Abstract
Telomeric regions in Drosophila are composed of three subdomains. A chromosome cap distinguishes the chromosome end from a DNA double-strand break; an array of retrotransposons, HeT-A, TART, and TAHRE (HTT), maintains telomere length by targeted transposition to chromosome ends; and telomere-associated sequence (TAS), which consists of a mosaic of complex repeated sequences, has been identified as a source of gene silencing. Heterochromatin protein 1 (HP1) and HP1-ORC-associated protein (HOAP) are major protein components of the telomere cap in Drosophila and are required for telomere stability. Besides the chromosome cap, HP1 is also localized along the HTT array and in TAS. Mutants for Su(var)205, the gene encoding HP1, have decreased the HP1 level in the HTT array and increased transcription of individual HeT-A elements. This suggests that HP1 levels directly affect HeT-A activity along the HTT array, although they have little or no effect on transcription of a white reporter gene in the HTT. Chromatin immunoprecipitation to identify other heterochromatic proteins indicates that TAS and the HTT array may be distinct from either heterochromatin or euchromatin.
TELOMERES are nucleoprotein structures at the ends of eukaryotic chromosomes with important roles in chromosome replication, stability, segregation, and position within the nucleus (Hochstrasser et al. 1986; Blackburn 1991; Hari et al. 2001; Chan and Blackburn 2002; Biessmann and Mason 2003; Abad et al. 2004). In most eukaryotes, chromosomes terminate in an array of simple repeats that is synthesized by telomerase (Blackburn 1991). The terminal arrays at Drosophila telomeres, however, are composites of three telomere-specific nonlong terminal repeat (non-LTR) retrotransposons, HeT-A, TAHRE, and TART (Mason and Biessmann 1995; Mason et al. 2008), whose stochastic transposition creates an array (HTT) that differs in length at different chromosomal ends in a range of 147–26 kb in one stock (Abad et al. 2004). Telomeric retrotransposons maintain chromosome length by targeted transposition to chromosome tips and by terminal recombination/gene conversion (Kahn et al. 2000; Biessmann and Mason 2003). The attachment of the elements by their 3′ oligo (A) tails to the chromosome end probably occurs via target-primed reverse transcription (Luan et al. 1993) and does not depend on the DNA sequence at the terminus (Biessmann et al. 1992; Biessmann and Mason 2003). The HeT-A element is the most abundant telomeric retroelement; it has a promoter located at its 3′ end that directs transcription of a downstream sequence (Danilevskaya et al. 1997; Capkova Frydrychova et al. 2007).
The terminal part of the HTT array is covered by protein complex, termed the chromosome cap, that protects chromosome ends from telomeric fusions. The telomere capping complex is formed by a special interaction of heterochromatin protein 1(HP1) with the HP1/ORC-associated protein (HOAP) (Cenci et al. 2005). The formation of the cap is mediated by a sequence-independent mechanism regardless of the presence of telomeric retroelements (Biessmann and Mason 1988; Biessmann et al. 1990). Analysis of chromosome ends broken within the yellow upstream region suggested that there is a special chromatin structure that interferes with enhancer function when the chromosome end is within ∼4 kb of the enhancer (Mikhailovsky et al. 1999; Melnikova et al. 2004), suggesting that the chromosome cap may extend up to this distance from the chromosome end.
To date, mutations in several genes have been implicated in the control of telomere elongation: the HP1-encoding gene Su(var)205, Tel, E(tc), spn-E, aub, and the Drosophila orthologs of Ku70 and Ku80 (Melnikova and Georgiev 2002; Savitsky et al. 2002, 2006; Cenci et al. 2005; Melnikova et al. 2005). Although all these genes act as negative regulators of telomere length, so far only mutations in Su(var)205, spn-E, and aub have been shown to increase retroelement transcripts and transposition of the retroelements to chromosome ends (Savitsky et al. 2002, 2006). Su(var)205, Tel, and E(tc) regulate telomere length by controlling terminal gene conversion (Melnikova and Georgiev 2002; Savitsky et al. 2002; L. Melnikova and P. Georgiev, personal communication).
The terminal retrotransposon arrays are adjacent to the subterminal telomere-associated sequence (TAS), which in turn borders euchromatic transcribed genes (Karpen and Spradling 1992; Abad et al. 2004). The TAS region covers ∼20 kb and consists of several kilobases of complex satellite sequences, which, in spite of some sequence similarities, vary among telomeres (Mason et al. 2008). Drosophila telomeres have been considered heterochromatic, as they contain repetitive DNA sequences and have the ability to repress gene activity (Gehring et al. 1984; Karpen and Spradling 1992; Wallrath and Elgin 1995; Cryderman et al. 1999; Zhimulev and Belyaeva 2003). However, recent detailed genetic analysis of white (w) transgenes inserted into distal and proximal sites within a telomere region identified TAS as the primary source of telomeric silencing (Mason et al. 2003; Biessmann et al. 2005a,b). TAS-induced silencing is unidirectional (Kurenova et al. 1998) toward the chromosome end and shows decreasing effect with increasing distance. Transgenes in TAS or the HTT array close to TAS displayed repressed and variegated expression, whereas expression of transgenes inserted into HTT >10 kb from TAS was comparable to that of control euchromatic insertions (Biessmann et al. 2005a,b). As gene silencing is considered to be a feature of closed chromatin and telomeric retroelements seem to lack silencing potential, TAS and the HTT array may be two distinct chromatin domains resembling closed chromatin and open chromatin, respectively (Biessmann et al. 2005a,b; Mason et al. 2008). These genetic results agree with immunostaining data that indicate distinct protein components in the chromosome cap, and the HTT and TAS arrays of Drosophila polytene chromosomes (Andreyeva et al. 2005), with proteins associated with interband regions found at HTT and Polycomb group proteins found at TAS.
HP1 is a chromosomal protein that is predominantly associated with heterochromatin. It has been shown that HP1 is a component of the telomere capping complex and is required for telomere elongation and transcriptional repression of telomeric retrotransposons (Fanti et al. 1998; Savitsky et al. 2002; Cenci et al. 2003, 2005; Perrini et al. 2004). On the basis of several studies it has been proposed that heterochromatin formation and epigenetic gene silencing is a result of a multistep process including replacement of histone H2A with the histone variant H2A.v, deacetylation, and subsequent methylation of Lys9 on histone H3, and binding of HP1 (Nakayama et al. 2001; Volpe et al. 2002; Schotta et al. 2003, 2004; Verdel et al. 2004; Swaminathan et al. 2005).
Using ChIP analysis, we show the presence of HP1 at the promoter of w transgenes inserted into the HTT array and TAS. We mapped the effect of HP1 mutations on the transcriptional activity of individual HeT-A elements located along the HTT array. Transcription at three specific sites in the HTT array was measured by quantification of readthrough transcripts that were transcribed from a HeT-A element into the adjacent P-element insertion. In HP1 mutants we observed elevated levels of the readthrough transcripts. These data suggest that the presence of HP1at telomeres and HP1 regulation of HeT-A transcription are not restricted to a specific region, such as the chromosome cap, but rather extend along the whole length of the HTT array. A mutation in caravaggio (cav), the gene encoding HOAP, however, does not affect transcriptional activity of HeT-A elements located along the HTT array, suggesting that the cap itself has no role in the regulation of telomere elongation via retroelement transcription.
MATERIALS AND METHODS
Drosophila stocks:
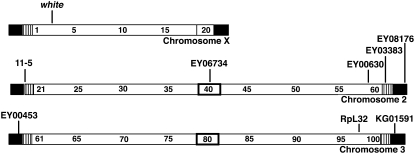
Drosophila stocks were raised and crosses performed at 25° on cornmeal-molasses medium with dry yeast added to the surface. Stocks were obtained from a P-element mobilization screen by the Berkeley Drosophila Genome Project described previously (Bellen et al. 2004; Biessmann et al. 2005a; Capkova Frydrychova et al. 2007) and from the Bloomington Stock Center. All original stocks were converted into similar y w67c23 genetic backgrounds by crossing with y w67c23; Sco/SM1; Sb/TM6 and then with control y w67c23; Sco/SM1 or y w67c23; Sb/TM6 before establishing new stocks. P{w+}EY00453 (hereafter EY00453) carries the Epgy2 element at the 3′ end of a TART element in the telomere at the left end of chromosome 3 (3L), 656 bp from its (A) tail and >20 kb from TAS. P{w+}EY03383 (hereafter EY03383) has an Epgy2 inserted into 2R TAS (Biessmann et al. 2005a). As controls, P{w+}EY00630 and P{w+}EY06734 (hereafter EY00630 and EY06734) carry an Epgy2 element in euchromatin at 59D8 or in 2R pericentric heterochromatin, respectively. P{wvar}11-5 (hereafter 11-5) has a copy of the genomic w gene inserted between the HTT and TAS arrays on 2L (Golubovsky et al. 2001; Capkova Frydrychova et al. 2007). P{w+}KG01591 (hereafter KG01591) carries a SuPor-P element inserted into a HeT-A element 5 kb from 3R TAS.
Micrococcal nuclease digestion:
Nuclei were isolated from third instar larvae and treated with 0.1, 0.2, and 0.3 units of MNase as previously described (Cryderman et al. 1998). The DNA was purified, separated on a 1.5% agarose/TAE gel, transferred to a nylon membrane, and hybridized to a DNA probe labeled by PCR with [32P]dCTP.
Chromatin immunoprecipitation (ChIP):
Drosophila HP1 polyclonal antibody was purchased from Covance (cat. no. PRB-291C), the other antibodies from Abcam: rabbit polyclonal antibody to histone H2A.Z (cat. no. ab4174), rabbit polyclonal antibody to histone H2A (cat. no. ab13923), rabbit polyclonal antibody to tri-methyl K9 of histone H3 (cat. no. ab8898), rabbit polyclonal antibody to di-methyl K9 of histone H3 (cat. no. ab7312). Specificity of the antibodies was checked by Western blot. For the ChIP assay we used nuclei isolated from 100 mg of third instar larvae and followed a protocol described by Upstate Biotechnology. Crosslinking reactions were performed by 1% formaldehyde, nuclei were lysed, the DNA was fragmented by sonication, and 50 μl of the chromatin solution was saved as input. Five microliters of each antibody were added to tubes containing 1000 μl of chromatin solution. Following incubation, the antibody complexes were captured using protein A-agarose beads. The beads were pelleted and washed. The chromatin was extracted and reverse crosslinked, and the DNA was purified using phenol-chloroform. Samples were analyzed using real-time PCR. Threshold cycle (Ct) was used for assessing the relative level of each amplification product vs. the amplification product of 5% of input DNA.
RNA isolation and cDNA synthesis:
RNA samples were made using RNasy mini kit (QIAGEN) according to the manufacturer's instructions and reverse transcribed using oligo dT and the SuperScript first-strand synthesis system for RT–PCR (Invitrogen).
Real-time PCR:
Quantitation was performed in two independent experiments of three samples for each strain. Relative levels of transcripts were compared by real-time PCR using an Mx3000P real-time PCR system. The reactions were prepared using SYBR Green PCR master mix (Stratagene) and Ct was used to assess relative levels of target transcripts vs. reference RpL32 transcripts. Normalization of HeT-A transcript levels was done by calculating mean transcript levels and dividing by mean HeT-A copy number. A reverse transcriptase-minus control was included for each sample; in all cases the control gave undetectable Ct value. Primer sequences are given in supplemental Table S1.
RESULTS
HP1 has been shown to play a role in the control of telomere length via regulation of gene conversion and transcription of telomeric retroelements. HP1 has repressive effect on the telomeric retroelements and its mutations lead to dramatic increase in transcriptional activity of the elements (Savitsky et al. 2002; Perrini et al. 2004).
Despite the role of HP1 in transcriptional regulation of telomeric retroelements, immunostaining of polytene chromosomes of the Tel mutant in previous studies surprisingly failed to reveal localization of HP1 along the HTT array and showed HP1 localized only at chromosome cap, i.e., in a region at the extreme chromosome ends (Siriaco et al. 2002; Andreyeva et al. 2005). This led us to three alternative hypotheses. First, localization of HP1 specifically to the chromosome cap may indicate that only the retroelements under telomere cap are affected by HP1 and that these retroelements make the major contribution to the increase in overall retroelement transcription and telomere elongation seen in HP1 mutants. Second, the cap is a structure with extensive repressive effect on transcriptional activity of retroelements located along the HTT array both inside and by some unknown mechanism outside of the telomere cap. Finally, we could not exclude the possibility that immunostaining of polytene chromosomes might not reflect the general telomeric localization of HP1, perhaps because of the character of polytene chromosomes or the unusual features of exceedingly long HTT arrays in the Tel mutant or because of low sensitivity of immunostaining. Thus HP1 might be present along the HTT array outside chromosome cap. This led us to retest for the presence of HP1 at telomeric retroelements located in the HTT array outside of the chromosome cap using a ChIP assay performed on nuclei isolated from whole third instar larvae.
Presence of HP1 at Drosophila telomeres:
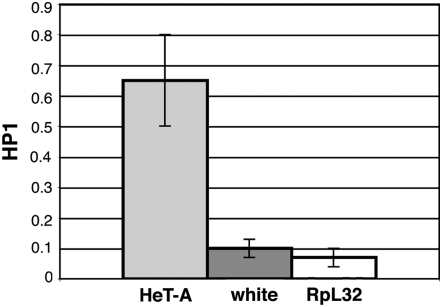
We first measured the average level of HP1 located at the 5′ end of the HeT-A elements in larvae of wild-type Oregon R with quantification of coprecipitating DNA by real-time PCR. As controls we used primers to the promoter of the w gene residing in its nontelomeric, euchromatic position on the X chromosome and primers to the coding sequence of the ribosomal protein gene RpL32. We found a 7-fold enrichment of HP1 at HeT-A compared to w and a 12-fold enrichment compared to RpL32 (Figure 1).
Figure 1.—
Chromatin immunoprecipitation (ChIP) analysis of HP1 at the 5′ end of HeT-A, the white promoter, and coding sequence of RpL32 in Oregon R. Quantitation of HP1 was performed using real-time PCR. ChIP samples were normalized to 5% of input DNA. Error bars represent standard deviations.
HP1 binding in the HTT array:
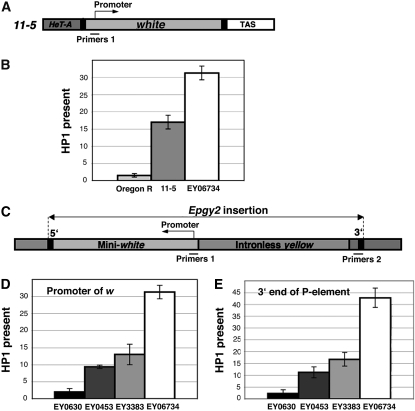
To distinguish between HP1 associated with retroelements in the chromosome cap and retroelements located outside of the cap, we could not probe any DNA sequence that is common in telomeric retroelements and we needed to test some unique sequence in telomeres. Assuming that HP1 can spread into adjacent transgenes (Danzer and Wallrath 2004) we looked for HP1 at P elements inserted into specific telomeric regions outside of the telomere cap (Bellen et al. 2004; Biessmann et al. 2005a,b). First, we compared HP1 at the w promoter of an insertion line 11-5 (Figures 2 and 3A), which has a copy of the genomic w gene inserted precisely between the HTT array and TAS at the 2L telomere (Golubovsky et al. 2001; Capkova Frydrychova et al. 2007), and at the wild-type w promoter of Oregon R. The distance between the w promoter of 11-5 and the chromosome end is estimated to be at least 30 kb on the basis of a correlation of P{wvar} variant eye color with HTT length (Golubovsky et al. 2001; Mason et al. 2003). HP1 showed a ninefold higher level at the telomeric w of 11-5 compared to the nontelomeric w gene of Oregon R (Figure 3B).
Figure 2.—
Schematic map showing locations of the wild-type white gene, the RpL32 gene, and the insertion sites of P elements used here. Boxes around 20, 40, and 80, indicate pericentric heterochromatin; striped boxes, TAS; filled boxes, HTT. Numbers indicate cytological map positions.
Figure 3.—
HP1 is found at telomeric insertions. (A) 11-5 bears a complete white gene inserted between the terminal retrotransposon array and TAS at the 2L telomere. Primers 1 used in the ChIP experiment surround the promoter. (B) ChIP analysis of HP1 at the w promoter of Oregon R, 11-5, and EY06734. Graphs represent real-time PCR results obtained after ChIP. HP1 measurements were normalized to 5% of input DNA and further normalized to the RpL32 locus. Error bars represent standard deviations. (C) The EPgy2 construct of EY insertions has a mini-white gene (mini-w) and an intronless yellow gene. Primers 1 and Primers 2, which correspond to the mini-w promoter and the 3′ end of the P-element insertion, respectively, were used for PCR analysis after ChIP. (D and E) The level of HP1 binding at the w promoter (D) and the 3′ end of the P element of EY insertions (E) analyzed by ChIP. Error bars represent standard deviations.
We also compared the HP1 level at the w promoter in 11-5 with HP1 at the promoter of a mini-w reporter transgene of the EY06734 insertion in pericentric heterochromatin of 2L (Figures 2 and 3B). The HP1 level at 11-5 was approximately twofold lower than at mini-w of the pericentric insertion. Further analyses were performed at EPgy2 elements inserted into the HTT array, TAS, euchromatin, and pericentric heterochromatin (Figure 2) to examine HP1 levels at the promoter of a mini-w reporter transgene and at the 3′ end of EPgy2 insertions immediately adjacent to the insertion site (Figure 3, C–E). In EY00453, the distance between the chromosome end and the w promoter is estimated to be at least 6.6 kb (including 3.8 kb between the w promoter and the 5′ end of the P element), and the distance between the chromosome end and the 3′ end of the P element is estimated to be at least 12.3 kb. The length estimation was based on a 2.8-kb PCR product generated with primers to HeT-A coding sequence and the P-element 5′ end (Het_seq1F, Car1P5_seq1B primers; specificity of the PCR product was checked by sequencing). Consistently, at both the w promoter and the 3′ end of the P element we found distinct HP1 levels showing an increase in the direction of EY00630 (euchromatin) < EY00453 (HTT) < EY03383 (TAS) < EY06734 (pericentric heterochromatin) (Figure 3, D and E), although HP1 levels at EY00453 and EY03383 are not significantly different from each other at either site. That is, HP1 is present in the HTT array and TAS, and the levels of HP1 in these regions are intermediate between euchromatin and pericentric heterochromatin.
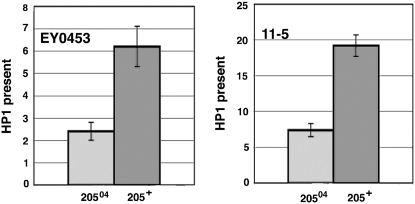
Mutations in Su(var)205 decrease HP1 levels in EY00453 and 11-5:
By genetic crosses we introduced Su(var)20504, which encodes a truncated HP1 protein that lacks part of the domain required for its nuclear localization (Powers and Eissenberg 1993), into the EY00453 and 11-5 insertion lines and quantified HP1 at the w promoter. Larvae heterozygous for the Su(var)20504 mutation showed a 2.5-fold decrease in HP1 at both insertions compared to wild-type larvae (Figure 4). These results indicate that the HP1 level in the HTT array is affected by the Su(var)20504 mutation, and it confirmed that HP1 is present in the internal region of the telomere and is not limited only to the telomere cap.
Figure 4.—
HP1 levels at the w the promoter in EY00453 and 11-5 insertion lines bearing a wild type or mutant Su(var)205 gene. Graphs represent real-time PCR results after ChIP. Measurements for each antibody were normalized to 5% of input DNA and further normalized to the results from the RpL32 locus. Error bars represent standard deviations.
Mutations in HP1 stimulate HeT-A transcription along the HTT array:
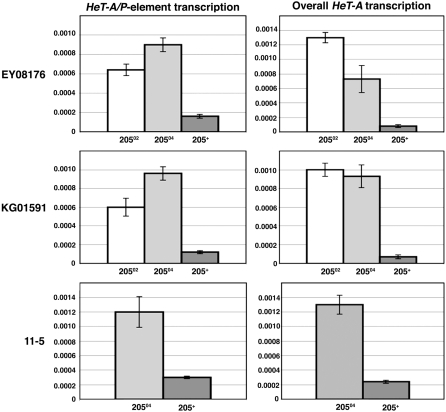
Localization of HP1 in the internal region of the HTT array suggests that expression of telomeric retroelements is regulated by local binding of HP1 to these elements. This led us to investigate the impact of Su(var)205 mutations on the transcriptional activity of individual HeT-A retroelements located in different positions of the HTT array. Promoter activity at the 3′ end of each HeT-A element may result in transcription into a downstream P-element insertion, which can be identified as a HeT-A/P-element readthrough transcript (Capkova Frydrychova et al. 2007). This allows us to measure transcriptional activity of individual HeT-A elements by quantitative real-time PCR with primers specific to a HeT-A/P-element transcript. We used the 11-5, KG01591, and EY08176 lines with P-element insertions in or adjacent to the HTT array (Figures 2 and 5) and compared levels of the HeT-A/P-element readthrough transcript between the Su(var)205 mutant and Su(var)205+ control flies (Figure 6). For the test we used two Su(var)205 mutants: Su(var)20502, with a point mutation in the conserved chromodomain and Su(var)20504 (Eissenberg et al. 1992; Platero et al. 1995). The stocks were kept for two generations before they were analyzed. Compared to the Su(var)205+ controls, larvae that were heterozygous for Su(var)205 displayed significantly increased levels of HeT-A/P-element readthrough transcripts compared to Su(var)205+ (Figure 6). Su(var)20502 exhibited a fourfold increase of the readthrough transcript in EY08176 and a fivefold increase in KG01591. The increase in the transcript level in Su(var)20504 was sixfold in EY08176, eightfold in KG01591, and fourfold in 11-5. As the same degree of increase was seen for all three of the insertions independent of position, it appears that upregulation of HeT-A transcription by Su(var)205 mutations is spread along the HTT array and is not limited to one specific region of the array.
Figure 5.—
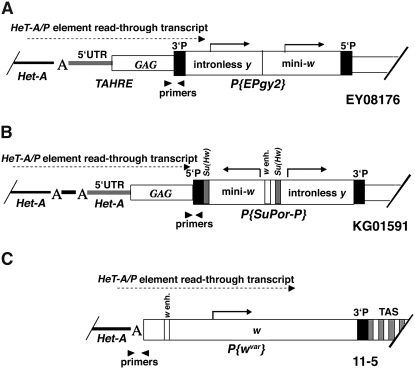
Diagram showing the structure of P-element insertions used in the experiment to measure HeT-A/P-element readthrough transcript levels. (A) EY08176 has a single EPgy2 element, containing an intronless yellow (y) gene and a mini-white (mini-w) gene. The EPgy2 construct is inserted in inverted orientation into the GAG open reading frame (ORF) of a TAHRE element >8 kb from the 2R chromosome end and >15 kb from TAS. The TAHRE bearing the insertion is bordered by an upstream HeT-A element. (B) KG01591 carries a SuPor-P element with a mini-w gene containing the w enhancer and an intronless y gene inserted 5 kb from 3R TAS. The mini-w is bordered by Su(Hw) insulators. The SuPor-P is inserted into the ORF of a HeT-A element. Directly upstream of this HeT-A lies a 168-bp HeT-A fragment with an oligo (A) tail followed by a 3′ HeT-A UTR with another oligo (A) tail. (C) 11-5 contains P{wvar} carrying a w transgene inserted between the HTT array and a truncated 2L TAS region. The P-element construct lacks its 5′ end (Biessmann et al. 2005a; Capkova Frydrychova et al. 2007). Arrowheads indicate the positions of primers used to quantify HeT-A/P-element readthrough transcript. “A” indicates the HeT-A oligo(A) tail. The presence of the HeT-A/P-element readthrough transcripts in all three insertions was reported previously (Capkova Frydrychova et al. 2007).
Figure 6.—
Levels of HeT-A and HeT-A/P-element readthrough transcripts are increased in Su(var)205 mutants. Levels of HeT-A/P-element and HeT-A transcripts in EY08176, KG01591, and 11-5 insertions heterozygous for Su(var)20502 or Su(var)20504 were compared with a Su(var)205 wild type. The transcript levels were normalized to RpL32 transcripts and insertion copy number. Overall HeT-A transcription was further normalized to genomic HeT-A copy number of each tested genotype. Error bars represent standard deviations.
Using primers specific to the HeT-A coding sequence, we also measured overall HeT-A transcript level and HeT-A genomic copy number, allowing us to calculate HeT-A transcript per genomic element. Comparison of genomic HeT-A copy numbers showed almost no differences between Su(var)205 mutants and the Su(var)205+ control (supplemental Table S2). This was probably due to the low number of generations since the Su(var)205 mutations were introduced into the insertion lines. When we analyzed the same stocks of EY08176; Su(var)20502 and EY08176; Su(var)20504 after 24 generations, we found twice the genomic HeT-A copy number compared to the EY08176 Su(var)205+ control. Despite little difference in genomic HeT-A copy number we found the levels of overall HeT-A transcript elevated 7.5- to 16-fold in mutant flies compared to the Su(var)205+ controls (Figure 6). The increase in overall HeT-A transcript in Su(var)205 mutants is more than the increase we observed in individual HeT-A/P-element transcript levels (Figure 6), which may indicate that the effect of Su(var)205 mutations on HeT-A transcription varies in different positions of the HTT array. These data suggest that regulation of HeT-A transcriptional activity by HP1 is not restricted to the telomere cap or any specific region of the HTT array, but affects the transcription of HeT-A elements along the HTT array.
Although we saw stimulation of HeT-A transcriptional activity in the presence of a mutation in Su(var)205, the mutation had no effect on transcription of the w transgene in any tested genotype. The w transcript was measured using real-time PCR with primers to coding sequence of the w gene (supplemental Table S3).
The capping complex has no significant effect on overall HeT-A transcription:
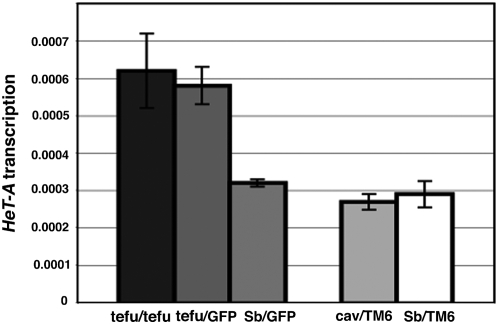
The telomere-capping complex is comprised of HP1 and HOAP. Mutants for the HOAP-encoding gene, cav, display a telomere fusion phenotype and a defect in HP1 localization at telomeres (Cenci et al. 2003). Formation of the telomere-capping complex may be disrupted by mutations in several telomere protective genes, such as tefu, which encodes the ATM kinase. ATM plays a role in DNA repair and telomere function and is required to recruit or maintain HP1 and HOAP at chromosome ends. Loss of ATM leads to telomeric fusions and significant reduction of HP1 and HOAP association with telomeres (Oikemus et al. 2004; Bi et al. 2005; Cenci et al. 2005). As HP1 acts as a repressor of transcription of telomeric retroelements, we asked whether cav and tefu mutations, through their effect on formation of the capping complex and association of HP1 with telomeres, lead to an increase in HeT-A transcriptional activity. As tefu homozygotes are viable during the third larval instar, and as the loss of ATM has been reported to reduce HP1 and HOAP localization at telomeres, we measured HeT-A transcript levels in tefu homozygotes. To distinguish homozygous larvae, we balanced tefu with the TM3 balancer chromosome marked with GFP. We simultaneously measured the HeT-A transcript level and HeT-A genomic copy number using the same set of primers specific to coding sequence of HeT-A and calculated HeT-A transcription per genomic element. tefu homozygotes showed no difference in normalized HeT-A transcription from tefu/TM3, GFP heterozygotes (Figure 7). As we saw an 8- to 10-fold increase in HeT-A transcriptional activity per HeT-A element caused by Su(var)205 mutations (Figure 6) the lack of an effect exhibited by the tefu mutant may indicate first, that ATM within the telomere interacts solely with the telomere cap and binding of HP1 in the HTT array outside of the telomere cap is ATM independent, and second, that a change in HP1 level in the cap due to loss of ATM is limited in distance and thus has no or a limited effect on retroelement activity in the HTT array outside of the telomere cap. We did not observe a significant change in HeT-A transcript level due to mutation in the cav heterozygote compared to a control (Figure 7), which is consistent with the idea that formation of the capping complex and association of HP1 at the end of telomeres do not have extensive effects on the overall level of transcriptional activity of telomeric retroelements.
Figure 7.—
Relative levels of HeT-A transcripts in homozygous and heterozygous tefu and heterozygous cav larvae. The levels of HeT-A transcript were normalized to transcript levels of RpL32 and to genomic HeT-A copy number of each tested strain. To minimize effects due to different genetic backgrounds, original mutant strains were crossed into the same y w67c23 background. Error bars represent standard deviations.
Chromatin domains in Drosophila telomeres:
Expression analysis of telomeric w transgenes suggested two distinct chromatin domains in Drosophila telomeres: the heterochromatic TAS and the euchromatic HTT array (Biessmann et al. 2005a,b). In our study, however, HP1 levels at the w promoter in the EY00453 and EY03383 insertions, in the HTT array, and TAS, respectively, revealed no significant differences and are intermediate compared to HP1 levels at EPgy2 elements located in euchromatin (EY00630) and pericentric heterochromatin (EY06734). This led us to look at levels of other chromatin proteins, histone modifications, and nucleosome organization at the w promoter and the 3′ end of these insertions (Figure 8) to better understand any difference between expression data and the presence of HP1 at tested transgenes.
Figure 8.—
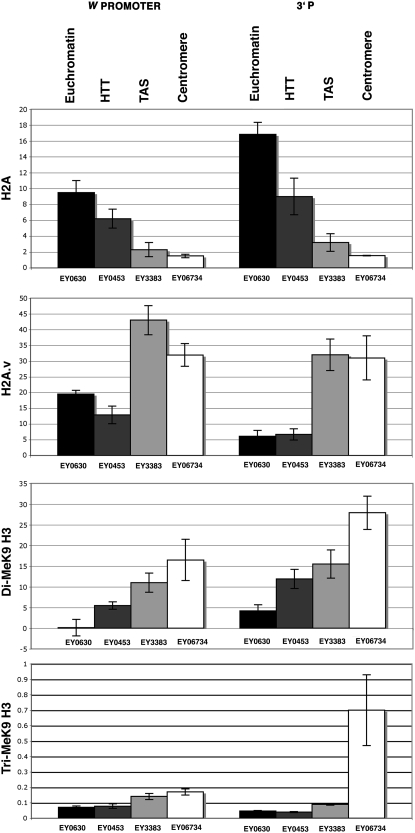
ChIP analysis of histones and histone modifications at EPgy2 insertion sites. Antibody quantification was performed using real-time PCR with primers specific to the promoter of the mini-w transgene and the 3′ end of the EPgy2 insertion. Graphs show real-time PCR results. The level of each antibody was normalized to 5% of input DNA and further normalized to the RpL32 locus. The data were obtained from four to six independent experiments, each of which included all of the strains compared. EY00630 carries an insertion in euchromatin of chromosome 2R. The EY00453 insertion is located in 3L HTT, EY03383 carries an insertion in 2R TAS, and the EY06734 insertion is located in 2R pericentric heterochromatin. Error bars represent standard deviations.
Mutations in His2Av are dominant suppressors of PEV in Drosophila, and exchange of histone H2A for H2A.v is implicated in heterochromatin formation (Swaminathan et al. 2005). Histone H2A and H2A.v levels did not show significant differences between TAS-located EY03383 and pericentric EY06734, with the exception of a slightly lower level of H2A.v in the w promoter region of EY06734 compared to EY03383 (Figure 8). H2A at these two insertions showed significantly lower levels in comparison to both euchromatic EY00630 and HTT-located EY00453. In contrast, H2A.v levels show significant elevation in EY03383 and EY06734. EY00453 shows a lack of proportionality in the transition between levels of H2A and H2A.v. Although H2A.v levels in EY00630 and EY00453 are comparable, H2A in EY00453 is intermediate between EY00630 and EY03383 (Figure 8). These data indicate that the TAS and pericentric domains contain H2A/H2A.v levels that are similar to each other, but distinct from those in HTT and the euchromatic control.
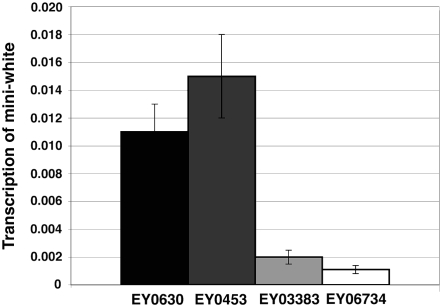
We tested levels of di- and trimethylated histone H3 at Lys9 (Me2K9H3 and Me3K9H3), as histone H3-Lys9 methylation plays a role in gene silencing (Schotta et al. 2003; Ebert et al. 2006). In both the w promoters and the 3′ ends of the insertions, the levels of Me2K9H3 resemble HP1 levels in that they show an increase in the direction of EY00630 < EY00453 < EY03383 < EY06734, and at the 3′ ends of these insertions the level of Me2K9H3 in EY00453 is comparable to that of EY03883. Me3K9H3 levels, on the other hand, did not show a significant difference between EY00630 and EY00453, or between EY03383 and EY06734 at the w promoter and only a relatively minor twofold difference between the latter pair and the former. More strikingly, the levels of Me3K9H3 at the 3′ end of these insertions was found to be similar in EY00630, EY03383, and EY00453, while EY06734 showed an approximately sevenfold increase relative to the others (Figure 8). Thus, although levels of HP1 do not distinguish the HTT array from TAS, other chromatin markers show that HTT more closely resembles open chromatin, while TAS resembles more closed chromatin. Of the chromatin marks examined here, the level of histone H2A.v, most closely (inversely) corresponds to the expression of the tested w transgenes as assayed by transcript levels (Figure 9) or by eye color (Biessmann et al. 2005a).
Figure 9.—
Levels of mini-w transcripts in EY00630 (euchromatin), EY00453 (3L HTT), EY03383 (2R TAS), and EY06734 (2R pericentric heterochromatin). Mini-w transcript was measured by real-time PCR with primers specific to coding sequence of the w gene. The levels of the transcript were normalized relative to transcript levels of RpL32. Error bars represent standard deviations.
Nucleosome organization at telomeres:
To examine a possible difference between the EY00453, EY03383, and EY00630 transgenes at the level of nucleosome organization we treated nuclei from third instar larvae with microccocal nuclease, an enzyme that preferentially cuts between nucleosomes. DNA fragments were analyzed using Southern hybridization with probes to the w promoter and the 3′ end of the insertion. The probe to the 3′ end was used to study nucleosome organization in regions adjacent to the insertions. Hybridization showed regular nucleosome spacing without significant differences among the different insertions (supplemental Figure S1). Thus, the functional differences in HP1 binding and white gene transcription between the TAS and HTT do not lie at the level of nucleosome organization.
DISCUSSION
On the basis of expression of telomeric white and yellow transgenes Drosophila telomeres have been proposed to have two distinct domains: TAS, which resembles heterochromatin and the HTT array, which behaves like euchromatin (Biessmann et al. 2005a,b). According to the pattern of chromatin proteins revealed by immunostaining of extended polytene chromosomes in a Tel mutant, telomeres consist of three distinct and nonoverlapping domains: the chromosome cap, the HTT array, and TAS (Andreyeva et al. 2005). The immunostaining results indicate that HP1 in telomeres is restricted to the cap region (Siriaco et al. 2002; Andreyeva et al. 2005).
Using ChIP, we show here that HP1 is also present along the HTT array outside of the cap as well as in TAS. The difference between our observations and previous reports might be due to a higher abundance of HP1 in the telomere cap than in the internal HTT region or better accessibility of antibodies to the telomere cap, and thus the difference in the reports may be explained by higher sensitivity of ChIP compared to immmnostaining of polytene chromosomes. The difference may be caused also by different properties of long telomeres of a Tel mutant or different biological properties of polytene salivary chromosomes compared to diploid or other polyploid cells. In any case, ChIP data on whole animals are more likely to be generalizable than immunostaining data on a specific cell type.
Su(var)205 belongs to a group of suppressor of variegation [Su(var)] genes, many of which encode chromosomal proteins or modifiers of chromosomal proteins. Mutations in Su(var) genes lead to suppression of position-effect variegation (PEV), which is repressed and variegated expression of genes placed in or near pericentric heterochromatin (Ebert et al. 2006). Despite phenotypic similarities between PEV and telomere position effect (TPE), TPE does not respond to Su(var) mutations (Cryderman et al. 1999; Mason et al. 2004). Although TAS was identified as a source of telomeric silencing, and the retrotransposon array genetically resembles euchromatin (Biessmann et al. 2005a,b), we found comparable levels of HP1 at transgenes inserted in these two telomeric domains. The levels of other marks for silent chromatin, such as histone H2A.v and MeK9H3, however, did vary between these two regions in a manner consistent with proposals in previous reports that HTT is associated with open chromatin and TAS is associated with closed chromatin. TPE may thus be caused by a silencing system different from HP1-mediated heterochromatin. One candidate is Polycomb silencing, as Polycomb group proteins were found associated with TAS (Boivin et al. 2003; Andreyeva et al. 2005). As levels of the chromatin markers in all tested regions, including euchromatin and pericentric heterochromatin, showed significant differences, interpretation of HTT and TAS as either heterochromatin or euchromatin is rather difficult. It may suggest that HTT and TAS are in a category of some transitional type of chromatin between euchromatin and heterochromatin, such as closed/inactive euchromatin, or it suggests the existence of additional chromatin types.
The relatively high level of HP1 on a transgene inserted into pericentric heterochromatin compared with transgenes in either HTT or TAS may suggest that failure of telomeric HP1 to silence telomeric transgenes is caused by its relative paucity. HP1, however, is a negative regulator of telomere length; its mutations lead to an increase in the transcriptional activity of HeT-A and TART, as well as an accumulation of these elements at the chromosome end (Savitsky et al. 2002; Perrini et al. 2004). We showed previously that the promoter activity of a telomeric w transgene inserted between the HTT array and TAS significantly exceeds the activity of a single HeT-A promoter (Capkova Frydrychova et al. 2007). Here we show that Su(var)205 mutations lead to a severalfold increase in the transcriptional activity of HeT-A, however we did not see any increase in transcription of a w gene inserted into the HTT array. In particular, using HeT-A/P-element readthrough transcripts in three P-element insertion lines, we found that Su(var)205 mutations lead to stimulation of HeT-A elements along the HTT array in all regions assayed. With regard to the low level of HP1 in telomeric regions compared to pericentric heterochromatin, as observed by ChIP experiments, it is conceivable that the relatively weak HeT-A promoter is more sensitive to HP1concentration than the more robust w promoter. However, HP1 per se cannot be considered as a signal for silencing. An analysis of genomewide correlations between the HP1 binding pattern and the pattern of gene expression revealed that recruitment of the protein is not sufficient to repress transcription completely (Greil et al. 2003). Moreover, some euchromatic genes in Drosophila are activated by the presence of HP1 (Cryderman et al. 2005). With respect to these observations, it is difficult to predict the effect of HP1 recruitment on the transcription pattern in any specific region.
HP1, by interaction with HOAP, forms capping complexes at the ends of Drosophila chromosomes (Cenci et al. 2005). Formation or maintenance of the HP1-HOAP capping complex requires ATM. Loss of ATM reduces localization of HP1 and HOAP at telomeres and leads to frequent telomeric fusions (Oikemus et al. 2004). tefu and cav mutations, however, did not lead to a profound increase in HeT-A transcription, as was observed in Su(var)205 mutants. This suggests that HP1 presence in the cap does not significantly participate in overall HeT-A transcriptional activity, and that HeT-A transcription is regulated mainly by HP1 in the HTT array outside the cap. Our data are consistent with Perrini et al. 2004, who suggested two distinct mechanisms for HP1 control of telomere capping and telomere elongation by retroelement transcription. They proposed that the capping function of HP1 is due to its direct binding to telomeric DNA, while the silencing of telomeric sequences and control of transcription of telomeric retroelements is due to interaction of HP1 with MeK9H3 and spreading of HP1 and repressive chromatin along the telomere.
Collectively, our data show that HP1 is present along the HTT array as well as in TAS and plays a role as a negative regulator of transcription of telomeric retroelements. The present data also support the observation that the HeT-A promoter is relatively weak compared with a mini-w promoter and more sensitive to local HP1 concentration and suggest that telomeric chromatin in Drosophila may be distinct from either euchromatin or heterochromatin.
Acknowledgments
We thank Karen Adelman and Daniel Menendez for critical reading of the manuscript and Harald Biessmann for providing sequences of EY and KG insertions and comments to the manuscript. This research was supported by the Intramural Research Program of National Institutes of Health National Institute of Environmental Health Sciences.
References
- Abad, J. P., B. de Pablos, K. Osoegawa, P. J. de Jong, A. Martin-Gallardo et al., 2004. Genomic analysis of Drosophila melanogaster telomeres: Full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 21 1613–1619. [DOI] [PubMed] [Google Scholar]
- Andreyeva, E. N., E. S. Belyaeva, V. F. Semeshin, G. V. Polkholkova and I. F. Zhimulev, 2005. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J. Cell Sci. 118 5465–5477. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, X. L., D. Srikanta, L. Fanti, S. Pimpinelli, R. Badugu et al., 2005. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc. Natl. Acad. Sci. USA 102 15167–15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., S. B. Carter and J. M. Mason, 1990. Chromosome ends in Drosophila without telomeric DNA sequences. Proc. Natl. Acad. Sci. USA 87 1758–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., L. E. Champion, M. O'Hair, K. Ikenaga, B. Kasravi et al., 1992. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 11 4459–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., and J. M. Mason, 1988. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 7 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., and J. M. Mason, 2003. Telomerase-independent mechanisms of telomere elongation. Cell. Mol. Life Sci. 60 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., S. Prasad, V. E. Semeshin, E. N. Andreyeva, Q. Nguyen et al., 2005. a Two distinct domains in Drosophila melanogaster telomeres. Genetics 171 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., S. Prasad, M. F. Walter and J. M. Mason, 2005. b Euchromatic and heterochromatic domains at Drosophila telomeres. Biochem. Cell Biol. 83 477–485. [DOI] [PubMed] [Google Scholar]
- Blackburn, E. H., 1991. Structure and function of telomeres. Nature 350 569–573. [DOI] [PubMed] [Google Scholar]
- Boivin, A., C. Gally, S. Netter, D. Anxolabehere and S. Ronsseray, 2003. Telomeric-associated sequences of Drosophila recruit Polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capkova Frydrychova, C. R., H. Biessmann, A. Y. Konev, M. D. Golubovsky, J. Johnson et al., 2007. Transcriptional activity of the telomeric retrotransposon HeT-A in Drosophila melanogaster is stimulated as a consequence of subterminal deficiencies at homologous and nonhomologous telomeres. Mol. Cell. Biol. 27 4991–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci, G., G. Siriaco, G. D. Raffa, R. Kellum and M. Gatti, 2003. The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5 82–84. [DOI] [PubMed] [Google Scholar]
- Cenci, G., L. Ciapponi and M. Gatti, 2005. The mechanism of telomere protection: A comparison between Drosophila and humans. Chromosoma 114 135–145. [DOI] [PubMed] [Google Scholar]
- Chan, S. W., and E. H. Blackburn, 2002. New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene 21 553–563. [DOI] [PubMed] [Google Scholar]
- Cryderman, D. E., M. H. Cuaycong, S. C. R. Elgin and L. L. Wallrath, 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107 277–285. [DOI] [PubMed] [Google Scholar]
- Cryderman, D. E., S. K. Grade, Y. Li, L. Fanti, S. Pimpinelli et al., 2005. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 232 767–774. [DOI] [PubMed] [Google Scholar]
- Cryderman, D. E., E. J. Morris, H. Biessmann, S. C. R. Elgin and L. L. Wallrath, 1999. Silencing at Drosophila telomeres: Nuclear organization and chromatin structure play critical roles. EMBO J. 18 3724–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya, O. N., I. R. Arkhipova, K. L. Traverse and M. L. Pardue, 1997. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell 88 647–655. [DOI] [PubMed] [Google Scholar]
- Danzer, J. R., and L. L. Wallrath, 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131 3571–3580. [DOI] [PubMed] [Google Scholar]
- Ebert, A., S. Lein, G. Schotta and G. Reuter, 2006. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 14 377–392. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J. C., G. D. Morris, G. Reuter and T. Hartnett, 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti, L., G. Giovinazzo, M. Berloco and S. Pimpinelli, 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2 527–538. [DOI] [PubMed] [Google Scholar]
- Gehring, W. J., R. Klemenz, U. Weber and U. Kloter, 1984. Functional analysis of the white+ gene of Drosophila by P-factor-mediated transformation. EMBO J. 3 2077–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky, M. D., A. Y. Konev, M. F. Walter, H. Biessmann and J. M. Mason, 2001. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics 158 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil, F., I. van der Kraan, J. Delrow, J. F. Smothers, E. de Wit et al., 2003. Distinct HP1 and SU(VAR)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 15 2825–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari, K. L., K. R. Cook and G. H. Karpen, 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15 1334–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M., D. Mathog, Y. Gruenbaum, H. Saumweber and J. W. Sedat, 1986. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J. Cell Biol. 102 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, T., M. Savitsky and P. Georgiev, 2000. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol. Cell. Biol. 20 7634–7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G. H., and A. C. Spradling, 1992. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P-element insertional mutagenesis. Genetics 132 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurenova, E., L. Champion, H. Biessmann and J. M. Mason, 1998. Directional gene silencing induced by a complex subtelomeric satellite from Drosophila. Chromosoma 107 311–320. [DOI] [PubMed] [Google Scholar]
- Luan, D. D., M. H. Korman, J. L. Jakubczak and T. H. Eickbush, 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72 595–605. [DOI] [PubMed] [Google Scholar]
- Mason, J. M., and H. Biessmann, 1995. The unusual telomeres of Drosophila. Trends Genet. 11 58–62. [DOI] [PubMed] [Google Scholar]
- Mason, J. M., R. Capkova Frydrychova and H. Biessmann, 2008. Drosophila telomeres: An exception providing new insights. BioEssays 30 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, J. M., A. Y. Konev, M. D. Golubovsky and H. Biessmann, 2003. Cis- and trans-acting influences on telomeric position effect in Drosophila melanogaster detected with a subterminal transgene. Genetics 163 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, J. M., J. Ransom and A. Y. Konev, 2004. A deficiency screen for dominant suppressors of telomeric silencing in Drosophila. Genetics 168 1353–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova, L., and P. Georgiev, 2002. Enhancer of terminal gene conversion, a new mutation in Drosophila melanogaster that induces telomere elongation by gene conversion. Genetics 162 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova, L., H. Biessmann and P. Georgiev, 2004. The vicinity of a broken chromosome end affects P-element mobilization in Drosophila melanogaster. Mol. Genet. Genomics 272 512–518. [DOI] [PubMed] [Google Scholar]
- Melnikova, L., H. Biessmann and P. Georgiev, 2005. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics 170 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailovsky, S., T. Belenkaya and P. Georgiev, 1999. Broken chromosomal ends can be elongated by conversion in Drosophila melanogaster. Chromosoma 108 114–120. [DOI] [PubMed] [Google Scholar]
- Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis and S. I. Grewal, 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292 110–113. [DOI] [PubMed] [Google Scholar]
- Oikemus, S. R., N. McGinnis, J. Queiroz-Machado, H. Tukachinsky, S. Takada et al., 2004. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 18 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini, B., L. Piacentini, L. Fanti, F. Altieri, S. Chichiarelli et al., 2004. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell 15 467–476. [DOI] [PubMed] [Google Scholar]
- Platero, J. S., T. Hartnett and J. C. Eissenberg, 1995. Functional analysis of the chromo domain of HP1. EMBO J. 14 3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, J. A., and J. C. Eissenberg, 1993. Overlapping domains of the heterochromatin-associated protein HP1 mediate nuclear localization and heterochromatin binding. J. Cell Biol. 120 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky, M., O. Kravchuk, L. Melnikova and P. Georgiev, 2002. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol. Cell. Biol. 22 3204–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky, M., D. Kwon, P. Georgiev, A. Kalmykova and V. Gvozdev, 2006. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert and G. Reuter, 2003. SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing. Genetica 117 149–158. [DOI] [PubMed] [Google Scholar]
- Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta et al., 2004. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 18 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriaco, G. M., G. Cenci, A. Haoudi, L. E. Champion, C. Zhou et al., 2002. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics 160 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, J., E. M. Baxter and V. G. Corces, 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. P. Gygi et al., 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal et al., 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wallrath, L. L., and S. C. R. Elgin, 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9 1263–1277. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., and E. S. Belyaeva, 2003. Intercalary heterochromatin and genetic silencing. BioEssays 25 1040–1051. [DOI] [PubMed] [Google Scholar]