1 lists key features of each construct (including promoters, reporters, 3′ ends, transformation markers, and Gateway sites). See supplemental Toolkit Documentation file and supplemental table for additional useful vectors and detailed strategies for their use. Construction details for all plasmids not listed below can be found in supplemental Methods. Entry clones and donor vectors that are used in plasmid constructions described below but that are not listed in the tables are underlined.
TABLE 1.
FLP recombinase expression constructs and FLP-out targets
| A. FLP recombinase expression constructs | |||
|---|---|---|---|
| Plasmid | Promoter | Genea | unc-119(+)b |
| pGC94 | hsp-16.2 | FLP | No |
| pGC95 | hsp-16.41 | FLP | No |
| pGC133 | hsp-16.41 | FLP | Yes |
| pGC146 | hsp-16.2 | FLP | Yes |
| pGC158 | lag-2 | FLP | No |
| B. FLP recombinase Gatewayd destination vectors to generate expression constructs in the form [promoter-FLP recombinase] | |||||
| Plasmid | attd | attd | iAc | Genea | unc-119(+)b |
| pGC180 | R1 | R2 | None | FLP | Yes |
| pGC181 | R2 | R1 | None | FLP | Yes |
| pGC267 | R1 | R2 | Yes | FLP | Yes |
| C. FRT-containing full FLP-out target cassettes in the form [promoter<GFP<gene/reporter/hairpin] | ||||||||||||||
| Plasmid | att | Promoter | att | FRT | iAc | Genea | unc-119(+)b | FRT | iAc | att | Gene | att | 3′end | unc-119(+)b |
| pGC185 | B1 | rpl-28 | B2 | Yes | No | 4xNLS-GFP | Yes | Yes | No | No | 4xNLS-tdimer2(12) | No | let-858 | No |
| pGC200 | B1 | pro-1 | B2 | Yes | No | 4xNLS-GFP | Yes | Yes | Yes | No | 1xNLS-lacZ | No | unc-54 | No |
| pGC219 | B1 | hlh-12 | B2 | Yes | Yes | 4xNLS-GFP | No | Yes | Yes | B4 | hlh-12(R15K) | B3 | let-858 | Yes |
| pGC220 | B1 | hlh-12 | B2 | Yes | Yes | 4xNLS-GFP | No | Yes | Yes | B4 | hlh-12(R25K) | B3 | let-858 | Yes |
| pGC240 | B1 | lim-7 | B2 | Yes | Yes | 4xNLS-GFP | No | Yes | Yes | No | 4xNLS-tdimer2(12) | No | let-858 | Yes |
| pGC452 | B4 | hlh-12 | B3 | Yes | Yes | 4xNLS-GFP | No | Yes | Yes | B1 | hairpin_hlh-12 | B1 | let-858 | Yes |
| D. FRT-containing Gateway destination vectors in the form [Gatewayd (R1-R2)<reporter<reporter] for insertion of promoter upstream of FLP-out cassette containing a reporter in the cassette and a reporter downstream of the cassette | |||||||||||||
| Plasmid | attd | attd | FRT | iA | Genea | unc-119(+)b | FRT | iA | att | Gene | att | 3′end | unc-119(+)b |
| pGC93 | R1 | R2 | Yes | No | 4xNLS-GFP | Yes | Yes | No | No | 1xNLS-lacZ | No | unc-54 | No |
| pGC97 | R1 | R2 | Yes | No | 4xNLS-GFP | Yes | Yes | No | No | tdimer2(12) | No | let-858 | No |
| pGC183 | R1 | R2 | Yes | No | 4xNLS-GFP | Yes | Yes | No | No | 4xNLS-tdimer2(12) | No | let-858 | No |
| pGC238 | R1 | R2 | Yes | Yes | 4xNLS-GFP | No | Yes | Yes | No | 4xNLS-tdimer2(12) | No | let-858 | Yes |
| pGC160 | R1 | R2 | Yes | No | tdimer2(12) | No | Yes | No | No | 4xNLS-GFP | No | let-858 | Yes |
| E. FRT-containing Gateway destination vectors in the form [Gatewayd (R1-R2)<reporter<Gatewayd (R4-R3)] for insertion of promoter upstream of FLP-out cassette and gene of interest downstream of FLP-out cassette | ||||||||||||
| Plasmid | attd | attd | FRT | iA | Genea | unc-119(+)b | FRT | iA | attd | attd | 3′end | unc-119(+)b |
| pGC162 | R1 | R2 | Yes | No | 4xNLS-GFP | Yes | Yes | No | R4 | R3 | let-858 | No |
| pGC247 | R1 | R2 | Yes | Yes | 4xNLS-GFP | No | Yes | No | R4 | R3 | let-858 | Yes |
| pGC163 | R1 | R2 | Yes | No | tdimer2(12) | No | Yes | No | R4 | R3 | let-858 | Yes |
| pGC225 | R1 | R2 | Yes | No | 4xNLS-tdimer2(12) | No | Yes | No | R4 | R3 | let-858 | Yes |
| pGC276 | R1 | R2 | Yes | Yes | 4xNLS-tdimer2(12) | No | Yes | Yes | R4 | R3 | let-858 | Yes |
| F. FRT-containing Gateway destination vectors in the form [Gatewayd (R4-R3)<reporter<Wormgatee] for insertion of promoter upstream of FLP-out cassette and RNAi hairpin downstream of FLP-out cassette | |||||||||||||
| Plasmid | attd | attd | FRT | iA | Genea | unc-119(+)b | FRT | iA | att | att | 3′end | unc-119(+)b | |
| pGC245 | R4 | R3 | Yes | Yes | 4xNLS-GFP | No | Yes | Yes | R1 | Wormgatee | R1 | let-858 | Yes |
| pGC320 | R4 | R3 | Yes | Yes | 4xNLS-mCherry | No | Yes | Yes | R1 | Wormgatee | R1 | let-858 | No |
| G. Donor vector for Gateway(R4-R3) cassette |
| Plasmid |
| pGC188f |
See materials and methods or supplemental Methods for plasmid construction details. See also supplemental Toolkit Documentation and supplemental table for strategies and additional constructs to facilitate construction of FLP- and FRT-containing plasmids.
All plasmids contain the let-858 3′end after the FLP recombinase and all reporters [GFP, tdimer2(12), or mCherry]. This sequence adds the let-858 3′-UTR onto the transcript.
C. briggsae unc-119(+); see text for details.
Intron A from Fire vectors.
Whole Gateway cassettes (with ccdB gene and chloramphenicol resistance gene; Invitrogen) are between attR sites unless otherwise indicated.
Wormgate (Johnson et al. 2005) consists of two reciprocally oriented Gateway cassettes flanking an intron. This system accepts cDNA clones directly from the ORFeome library.
See materials and methods and supplemental Toolkit Documentation for details on pGC188.
The orientation and type of att sites in the Gateway cassettes are given as “Gateway(R1-R2)” to indicate a Gateway cassette in the attR1 and attR2 orientation. NLS means nuclear localization signal. The < symbol is used to represent the FRT sequence. In oligo sequences, Gateway attB overhangs are indicated as uppercase letters while the gene-specific sequences are indicated as lowercase letters.
The starting point for construction of worm-compatible FLP/FRT vectors were the existing “Fire vectors” created in Andrew Fire's laboratory (http://www.addgene.org/Andrew_Fire) and the FLP/FRT vectors made in Gary Struhl's laboratory that have been widely used in the fly community, FL119 [Adh-Flp-Adh(3′), C20NX backbone] and J33R [>hsp70(3′)>, pUC19 backbone] (Struhl and Basler 1993).
FLP recombinase expression plasmid construction:
The FLP recombinase cDNA from plasmid FL119 (Struhl and Basler 1993) is the wild-type FLP recombinase. We chose this FLP recombinase over high-temperature forms used for mammalian applications such as FLP-F70L (pOG44, Stratagene) or FLPe (Buchholz et al. 1998), since the original wild-type FLP activity optimum is at 23° (Buchholz et al. 1996), a temperature compatible with worm husbandry. This becomes important when tissue-specific promoters are used to drive FLP expression (see results).
Heat-shock-driven FLP recombinase plasmids pGC94 [Phsp16.2∷FLP∷let-858(3′)] and pGC95 [Phsp16.41∷FLP∷let-858(3′)]:
The FLP cDNA in plasmid FL119 (Struhl and Basler 1993) is flanked by KpnI and BamHI restriction sites, with which it was excised and ligated to a similarly digested pBluescript SK+ (Stratagene) backbone, resulting in plasmid pGC92. This was done to facilitate use of the XbaI restriction site that follows the FLP cDNA in pGC92. Using KpnI and XbaI, the FLP recombinase cDNA fragment was excised from pGC92 and ligated to KpnI/NheI-digested pPD118.26 or pPD118.28 (A. Fire, S. Xu, J. Fleenor, J. Ahnn and G. Seydoux, personal communication; http://www.addgene.org/1594 and http://www.addgene.org/1595), which are Phsp16.2∷GFP∷let-858(3′) and Phsp16.41∷GFP∷let-858(3′), respectively. We thereby replaced the GFP with the FLP cDNA resulting in pGC94 [Phsp16.2∷FLP∷let-858(3′)] and pGC95 [Phsp16.41∷FLP∷let-858(3′)].
Bombardable heat-shock-driven FLP recombinase plasmids pGC133 [Phsp16.41∷FLP∷let-858(3′)-(Cb)unc-119(+)] and pGC146 [Phsp16.2∷FLP∷let-858(3′)-(Cb)unc-119(+)]:
We generated additional Phsp∷FLP constructs that can be used in microparticle bombardments by adding the C. briggsae unc-119 rescuing fragment [(Cb)unc-119(+)] in their backbones. We used the C. briggsae unc-119 gene rather than the C. elegans unc-119 because it is smaller (∼2 kb vs. ∼5.7 kb). This insertion was done by digesting both pGC94 and pGC95 with SbfI and NgoMIV and ligating each of the resulting fragments, Phsp16.2∷FLP or Phsp16.41∷FLP, to a SbfI/NgoMIV-digested vector derivative of pPD117.01 (A. Fire, S. Xu, J. Fleenor, J. Ahnn and G. Seydoux, personal communication; http://www.addgene.org/1587) containing (Cb)unc-119(+) (pPD117.01GtwyGFP_S65T_CbUnc), which was generously given to us by Barth Grant. This produced pGC133, [Phsp16.41∷FLP∷let-858(3′)-(Cb)unc-119(+)] and pGC146 [Phsp16.2∷FLP∷let-858(3′)-(Cb)unc-119(+)].
lag-2-promoter driven FLP recombinase plasmid pGC158 [Plag-2∷FLP∷let-858(3′)]:
We created a lag-2-driven FLP, pGC158, by replacing GFP∷unc-54(3′) from pJK590 (Blelloch et al. 1999; Mathies et al. 2003) between XmaI and ApaI with the FLP∷let-858(3′) fragment cut from pGC94 with the same restriction enzymes.
Gateway-compatible vectors pGC180, pGC181, and pGC267 that facilitate construction of [Ppromoter-of-interest∷FLP-recombinase]:
See supplemental Methods.
FRT-containing target plasmid and related vector construction:
“Full” FRT-containing FLP-out targets in the form [Pubiquitous<GFP<reporter]:
pGC93 and pGC183 were used as destination vectors in an LR reaction to insert pro-1 and rpl-28 promoters from entry vectors pGC22 (Killian and Hubbard 2004) and pGC157, respectively; the resulting plasmids are pGC200 and pGC185, respectively. The rpl-28 promoter was PCR amplified with Gateway primers GGGGACAAGTTTGTACAAAAAAGCAGGCTctgcagtttgtgcaacaaattgag and GGGGACCACTTTGTACAAGAAAGCTGGGTcacgagagcgtcggatattttacc and inserted in pDONR221 by BP reaction, resulting in pGC157.
pGC188, a new P4-P3 donor vector:
We generated pGC188, a new P4-P3 donor vector that is compatible with Gateway(R4-R3) cassette. To make pGC188, we first PCR amplified the attP4 site from pDONRP4-P1R (Invitrogen) with the following primers: aagctcgggcccgcgttaac and aaggctgtcggtcgacctcg. Then, we replaced the P2R site in pDONRP2R-P3 (Invitrogen) with the P4 PCR product after cutting it with ApaI and SalI, thus generating pGC188.
lim-7-promoter driven FLP-out construct (pGC240):
pGC238 was used as a destination vector in an LR reaction to introduce the lim-7 promoter from the pGC235 entry clone. pGC235 was created by PCR amplification of the first intron of lim-7 (sequences that drive expression in the gonadal sheath; R. Voutev, R. Keating, E. J. Hubbard and L. G. Vallier, unpublished results) with Gateway primers GGGGACAAGTTTGTACAAAAAAGCAGGCTacttgtgccttgattctc and GGGGACCACTTTGTACAAGAAAGCTGGGTcggtggttggtgctgacg and inserted in pDONR221 by BP reaction. An LR reaction between pGC235 and pGC238 resulted in pGC240.
hlh-12-promoter driven FLP-out constructs:
pGC247 was used as a destination vector to insert in a single LR reaction Phlh-12 and hlh-12(R25K) or hlh-12(R15K) into Gateway(R2-R1) and Gateway(R4-R3), respectively. First we created a Phlh-12 entry clone (pGC291) by PCR amplification of the ∼4-kb upstream region of hlh-12 and inserted this region into pDONR221 by a BP reaction. We used the following primers for the PCR: GGGGACAAGTTTGTACAAAAAAGCAGGCTgcggcgaggtcggcggtacgggcg and GGGGACCACTTTGTACAAGAAAGCTGGGTaataaaattgtgtaagatgacgc. Then we made hlh-12(R15K) and hlh-12(R25K) entry clones by PCR amplification from pGC85 and pGC86, respectively (see “hlh-12 constructs and site-directed mutagenesis” below) using the following primers: GGGGACAACTTTGTATAGAAAAGTTGatggcgaagaaaccgagag and GGGGACAACTTTGTATAATAAAGTTGcaactcaaatacaaactc. The PCR products were inserted into pGC188 by a BP reaction resulting in pGC192 and pGC193, respectively. Last, an LR reaction was performed between pGC291, pGC193, and pGC247 to create pGC220 [Phlh-12<iA-4xNLS-GFP∷let-858(3′)<iA- hlh-12(R25K)∷let-858(3′)-(Cb)unc-119(+)]. In a separate LR reaction pGC291, pGC192, and pGC247 were recombined to create pGC219, [Phlh-12<iA-4xNLS-GFP∷let-858(3′)<iA- hlh-12(R15K)∷let-858(3′)-(Cb)unc-119(+)].
hlh-12-promoter driven FLP-out RNAi-hairpin construct (pGC452):
We used pGC245 as a destination vector to create Phlh-12<GFP<hairpin_hlh-12. First we PCR amplified the hlh-12 promoter from pGC81 with primers: GGGACAACTTTGTATAGAAAAGTTGgcggcgaggtcggcggtacgggcg and GGGGACAACTTTGTATAATAAAGTTGaataaaattgtgtaagatgacgc. Then we inserted Phlh-12 into pGC188 by BP reaction to create pGC450. We PCR amplified the hlh-12 coding region from pGC81 with primers: GGGGACAAGTTTGTACAAAAAAGCAGGCTatggcgaagaaaccgagag and GGGGACCACTTTGTACAAGAAAGCTGGGTgcaatataaacattggtttggggc and used the PCR product in a BP reaction inserting it into pDONR221, yielding pGC451. Next we performed a single LR reaction using pGC450 and pGC451 as entry clones and pGC245 as a destination vector to create pGC452 [Phlh-12<iA-4xNLS-GFP∷let-858(3′)<iA-hairpin_hlh-12∷let-858(3′)-(Cb)unc-119(+)].
hlh-12 genomic region constructs and site-directed mutagenesis:
A ∼7.6-genomic region of hlh-12 was PCR amplified using the following primers: atgcgtgttgtcatagcctatattgg and catcacttgaatgttcacagattccg. The PCR product was TA cloned into PCR-XL-TOPO (Invitrogen) to create pGC81 (the insert went into reverse orientation into the vector). hlh-12(R15K) and hlh-12(R25K) were made by introducing point mutations in pGC81 using QuikChange II site-directed mutagenesis kit (Stratagene). The following primers were used for the site-directed mutagenesis, creating pGC85[hlh-12(R15K)] and pGC86[hlh-12(R25K)], respectively: ccaagctgaatacggatcgaaaatcgagagcaaacgagtacgttc gaacgtactcgtttgctctcgattttcgatccgtattcagcttgg and cattgtaaactttcagacgagaacgacagaaagtttccgagatg catctcggaaactttctgtcgttctcgtctgaaagtttacaatg.
Additional molecular methods
PCR amplifications:
All PCR amplifications were performed using the Expand Long Template PCR system (Roche). hlh-12 coding region and intron regions in pGC81, pGC85, pGC86, pGC192, and pGC193 were examined by sequence analysis. The promoter-containing constructs that were constructed using PCR fragments (Plim-7, Prpl-28, and Phlh-12) were not sequenced but were assessed functionally by examining the expression pattern of the respective promoter–reporter fusions.
Gateway recombination reactions:
All Gateway recombination reactions were performed according to the recommendations in the Invitrogen manuals: Gateway Technology with Clonase II (Invitrogen) and MultiSite Gateway three-fragment vector construction kit (Invitrogen) except that LR Clonase II was used in all LR reactions. LR Clonase Plus is recommended in Multisite Gateway reactions, but we did not detect a difference in obtaining colonies when using either LR clonase II or LR Clonase Plus II. We used One Shot TOP10 chemically competent cells (Invitrogen) when performing LR reaction with FLP-out constructs containing double and triple Gateway cassettes and (rubidium chloride competent) DH5α when performing LR reactions with single Gateway cassettes. Constructs containing Gateway cassettes were grown in DB3.1 cells (Invitrogen).
Worm handling and strains
Strains:
Preexisting strains used in this study were N2, him-5(e1490) (Brenner 1974) and unc-119(ed3) (Maduro and Pilgrim 1995). Transgenic lines are listed and described in Table 2. We used two different unc-119(ed3) background strains (GC729 and DP38) for injections and bombardment as indicated in Table 2. naIs3, naIs6, naIs7, naIs35, and naEx75 were obtained by microparticle bombardment (Praitis et al. 2001) of the respective constructs into unc-119(ed3) worms. In the case of naIs6, naIs7, and naEx75, pCW2.1 was co-bombarded together with the construct, however naIs7 did not show expression of Pceh-22∷GFP.
TABLE 2.
C. elegans strains used in this study
| A. Strains carrying FLP recombinase expression constructs, listed in order of pGC FLP-bearing plasmid | |||||
|---|---|---|---|---|---|
| Plasmid | Method | Alleleabc | Strain name | Background straind | Cotransformation markerb |
| pGC95a | Injection | naEx40 | GC729 | CB4088 | pCW2.1 |
| pGC133 | Bombard | naIs3 | GC773 | GC769 | no |
| pGC146 | Bombard | naIs6 | GC817 | GC769 | pCW2.1 |
| pGC146e | Bombard | naIs7e | GC827 | GC769 | pCW2.1e |
| pGC146b | Bombard | naEx75b | GC822 | GC769 | pCW2.1 |
| pGC158a | Injection | naEx57 | GC771 | CB4088 | pCW2.1 |
| B. Strains carrying FLP-out target constructs, listed in order of pGC FRT-bearing plasmid | |||||
| Plasmid | Method | Alleleabc | Strain name | Background straind | Cotransformation markerb |
| pGC185 | Injection | naEx56 | GC772 | GC769 | No |
| pGC200 | Injection | naEx64 | GC801 | GC769 | No |
| pGC200 | Bombard | naIs35 | GC1020 | DP38 | No |
| pGC219 | Injection | naEx167 | GC1010 | GC769 | pGC204 |
| pGC220 | Injection | naEx164 | GC987 | GC769 | pGC204 |
| pGC240 | Injection | naEx66 | GC804 | GC769 | pGC204 |
| pGC452 | Injection | naEx168 | GC1011 | GC817 | pGC204 |
| C. Strains carrying hlh-12 constructs | |||||
| Plasmid | Method | Alleleab | Strain name | Background straind | Cotransformation markerb |
| pGC81a | Injection | naEx161 | GC984 | N2 | pRF4 |
| pGC85a | Injection | naEx162 | GC985 | N2 | pRF4 |
| pGC86a | Injection | naEx163 | GC986 | N2 | pRF4 |
With the exception of pGC81, pGC85, pGC86, pGC95, and pGC158, all pGC constructs carry the Cb[unc-119(+)] as a transformation marker on the pGC construct.
With the exception of naEx75 that was generated by microparticle bombardment (Praitis et al. 2001) but is not integrated, all naEx allele-bearing strains were generated by injection (Mello et al. 1991) with the pGC plasmid at 10 ng/μl and the cotransformation maker at 20 ng/μl (pCW2.1 or pGC204) or 100 ng/μl (pRF4). Injection mixes also included pBluescript DNA for naEx40, naEx57, naEx66, naEx164, and naEx167 (70 ng/μl), naEx56 and naEx64 (90 ng/μl), and naEx168 (100 ng/μl). pCW2.1 carries the ceh-22∷GFP pharyngeal marker (Okkema et al. 1997); pGC204 carries Pceh-22∷tdimer2(12); pRF4 carries DNA encoding the dominant rol-6(su1006) allele (Mello et al. 1991). All remaining naEx alleles were selected using the pGC plasmid-borne Cb[unc-119(+)] as a transformation marker.
naIs alleles were generated by microparticle bombardment (Praitis et al. 2001) with the pGC plasmid at 10 μg and, where included as a co-bombarded marker, pCW2.1 at 10 μg.
Genotypes of background strains for injection and bombardment: CB4088 him-5(e1490); DP38 unc-119(ed3); GC769 unc-119(ed3) (note: same genotype as DP38 but healthier and somewhat less Unc); GC817 naIs6; N2 wild type.
Expression of the co-bombarded marker is not observed in naIs7 strain although expression of the pGC146 plasmid-borne Cb[unc-119(+)] phenotype is observed.
Temperature regimes for heat-shock inducible FLP-out and detection of lacZ induction:
We tested several temperature conditions for FLP recombinase expressed from different promoters, one on an array and one integrated (naEx40[Phsp-16.41∷FLP] and naIs6[Phsp-16.2∷FLP]). First, we determined that the progeny of worms grown at 15° or 25° (either prior to or following the heat shock) did not inappropriately induce FLP-out, indicating that the heat-shock promoters were not leaky throughout development and that the system is compatible with strains that must be maintained at these temperatures. Second, we observed successful FLP-out under several heat-shock protocols including 33° for 2, 3, and 4 hr or 37° for 40 min. We did not use prewarmed plates in these experiments. Unless otherwise specified, we used 33° as heat-shock temperature since 33° provides optimal heat shock with the promoters we use (Stringham et al. 1992). For experiments where lacZ was induced by FLP-out, β-gal activity was assayed after acetone fixation and X-gal staining for ∼6–24 hr.
RESULTS
Temporal control of gene expression by FLP/FRT:
To determine whether the yeast FLP recombinase is active in C. elegans, we first tested whether it could direct intramolecular excision of cassettes flanked by tandem unidirectional FRT sites (FLP-out cassettes) introduced as transgenes on “simple” extrachromosomal arrays (Stinchcomb et al. 1985; Mello et al. 1991; Kelly et al. 1997). Simple arrays are the easiest and most common method used to generate transgenic C. elegans. In theory, upon induction of the FLP recombinase, expression of the reporter between the FRT sites (reporter 1) would be lost while the reporter gene following the FLP-out cassette (reporter 2) would be expressed (Figure 1A). Because the target constructs are present in multiple copies, depending on the efficiency of the FLP-out reaction, several possible results were anticipated depending on the efficiency of the reaction. If no recombination occurred we would expect to see only reporter 1 expression. If a subset of the target copies were excised we would expect to see expression of reporter 1 both before and after induction and reporter 2 only after induction. Finally, if every target copy in the array is excised, reporter 1 expression should be abolished and only reporter 2 expression should remain after induction.
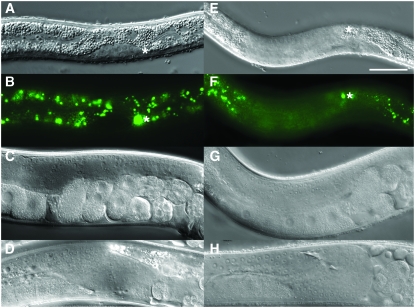
We generated and crossed two strains of worms, one carrying a heat-inducible FLP recombinase and the other carrying the target construct: a ubiquitous promoter driving expression of one reporter embedded in the FLP-out cassette and another reporter following the FLP-out cassette (Figure 1A, see materials and methods). We found that expression of the FLP recombinase from the heat-shock promoter Phsp-16.41 resulted in robust induction of the FLP recombinase as assayed by ubiquitously expressed reporters both within and following the FLP-out cassette (Figure 1, F–I). Specifically, worms that carried both the FLP-out cassette, Ppro-1<GFP<lacZ (pGC200, naEx64; Tables 1 and 2), and the heat-inducible FLP recombinase (pGC95, naEx40; Tables 1 and 2) were heat-shocked for 2 hr at 30°, returned to 20°, and assayed for β-galactosidase (β-gal) activity 24 hr later. Successful FLP-out of the FRT-flanked GFP (“<GFP<”) cassette was revealed by expression of β-gal (Figure 1I). In contrast to the non-heat-shocked control worms that expressed only GFP (Figure 1, B–E), the heat-shocked worms expressed both GFP and β-gal (Figure 1, F–I). We conclude that the FLP-out excision reaction occurred on a subset of the target cassettes on the transgenic array.
In rare cases, we observed apparently complete FLP-out. For example, in a worm expressing the FLP recombinase from the hsp-16.41 promoter (naEx40[Phsp-16.41∷FLP]) and the FLP-out target from the ubiquitous rpl-28 promoter (naEx56[Prpl-28<GFP<tdimer2(12)]) (pGC185, naEx56; Tables 1 and 2), an apparently complete FLP-out was observed in some cells since the otherwise robust GFP signal completely disappeared after heat-shock induction of the tdimer2(12) reporter (Figure 2).
Figure 2.—
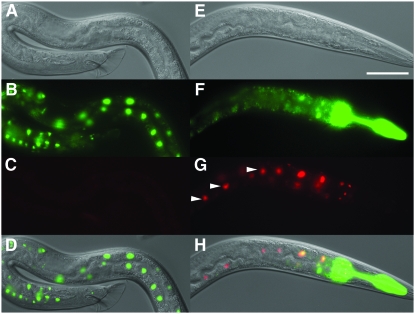
Highly efficient temporal control of FLP-out. Heat-induced FLP-out as indicated by GFP-to-tdimer2(12) switch in a male worm of the genotype naEx40[Phsp-16.41∷FLP, Pceh-22∷GFP]; naEx56 [Prpl-28<GFP<tdimer2(12)] (see Tables 1 and 2 and materials and methods for details on constructs and strains). (A–D) Worm before heat shock. Worm was raised at 25°, heat-shocked for 2 hr at 33°, and returned to 25° for 24 hr. (E–H) Same individual worm as in A–D after heat shock. All images were captured at 400× magnification. A and E, DIC; B and F, green channel; C and G, red channel; D and H, merges of DIC, green, and red panels above. Cells that no longer express GFP and only express tdimer2(12) are indicated by arrowheads in G. Bar, 50 μm. Note that tdimer2(12) was readily visible in worms grown at 25° but variably visible in worms grown at 15° and 20°.
Spatial control of gene expression:
Next, we asked whether we could use this system for spatial control of gene expression. We placed the FLP recombinase under the control of a tissue-specific promoter, Plag-2, which is strongly expressed in the DTCs (Henderson et al. 1994). We found that FLP-out occurs appropriately in the DTC, as read out from the ubiquitously expressed Prpl-28<GFP<tdimer2(12) or Ppro-1<GFP<lacZ (Figure 3, A–D and Figure 4, respectively). Using Plag-2∷FLP (pGC158, naEx57; Tables 1 and 2) directed FLP-out of the Ppro-1<GFP<lacZ array, FLP-out occurred in 16% of DTCs (n = 32 gonad arms).
Figure 3.—
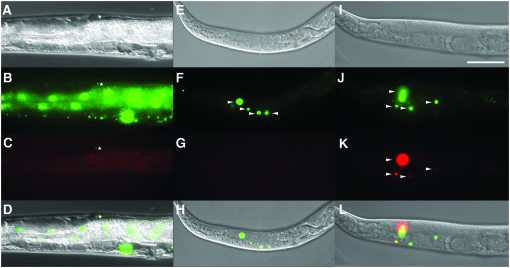
Spatial and spatio-temporal control of gene expression. (A–D) Tissue-specific FLP-out visualized by tdimer2(12) expression in DTC of worms carrying naEx57[Plag-2∷FLP, Pceh-22∷GFP] and naEx56[Prpl-28<GFP<tdimer2(12)] (see Tables 1 and 2 and materials and methods for details on constructs and strains). Worms were raised at 20°. The DTC is indicated with an asterisk. (E–L) Spatio-temporal control of FLP-out that induces GFP-to-tdimer2(12) switch in gonadal sheath cells of a worm with genotype naEx75[Phsp16.2∷FLP, Pceh-22∷GFP]; naEx64[Plim-7<GFP<tdimer2(12)]. (E–H) worm prior to heat shock; (I–L) Same individual worm as in E–H 24 hr after a 2-hr heat shock at 33° followed by return to 20°. All images were captured at 400× magnification. A, E, and I, DIC; B, F, and J, green channel; C, G, and K, red channel; D, H, and L, merges of DIC, green, and red panels above. Nuclei of gonadal sheath cells are indicated by arrowheads. Bar, 50 μm.
Figure 4.—
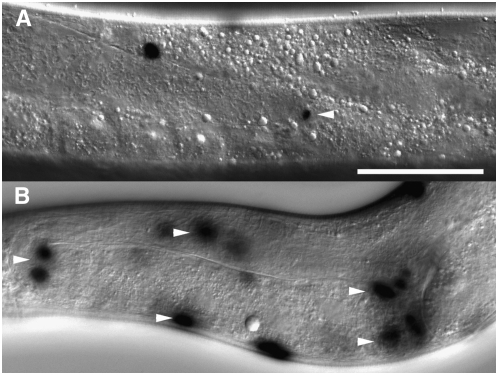
Plag-2-induced FLP-out induces lacZ expression in the somatic gonad lineage. (A) β-gal expression in the DTC only; FLP-out occurred after the DTC was born. (B) β-gal expression in the entire somatic gonad; FLP-out occurred in the early L1 stage (prior to the division of Z1 or Z4). Worms were of the genotype naEx57[Plag-2∷FLP, Pceh-22∷GFP]; naEx64[Ppro-1<GFP<lacZ]. Both images were captured under DIC optics, and worms were stained with X-gal staining solution to visualize β-gal. Arrowheads indicate the DTC (A) and gonadal sheath cells (B) where FLP-out occurred. Bar, 50 μm.
We note that in experiments of this type where the recombinase is driven from a non-heat-shock promoter, the FLP recombinase must be active at normal growth temperatures for the worm; the form of the FLP recombinase we used is optimally active at 23° (Buchholz et al. 1996, see materials and methods).
Combined temporal and spatial control of gene expression:
We further tested whether the system could be used to combine both spatial and temporal control of gene expression. We asked whether transgenic worms carrying two constructs, Phsp-16.2∷FLP (pGC146, naEx75; Tables 1 and 2) and Plim-7<GFP<tdimer2(12) (pGC240, naEx66; Tables 1 and 2; materials and methods), could be induced to express tdimer2(12) specifically in the gonadal sheath cells after heat shock. We indeed detected tdimer2(12) in gonadal sheath cells only after heat shock and not prior to heat shock (Figure 3, E–L).
Taken together, these results demonstrate that the FLP/FRT system can be used effectively for spatial and temporal control of gene expression in C. elegans when expressed from simple arrays.
The FLP/FRT system as lineage tracing tool:
The Plag-2∷FLP directed FLP-out also provided a test of the system as a cell lineage tracer. Cell lineage markers are useful for analysis of development. In particular, cell fate-independent lineage tracers are useful under experimental conditions that may alter cell fate during development. Plag-2 is expressed in L1 larvae in Z1 and Z4 (Henderson et al. 1994), cells that give rise to the mirror-symmetric anterior and posterior parts of the hermaphrodite somatic gonad, respectively. Subsequently, these cells divide and only their distal granddaughters, the DTCs (Z1.aa and Z4.pp), maintain Plag-2 expression, while other somatic gonad cells do not express Plag-2. Therefore, if FLP-out occurs in Z1 and Z4 prior to their division in the early L1 larval stage, the entire somatic gonad lineage should express the reporter downstream of the FRT cassette. If FLP-out occurs in only Z1 or only Z4 (but not both), either the entire anterior half or posterior half of the somatic gonad would be marked. Finally, if FLP-out occurs after the division of Z1 and Z4, a subset of somatic gonad cells would be marked, depending on the timing of the FLP relative to the lineage (that is, either all descendents of Z1.a and/or Z4.p or the DTCs Z1.aa and/or Z4.pp). In 3/32 animals carrying Plag-2∷FLP and the ubiquitously expressed Ppro-1<GFP<lacZ (both on extrachromosomal arrays), we observed lacZ expression in all descendents of Z1 or Z4, while in 2/32 we observed expression only in the DTC (Figure 4). We interpret the former as FLP-out in Z1 or Z4 and the latter as FLP-out in Z1.aa or Z4.pp. The remaining 27 animals did not express lacZ in the somatic gonad. In this case, because both the FLP recombinase and the target were present on transgenic arrays, the relatively low percentage of worms in which FLP-out occurred in the gonad may have been the result of mosaicism. An integrated FLP recombinase provided more robust results (see below). We conclude that the FLP-out technique can be used to mark cells and cell lineages in C. elegans.
Efficacy of FLP-out from an integrated Phsp-16.2∷FLP transgenic line:
To evaluate in a more systematic way the efficiency and possible cell-type bias of FLP-out, we carried out further experiments using a transgenic line carrying integrated Phsp-16.2∷FLP (pGC146, naIs6; Tables 1 and 2). Integration eliminates mosaicism, a characteristic of extrachromosomal arrays (Praitis et al. 2001). We compared the viability and brood size of this strain with that of N2, both with and without heat shock, and found that they did not exhibit defects (Table 3).
TABLE 3.
Test for viability/fertility of FLP-expressing worms
| Genotype | Heat shocka | Survival (%) | n |
|---|---|---|---|
| N2 | — | 97 | 100 |
| naIs6b | — | 99 | 100 |
| N2 | + | 85 | 100 |
| naIs6b | + | 91 | 100 |
| Genotype | Heat shockc | Average brood | nd |
| N2 | — | 226 | 7 |
| naIs6b | — | 234 | 8 |
| N2 | + | 82 | 10 |
| naIs6b | + | 86 | 11 |
L1 worms were heat-shocked for 4 hr at 33° and put at 25° until adulthood when survival was scored. The non-heat-shocked worms were kept at 25° and scored at the same time as the heat-shocked worms.
Full genotype: naIs6[Phsp-16.2∷FLP-(Cb)unc-119(+), Pceh-22∷GFP]
L4 animals were heat-shocked (where indicated) for 2 hr at 33° and then transferred to 25° for 72 hr. Non-heat-shocked worms were kept at 25° throughout.
n is the number of broods counted.
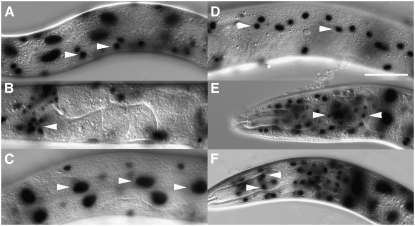
To assess tissue distribution of FLP-out, we introduced a FLP-recombinase target Ppro-1<GFP<lacZ extrachromosomal array (naEx64; Table 2) in the background of the Phsp-16.2∷FLP integrated line and heat-shocked L4 animals for 2 hr at 33° followed by return to 20° for 24 hr. We observed FLP-out activity in head neurons, pharyngeal muscle, intestine, hypodermis, somatic gonad, and body wall muscle in 97, 81, 90, 94, 45, and 97% of animals, respectively (n = 64, Figure 5, A–F). In addition Phsp-16.2-directed FLP-out was observed in late embryos but not in early embryos (data not shown). The latter observation is consistent with the results for Phsp-16.2 induction previously described (Stringham et al. 1992).
Figure 5.—
FLP-out in different tissues and cell types. Expression in (A) body wall muscles, (B) somatic gonad (spermatheca), (C) intestine, (D) hypodermis, (E) neurons, and (F) pharyngeal muscle cells. All images were captured under DIC. Genotype of all worms was naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx64[Ppro-1<GFP<lacZ]. Arrowheads in A–F indicate examples of the respective cell types scored. FLP-out also occurred in additional tissues in each worm shown. Bar, 50 μm.
Using the same strain, we wished to evaluate the overall efficiency of FLP-out. We first tried quantitative PCR, using multiple primer sets. However, we did not detect a significant change in the amount of FLPed cassettes in the heat-shocked worms vs. non-heat-shocked worms (data not shown). We do not have a satisfactory explanation for these results, but they suggest that, on average, few targets undergo excision or that the complex arrangement of the arrays and/or perdurance of excised fragments permits priming.
In the absence of a reliable molecular readout, we measured the efficiency of FLP-out by assessing a more experimentally relevant readout: the efficiency of FLP-out in single cell. First, we tested several heat-shock conditions and found that for the recombinase to drive FLP-out from naEx66[Plim-7<GFP<tdimer2(12)], 3 hr of heat shock at 33° was more effective than 2 or 4 hr (Table 4). Next we assessed the frequency of FLP-out reporter induction in a single cell (dorsal cell of sheath pair one) and found it was 20% for this set of reagents and conditions (Table 4).
TABLE 4.
FLP-out rate in dorsal cell of gonadal sheath pair 1
| Heat-shocka length (hr) | Gonad armsb with FLP-out (%) | Dorsal Sh1 with FLP-out (%) | nc |
|---|---|---|---|
| 2 | 56 | 11 | 18 |
| 3 | 65 | 20 | 20 |
| 4 | 58 | 17 | 12 |
Early adult animals with genotype naIs6[Phsp-16.2∷FLP-(Cb)unc-119(+), Pceh-22∷GFP]; naEx66[Plim-7<GFP<tdimer2(12)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)] were heat-shocked for the specified time at 33° and put at 25° for 24 hr, after which they were examined for t-dimer2(12) expression in the gonadal sheath.
Percentage of gonad arms in which tdimer2(12) was expressed in any of the 10 cells of the somatic gonad sheath.
Number of gonad arms examined.
Conditional expression of dominant-negative hlh-12 using FLP/FRT:
To test a biologically relevant phenotypic scenario, we asked if conditional expression using the FLP-out system could facilitate analysis of a transgene predicted to interfere with fertility. Transgenes that interfere with fertility pose difficulties using traditional nonconditional expression technologies. Even low-level sterility caused by an extrachromosomal array that is, itself, inherited at a low frequency can significantly interfere with maintenance of the transgenic line.
Mutations in hlh-12 cause early DTC migration defects that secondarily cause proximal germline tumor formation (Pro phenotype) and thereby reduce fertility. hlh-12 encodes a class II (Massari and Murre 2000) or group A (Atchley and Fitch 1997) bHLH transcription factor (see supplemental Results for further consideration of HLH-12 family assignment). HLH-12 is expressed in the DTC and forms heterodimers with HLH-2 to activate transcription of gon-1, and reduction of hlh-12 activity produces Mig phenotype (Tamai and Nishiwaki 2007).
We used the FLP-out system to assess the activity of a putative dominant-negative form of hlh-12. To generate a dominant-negative allele of hlh-12, we reasoned that interfering with the ability of HLH-12 to bind DNA without interfering with its ability to bind its partner HLH-2 (daughterless ortholog) could generate inactive HLH-12/HLH-2 heterodimers (Tamai and Nishiwaki 2007) and cause a dominant-negative phenotype. Previously, it was shown that R-to-K substitutions of the highly conserved arginine residues at the beginning and the end of the basic region of the mammalian daughterless ortholog E47, abolishes DNA binding (Voronova and Baltimore 1990). On the basis of these findings and the identical position of these arginines in the basic domain sequence (see supplemental Results), we tested two different HLH-12 mutant constructs: hlh-12(R15K) and hlh-12(R25K) to see if they would produce a dominant-negative effect in vivo.
When we expressed each of these mutant forms in the context of genomic hlh-12 from conventional arrays, we found that one of the substitutions, HLH-12(R25K) indeed produced a penetrant dominant-negative defect (Table 5). These data suggest that HLH-12(R15K) likely retains its wild-type DNA binding capability, whereas HLH-12(R25K) does not. Consistent with our prediction, the hlh-12(R25K) transgenic line was, in fact, difficult to maintain since nearly 25% of the worms that inherited the array were sterile.
TABLE 5.
Penetrance of Mig in nonconditional and conditional hlh-12 mutants and RNAi
| Genotype | Heat shocka | Phenotype (% Mig) | N (gonad arms) |
|---|---|---|---|
| naEx161[hlh-12(WT)]b | — | 2.1 | 140 |
| naEx162[hlh-12(R15K)b | — | 2.8 | 108 |
| naEx163[hlh-12(R25K)b | — | 24.2*** | 132 |
| hlh-12(RNAi)c | — | 50 | 300 |
| L4440c | — | 0 | 300 |
| naIs6d | + | 0 | 140 |
| naIs6d; naEx164e | + | 7.7* | 78 |
| naIs6d; naEx164e | — | 0 | 84 |
| naIs6d | + | 0 | 110 |
| naIs6d; naEx167f | + | 0 | 87 |
| naIs6d; naEx167f | — | 0 | 36 |
| naIs6d | + | 1.5 | 136 |
| naIs6d; naEx168g | + | 8.9** | 158 |
| naIs6d; naEx168g | — | 1.7 | 120 |
A Fisher exact test (one-sided) was used to test the null hypothesis that there is no difference between the percentage of animals that display the Mig phenotype among animals of the indicated genotype compared to the genotype directly below it. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001.
+ indicates L1 larvae were heat-shocked for 3 hr at 33° and afterwards grown at 25° to adulthood; − indicates non-heat-shocked worms that were grown at 25°.
naEx161, naEx162, and naEx163 express hlh-12 from its native promoter and 3′-UTR (see materials and methods for details).
hlh-12(RNAi) was induced by feeding L4 animals (Ahringer library clone IV-4G20; Kamath et al. 2003, which we validated by sequence analysis) and scoring their progeny for the Mig phenotype; L4440 is the double T7-vector control for the RNAi feeding plasmid (Timmons and Fire 1998). Feeding L1 larvae within 2 hr of hatching produced similar results (44%, n = 102 with 0% in parallel control, n = 100).
naIs6[Phsp-16.2∷FLP-(Cb)unc-119(+), Pceh-22∷GFP]. For each set of three strains, the first and third lines are control siblings segregating from mothers that gave rise to all three strains. That is, animals labeled naIs6 in the first set are siblings of naIs6; naEx164[Phlh-12<GFP<hlh-12(R25K)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)] that appear to have lost the array by virtue of their failure to express Pceh-22∷tdimer2(12).
naEx164[Phlh-12<GFP<hlh-12(R25K)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)]. For each set of three strains, the first and third lines are control siblings segregating from mothers that gave rise to all three strains. That is, animals labeled naIs6 in the first set are siblings of naIs6; naEx164[Phlh-12<GFP<hlh-12(R25K)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)] that appear to have lost the array by virtue of their failure to express Pceh-22∷tdimer2(12).
naEx167[Phlh-12<GFP<hlh-12(R15K)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)]. For each set of three strains, the first and third lines are control siblings segregating from mothers that gave rise to all three strains. That is, animals labeled naIs6 in the first set are siblings of naIs6; naEx164[Phlh-12<GFP<hlh-12(R25K)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)] that appear to have lost the array by virtue of their failure to express Pceh-22∷tdimer2(12).
naEx168[Phlh-12<GFP<hairpin_hlh-12-(Cb)unc-119(+), Pceh-22∷tdimer2(12)]. For each set of three strains, the first and third lines are control siblings segregating from mothers that gave rise to all three strains. That is, animals labeled naIs6 in the first set are siblings of naIs6; naEx164[Phlh-12<GFP<hlh-12(R25K)-(Cb)unc-119(+), Pceh-22∷tdimer2(12)] that appear to have lost the array by virtue of their failure to express Pceh-22∷tdimer2(12).
Next we expressed HLH-12(R25K) conditionally via the FLP/FRT system. To create Phlh-12<GFP<hlh-12(R25K) (pGC220; Table 1), we used one of our Gateway compatible FLP-out constructs that permit efficient insertion of promoters and genes of interest in one step (see Table 1, materials and methods, discussion, and supplemental Toolkit Documentation). Worms carrying Phlh-12<GFP<hlh-12(R25K) expressed GFP in the DTC during migration (Figure 6, A and B). We introduced naEx164[Phlh-12<GFP<hlh-12(R25K)] in the background of naIs6[Phsp-16.2∷FLP] (see Table 2) and heat-shocked L1 larvae for 3 hr at 33°. We detected FLP-out in 7.7% of the gonad arms examined as readout by the Mig phenotype (Table 5). The Mig phenotype was similar to that observed in the strain bearing the original non-FLP transgene carrying hlh-12(R25K) (Figure 6, C and D). Although this overall penetrance of Mig after induction of FLP is relatively low, we note that the penetrance of the Mig phenotype using the non-FLP transgene is only 24% (Table 5), suggesting that the FLP-out strain was roughly one-third as effective as the normal transgene.
Figure 6.—
Conditional expression of HLH-12(R25K) and induction of hlh-12(RNAi) by FLP-out. (A) DIC image of an L2 worm of the genotype naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx164[Phlh-12<GFP<hlh-12(R25K)] . (B) GFP image of the same worm as in A. GFP expression driven by Phlh-12 in the DTC (asterisk). (C) Non-heat-shocked naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx164 [Phlh-12<GFP<hlh-12(R25K)] worm that did not display the Mig phenotype. (D) Heat-shocked naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx164 [Phlh-12<GFP<hlh-12(R25K)] worm that exhibited the Mig phenotype. (E) DIC image of an L4 worm of the genotype naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx168[Phlh-12<GFP<hairpin_hlh-12)]. (F) GFP image of the same worm as in E; GFP expression driven by Phlh-12 in the DTC (asterisk). (G) Non-heat-shocked naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx168[Phlh-12<GFP<hairpin_hlh-12)] worm that did not display the Mig phenotype. (H) Heat-shocked naIs6[Phsp-16.2∷FLP, Pceh-22∷GFP]; naEx168[Phlh-12<GFP<hairpin_hlh-12)] worm that exhibited the Mig phenotype. Bar, 50 μm.
Conditional expression of an RNAi-inducing hairpin using the FLP/FRT system:
We next asked whether we could conditionally induce the hlh-12 Mig phenotype by FLP-mediated expression of a hlh-12(RNAi) from an hlh-12 hairpin [inverted hlh-12 sequences separated by an intron to produce an RNA “hairpin” (Tavernarakis et al. 2000; Johnson et al. 2005)]. For this experiment we created a Phlh-12<GFP<hairpin_hlh-12 construct using the advantage of pGC245, a Gateway/Wormgate FLP-out destination construct we created (materials and methods; Figure 7C; Table 1; supplemental Methods). We observed a 7% increase in the percentage of worms displaying the Mig defect (Table 5) in heat-shocked animals (over the background <2% in siblings that had lost the array—as judged by the pharyngeal marker—and non-heat-shocked siblings that carried the array; Figure 6, E–H). These results are comparable to what we observed with the dominant-negative allele. The penetrance is lower than the ∼50% Mig observed in worms fed hlh-12(RNAi) continuously from the previous generation or from the L1 stage (Table 5). That we observed a lower penetrance of the phenotype is not surprising given that our assay was quite stringent and required several independent events (all of which occur with <100% probability) to occur simultaneously in a single cell. For example, the FLP recombinase had to be sufficiently induced, the array carrying the FRT target had to be inherited by the DTC, the FLP-out reaction had to occur, and the RNAi had to be effective. We conclude that the FLP-out system allows conditional induction of RNAi that results in dominant depletion of gene activity.
Figure 7.—
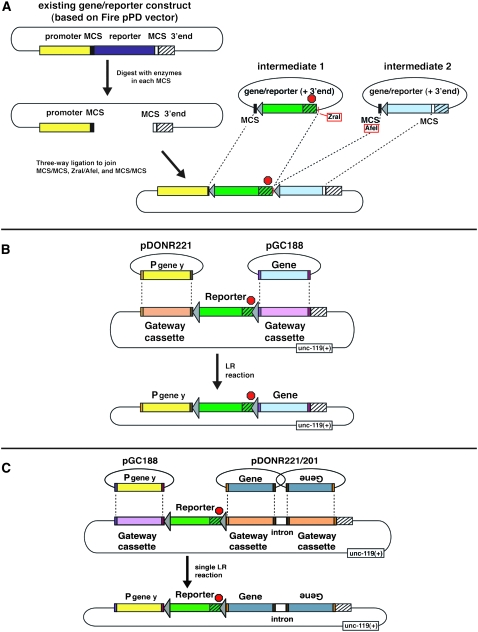
Schematic of three strategies to generate FLP-out target constructs. See supplemental Toolkit Documentation for full details on plasmids and strategies for their use in customizing both FLP recombinase-containing and FRT-containing reagents. (A) Converting a preexisting Fire vector-based promoter/reporter (or promoter/gene) construct into an FRT-bearing FLP-out construct. Any existing promoter/reporter fusion can be digested in the resident multiple cloning sites (MCS, two different MCS sequences are indicated by black and white boxes, respectively) and then ligated to fragments from FRT intermediate vectors. In this strategy, corresponding restriction enzymes are used to prepare the inserts and vector backbone. An additional convenient aspect of the strategy is the ZraI site in the FRT intermediate vector and the MCS-resident AfeI site (both of which result in blunt ends after digestion, indicated in red boxes). These sites facilitate three-way ligation to complete the FRT-containing vector construction in one step. Other compatible sites could be used for this junction. (B) Gateway-mediated insertion of promoter and gene of interest to generate a final promoter<reporter<gene-of-interest FLP-out target product in a single LR reaction. Sequences in the two donor vectors can be recombined to any destination FLP-out cassette vector that follows the formula Gateway(attR1-attR2)<reporter<Gateway(attR4-attR3). (C) Generation of a final promoter<reporter<RNAi-hairpin product in a single LR reaction from two donor vectors to any FLP-out cassette destination vector that follows the formula Gateway(attR4-attR3)<reporter<Gateway(attR1-attR2)-intron-Gateway(attR2-attR1). This strategy takes advantage of the Wormgate system and ORFome clones (Johnson et al. 2005). In A–C, the red stop sign indicates a required transcriptional stop and 3′ end sequences are indicated by hatched boxes. See materials and methods, Table 1, and supplemental Toolkit Documentation for details.
DISCUSSION
Here, we examine combinations of inducible, ubiquitous, and tissue-specific promoter FLP expression and FRT-construct targets, several heat-shock promoters and induction conditions, and the behavior of an integrated heat-shock FLP. In addition, we demonstrate the utility of the system for cell lineaging, and for expression of both dominant-negative and RNAi-hairpin forms of a gene to produce conditional reduction of gene expression. We introduce useful strains and constructs (both FLP recombinase and FRT-bearing components), as well as a vector “kit” to facilitate construction of additional plasmids of interest (both de novo and adaptations of preexisting Fire-vector promoter-reporter constructs; see supplemental Toolkit Documentation). Our analysis extends the recent Davis et al. (2008) report of a similar FLP/FRT-based system that they validated using reporters and an inducible tetanus toxin to interfere with neuronal transmission (Davis et al. 2008).
Below, we further discuss general features of recombinase-mediated gene expression in C. elegans compared to other organisms. We then discuss the uses, caveats, and possible extensions to this technology in C. elegans as they apply to lineaging and to control of gene expression (including RNAi), noting useful reagents for each of these applications. Finally, we present an overview of strategies by which our vectors can be used to generate customized FLP-out target cassettes.
FLP-out technology is applicable to C.elegans:
In other model organisms, the utility of the FLP/FRT and other recombinase-mediated systems are well established for lineage marking and temporal and spatial control of gene expression (see Branda and Dymecki 2004, for review; McGuire et al. 2004). Certain features of C. elegans biology both limit and extend the application of this technology.
While we have shown that FLP-out is useful in C. elegans, we note that FLP/FRT-mediated mitotic recombination is not applicable. Recombinase-mediated gene expression systems are widely used in Drosophila in two capacities: to generate mosaics by crossover between chromatids in mitosis and for intramolecular excision (e.g., FLP-out). Unlike Drosophila, C. elegans homologous chromosomes do not pair during mitosis, therefore the FLP/FRT system cannot be used to efficiently generate mosaic “clones” by mitotic crossover as it is in Drosophila (Golic 1991). Because techniques for generating genetic mosaics by mitotic loss of free duplications or extrachromosomal arrays have long been part of the worm “toolkit” (Herman 1984; Yochem and Herman 2005), the lack of a recombinase-mediated system to generate mosaics is not at issue. Nevertheless, the FLP-out technique does generate a permanent change in gene expression within a cell lineage, in effect providing an alternate way to generate mosaics.
We focused our attention on the other commonly used method to alter gene expression with recombinase-mediated technology: the intramolecular excision or FLP-out application of the FLP/FRT recombination system (Figure 1A). In this system, as in other similar systems like Cre-lox, an intervening sequence that limits or prevents gene expression (reporter 1, Figure 1A) is excised to permit expression of a second gene (reporter 2, Figure 1A). A critical feature of this system is that prior to activation of the FLP recombinase, the excision cassette must prevent transcriptional readthrough into the second gene downstream of the FRT sequences.
In each of our excision cassettes we use DNA sequences 3′ to the let-858 gene (http://www.addgene.org/Andrew_Fire) and have not observed readthrough in any case that we tested. This sequence contains several possible transcriptional stop sites (Blumenthal and Steward 1997) but EST evidence shows that one of them is used preferentially (see supplemental Toolkit Documentation for details). The commonly used unc-54 3′ end also contains several consensus transcriptional stop sites, so we predict that it would also work well (see supplemental Toolkit Documentation for details). Nonetheless, even if predicted transcriptional terminators are present in an excision cassette, they should be tested for readthrough in vivo.
In our studies, we excised both a reporter and a transcriptional termination site. In this way, we could monitor expression both before (reporter 1 expression) and after (reporter 2 expression) excision (Figure 1A). If alternative sequences within the FLP-out cassettes are used, it would be imperative to first assess expression of the downstream of the FRT cassette when the FLP recombinase is absent. Nonrecombinase-induced expression could result from readthrough or from unanticipated rearrangements of the transgenes during the formation of the FRT-cassette-bearing transgene (see below).
Although our studies focused on the FLP-out technology where the FLP recombinase and the FLP-out target genes are driven from specific promoters, the role of 3′-UTRs in gene expression must also be taken into account in the experimental design.
Finally, we note that to promote FLP-out, the FRT target sequences must be in tandem (that is, the same orientation) to precisely excise the intervening sequences. This consideration becomes important below when we consider the arrangement of transgenes in C. elegans.
How C.elegans-specific transgenic methods influence FLP-out technology:
The ways in which transgenic worms are generated influences how the FLP-out technology can be applied in C. elegans. Transgenes are most often carried in multiple copies on extrachromosomal arrays that are generated by injection of DNA into the germ line. These arrays are formed by recombination in the worm and therefore carry the injected DNA in an unpredictable order, orientation, and copy number (Mello et al. 1991). In addition, while transgenic arrays are relatively stable and can be selected for retention in meiosis by dominant markers in progeny, they can also be lost during mitosis within an individual worm. Transgenes can also be integrated into the chromosome in multiple, low, or (rarely) single copy by induction of double-strand breaks (to insert extrachromosomal arrays; Mello and Fire 1995) or by microparticle bombardment (Praitis et al. 2001). Thus, unlike transposon-mediated insertion of single-copy trangenes in Drosophila, transgenes in C. elegans are usually present in multiple copies in an unpredictable arrangement.
FRT targets are borne on transgenes and C. elegans transgenes are not present in single copy. In addition, FLP-out is rarely 100% efficient. Taken together, these considerations mandate that the desired effect of the FLP-out must be dominant. Therefore, FLP-out technology is good for controlled expression of dominantly acting genes such as (1) exogenous transgenes with dominant effects such as reporters, poisons (Davis et al. 2008), or cell-death inducers (Chelur and Chalfie 2007), (2) dominant forms of genes (e.g., dominant-negative, hypermorphic, neomorphic), including wild-type gene expression in an otherwise mutant background, and (3) dominant knockdown such as RNAi.
Should technology become available to reliably produce single-copy insertions of transgenes in worms, these considerations would change. A promising technology for single-copy insertions is being developed (E. Jorgensen, personal communication). Completely efficient FLP-out would be possible in this case. For example, the provision of a single wild-type copy of a gene on a FLP-out cassette could be reversed by FLP-out, generating a mosaic. However, because RNAi acts in a dominant fashion, multiple copy transgenesis does not prevent use of the FLP-out technology to reduce gene function in a spatially and temporally controlled manner, provided the gene in the targeted tissue is sensitive to RNAi.
Regarding efficiency of FLP-out, our observation is that excision of all copies of the FLP-out cassettes on transgenic arrays is rare. While it is likely that improvements in recombinase efficiency will be made with further extension of the technology (e.g., by more efficient recombination systems, stronger promoters driving FLP recombinase, and recombination-efficient genetic backgrounds), the presence of residual unexcised FLP-out cassettes does not interfere with the utility of the technology, provided the effect of the conditional gene expression is dominant.
We also observed that the integrated hs-FLP recombinase (naIs6 in strain GC817) provided more uniform and higher efficiency FLP activity than did the same construct on an extrachromosomal array. It is likely that promoter–recombinase fusions will behave similarly. We found that overall efficiency was reduced when the recombinase construct and the target construct are on separate arrays. Not only was it rare to find individuals that inherited both arrays, efficiency was reduced by array mosaicism and by general health problems associated with multiple markers. Integration may also improve transcriptional output. Therefore, if one recombinase line will be used in multiple experiments, generating an integrated line will be worth the effort. Alternatively the recombinase-bearing construct can be co-injected with the target such that they are expressed off the same array. Additional differences in efficiency were noted with different target lines, likely due to different degrees of mosaicism among extrachromosomal arrays.
Integration of FRT target sequences poses other potential problems. Because transgenes are not usually generated in predictable single copy the orientation of the FRT sequences is not always predictable. For FLP-out to occur, the FRT sequences must be oriented in the same direction. However, the unpredictable orientation of transgenes in arrays or insertions means that nontandem FRT sequences may arise on an array. While this is not a limitation for the technology when FRT targets are on extrachromosomal arrays, the presence of nontandem FRTs in an integrated line could possibly result in sequence inversions or other aberrations within or neighboring the FRT target sites. However, because extrachromosomal arrays are relatively easy to generate, and post-FLP arrays are not maintained over generations, sequence inversions or other abnormalities on these arrays do not pose a problem. Although the degree to which target integration would interfere with the utility of the technology has not been rigorously assessed, we tested one integrated target (naIs35) and found that it responded appropriately to FLP recombinase and did not cause obvious lethality (data not shown).
In summary, features of C. elegans transgenes, especially their multi-copy content and optional extrachromosomal location, pose limitations and provide opportunities for the application of FLP-out technology.
Lineaging applications:
Perhaps the greatest feature of C. elegans as a model system is the feasibility of following development in real time at the level of individual cells. Although the entire somatic cell lineage has been mapped in real time (Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983), following a cell lineage is often an arduous task that is made more difficult when mutant conditions perturb landmarks or alter cell identities. Our application of the FLP-out technology to lineaging offers an alternative whereby a cell lineage can be followed by virtue of an independent and permanent lineage mark. This application has gained wide use in other organisms. We examined the somatic gonad lineage in wild-type worms as a test case and found that in a subset of animals, the entire lineage was labeled by FLP recombinase activity in the appropriate founder cells (Figure 4).
To monitor a cell lineage using this system, there are several options for driving expression of the recombinase. One option is to use a well-characterized promoter driving the FLP recombinase. In this case, it is vital that the promoter is well characterized, preferably with a reporter that does not perdure so that its full temporal and spatial effect on the lineage of interest is known. Ideally, the promoter would be active for a short time in early cells of the lineage and would subsequently be restricted to a small subset of the lineage or would turn off altogether. Continued expression of the promoter within the lineage of interest could interfere with labeling since the FLP-out could occur during the course of development. The strength of the promoter would likely dictate the percentage of animals in which the lineage would be marked. Mosaicism of the target array could also reduce efficiency, but because the activity of FLP-out is a traceable positive result that is marked by a reporter, inefficiency of the system is unlikely to interfere with interpretation of the results.
Nevertheless, lineaging with a specific FLP-out strategy should be tested in the wild type prior to use in a mutant condition. Tests should be designed to assess whether the presence of the array interferes with the normal lineage and to measure the background rate at which the target array is lost within the lineage. Coexpression of a Promoter–reporter fusion together with the Promoter–FLP fusion would provide a means to monitor the activity of the recombinase in the desired lineage. For example, in situations in which a suitable cell-specific promoter is not available, an alternative strategy is to use heat shock to induce the recombinase randomly and to select worms in which the FLP-out has occurred in the cells of interest. Again, when lineaging is conducted in mutants, marking strategies should be compared with wild-type controls. The ultimate strategy that takes full advantage of C. elegans would be induction of gene expression within a single cell (e.g., with laser-induced heat shock, e.g., Stringham and Candido 1993) to follow directly a single cell's lineage by the alteration in reporter expression caused by FLP-out excision.
We generated several reagents that are particularly useful for lineaging. Worm strains that carry integrated transgenes with the FLP recombinase driven by the heat-shock promoter can be used with two reporter FLP-out targets (in the form [Promoter-of-interest<reporter 1<reporter 2]) to follow cell lineage after heat-shock-induced excision of the reporter 1 FLP-out cassette. In addition to the variety of FLP-out target vectors we made, we have generated useful constructs and strategies to generate promoter–FLP recombinase fusions of choice (see supplemental Toolkit Documentation). Should other reporters and/or promoters be required for the FRT target, our half-cassette vectors (in the form [<reporter]) and Gateway-compatible destination vectors provide easy ways to build appropriate targets. See supplemental Toolkit Documentation for details.
Gene expression applications:
Tools that allow temporally controlled or spatially controlled gene expression are in wide use in C. elegans (e.g., using heat-shock or tissue-specific promoters). RNAi can be similarly controlled by tissue-specific or tissue-restricted expression or activity (e.g., Tavernarakis et al. 2000; Sijen et al. 2001) or by timing of RNAi feeding (Karp and Greenwald 2003). These strategies permit temporal or spatial control of gene expression, but not both. Bacaj and Shaham (2007) describe a useful method to circumvent this limitation by using cell-specific rescue of heat-shock factor-1 (hsp-1) to restrict heat-shock-induced expression of a gene of interest to cells that express active hsf-1 (Bacaj and Shaham 2007). A similar strategy can be applied to rescue of genes required for RNAi, such as rde-1 (Tabara et al. 1999) to restrict RNAi to certain times and/or places (Qadota et al. 2007). While these strategies are useful, expression of the rescuing gene (e.g., hsf-1) is dependent upon continued expression from the promoter driving it and/or its activity (e.g., binding and activating the heat-shock promoter). Moreover, experiments must be conducted in the relevant mutant background (hsf-1, rde-1, etc.), which may have unanticipated effects.
The FLP-out system offers additional and complementary approaches to those already available in the C. elegans molecular-genetic toolkit. The major advantage of two-component systems such as FLP-out over single-component systems is that control can be exerted over both spatial and temporal aspects of gene expression. We demonstrated that FLP recombinase activity can be controlled temporally by heat shock or by a tissue-specific promoter, while target activity can be ubiquitously expressed or tissue restricted.
FLP-out or similar recombinase-mediated systems are particularly useful in circumstances where a cell-heritable and permanent change in gene expression is desired. (We note that in this case, if the target sequences are present on an extrachromosomal array, the possibility of loss of the array must be taken into account by monitoring the reporter in the excision cassette.) The possibility of driving both components of the system with promoters is advantageous in conditions where heat-shock treatment is incompatible with the experiment. Perhaps the most useful application of this system will be in situations where the desired manipulation of gene expression in a single-component system would cause lethality, sterility, or pleiotropic defects. In addition, because of the temporal control it affords, this system will be useful in cases where gene expression is required in multiple cell types and/or iteratively during development.
Examples of alteration in gene expression that can be spatially and/or temporally manipulated is the induction of wild-type gene expression in a mutant background, of dominant forms of gene products, or ectopic expression. These apply to protein-coding genes as well as genes that produce noncoding RNAs.
Most of the worm and plasmid reagents we have generated are useful in the context of controlled gene expression. We built plasmids to facilitate the construction of both FLP recombinase and the FLP-out target components of the technology. These are described in detail together with strategies for their use in the supplemental Toolkit Documentation. In particular, we note that previously characterized Fire vector-based constructs can be easily transformed into FLP recombinase or FLP-out targets. We also provide Gateway-based vectors to facilitate plasmid construction. Destination vectors of particular use in generating promoter–FLP recombinase fusions and FLP-out targets of the form [promoter-of-interest<reporter<gene-of-interest] are listed in Table 1. See supplemental Methods for details.
RNAi applications:
Because RNAi causes a dominant loss-of-function effect, FLP-out technology can be used in worms to control reduction of gene function. Application of the FLP-out technology to drive RNAi in a temporally and/or spatially controlled way is potentially quite useful, especially for the analysis of genes for which systemic loss of the gene product is pleiotropic or causes lethality or sterility. Another useful application for this technology is in situations where gene function is required reiteratively. In these circumstances, temporal control of RNAi induction allows stage-specific removal of gene function. Finally, there may be situations where inducing RNAi in specific cell types is difficult, such as in the embryo or neurons.
Special considerations must be made in the use of this technology for induction of RNAi. First, the FLP-out must be performed under conditions in which the gene/tissue of interest is RNAi sensitive. If RNAi does not produce a phenotype in the wild type, it can be assayed in an RNAi-sensitive genetic background such as rrf-3 (Simmer et al. 2002). In this case, the FLP-out should also be performed in the same RNAi-sensitive background. We have generated an rrf-3; heat-shock-FLP strain (strain GC1021) that will facilitate this use in these circumstances.
Even if a gene is sensitive to systemic RNAi, efficient RNAi from a cell-specific hairpin construct after FLP-out may induce RNAi less efficiently. Indeed, in our test case, hairpin-hlh-12, RNAi feeding starting in the L1 produced the DTC Mig defect at a 50% penetrance. In a strain bearing an integrated hs-FLP and the hlh-12 RNAi-hairpin FRT target, the phenotype was observed less frequently (∼7%). However, our test case was particularly stringent since we started with an incompletely penetrant RNAi defect, performed an L1 heat shock, and scored for a phenotype that required maintenance of an array and loss of hlh-12 function in one cell. That we were still able to observe the expected phenotype is encouraging for other less stringent applications. Moreover, our results suggest that the rate of false-positive results for these experiments will be negligible. See below for additional discussion of monitoring FLP-out efficiency.
Another concern for RNAi is the possibility of RNAi spreading between cells and tissues within the worm (systemic RNAi, Jose and Hunter 2007), that is, where expression of an RNAi hairpin in one tissue causes silencing of the same gene in other tissues within the individual. For experiments where spreading could pose a problem, the FLP-out experiment could be performed in a spreading-resistant strain such as sid-1 mutants (Winston et al. 2002). An alternative strategy to limit the effect of RNAi to desired cell type would be to perform the experiment in an RNAi-resistant background and replace RNAi-susceptibility only in the cells of interest. For example, a transgene that drives expression of rde-1(+) from a promoter of interest in an rde-1(-) background could be co-injected with the FLP-out plasmid-bearing array. In this case, FLP-out of an RNAi hairpin could occur in all cells, but only those expressing the rde-1(+) transgene would be sensitive to the RNAi. The same result (cell-specific RNAi without effects of spreading) could be achieved by conditionally expressing rde-1(+) by FLP-out while feeding RNAi directed against a gene of interest.
We have generated several reagents that are particularly useful for RNAi hairpin application of the FLP-out technology. As noted above, we crossed our integrated heat-shock FLP recombinase into an rrf-3(pk1426) RNAi hypersensitive background. Second, we generated a double-Gateway Wormgate (Johnson et al. 2005) destination FLP-out vector that can accept—in one reaction—a promoter of interest and an ORFeome clone such that the ORFeome cDNA is inserted into the vector as an RNAi-inducing hairpin (Table 1; materials and methods). See supplemental Toolkit Documentation, table, and Methods for additional details on plasmids and FRT target constructions.
General strategies to monitor FLP-out:
Having noted that it is not necessary for all copies of the FLP-out cassette on an array to undergo excision for the applications we describe, it is, nonetheless, vital that there is no transcriptional readthrough prior to FLP-out. In other organisms, non-reporter-bearing minimal sequences are often used in the excision cassette (Struhl and Basler 1993). However, given that FLP-out does not occur in every copy, reporters are a convenient way to monitor several aspects of FLP-out. Our results suggest that highly efficient FLP-out is possible although rare. Where this level of efficiency is required, animals can be screened for loss of expression of the excised reporter.
In cases where FLP-out causes a change in native gene function (as opposed to activation of exogenous reporter expression), and where the outcome of the FLP experiment is not known a priori, a control target construct should be tested in parallel. This control should be identical to the test construct. The former would be in the form [Promoter<gene/reporter<gene/reporter] while the latter would be in the form [Promoter<reporter<reporter]. The control plasmid can be tested in parallel in the same FLP recombinase-bearing strain or co-injected so that it is on the same array as the experimental target. The control target will allow the user to determine (a) expression pattern from the promoter before and after excision (especially important if the promoter was not previously characterized without the intervening FRT), (b) whether the downstream gene is inappropriately expressed prior to expression of the recombinase, and (c) whether the conditions for excision are working. Where possible, the best alternative is to monitor FLP-out efficiency directly using a gene/reporter fusion in the downstream part of the target. A strategy for construction of these types of fusions is presented in supplemental Figure 3 in the supplemental Toolkit Documentation.
Combining FLP-out with other technologies:
There are additional possibilities of controlling gene expression using combinations of this technology with other technologies that limit gene expression. Directed expression of hsf-1 (Bacaj and Shaham 2007, see above) could be used to limit the effect of the heat shock to a subset of cells of interest. It may also be possible to combine this technology with other inducible/repressible promoter systems such as those that respond to small molecules (e.g., tetracycline or tamoxifen) or temperature (e.g., temperature-sensitive forms of transcriptional regulators such as GAL80) (see Branda and Dymecki 2004, for reviews; McGuire et al. 2004).
Additional general information regarding useful strains and constructs:
Davis et al. (2008) introduce useful constructs that are compatible with promoterome (Dupuy et al. 2004) and ORFeome (Reboul et al. 2003) reagents; we expand the set of useful FLP-out technology reagents by introducing a variety of additional plasmids for constructing specific FLP recombinase and FLP-out target vectors of choice by both traditional methods and by Gateway cloning methods. The general strategies presented below were used to build plasmids used in this study. Here, we give the highlights of these reagents and strategies (Figure 7); full details are presented in the supplemental Toolkit Documentation file and supplemental figures therein.
To build “promoter-of-interest driving FLP recombinase” plasmids, we suggest three general strategies. The first two strategies facilitate modification of existing Fire vector-based constructs: (1) replacement of expressed sequences within an existing Fire vector promoter-gene construct with FLP cDNA from pGC92, pGC94, or pGC95 and (2) insertion of a promoter or 3′end of interest in pGC94 or pGC95. The third strategy is to insert a promoter of interest into a Gateway-based FLP recombinase destination vector (pGC180, pGC181, or pGC267). Davis et al. (2008) provide a similar Gateway strategy, but with different Gateway acceptor sites. If the starting sequences are in pDONR221 (Invitrogen), our destination vectors are most useful; if the starting sequences are from the promoterome (Dupuy et al. 2004) or ORFeome (Reboul et al. 2003) reagents, the Davis et al. (2008) Gateway destination vectors may be more convenient.
To build FLP-out targets in the form [promoter-of-interest<reporter 1<reporter 2] (where < refers to an FRT site), we provide two general strategies: traditional cloning (facilitated by available intermediates) and Gateway-based strategies. The first consists of three components, a promoter (from a preexisting vector with suitable restriction sites) and two “half cassettes” in the form [<reporter]. If the starting material is a Fire vector-based construct, the half-cassette strategy enables construction of a full FLP-out cassette with a single three-way ligation. The half cassettes are, themselves, very flexible since they derive from the Fire vector kit. In the supplemental Toolkit Documentation, we provide useful reagents and strategies for modifying the half cassettes to generate a large variety of reagents including gene∷reporter fusions. The second general strategy uses Gateway technology to introduce a promoter of interest into a destination vector in the form [Gateway<reporter<reporter].
To create FLP-out targets in the form [promoter-of-interest<reporter<gene-of-interest], we again provide traditional (see above) and Gateway options. In this case, the latter includes a double-Gateway destination vector in the form [Gateway<reporter<Gateway] so that a promoter of interest and gene of interest can be inserted simultaneously. Again, depending on the starting material, the Davis et al. (2008) vectors may be more convenient.
Finally, to generate FLP-out targets in the form [promoter-of-interest<reporter<RNAi-inducing hairpin-of-interest], we provide a triple Gateway destination vector that simultaneously accepts a promoter of interest and, using Wormgate technology (Johnson et al. 2005), generates an RNAi hairpin from an ORFeome (Reboul et al. 2003) entry clone.
We have demonstrated that FLP-out technology can be used in C. elegans to control gene expression and facilitate lineage tracing. This technology can also be used in conjunction with other existing and emerging technologies to further manipulate this powerful model organism.
Acknowledgments
We thank Gary Struhl, Andy Fire, Oliver Hobert, Barth Grant, Roger Tsien, and Scott Clark for reagents; Darrell Killian, John Maciejowski, Jim Lai, and James Ahn for discussion, reagents, and technical support. Thanks also go to Jeremy Nance and Dorian Anderson and other members of the Hubbard, Fitch, and Nance labs for helpful discussions. This work was supported by National Institutes of Health grant GM-61706.
References
- Atchley, W. R., and W. M. Fitch, 1997. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 94 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj, T., and S. Shaham, 2007. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics 176 2651–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch, R., S. S. Anna-Arriola, D. Gao, Y. Li, J. Hodgkin et al., 1999. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev. Biol. 216 382–393. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., and K. Steward, 1997. RNA processing and gene structure, pp. 117–145 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Branda, C. S., and S. M. Dymecki, 2004. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6 7–28. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz, F., L. Ringrose, P. O. Angrand, F. Rossi and A. F. Stewart, 1996. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 24 4256–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz, F., P. O. Angrand and A. F. Stewart, 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16 657–662. [DOI] [PubMed] [Google Scholar]
- Chelur, D. S., and M. Chalfie, 2007. Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. USA 104 2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M. W., J. J. Morton, D. Carroll and E. M. Jorgensen, 2008. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 4 e1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy, D., Q. R. Li, B. Deplancke, M. Boxem, T. Hao et al., 2004. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 14 2169–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Golic, K. G., 1991. Site-specific recombination between homologous chromosomes in Drosophila. Science 252 958–961. [DOI] [PubMed] [Google Scholar]
- Golic, K. G., and S. Lindquist, 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59 499–509. [DOI] [PubMed] [Google Scholar]
- Henderson, S. T., D. Gao, E. J. Lambie and J. Kimble, 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120 2913–2924. [DOI] [PubMed] [Google Scholar]
- Herman, R. K., 1984. Analysis of genetic mosaics of the nematode Caenorhabditis elegans. Genetics 108 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, N. M., C. A. Behm and S. C. Trowell, 2005. Heritable and inducible gene knockdown in C. elegans using Wormgate and the ORFeome. Gene 359 26–34. [DOI] [PubMed] [Google Scholar]
- Jose, A. M., and C. P. Hunter, 2007. Transport of sequence-specific RNA interference information between cells. Annu. Rev. Genet. 41 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Karp, X., and I. Greenwald, 2003. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W. G., S. Xu, M. K. Montgomery and A. Fire, 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian, D. J., and E. J. Hubbard, 2004. C. elegans pro-1 activity is required for soma/germline interactions that influence proliferation and differentiation in the germ line. Development 131 1267–1278. [DOI] [PubMed] [Google Scholar]
- Kimble, J., and D. Hirsh, 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70 396–417. [DOI] [PubMed] [Google Scholar]
- Maduro, M., and D. Pilgrim, 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari, M. E., and C. Murre, 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies, L. D., S. T. Henderson and J. Kimble, 2003. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development 130 2881–2892. [DOI] [PubMed] [Google Scholar]
- McGuire, S. E., G. Roman and R. L. Davis, 2004. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 20 384–391. [DOI] [PubMed] [Google Scholar]
- Mello, C., and A. Fire, 1995. DNA transformation. Methods Cell Biol. 48 451–482. [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema, P. G., E. Ha, C. Haun, W. Chen and A. Fire, 1997. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development 124 3965–3973. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., M. Inoue, T. Hikita, M. Koppen, J. D. Hardin et al., 2007. Establishment of a tissue-specific RNAi system in C. elegans. Gene 400 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul, J., P. Vaglio, J. F. Rual, P. Lamesch, M. Martinez et al., 2003. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34 35–41. [DOI] [PubMed] [Google Scholar]
- Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish et al., 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465–476. [DOI] [PubMed] [Google Scholar]
- Simmer, F., M. Tijsterman, S. Parrish, S. P. Koushika, M. L. Nonet et al., 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12 1317–1319. [DOI] [PubMed] [Google Scholar]
- Stinchcomb, D. T., J. E. Shaw, S. H. Carr and D. Hirsh, 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham, E. G., and E. P. Candido, 1993. Targeted single-cell induction of gene products in Caenorhabditis elegans: a new tool for developmental studies. J. Exp. Zool. 266 227–233. [DOI] [PubMed] [Google Scholar]
- Stringham, E. G., D. K. Dixon, D. Jones and E. P. Candido, 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell 3 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, G., and K. Basler, 1993. Organizing activity of wingless protein in Drosophila. Cell 72 527–540. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100 64–119. [DOI] [PubMed] [Google Scholar]
- Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok et al., 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123–132. [DOI] [PubMed] [Google Scholar]
- Tamai, K. K., and K. Nishiwaki, 2007. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev. Biol. 308 562–571. [DOI] [PubMed] [Google Scholar]
- Tavernarakis, N., S. L. Wang, M. Dorovkov, A. Ryazanov and M. Driscoll, 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24 180–183. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395 854. [DOI] [PubMed] [Google Scholar]
- Voronova, A., and D. Baltimore, 1990. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc. Natl. Acad. Sci. USA 87 4722–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, W. M., C. Molodowitch and C. P. Hunter, 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295 2456–2459. [DOI] [PubMed] [Google Scholar]
- Yochem, J., and R. K. Herman, 2005. Genetic mosaics, pp. 1–6 in WormBook, edited by The C. elegans Research Community. http://www.workbook.org. [DOI] [PMC free article] [PubMed]