Abstract
Selection of spontaneous, loss-of-function mutations at two chromosomal loci (pyrF and pyrE) enabled the first molecular-level analysis of replication fidelity in the extremely thermophilic bacterium Thermus thermophilus. Two different methods yielded similar mutation rates, and mutational spectra determined by sequencing of independent mutants revealed a variety of replication errors distributed throughout the target genes. The genomic mutation rate estimated from these targets, 0.00097 ± 0.00052 per replication, was lower than corresponding estimates from mesophilic microorganisms, primarily because of a low rate of base substitution. However, both the rate and spectrum of spontaneous mutations in T. thermophilus resembled those of the thermoacidophilic archaeon Sulfolobus acidocaldarius, despite important molecular differences between these two thermophiles and their genomes.
EXPERIMENTAL study of genetic stability in prokaryotes has focused historically on mesophilic bacteria that allow facile genetic manipulations. However, diverse prokaryotes adapted to extreme environments may also provide strategic insights into mechanisms of genetic stability, particularly with respect to environmental stresses exerted on DNA structure, replication, and repair. For instance, Deinococcus radiodurans, which belongs to a basal lineage of bacteria, exhibits extreme resistance to ionizing radiation and desiccation and has been used as a model system to understand the molecular basis of highly efficient repair of DNA double-strand breaks (Cox and Battista 2005).
Species of the related genus Thermus show a corresponding potential for the study of cellular and molecular adaptations of bacteria to extremely high temperature. This potential is reinforced by progress in the genetic analysis of Thermus thermophilus supported, in turn, by convenient growth and transformation conditions, cloning vectors, selectable markers, and complete genome sequences for two strains (Henne et al. 2004; Brüggemann and Chen 2006). One of these T. thermophilus strains (HB27) lacks a functional reverse gyrase, which is otherwise highly conserved among extremely thermophilic bacteria and archaea (Henne et al. 2004). In addition, both strains appear to lack family-Y DNA polymerases (Henne et al. 2004; Brüggemann and Chen 2006), which generate many of the mutations induced by DNA damage and may also contribute to spontaneous mutation (Goodman 2002). The apparent lack of these enzymes, combined with the ability to grow >80°, raises questions about the accuracy of genome replication in T. thermophilus. At the same time, genetic manipulations made possible by various T. thermophilus vectors and selections suggest ways to investigate the mechanisms affecting replication accuracy in vivo. Both considerations provide incentives to examine the rate and molecular nature of replication errors.
Errors that occur in the course of normal DNA replication and escape correction can be detected in descendant genomes as spontaneous mutations, provided the mutations do not impair survival. The errors include unforced misincorporations by replicative polymerases as well as the inaccurate insertion of bases opposite template damage, often by specialized family-Y polymerases (Goodman 2002). Despite the presumably chaotic nature of such replication errors, distinct patterns emerge from analysis of the resulting mutations among diverse organisms. For example, microorganisms with DNA genomes (including DNA viruses, prokaryotes, and eukaryotes) display very similar genomic mutation rates that cluster around 0.003 mutations per genome per replication despite tremendous differences in genome size and error rates per base pair (Drake et al. 1998). Furthermore, base-pair substitutions (BPSs) tend to outnumber insertion and deletion mutations (indels) in these genomes (Grogan et al. 2001). The similar genomic rates and mutational spectra observed across such organismal diversity suggest that all these genomes are subject to similar selective forces. These forces can be interpreted formally as a balance between the costs of achieving replication accuracy (such as limits on the efficiency of DNA repair and the rate of DNA synthesis) and its benefits (such as maintaining the fitness of descendents over many generations). However, the precise nature of these costs and benefits remains unclear, and detailed analysis of spontaneous mutation across a range of microbial diversity offers a useful approach to identifying them in molecular terms.
The molecular analysis of replication errors in vivo requires target genes in which rare spontaneous mutations can be recovered efficiently for quantification and identification. The Saccharomyces cerevisiae orotidylate decarboxylase (URA3) gene provides an example of such a mutational target, and corresponding UMP biosynthetic genes have provided favorable mutational targets in archaea. Use of these related targets has yielded dramatically different results in different organisms, however. In yeast, the mutational spectrum was dominated by base-pair substitutions (86% of the total) (Lee et al. 1998) and yielded a genomic mutation rate similar to those of other mesophilic genomes (Drake et al. 1998). In the thermoacidophilic crenarchaeote Sulfolobus acidocaldarius (optimal growth near 80° and pH 3), loss-of-function mutations in the orotate:phosphoribosyl transferase gene, pyrE (Grogan et al. 2001), indicated a genomic mutation rate slightly below the consensus rate for diverse mesophilic genomes, whereas the orotidylate decarboxylase gene, pyrF, yielded an even lower estimate. In these targets, only about a third of the detected mutations were base-pair substitutions (BPSs) compared to about two-thirds in most other microbial DNA genomes (Grogan et al. 2001). The mesophilic, extremely halophilic euryarchaeote Haloferax volcanii yielded pyrE mutants at a still lower rate (Mackwan et al. 2007), although this probably resulted from long phenotypic lag or other effects of the unusually high copy number of its genome (Breuert et al. 2006). Furthermore, the mutations recovered from H. volcanii pyrE2, the orotate:phosphoribosyl transferase gene, were almost exclusively in-frame deletions templated by short direct repeats (Mackwan et al. 2007); this kind of mutation has not been observed in S. acidocaldarius (Grogan et al. 2001; Grogan and Hansen 2003). The fact that mutational patterns of the two archaea (one a thermoacidophile, the other an extreme halophile) differ from each other as well as from the norm of evolutionarily diverse mesophiles raises the possibility that patterns of spontaneous mutation in microorganisms may correlate more strongly with environmental parameters than with phylogeny. A similar hypothesis has drawn support from the observed base-substitution patterns of orthologous gene pairs within mesophilic vs. thermophilic genera of both archaea and bacteria (Friedman et al. 2004).
Quantitative molecular analysis of spontaneous mutation in the extremely thermophilic bacterium T. thermophilus would provide a test of the hypothesis that adaptation to high growth temperatures is accompanied by low BPS rates. We therefore established conditions for selecting loss-of-function mutations in two UMP biosynthetic genes, pyrE and pyrF, and used this selection to analyze spontaneous mutation both quantitatively and qualitatively. The results indicate that these T. thermophilus genes have BPS rates well below those typically observed in target genes of mesophiles but similar to those of the orthologous S. acidocaldarius genes.
MATERIALS AND METHODS
Growth conditions:
T. thermophilus strain HB27 (Oshima and Imahori 1974) was obtained from the American Type Culture Collection as BAA-163. Liquid growth medium contained (g/liter) NaCl, 3.0; sodium citrate, 0.3; CaCl2·2H2O, 0.03; MgSO4·7H2O, 0.2; K2HPO4, 0.17; and 0.1 ml/liter of trace-mineral solution (5% FeCl3 + 0.5% each CuCl2, CoCl2, MnCl2, and ZnCl2 in 1 m HCl). This mineral base was supplemented with 5 g/liter enzymatic digest of casein (Sigma) and the pH of the resulting complete medium was adjusted to 7.5 with NaOH. For plating, the tryptone medium was supplemented with an additional 33 mm MgSO4 and 4 mm CaCl2 and solidified with 7 g/liter Gellan gum. Uracil (20 mg/liter) was routinely added to support the growth of auxotrophs. In some experiments, the growth medium was further supplemented with 0.1% yeast extract (Sigma), with no measurable effect on mutation rates. All plates and liquid cultures were incubated aerobically at 76°.
Growth of liquid cultures was routinely monitored by turbidity measurements at 600 nm. Viable counts were determined by serial dilution in sterile SM buffer (Sambrook and Russell 2001) and plating on solid medium; colonies typically reached 1 mm diameter in 2–3 days. The minimal inhibitory concentration of 5-fluoroorotic acid (FOA) was determined by incubating serial 1:2 dilutions of FOA in liquid growth medium inoculated with ∼107 cells/ml. Under these conditions, the minimal inhibitory concentration was ≤1.25 g/liter. However, FOA concentrations of 0.5–0.6 g/liter blocked all growth and colony formation of wild-type HB27 on solid medium and were used in selective plates.
To determine whether mutants resistant to FOA grow more slowly than the wild type, we performed two tests on a set of mutants of independent origin. In a reconstruction test, eight binary mixtures, consisting of liquid cultures of one mutant mixed with a 10-fold excess of wild-type cells, were grown for several hours in nonselective medium, during which time the populations expanded by factors of ∼100. For each mixture, the numbers (N) of mutant and total cells were measured by plating before and after incubation. The ratio of growth-rate constants (km/kw) was calculated under the model of exponential growth, where the superscripts m and w represent mutant and wild-type strains, respectively. The ratios of growth-rate constants (averaged over several trials and arranged monotonically) were 0.83, 1.02, 1.15, 1.18, 1.21, 1.24, 1.36, and 1.62 (mean 1.20 ± 0.23). In a turbidometric test, nonselective medium (3.0 ml in 16-mm glass tubes) was inoculated to yield ∼107 cells/ml and incubated. Absorbance at 600 nm was read at hourly intervals and the resulting values (corrected for the absorbance of the growth medium) were used to calculate km/kw for each interval between two absorbance measurements. The wild-type strain was represented by three cultures whose absorbance values were averaged at each time point. Six independent comparisons (representing six 1-hr intervals of growth) were made of each of nine different mutant cultures to the triplicate average of the wild type, yielding an average relative growth rate of 0.98 ± 0.29. Thus, the mutants grew at rates indistinguishable from that of the wild type.
In efforts to measure a delay in the appearance of resistant clones (phenotypic lag), exponentially growing cultures were treated with a minimally toxic dose of mutagen and sampled frequently to detect any abrupt increase in mutant frequency. Among N-methyl N′-nitro N-nitroso guanidine (MNNG), butadiene diepoxide, mechlorethamine, N-nitroso-N-methyl urea, and cyclophosphamide, only MNNG appeared to stimulate forward mutation in preliminary tests. However, in experiments extending over several generations, MNNG concentrations that permitted uninterrupted exponential growth generated no detectable increases in mutant frequency (data not shown).
Mutation rates and spectrum:
Fluctuation tests used series of liquid cultures started from small inocula. A single wild-type colony was inoculated into nonselective medium and allowed to grow to ∼107 cells/ml. It was then diluted with fresh medium and dispensed into microdilution trays to yield 103 cells in 200 μl in 10–15 wells. The trays were incubated until the cultures reached a density of ∼108 cells/ml, and any (small) volume losses due to evaporation were replaced with distilled water. The average number of viable cells in 5 wells per series was then determined by plating on nonselective medium. To determine the number of FOA-resistant mutants, the remaining cultures were plated in their entirety as two aliquots (50 and 150 μl) on selective medium (uracil + FOA). The number of FOA-resistant colonies varied by >100-fold, and this variance was not affected by varying the size of the inocula by 10-fold, indicating that the inocula did not contain preexisting mutants (Luria and Delbrück 1943). The number of mutant colonies arising from the two aliquots of each culture showed no effect of cell density on the efficiency of recovering mutants, and the individual 50- and 150-μl counts were therefore combined. The cultures were ranked according to the number of mutants; the first, second, and third quartiles of the resulting distribution were then used to calculate three corresponding estimates of the mutation rates using the numerical solutions of Koch (1982).
The accumulation method (Drake et al. 1998; Mackwan et al. 2007) used 25-ml cultures, each started from separate small inocula and grown to total populations of ∼1010 cells. Several samples of each culture were plated on selective and nonselective media. The resulting colony counts were used to determine the mutant frequency (f) and the population size (N) from which the mutation rate was calculated as μ = f/ln(Nμ). The median value from this series was taken as the best estimate of the mutation rate (Rosche and Foster 2000).
Mutation rates are given ±1 standard deviation (SD). The SD for the median value from the accumulation test was calculated according to Maritz and Jarrett (1978).
FOA-resistant colonies were confirmed to be uracil auxotrophs by inoculation of liquid medium with and without uracil supplementation. Clonally pure derivatives were stored frozen at −70° in growth medium supplemented with dimethyl sulfoxide (9% w/v). Genomic DNAs, purified from liquid cultures essentially as described (Boom et al. 1990), served as templates in separate standard PCR reactions to amplify pyrF and pyrE, which are separated by a GGGGGG spacer [GenBank accession nos. AE017221 and AE017222 (Henne et al. 2004)]. The amplification primer upstream of pyrF was 5′-AAAGCGGGAGGAAGGAGGGGA-3′ and the primer downstream of pyrE was 5′-GAAGGGTGGGCAGGTGGAAGAGGA-3′. The PCR reaction was adjusted for a GC-rich template and included a 2-min denaturing step at 95° with Platinum Taq DNA polymerase, PCRx enhancer solution, MgSO4, and PCRx amplification buffer (Invitrogen, Carlsbad, CA). The reaction was then run for 35 cycles of 30 sec at 95°, 30 sec at 60°, and 168 sec at 68°, followed by a hold temperature of 4°. The PCR products were purified using the QIAquick purification kit (QIAGEN, Valencia, CA). The BigDye-Terminator cycle sequencing kit was used to sequence the purified PCR products using the primers (F, forward; R, reverse) F5′-GGTGCTTTCGGCGTTTT-3′, F5′-AGGGGGAGGGGCTTCTTT-3′, R5′-AAAGAAGCCCCTCCCCCT-3′ and R5′-AAGAAGGGCAGCGAGGAA-3′. When these were inadequate, we used the alternative primers F5′-CCCTGGCTCCGGTTTCC-3′, R5′-AACGCAGGAGGAAGTGGC-3′, F5′-GCCACTTCCTCCTGCGTT-3′F and F5′-CTTCTTCCAGGCCGCTT-3′. The sequencing cycle consisted of 2 min at 96° followed by 25 cycles of 10 sec at 96°, 5 sec at 60°, and 4 min at 60°, followed by a hold temperature of 4°.
RESULTS
Genic mutation rates:
Mutation-rate assays were based on the ability of growth medium containing FOA + uracil to select pyrimidine auxotrophs lacking either of two UMP biosynthetic enzyme activities, orotate:phosphoribosyl pyrophosphate transferase or orotidylate decarboxylase, encoded by the pyrE and pyrF genes, respectively (Yamagishi et al. 1996). Efforts to develop other selections for spontaneous T. thermophilus mutants were unsuccessful. For example, in preliminary tests, rifampicin, l-canavanine, l-ethionine, d-cycloserine, naladixic acid, novobiocin, and 5-fluorouracil either failed to inhibit growth at reasonable concentrations or failed to select spontaneous mutants with sufficiently elevated resistance to the inhibitor (R. R. Mackwan, S. Teredesai and D. W. Grogan, unpublished results). To our knowledge, therefore, the FOA selection provides the only quantitative assay of forward mutation currently feasible for Thermus spp.
To determine whether physiological properties of the FOA-resistant mutants would affect the mutation assays, control experiments evaluated the fitness of FOA-selected pyrimidine auxotrophs, whereas possible effects of phenotypic lag were evaluated by comparing different mutation-assay methods. By these criteria, the mutants showed no significant reproductive disadvantage (see materials and methods) and no detectable impact of phenotypic lag (see below). On average, 20% of the FOA-resistant colonies from selection plates scored as prototrophs (i.e., did not require uracil for growth). In S. solfataricus strain P2 and H. volcanii, all such FOA-resistant uracil prototrophs were confirmed to have intact pyrE and pyrF genes (Redder and Garrett 2006; Mackwan et al. 2007). Therefore, we based our mutation-rate calculations only on pyrimidine-auxotrophic mutations that could be localized to pyrE or pyrF by DNA sequencing (see below).
Fluctuation-test mutation rates are shown in Table 1. In Koch's quartile analysis (Koch 1982), a fluctuation test yields three estimates of the mutation rate, each derived from the corresponding quartile of the frequency distribution. This approach can reveal systematic distortions caused by processes such as phenotypic lag, to which the first quartile is most sensitive and the third quartile is least sensitive. The substantial variation in the rates among the 10 trials was consistent with the small numbers of cultures in each trial (5–10) assayed for mutants. The three quartiles yielded similar mean values, thus providing no hint of phenotypic lag. The genic rate for pyrE + pyrF, obtained by averaging across the three quartiles and 10 trials, was (4.34 ± 2.92) × 10−7 mutations per cell division.
TABLE 1.
Fluctuation-test mutation rates per 107 cell divisions
| Trial | Quartile 1 | Quartile 2 | Quartile 3 |
|---|---|---|---|
| 1 | 1.11 | 2.36 | 2.22 |
| 2 | 1.99 | 2.17 | 1.88 |
| 3 | 4.39 | 7.92 | 9.47 |
| 4 | 6.67 | 6.93 | 6.93 |
| 5 | 0.022 | 0.022 | 0.11 |
| 6 | 1.03 | 2.71 | 2.54 |
| 7 | 7.79 | 6.72 | 6.45 |
| 8 | 2.58 | 6.32 | 6.32 |
| 9 | 3.14 | 3.06 | 2.27 |
| 10 | 7.97 | 9.11 | 7.97 |
| Mean ± SD | 3.67 ± 2.91 | 4.73 ± 3.02 | 4.62 ± 3.16 |
The global mean ± SD for all 10 fluctuation tests was (4.34 ± 2.92) × 10−7.
From the accumulation method, mean mutation rates per 107 cell divisions for each of the six cultures (arranged monotonically) were 1.61, 1.80, 2.11, 2.41, 3.46, and 3.59. The appropriate measure of this distribution is the median (Drake et al. 1998; Rosche and Foster 2000), which was (2.26 ± 0.66) × 10−7. The accumulation method is intrinsically less sensitive to phenotypic lag than is the fluctuation test, so that the similarity of the rates determined by the two methods again suggests the absence of significant phenotypic lag. Because the fluctuation-test mean and the accumulation-method median produced statistically indistinguishable values, we adopted the mean of the two, (3.30 ± 2.12) × 10−7, as the most reliable first measure of the genic rate. This rate must be adjusted slightly to reflect the fact that 2 of the 75 sequenced FOA-resistant uracil auxotrophs contained no mutation in the target genes. Thus, the corrected genic rate for the combined target (pyrE + pyrF) is (73/75)(3.30 ± 2.12) × 10−7 = (3.21 ± 2.06) × 10−7.
Mutation spectrum:
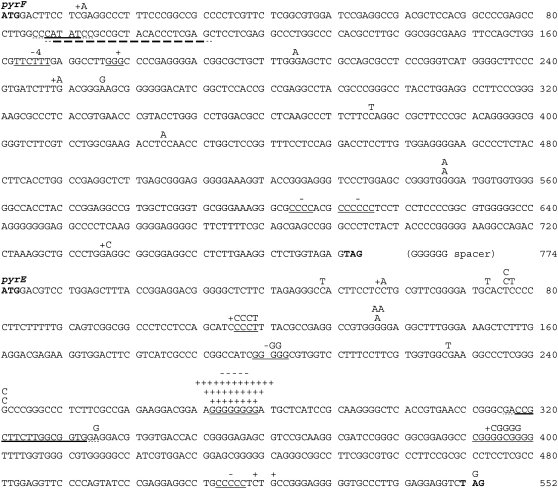
The mutational alterations in the sequenced mutants are listed in Table 2, their sequence contexts are shown in Figure 1, and the molecular nature of the mutations is summarized in Table 3. Half of the mutations arose at a single hotspot, a run of eight Gs in pyrE, which accounted for more mutations in the 552-nt pyrE gene (57) than in the 774-nt pyrF gene (26). Transitions outnumbered transversions by 16 to 3, a not particularly unusual bias. However, +1 indels outnumbered −1 indels by 37 to 10 including the hotspot, or 7 to 3 ignoring it, which represents a bias not commonly observed in vitro (Garcia-Diaz and Kunkel 2006). We observed no complex mutations, transposable-element insertions, or multiple mutations.
TABLE 2.
Molecular characteristics of spontaneous pyrE and pyrF mutations
| pyr | Positiona | Mutationb | No. | Notes |
|---|---|---|---|---|
| F | 12/13 | +A | 1 | |
| 79–186 | −108 | 1 | GGCCCCGAG(99)GGCCCCGAG | |
| 86–94 | TD7 | 1 | CCCATATCC | |
| 88–108 | TD20 | 1 | CATATCCGCCGCTACACCCTC | |
| 163–168 | −4 | 1 | TTCTTT → TT (three ways) | |
| 177–179 | +G | 1 | ||
| 204 | G → A | 1 | trp → UGA | |
| 249/250 | +A | 1 | ||
| 256 | A → G | 1 | lys → glu | |
| 376 | C → T | 1 | gln → UAG | |
| 425 | C → A | 1 | ser → tyr | |
| 547 | G → A | 2 | gly → glu | |
| 604–607 | –C | 1 | ||
| 611–616 | –C | 1 | ||
| 736/737 | +C | 1 | ||
| E | 49 | C →T | 1 | his → tyr |
| 57/58 | +A | 1 | ||
| 73 | C → T | 1 | his → tyr | |
| 76 | T → C | 2 | ser → pro | |
| 77 | C → T | 1 | ser → phe | |
| 116–119 | +CCCT | 1 | ||
| 137 | G → A | 3 | gly → glu | |
| 199–203 | –GG | 1 | ||
| 227 | C → T | 1 | ala → val | |
| 241 | G → C | 2 | ala → pro | |
| 272–279 | 8G → 9G | 31 | ||
| 272–279 | 8G → 7G | 5 | ||
| 316–335 | TD18 | 1 | GACCGCTTCTTGGCGGTGGA | |
| 335 | A → G | 1 | glu → gly | |
| 391–400 | TD5 | 1 | ||
| 513–517 | –C | 1 | ||
| 519 | C → C2C | 1 | ||
| 521 | G → GG | 1 | ||
| 551 | A → G | 1 | UAG → trp |
Relative to the first base of the initiation codon. / denotes an insertion between the indicated positions.
+ or − followed by a base indicates an insertion or deletion, respectively, within a short homopolymeric run or, followed by a number, indicates the number of bases inserted or deleted. TD followed by a number indicates a tandem duplication of that many bases. Flanking repeats are underlined in the last column, one copy of the repeat being included in the duplication or deletion. The deletion at pyrF 163–168 can be generated by removing any consecutive four bases from the indicated sequence.
Figure 1.—
Spectrum of pyrF and pyrE mutations. Initiating and terminating codons are in boldface type. Insertions between adjacent bases are written directly above the two bases. Insertions or deletions within mononucleotide repeats are shown as + or − above the lightly underlined repeat. Tandem duplications are heavily underlined, the flanking repeats have dashed underlines. The single large pyrF deletion is not shown; its flanking repeats are at positions 79–87 and 178–186.
TABLE 3.
Types of mutations arising spontaneously in T. thermophilus
| Type | pyrE | pyrF | Both |
|---|---|---|---|
| Total | 57 | 16 | 73 |
| BPSs | 13 | 6 | 19 |
| Transitions | 11 | 5 | 16 |
| Transversions | 2 | 1 | 3 |
| Indels | 44 | 10 | 54 |
| +1 | 34 | 4 | 38 |
| (HS)a | (31) | — | — |
| −1 | 6 | 2 | 8 |
| (HS)a | (5) | — | — |
| TDb | 2 | 2 | 4 |
| ±(2–108) | 2 | 2 | 4 |
The contribution from indels at the 8G hotspot.
Tandem duplications.
Genomic mutation rate:
Most mutations are deleterious, some are neutral, and few are advantageous. Because the load of deleterious mutations depends primarily upon the rate and kinds of spontaneous mutations, a primary goal of this study was to estimate the genomic mutation rate for T. thermophilus. The calculation involves correcting the genic rate for mutation-detection efficiency, followed by scaling the resulting value to the size of the entire genome. Most mutation-reporter genes detect indels with high efficiency, reflecting the gross disruption of the encoded protein, but they typically underestimate the frequencies of BPSs by severalfold, reflecting the predominance of phenotypically subtle or silent BPSs. This phenotypic underrepresentation requires some form of correction to estimate the total BPS frequency. The first approach uses an average correction factor of 4.726 (±2.778, 14 samples) determined historically for numerous genes (Grogan et al. 2001). One potential weakness of this approach is the large variance around the average, while another is the evidence that extreme thermophiles experience stronger purifying selection on BPSs than do the mesophiles from which this average is derived (Friedman et al. 2004). The latter result suggests that the average missense mutation is more deleterious in an organism growing at higher temperatures, and failure of the historical ratio to account for this stronger selection would cause the BPS rate in T. thermophilus to be overestimated. Another approach estimates the BPS rate from the number of chain-termination mutations (CTs) recovered. The total number of CTs is divided by the total number of pathways to CTs in the mutation-reporter sequence and is then multiplied by 3 because there are three possible base substitutions per base pair. However, this approach has two limitations. First, A·T → G·C mutations cannot generate CTs. Second, the number of CTs in a spectrum can be small; in the present case, for example, it was only 2 (Table 2).
Starting from the observed average mutation rate for pyrE + pyrF of (3.21 ± 2.06) × 10−7 based on 73 mutations (54 indels + 19 BPSs), the genic rate corrected using the historical factor is (3.21 ± 2.06)(10−7)(54 + 4.726 × 19)/73 = (6.32 ± 4.06) × 10−7 with a 95% confidence interval (CI) from (3.21 ± 2.06) × 10−7 to (11.0 ± 7.06) × 10−7. (Note that the interval boundaries themselves have SDs.) The mutation-reporter consists of 1326 nt, and the genome size, G, equals 2,127,482 nt, yielding a genomic rate of (6.32 ± 4.06)(10−7)(1/1326)(2.13 × 106) = 0.00101 ± 0.00065 with a 95% CI from 0.00052 ± 0.00033 to 0.00177 ± 0.00113. This overall rate is composed of an indel component of 0.00038 and a BPS component of 0.00063.
There are 103 pathways to CTs in the mutational target, including multiple pathways within a codon and multiple pathways at a site, and 2 CTs were observed. Thus, the BPS rate is (2/73)(3.21 ± 2.06)(10−7)(3/103) = (2.56 ± 1.64) × 10−10 per nt or (2.56 ± 1.64)(10−10)(2.13 × 106) = 0.00054 ± 0.00035 per genome, close to the rate calculated by the alternative method. Adding the indel genomic rate of 0.00038 gives a total genomic rate of 0.00093.
Because of their close correspondence, we averaged the values derived by the two methods to obtain a consensus genomic rate of 0.00097 ± 0.00052 (with a 95% CI from 0.00073 ± 0.00034 to 0.00135 ± 0.00084) mutations per replication, representing an indel rate of 0.00038 and a BPS rate of 0.00059. This total genomic rate is distinctly lower than the value of 0.0035 ± 0.0007 observed for DNA genomes of diverse mesophiles (see discussion), primarily because of the low value for the BPS rate. Using a parametric bootstrap method, the thermophile–mesophile difference has P < 0.0001 for the lower CI boundary, P = 0.0002 for the central value, and P < 0.005 for the upper CI boundary.
DISCUSSION
To our knowledge, this study provides the first quantitative molecular assessment of DNA replication accuracy in vivo at an extremely high temperature in a bacterium (as opposed to an archaeon). Furthermore, use of the same selection and orthologous mutational targets in T. thermophilus and S. acidocaldarius (Grogan et al. 2001) facilitates comparison of the mutational propensities of two highly divergent prokaryotes, each adapted to geothermal environments. The mutation assays gave no hint of phenotypic lag in T. thermophilus, neither in the rates estimated separately from the three quartiles in the fluctuation tests nor in the accumulation-test estimate, which was only slightly lower than the value determined from the fluctuation tests, rather than substantially higher as would have resulted from phenotypic lag. The forward (loss-of-function) mutations recovered using FOA selection included the types of events typical of spontaneous mutation in other systems: a minority of BPSs, many small indels, and a few larger tandem duplications and deletions. Most of the latter two types of mutations arose between short repeated sequences, which is seen in most target genes of mesophiles, although not those of the hyperthermophilic archaeon S. acidocaldarius (Grogan et al. 2001). This result can be compared with 86% BPSs among loss-of-function mutations of the S. cerevisiae URA3 gene recovered by the same selection (Lee et al. 1998).
The question sometimes arises of the reliability of a genomic mutation rate obtained by extrapolation from mutation reporters which may comprise ≤0.1% of the genome. Even in the most sophisticated genetic systems, few target genes can assay spontaneous forward mutation via selection, and the choices remain particularly limited in T. thermophilus. However, empirical data relate directly to the question of whether spontaneous mutations are expected to be significantly less frequent in the pyrEF genes than in the average genomic interval of similar size in T. thermophilus, which is the aspect of variation most relevant for interpretations of this study. First, the mutant reporter genes must be sequenced for the data to be considered robust, due to the influence of mutational hotspots on the genic rate and spectrum. These hotspots almost always consist of simple repeats, which are prone to slippage during DNA replication. As a consequence, contributions from these sites are readily distinguished from BPSs by sequence analysis, which allows BPS rates to be calculated with minimal distortion from the irregular distribution of hotspots among genes.
Second, the T. thermophilus rate is based on two genes with only one indel hotspot between them. This hotspot is moderate in strength and, as a mononucleotide run, conforms to the classical slipped-strand model of frameshift mutation (Streisinger and Owen 1985). The rates of spontaneous frameshifts at these runs have been analyzed in diverse mismatch-deficient and -proficient systems; the rates increase significantly with run length for a given mononucleotide, and are significantly higher for G(C) vs. A(T) runs of equal length (Sagher et al. 1999; Gragg et al. 2002). These intrinsic properties allow the quantitative impact of the pyrE hotspot to be compared to that of mononucleotide runs in the rest of the T. thermophilus genome. We analyzed the complete HB27 genome sequence and identified 1 occurrence each of 8A(T) and 9A(T), 174 occurrences of 8G(C), 5 of 9G(C), and 2 of 10G(C). Thus, A(T) runs represent a relatively minor source of spontaneous mutation for the T. thermophilus genome overall. Furthermore, only ∼9% of T. thermophilus genes have 8G(C) runs expected to match the mutational strength of the pyrE hotspot, and only ∼0.4% have longer runs expected to be more mutable than this. Corresponding repeats of dinucleotides, which do not occur in the pyrEF sequences, are only about one-tenth as abundant as mononucleotide runs throughout the T. thermophilus genome. Thus, the frameshift hotspot found in pyrE is one of the few in T. thermophilus having both the strength and abundance to affect the genomic rate of spontaneous mutation, and its distribution predicts that the target genes would tend to overestimate, rather than underestimate, this genomic rate.
Third, the set of mesophiles that forms a basis for comparison have genomic mutation rates of 0.0025, 0.0027, 0.0027, 0.0030, 0.0038, 0.0040, 0.0045, and 0.0046 (mean = 0.0035 ± 0.00097) for Escherichia coli, S. cerevisiae, Herpes simplex virus 1, Neurospora crassa, bacteriophage λ, bacteriophage T2/T4, Schizosaccharomyces pombe, and bacteriophage M13, respectively, estimated using the historical correction factor (Drake et al. 1998; Fraser et al. 2003; Drake and Hwang 2005). All but one of these values are based on only one or two reporter genes but the range of values resulting from the same methods of calculation is nevertheless less than twofold. The similarity of these individual genomic mutation rates drawn from diverse mesophiles, and the deviation of the corresponding T. thermophilus rate from this norm, thus provide empirical evidence for the robustness of the conclusion that the T. thermophilus genomic rate is lower than the characteristic mesophile rate.
Because indel hotspots were more the exception than the rule in the reporter genes used to obtain the mesophilic genomic rates listed above, it is informative to calculate the residual T. thermophilus mutation rates in the absence of the 8G hotspot. The genomic indel rate would decrease from 0.00038 to 0.000056. Using the historical factor to calculate the BPS rates, the total genomic rate would decrease from 0.00101 to 0.00069. Using the CT method, it would decrease from 0.00093 to 0.00060. The corresponding change in average genomic rate for the reference set of mesophiles is quite small, so that the contrast between the thermophilic and the average mesophilic genomic rates increases substantially when the thermophile hotspot is discounted.
The similarity of the mutational properties of T. thermophilus and S. acidocaldarius contrasts with the deep evolutionary and biochemical divergence between them. We note, for example, that T. thermophilus and S. acidocaldarius share aerobic and heterotrophic modes of growth, extremely high optimal growth temperatures, and genomes of ∼2.2 Mbp. However, they have dramatically different DNA base compositions (69 vs. 38 mol% G + C) and represent different domains of cellular life (bacteria vs. archaea), resulting in different molecular contexts of genome replication. Thus, T. thermophilus employs a family-A DNA polymerase for genome replication (Bullard et al. 2002), whereas Sulfolobus spp. and other archaea possess only family-B polymerases capable of genome replication (Kawarabayasi et al. 2001; She et al. 2001). Archaeal family-B polymerases have the peculiar characteristic of stalling just before template deoxyuridine residues (Lasken et al. 1996; Shuttleworth et al. 2004) although, to our knowledge, the functional significance of this property has not been confirmed in vivo. T. thermophilus and S. acidocaldarius have different types of DNA-binding proteins (Zierer and Choli 1990; Du and Pène 1999) and different numbers of replication origins (Lundgren et al. 2003; Henne et al. 2004). In addition, strain HB27 lacks an intact gene for reverse DNA gyrase, which is otherwise highly conserved among bacteria and archaea from geothermal habitats (Forterre 2002), including S. acidocaldarius and T. thermophilus strain HB8 (Henne et al. 2004; Chen et al. 2005). With respect to DNA repair, T. thermophilus encodes complete, bacterial-type systems for nucleotide excision repair (UvrABC) and DNA mismatch repair (MutHSL), whereas S. acidocaldarius encodes an apparently incomplete nucleotide-excision-repair system of the eukaryotic type (XPF and XPG) and no identifiable mismatch-repair genes (Grogan 2004). Conversely, T. thermophilus apparently lacks any DNA polymerase of the Y family, whereas S. acidocaldarius encodes at least one such enzyme, designated Dbh (Boudsocq et al. 2004). One predicted consequence of this latter difference is weaker induced mutation in T. thermophilus compared to S. acidocaldarius, which is consistent with our observation that chemical mutagenesis in T. thermophilus appeared to be less effective than similar treatment of S. acidocaldarius (Reilly and Grogan 2002).
In summary, quantitative molecular analysis of spontaneous mutations occurring in vivo indicates that DNA replication in T. thermophilus growing at 76°, far from being error prone (Castán et al. 2003), generates mutations at an unusually low rate per genome replication. Comparison of the gene inventories of T. thermophilus and S. acidocaldarius further suggests that these two thermophiles achieve their low mutation rates by distinct sets of biochemical strategies. Overall, the available data suggest that cellular organisms in geothermal environments may be subject to a selective pressure against substitution mutations which is more stringent than the corresponding selection exerted on mesophiles, with corresponding implications for the rates of molecular evolution in these lineages.
Acknowledgments
We thank Marilyn Diaz and Tom Kunkel for critical readings of the manuscript. This work was supported by National Science Foundation grants MCB 9733303 and MCB 0543910 to D.W.G. and by the Intramural Research Program of the National Institutes of Health National Institute of Environmental Health Sciences.
References
- Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen et al., 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, F., R. J. Kokoska, B. S. Plosky, A. Vaisman, H. Ling et al., 2004. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 279 32932–32940. [DOI] [PubMed] [Google Scholar]
- Breuert, S., T. Allers, G. Spohn and J. Soppa, 2006. Regulated polyploidy in halophilic archaea. PLoS ONE 1 e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann, H., and C. Chen, 2006. Comparative genomics of Thermus thermophilus: plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 124 654–661. [DOI] [PubMed] [Google Scholar]
- Bullard, J. M., J. C. Williams, W. K. Acker, C. Jacobi, N. Janjic et al., 2002. DNA polymerase III holoenzyme from Thermus thermophilus. Identification, expression, purification of components, and use to reconstitute a processive replicase. J. Biol. Chem. 277 13401–13408. [DOI] [PubMed] [Google Scholar]
- Castán, P., L. Casares, J. Barbé and J. Berenguer, 2003. Temperature-dependent hypermutational phenotype in recA mutants of Thermus thermophilus HB27. J. Bacteriol. 185 4901–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., K. Brugger, M. Skovgaard, P. Redder, Q. She et al., 2005. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 187 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, M. M., and J. R. Battista, 2005. Deinococcus radiodurans: the consummate survivor. Nat. Rev. Microbiol. 3 882–892. [DOI] [PubMed] [Google Scholar]
- Drake, J. W., and C. B. C. Hwang, 2005. On the mutation rate of herpes simplex virus type 1. Genetics 170 969–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., B. Charlesworth, D. Charlesworth and J. F. Crow, 1998. Rates of spontaneous mutation. Genetics 148 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., and J. J. Pène, 1999. Identification, cloning and expression of p25, an AT-rich DNA-binding protein from the extreme thermophile, Thermus aquaticus YT-1. Nucleic Acids Res. 27 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre, P., 2002. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18 236–237. [DOI] [PubMed] [Google Scholar]
- Fraser, J. L. A., E. Neill and S. Davey, 2003. Fission yeast Uve1 and Apn2 function in distinct oxidative damage repair pathways in vivo. DNA Repair 2 1253–1267. [DOI] [PubMed] [Google Scholar]
- Friedman, R., J. W. Drake and A. L. Hughes, 2004. Genomewide patterns of nucleotide substitution reveal stringent functional constraints on the protein sequences of thermophiles. Genetics 167 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz, M., and T. A. Kunkel, 2006. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 31 206–214. [DOI] [PubMed] [Google Scholar]
- Goodman, M. F., 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71 17–50. [DOI] [PubMed] [Google Scholar]
- Gragg, H., B. D. Harfe and S. Jinks-Robertson, 2002. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 22 8756–8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan, D. W., 2004. Stability and repair of DNA in hyperthermophilic Archaea. Curr. Issues Mol. Biol. 6 137–144. [PubMed] [Google Scholar]
- Grogan, D. W., and J. E. Hansen, 2003. Molecular characteristics of spontaneous deletions in the hyperthermophilic archaeon Sulfolobus acidocaldarius. J. Bacteriol. 185 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan, D. W., G. T. Carver and J. W. Drake, 2001. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 98 7928–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne, A., H. Brüggemann, C. Raasch, A. Wiezer, T. Hartsch et al., 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22 547–553. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi et al., 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8 123–140. [DOI] [PubMed] [Google Scholar]
- Koch, A. L., 1982. Mutation and growth rates from Luria-Delbrück fluctuation tests. Mutat. Res. 95 129–143. [Google Scholar]
- Lasken, R. S., D. M. Schuster and A. Rashtchian, 1996. Archaebacterial DNA polymerases tightly bind uracil-containing DNA. J. Biol. Chem. 271 17692–17696. [DOI] [PubMed] [Google Scholar]
- Lundgren, M., L. Chen, A. Andersson, P. Nilsson and R. Bernander, 2003. Three replication origins in Sulfolobus spp.: Synchronous initiation of replication and asynchronous termination. Proc. Natl. Acad. Sci. USA 101 7046–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G. S., E. A. Savage, R. G. Ritzel and R. C. von Borstel, 1998. The base-alteration spectrum of spontaneous and ultraviolet-induced forward mutations in the URA3 locus of Saccharomyces cerevisiae. Mol. Gen. Genet. 214 396–404. [DOI] [PubMed] [Google Scholar]
- Luria, S. E., and M. Delbrück, 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackwan, R. R., G. T. Carver, J. W. Drake and D. W. Grogan, 2007. An unusual pattern of spontaneous mutations recovered in the halophilic archaeon Haloferax volcanii. Genetics 176 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritz, J. S., and R. G. Jarrett, 1978. A note on estimating the variance of the sample median. J. Am. Stat. Assoc. 73 194–196. [Google Scholar]
- Oshima, T., and K. Imahori, 1974. Physiochemical properties of deoxyribonucleic acid from an extreme thermophile. J. Biochem. 75 179–183. [DOI] [PubMed] [Google Scholar]
- Redder, P., and R. A. Garrett, 2006. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 188 4198–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly, M. S., and D. W. Grogan, 2002. Biological effects of DNA damage in the hyperthermophilic archaeon Sulfolobus acidocaldarius. FEMS Microbiol. Lett. 208 29–34. [DOI] [PubMed] [Google Scholar]
- Rosche, W. A., and P. L. Foster, 2000. Determining mutation rates in bacterial populations. Methods 20 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagher, D., A. Hsu and B. Strauss, 1999. Stabilization of the intermediate in frameshift mutation. Mutat. Res. 432 73–77. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual, Vol. 3, p. A2.8. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard et al., 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuttleworth, G., M. J. Fogg, M. R. Kurpiewski, L. Jen-Jacobson and B. A. Connolly, 2004. Recognition of the pro-mutagenic base uracil by family B DNA polymerases from archaea. J. Mol. Biol. 337 621–634. [DOI] [PubMed] [Google Scholar]
- Streisinger, G., and J. Owen, 1985. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109 633–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, A., T. Tanimoto, T. Suzuki and T. Oshima, 1996. Pyrimidine biosynthesis genes (pyrE and pyrF) of an extreme thermophile, Thermus thermophilus. Appl. Environ. Microbiol. 62 2191–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierer, R., and D. Choli, 1990. The primary structure of DNA binding protein II from the extreme thermophilic bacterium Thermus thermophilus. FEBS Lett. 273 59–62. [DOI] [PubMed] [Google Scholar]