Abstract
In Drosophila, sexual differentiation, physiology, and behavior are thought to be mediated by numerous male- and female-specific effector genes whose expression is controlled by sex-specifically expressed transcriptional regulators. One such downstream effector gene, sex-specific enzyme 1 (sxe1, cyp4d21), has been identified in a screen for genes with sex-biased expression in the head. Sxe1 was also identified in another screen as a circadian regulated gene. Here, we analyzed the spatial and temporal regulation of sxe1 and identified a function for this gene in male courtship. We show that male-specific transcriptional regulator DSXM and the clock genes are necessary for cycling of sxe1 mRNA during the diurnal cycle. Similar to sxe1 mRNA, expression of SXE1 protein oscillates in a diurnal fashion, with highest protein levels occurring around midnight. SXE1 protein expression is restricted to nonneuronal cells associated with diverse sensory bristles of both the chemo- and mechanosensory systems. Suppression or knockout of sxe1 significantly reduces mating success throughout the diurnal cycle. Finally, the metabolomic profile of wild-type and sxe1 mutant males revealed that sxe1 likely functions as a fatty acid ω-hydroxylase, suggesting that male courtship and mating success is mediated by small compounds generated by this enzyme.
DEVELOPMENT and differentiation of sex-specific structures and organs, and the establishment of sex-specific physiological/neuroanatomical traits, are essential processes common to virtually all animals. In Drosophila melanogaster, somatic differentiation of male and female flies is regulated by sex-specific splicing regulators including SEX-LETHAL (SXL) and TRANSFORMER (TRA) that lead to the expression of male- and female-specific spliced isoforms of the transcription factors, FRUITLESS (FRUM) and DOUBLESEX (DSXM and DSXF). DSXF, DSXM, and FRUM are thought to control the expression of numerous downstream effector genes both in the nervous and other organ systems, and ultimately establish the sex-specific properties and features of the adult fly (Burtis and Baker 1989; Coschigano and Wensink 1993; Ito et al. 1996; Ryner et al. 1996; Demir and Dickson 2005). These effector genes are thought to fall into two broad categories: (1) genes expressed during critical stages of development (pupae stage) and required for the differentiation of sex-specific phenotypical traits (male and female genitalia, male sex-comb, coloration of abdominal cuticle) and wiring of sex-specific neural circuits (male courtship, female egg laying, and mating/rejection behavior) and (2) genes required after development to assure sex-specific physiological properties and maintenance of sex-specific behaviors. While microarray and other large scale expression screens have led to the identification of hundreds of putative effector genes that are differentially expressed in males and females (Arbeitman et al. 2002, 2004; Dauwalder et al. 2002; Fujii and Amrein 2002; Parisi et al. 2004), the specific functions of virtually all of them remain unknown.
Courtship is an innate behavior that is characterized by manifestation of several behavioral displays executed in a characteristic sequential fashion by a male when encountering a female (Hall 1994). The appropriate execution of the male courtship sequence is genetically controlled mainly by FRUM, and to a lesser degree DSXM (Demir and Dickson 2005; Shirangi et al. 2006), and by inference, the effector genes regulated by these transcription factors.
We have previously identified 57 sex-biased and sex-specific candidate genes in adult heads using serial analysis of gene expression (SAGE) (Fujii and Amrein 2002). Nine of these genes were virtually sex specific, including all 5 genes known at the time to be expressed in the head in a sex-specific manner: the 3 female-specific yolk protein (yp1, 2, and 3) genes and the male-specific RNAs roX1 and roX2 (RNA on the X 1 and 2). The four novel genes included 3 male-biased genes sex-specific enzyme 1 and 2 (sxe1, sxe2) and turn on sex-specificity (tsx) and a female-specific gene female independent of transformer (fit). The common feature of these 4 novel sex-biased genes is that their expression is nonneuronal, yet is largely restricted to the head, and expression (sex-biased expression in the case of tsx) is restricted to the adult stage (Fujii and Amrein 2002). This suggests that the main function of these genes is necessary in the adult physiology and/or behavior of male or female flies.
Sxe1 encodes one of >80 cytochrome P450 enzymes, which are crucial components in many biochemical pathways (Simpson 1997). We have previously shown that sxe1 is a downstream target of dsx (Fujii and Amrein 2002). Specifically, XX; dsxD/Df(dsx) flies, which only express DSXM protein, show robust expression of sxe1, as do XX; tra/tra flies, which express both DSXM and FRUM (Fujii and Amrein 2002). Expression of sxe1 mRNA was detected in the head and at reduced levels in the thorax, but not the abdomen. Within the head, sxe1 mRNA is mostly found in carcasses (cuticle) and possibly fat cells, but is absent in the brain. Sxe1 has also been identified in a screen for genes expressed in the head of adult flies that oscillate in a circadian fashion using microarray analysis (Claridge-Chang et al. 2001; McDonald and Rosbash 2001; Lin et al. 2002), suggesting that this gene may also be a downstream effector of the clock genes. Indeed, oscillation of sxe1 is dependent on Clock (Clk), period (per), and timeless (tim), the main regulators required for oscillation of downstream effectors (Hardin 2005). Interestingly, takeout (to), which encodes a pheromone binding protein and is male-biased expressed in the fat body (Dauwalder et al. 2002), also oscillates with a 24-hr period (Sarov-Blat et al. 2000).
Circadian locomotor behavior was recently found to exhibit sex-specific features (Fujii et al. 2007), albeit both males and females are largely active during the day and rest during the night. Specifically, the activity of females, after an initial steep rise after dawn, continues to increase throughout the day, ending in a single peak before dusk, while males show two distinct activity peaks, one after dawn and a second one at dusk (coinciding with the single female peak). Surprisingly, circadian activity is reset when a male and a female cohabitate the same arena, with a drastic increase in nocturnal activity (Fujii et al. 2007). This activity shift is driven by male courtship and is dependent on the male's circadian system. Together, these observations suggest that the circadian clock of the two sexes may be regulated by distinct sex-specific factors.
Here, we determined SXE1 expression using sxe1-Gal4 drivers as well as anti-SXE1 antiserum and found that SXE1 is expressed in nonneuronal tissues of the chemo- and mechanosensory systems. Molecular genetic analyses revealed that circadian regulation for sxe1 is dependent on the male-specific transcription factor DSXM. We employed RNA interference and gene targeting to show that sxe1 is required for high mating success in males. Finally, we investigated the molecular function of SXE1 as a fatty acid ω-hydroxylase by analyzing the metabolomic profile of wild-type and sxe1 mutant males and we show that this protein is essential for the production of many small molecules in the male, which may function as male- and species-specific neuromodulators.
MATERIALS AND METHODS
Fly strains:
The following fly strains were used: Ore-R, w1118, per01, yw; tim01, ClkJRK, cyc01 ry, BsY; dsx1/TM3, BsY; hs->traf Df[3L] st tra pp dsxD Sb e/TM6, BsY; dsxDf/TM3, p[ELAV]GAL4C155, UAS-y_ds (R. Costa), UAS-mCD8-GFP, w1118; 70FLP, 70I-SceI, Sco/CyO, and w1118; 70FLP (K. Golic). All stocks were maintained under a 12-hr light:12-hr dark cycle at 25°.
Over 80% of homozygous dsx1 flies die <4 days after eclosion with standard food. To elongate their longevity and entrain them for LD cycles, we collected flies within 8 hr after eclosion and kept them in vials with food lacking yeast extract for 4 days. Oregon-R, Clkjrk, cyc01, per01, and tim01 males were entrained for 7–10 days, whereas all flies carrying the dsx1 allele were entrained for 4 days.
Immunochemistry:
Whole mount LacZ and antibody staining were carried out as described (Bray and Amrein 2003), with some modifications. A rabbit polyclonal antibody was generated against synthetic peptide STGNNVGLKPRTRVK, corresponding to amino acid 497 to 511 of SXE1 and affinity purified (AnaSpec, San Diego). The antiserum was used at 1:800 dilution for Western blotting and whole mount antibody staining. For Western blotting, ECL anti-rabbit IgG, HRP-linked F(ab′)2 fragment and ECL Plus detection kit (Amersham Biosciences) were used. Western blots were repeated between five and nine times using freshly prepared protein extract for each genotype/time point.
Monoclonal anti-GFP (Molecular Probes), anti-α-tubulin (Sigma-Aldrich), anti-ELAV antibody and TOTO3 (Molecular Probes) were used at 1:1000, 1:50,000, 1:10, and 1:1000, respectively. Immunofluorescence imaging was performed using a Leica TCS5 confocal microscope as described previously (Thorne et al. 2004).
Northern blot analysis:
Total RNA from heads was isolated for all Northern blot analysis using a TRIsol (Life Technologies). The coding region of sxe1 cDNA was labeled with [32P]-dCTP using Random Primer labeling kit (Stratagene). Northern blotting and hybridization was performed as previously described (Fujii and Amrein 2002). A Typhoon Imager (Amersham Biosciences) was used for measurement of signal strength. Northern blots in Figure 2A were repeated four to five times, each time with a new RNA sample for each genotype/time point, except for Ore-R females (performed once). Northern blots in Figure 2C were repeated twice, each time with a new RNA sample for each genotype/time point.
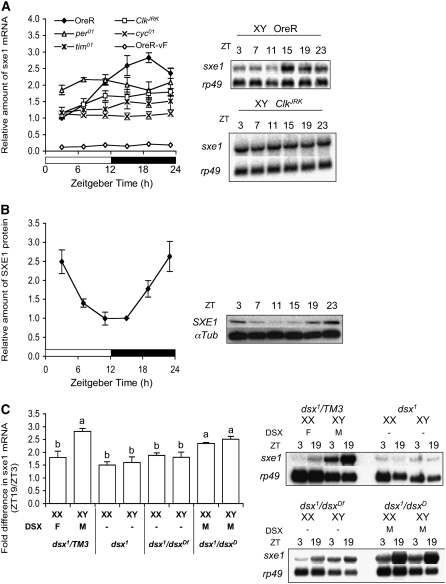
Figure 2.—
Daily oscillation of sxe1 depends on DSXM. (A) Relative amount of sxe1 expression in wild-type males and virgin females (OreR-vF) and various clock mutant males is plotted at various Zeitgeber times (ZT; left). Signal intensity of Northern blot for sxe1 was normalized by that of rp49, and then all data were converted to relative amount to OreR males signal at ZT 3 (value 1). (n = 4–5 for each genotype and time point. n = 1 for OreR-vF). Only OreR showed a significant difference (P < 0.0001) in time by one-way ANOVA. Error bar represents SEM. Two representative Northern blots of OreR and ClkJRK mutant males are shown on the right. An rp49 probe was used as a control for RNA loading. (B) SXE1 protein expression in the head of OreR males (n = 9–10 for each time point) is plotted at various ZTs. Signal intensity of Western blot for SXE1 at ZT 15 was converted to 1. Difference in time is significant (P < 0.0001) by one-way ANOVA. A Western blot is shown on the right. An α-Tub was visualized with a specific antibody monitor sample loading. (C) The relative amount of sxe1 mRNA at ZT 19 as fold increase of levels at ZT 3 is shown for various genotypes. The genotypes expressing DSXM show a significant higher elevation of sxe1 mRNA at ZT 19 (a, P < 0.05), as compared to genotypes lacking DSXM (b), as shown by one-way ANOVA followed by post hoc each pair Student's t-test. The Northern blots are shown on the right. An rp49 probe was used as a control for RNA loading.
Transgenic fly construction:
For the p[sxe1]GAL4 driver, a 3.0-kb fragment upstream of the start codon was amplified from genomic DNA, sequenced, and cloned into the GAL4 transformation vector SM1 (Dunipace et al. 2001). Transgenic flies were generated as described previously (Dunipace et al. 2001).
For the UAS-sxe1_ds reporter, the entire coding region of sxe1 was amplified from cDNA, cloned into pZero1 vector (Invitrogen), and sequenced. Two copies of this fragment were ligated sense and antisense (head-to-head) orientation, separated by a spacer (partial EGFP cDNA), and subsequently cloned into the pUAST vector (Brand and Perrimon 1993).
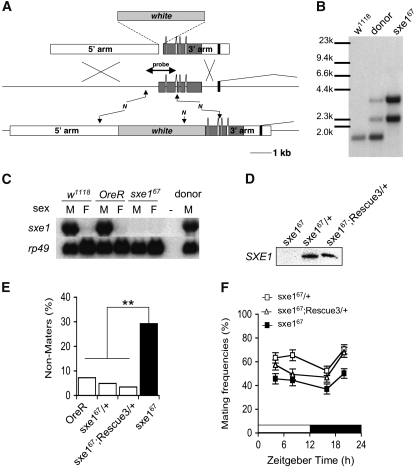
For the sxe1 targeting construct, a 4.873-kb fragment (corresponding to nucleotides 7,600,804–7,605,677 on chromosome arm 2L; release 5.3 in FlyBase) was amplified to generate the 5′ arm (see Figure 4) using primers CCGGCGCGCCCACTTCGAGTGCTGATCGCAATTGG and GGCCTAGGTATTCTTACGGATAAATCGGCTTTACG, and a 3.619-kb fragment (nucleotides 7,605,736–7,609,355) was amplified to generate the 3′ arm using primers CCCGGTCCGAGTCGCAAACAATGGAGAGTTAACAC and CCGCTAGCGCCTTAAAAAAGATCTCAGAATGGTTC. Fragments were subcloned into CMC105 vector (C.-M. Chen and G. Struhl, personal communication). The resulting targeting construct created a sxe1 mutation in which the 58-bp DNA region starting at the −5 position of the translational start was replaced by the white minigene, creating a null allele for sxe1. Virgin female flies carrying a single targeting construct on the third chromosome (donor) were crossed to w1118;70FLP, 70I-SceI/TM3, and 3- to 4-day-old progeny were heat-shocked at 38° for 60 min. Approximately 3000 white-eyed adult virgin females were crossed to w1118;70FLP males and progeny were heat-shocked as described above. Four red-eyed flies were obtained in which the targeting construct relocated to the second chromosome, and a homozygous line for each of them was established. Genomic DNA of these lines was digested with NcoI, analyzed by Southern blotting using a DNA probe corresponding to the 5′ region of sxe1 (see Figure 4). One line, sxe167, representing a precise recombination event, was backcrossed five times to Oregon-R and used for further investigation. A genomic rescue fragment was amplified with the most 5′ and the most 3′ primers used for the targeting construct, sequenced, cloned into pCsper4 transformation vector, and used to generate three independent transgenic rescue lines. Initial rescue experiments revealed varying degrees of rescue in mating assays; line 3, which conferred the strongest rescue, was backcrossed five times to OreR and used for further investigations as a rescue line.
Figure 4.—
Sxe1 mutant males exhibit reduced mating success. (A) Diagram of the sxe1 genomic region prior to (middle) and after (bottom) homologous recombination of the sxe1 targeting construct (top). The mutant sxe1 gene lacks the 5′ part (58 bp) of the sxe1 gene including the start codon. N's indicate NcoI sites. (B) Genomic Southern analysis of DNA from control flies, flies carrying the targeting construct, and flies homozygous for the mutant sxe167 allele. The 1.5-kb NcoI fragment present in the sxe1 gene, which also served as a probe in Figure 4A, is replaced by the white minigene and adjacent sxe1 sequence, represented by two fragments of 3.3 and 2.3 kb in the sxe167 mutant allele. (C and D) Expression of sxe1 mRNA (C) and SXE1 protein (D) is completely abolished in homozygous sxe167 mutant flies. (E) Frequencies of nonmaters in single pair mating assays of sxe167 mutant and control males (n = 55, 41, 29, or 89 for OreR, sxe167/+, sxe167;Rescue3/+ or sxe167, respectively). **P < 0.01 (chi-square test) for values compared with sxe167. (F) Mating frequencies at different ZT points. Five virgin males and five virgin females were placed in a vial and left for 15 min to copulate in the dark (<1 lux; n = 38–44 for each genotype and time point). Error bars represent SEM. Differences were analyzed by two-way ANOVA and were as follows for sxe167/+ vs sxe167: P < 0.0001 (genotype), P = 0.0036 (time), and P = 0.9068 (interaction of time and genotype). For sxe167/sxe167;Rescue3/+ vs. sxe167/sxe167, they were P = 0.0003 (genotype), P = 0.0008 (time) and P = 0.5158 (interaction of time and genotype).
Behavioral analysis:
All males used in behavioral assays were of w+ background to eliminate any effects due to impaired visual perception caused by different levels of eye pigmentation provided by the various w minigene-containing transgenes. w1118 virgin females were used as mating targets in all assays. CI and frequency of nonmaters were measured as described (Bray and Amrein 2003). Males were kept in isolation for 14 days, whereas ∼20 virgin females were kept in a vial together for 5–7 days.
Mating frequency assays were performed as described (Sakai and Ishida 2001), with the following modifications. Virgin flies were collected and sexed within 6 hr after eclosion, and animals of the same sex were kept for 1–2 weeks (∼20/vial). One day before the experiment, animals of the same sex were transferred into new vials in batches of 5/vial; the following day, they were placed in a light-protected box for 30 min before ZT 4, 8, 16, and 20. After letting all the flies adjust to darkness, 5 males and 5 females were combined into one vial and kept for 15 min under darkness (dim red light, <1 lux); they were then knocked out using CO2 to score actively mating (physically connected) pairs. Females not actively copulating were transferred into new vials individually, which were examined for larvae after 10 days to determine the female's mating status.
Locomotor activity:
Virgin males were kept individually for 14 days. Flies were transferred with a “fly aspirator” into a small Plexiglas mating chamber (4 × 10 × 30 mm), left to adjust to the new environment for 2 min, and then monitored for crossing the midline of the chamber for 4 min at ZT 2–4.
Taste preference assay was performed as described previously (Thorne et al. 2004). Preference of males for 25 mm trehalose vs. water was measured. Three independent experiments were performed with ∼40 flies per genotype per experiment.
Longevity of flies was measured as follows: Virgin males were collected within 6 hr after eclosion and kept at 10 flies per vial at 25°. Flies were transferred into new vials every 2 days, and dead flies were scored until all flies were dead. Three independent experiments were performed with 100 flies per genotype per experiment.
Fertility test:
A single male and single w1118 virgin female were kept for 4 days in a vial at 25°. Parents were discarded and the vial was kept until adults emerged. The number of adult flies was scored.
Quantification and analysis of fatty acids:
Roughly 1000 heads of sxe167/sxe167; Rescue3/+ and sxe167/sxe167 males of 5–10 days of age were collected, weighed, and provided to Lipomics Technologies for lipid analysis using the TrueMass protocol. All lipid species were quantified as nanomoles per gram of tissue. A molecular description of fatty acid species is provided at http://www.lipomics.com/fatty_acids.
Statistical analysis:
All analyses were performed using the JMP6 software suite (SAS).
RESULTS
Sxe1 is expressed in nonneuronal cells associated with chemo- and mechanosensory sensilla:
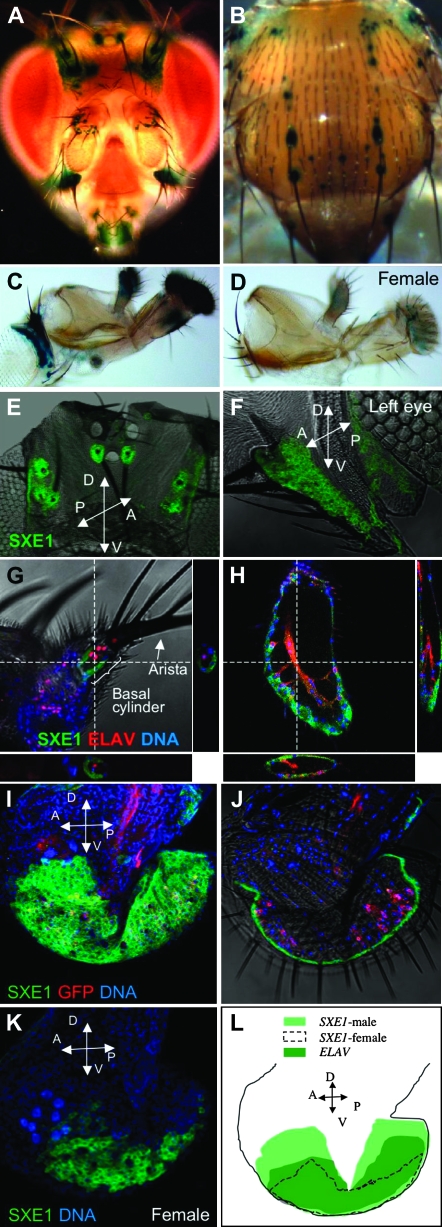
We previously analyzed the expression of sxe1 mRNA using Northern analysis, which revealed that the gene is abundantly expressed in the head and the thorax, but not in the abdomen. Dissection of various head parts further indicated that sxe1 is absent from the brain, but is mostly expressed in tissues associated with the head carcass (Fujii and Amrein 2002). To determine the cellular expression profile of sxe1, we generated flies containing a sxe1-Gal4 driver and a UAS-lacZ reporter (p[sxe1]GAL4/UAS-LacZ) and performed β-Gal staining. lacZ activity was observed in numerous sites, most notably the second antennal segment, at the base of the arista, in the labial palps, at the tip of maxillary palps, the vibrissae (ventral outside edge of compound eyes), and at the base of macrochaetae in the head and thorax (Figure 1, A–C). sxe1 is also expressed in the base of bristles of the male forelegs (data not shown). In females, expression is absent in most of these sites, with the exception of the labial and maxillary palps; there, expression is much weaker than in the corresponding male structures (Figure 1D). These findings are consistent with our previous Northern analyses, which revealed male-specific sxe1 expression in the head and thorax (Fujii and Amrein 2002).
Figure 1.—
Sxe1 expression in the head and thorax of adult males and females (A–D) Whole-mount LacZ staining of p[sxe1]GAL4/UAS-LacZ in a male (A–C) and a female (D). In the head and thorax, sxe1 is mainly expressed in cells associated with long mechanosensory bristles of males (macrochaetae, A and B), but absent in females (not shown). Sxe1 expression is also observed in cells associated with taste (labial palps) and olfactory sensilla (maxillary palps) of males (C); in females weak expression in the later is also observed. (E–K) Whole-mount antibody staining of p[elav]GAL4/UAS-mCD8GFP in a male (E–J) and a female (K) using anti-SXE1, anti-ELAV (only G), or anti-GFP primary (H, I, and J) antibodies. SXE1 antigen (visualized in green) is present in cells surrounding the base of the long bristles on the head and the vibrissae (E and F). SXE1 is clearly nonneuronal as no colocalization is observed with ELAV (red, labels nuclei of neurons in G) or GFP (red, labels entire neural network in H, I, and J) in the third antennal segment/arista (G), the maxillary (H), and the labial palp (I), but is confined to a cell sheet just underneath the cuticle (H and J). Only very weak staining is observed in the labial palps of the female (K). (L) Schematic map of sexually dimorphic SXE1 expression in labial palps.
To determine protein expression of SXE1, we generated antisera against this protein (see materials and methods) and performed Western analysis, which revealed a distinct band in wild-type but not sxe1 mutant flies of the expected molecular weight of 58 kDa (supplemental Figure 1). We then performed whole-mount antibody staining of p[elav]GAL4/UAS-mCD8-GFP flies using anti-SXE1, anti-ELAV, and anti-GFP antibodies, which allowed us to determine whether SXE1 was present in any neuronal cell compartment. SXE1 immunoreactivity was observed in the same tissues where lacZ activity was localized in sxe1-GAL4;UAS-lacZ flies (compare Figure 1, A–C, with Figure 1, E–I). Confocal microscopy revealed that SXE1 is expressed in the cytoplasm of nonneuronal cells associated largely with sensory bristles (Figure 1, G, H, and J). SXE1 immunoreactivity was also observed in the female labial palps, but expression is weaker and restricted to a smaller area when compared to males (compare Figure 1, I, K, and L). Taken together, these studies indicate that SXE1 is localized in cells mostly associated with sensory bristles and, while not entirely male specific, it is expressed with a strong male bias.
Clock and sex determination genes coregulate sxe1 expression:
Claridge-Chang et al. (2001) showed significant changes in sxe1 expression in the heads in Clk, per, and tim mutant flies using microarray analyses and suggested that the clock genes regulate sxe1 expression (McDonald and Rosbash 2001; Lin et al. 2002). To obtain a definite view of how expression of sxe1 is regulated, we performed a series of Northern and Western analyses with RNA and protein extracts from adult heads isolated at various time points during the day from flies mutant for individual clock and sex determination genes (Figure 2). First, we confirmed that the amount of sxe1 mRNA in wild-type males oscillates (one-way ANOVA, P > 0.0001), with a peak at Zeitgeber time (ZT) 19. The strongest Zeitgeber for most animals is light, and zeitgeber time refers to a specific time point during the 12-hr light:12-hr dark cycle (ZT 0/24, light on; ZT 6, midday; ZT 12, light off ; ZT 18, midnight). Mutations in all four major clock regulators abolish sxe1 oscillation in males, but the expression levels are still higher than those of wild-type females. To investigate whether the cycling of sxe1 was also observed at the protein level, we performed Western analyses of protein extracts from heads at different circadian times. Indeed, the abundance of SXE1 protein followed an oscillating pattern throughout the day (one-way ANOVA, P > 0.0001; Figure 2B) with peak time at ZT 23, corresponding to a 4-hr time shift of the mRNA peak.
We have previously shown that male-biased sxe1 expression is regulated by dsx (Fujii and Amrein 2002). We therefore determined the effects of the two DSX proteins on cycling of sxe1 mRNA at ZT 3 and ZT 19, the times of trough and peak, respectively, in XX and XY flies with various dsx alleles. Homo- and hemizygous dsx1 mutant flies (dsx1/dsx1 and dsx1/dsxDf), which show an intersexual (male and female characteristics) phenotype (Erdman and Burtis 1993), generate no functional DSX protein, regardless of their sex chromosomes; in contrast, the dsxD allele leads to TRA-independent splicing of dsxm mRNA (and production of DSXM protein), and therefore, X/X; dsx1/dsxD and X/Y; dsx1/dsxD flies are phenotypic male and express only DSXM, but not DSXF (Nagoshi and Baker 1990). As controls, we included XX and XY flies that contained one intact dsx allele (TM3/dsx1), which produce DSXF and DSXM and develop as normal females and males, respectively. RNA from all flies showed an increase of sxe1 mRNA from ZT 3 to ZT 19 in all genotypes, even in the absence of any DSX protein; however, only DSXM, but not DSXF leads to a significant increase in sxe1 mRNA at ZT 19 (Figure 2C; P < 0.05). Taken together, these experiments establish that the clock genes are necessary for cycling of sxe1 mRNA, and that DSXM is required for amplifying the increase of sxe1 expression at ZT 19.
Suppression of sxe1 decreases male courtship and mating success:
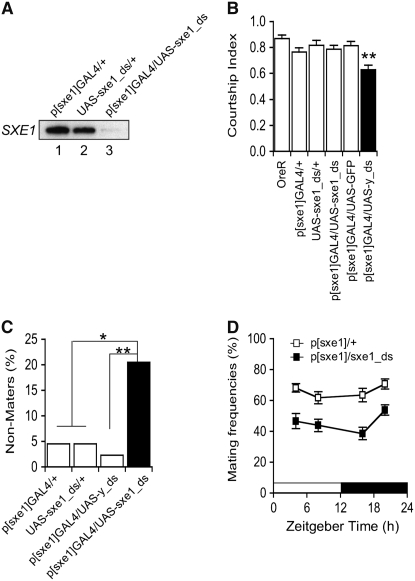
Male-specific expression of sxe1 in the adult suggests a role for this gene in male reproductive physiology and/or behavior. We therefore employed RNA interference (RNAi) to suppress SXE1 activity. We generated flies expressing double-stranded sxe1 mRNA under the control of the sxe1 promoter (sxe1-RNAi flies: p[sxe1]GAL4/UAS-sxe1_ds; for details see materials and methods) and performed both Northern and Western analyses. These experiments showed that both mRNA and protein expression levels were drastically reduced in sxe1-RNAi males, when compared to control males containing only the driver or the reporter (Figure 3A and data not shown). These flies are viable, healthy, and morphologically indistinguishable from control flies. Moreover, suppression of sxe1 in male flies did not affect locomotor activity (Table 1). To investigate whether sxe1 is involved in sexual behavior, we performed behavioral assays and determined the courtship index (CI) and mating success (see materials and methods). Indeed, both the CI (P < 0.001) and mating success (P < 0.05) was significantly reduced in sxe1-RNAi males compared to control males (Figure 3, B and C). Because SXE1 expression oscillates during the diurnal cycle (Figure 2, A and B) and because male courtship is also under circadian control (Hardeland 1972; Sakai and Ishida 2001; Tauber et al. 2003; Fujii et al. 2007), we wondered whether mating success was affected differently at various time points during the day and night. Thus, we measured the frequency of successful matings at ZT 4, 7, 16, and 19 among sexually naive males and females. Because visual cues may override subtle deficiencies in other sxe1 dependent sensory modalities, we performed these experiments under dim red light (<1 lux). Mating frequency of sxe1-RNAi males was significantly lower at all time points (Figure 3D). Two-way ANOVA revealed that there are significant differences between sxe1-RNAi males and control males (p[sxe1]GAL/+) in genotype (P < 0.0001) and time (P = 0.022), but not in the interaction of genotype and time (P = 0.72). Thus, our observations indicate that sxe1 is required throughout the day for high mating success and show that sxe1 is necessary in male courtship behavior and mating.
Figure 3.—
Suppression of sxe1 by RNAi decreases courtship and mating performance. (A) Western blot with anti-SXE1 of protein extracts from heads of two types of control males (lanes 1 and 2) and p[sxe1]GAL4/UAS-sxe1_ds males (RNAi males; lane 3). (B) Courtship index (CI) for control and RNAi males (n > 20 for OreR, UAS- p[sxe1]GAL4/UAS-y_ds, and p[sxe1]GAL4/UAS-GFP; n > 50 for p[sxe1]GAL4/+ and p[sxe1]GAL4/UAS-sxe1_ds). Error bars represent SEM. **P < 0.001 (Wilcoxon/Kruskal-Wallis tests, the value of p[sxe1]GAL4/UAS-sxe1_ds was compared with that of p[sxe1]GAL4/+). (C) Frequencies of nonmaters in single pair mating assay (n = 44 for each genotype; 30-min observation period). *P < 0.05 or **P < 0.01 (chi-square test) for values compared with p[sxe1]GAL4/UAS-sxe1_ds. (D) Mating frequencies at different ZT points. Five virgin males and five virgin females were placed in a vial and left for 15 min to copulate in the dark (<1 lux; n = 39–50 for each genotype and time point). Error bars represent SEM. Differences in genotype (P < 0.0001), time (P = 0.0223), and interaction of time and genotype (P = 0.7249) were analyzed by two-way ANOVA.
TABLE 1.
Suppression of sxe1 affected neither locomotor activity nor fertility
| Locomotiona (“no.” of events/4 min) | Tasteb | Life spanc (days) | Fertilityd | |
|---|---|---|---|---|
| Oregon-R | — | — | — | 69.3 ± 3.4 |
| p[sxe1]GAL4/+ | 39 ± 3.7 | — | — | — |
| UAS-sxe1_ds/+ | 37 ± 3.3 | — | — | — |
| p[sxe1]GAL4/UAS-y_ds | 34 ± 3.5 | — | — | — |
| p[sxe1]GAL4/UAS-sxe1_ds | 37 ± 3.2 | — | — | — |
| sxe167/+ | 38 ± 3.6 | 0.85 ± 0.06 | 48 ± 1.2 | 72.1 ± 3.3 |
| sxe167; rescue3/+ | 43 ± 2.6 | 0.86 ± 0.01 | 49 ± 1.9 | — |
| sxe167 | 40 ± 2.9 | 0.86 ± 0.01 | 48 ± 2.6 | 75.2 ± 2.7 |
Sxe1 is not required for locomotion, taste (sugar) perception, normal life span, or fertility.
Line crossing events were counted for 4 min (n = 14 for each genotype).
Taste preference was determined by using the two-choice preference assay for 25 mm trehalose vs. water (Thorne et al. 2004). Three independent experiments (n = 40/experiment) for each genotype were performed.
Life span indicates at which point 50% of flies have died. Three independent experiments per genotype were performed (n = 100/experiment).
A single male and a single w1118 virgin female were kept in a vial. After 4 days, they were removed and flies were counted after all progeny had hatched. The number indicates the average progeny per vial. The number of single crosses for each genotype was between 20 and 26.
Sxe1 mutant males show reduced mating efficiency:
Although RNAi is an effective tool for investigating gene function, downregulation is often incomplete; indeed SXE1 protein is not entirely depleted in sxe1 RNAi males (Figure 3A). Conversely, RNAi can cause off target effects, especially if the target gene belongs to a large gene family that has extensive DNA homology, as do many cyp genes. Thus, we generated a null mutation for sxe1 to verify the phenotypes observed with RNAi using homologous recombination (Gong and Golic 2003). Mapping and PCR analysis indicated that two potential sxe1 mutations were obtained (Figure 4A and data not shown), and Southern analysis indicated that one of these two alleles (sxe167) represented a precise recombination event in which the first 18 codons of the sxe1 gene were replaced by the mini-white gene (Figure 4B and materials and methods). To investigate whether sxe167 was a true null allele, we performed Northern and Western analyses, which revealed that neither RNA nor protein is produced in homozygous mutant males (Figure 4, C and D). We then performed the same behavioral experiments on wild-type, sxe167 homozygous and heterozygous males as we did on sxe1-RNAi males. While no effect on the courtship index of sxe167 homozygous mutant males compared to control males was apparent (data not shown), we observed a significant increase in the fraction sxe167 homozygous mutant nonmaters, similar to sxe1-RNAi males (compare Figures 4E to 3C). Moreover, this phenotype was displayed throughout the diurnal cycle as in sxe1-RNAi males (compare Figures 4F and 3D). Significantly, these phenotypes were rescued when a sxe1 transgene was introduced into a sxe167 homozygous mutant background (Figure 4, E and F). Two-way ANOVA indicates that there are significant differences in genotype (P < 0.0001) and time (P = 0.0036), but not in the interaction of genotype and time (P = 0.9068), between homozygous (sxe167/sxe167) and heterozygous (sxe167/+) males. Genotype (P = 0.0003) and time (P = 0.0008), but not the interaction of genotype and time (P = 0.5158) are also significantly different between homozygous (sxe167/sxe167) males and the same males carrying a rescue transgene (sxe167/sxe167;Rescue3/+). We tested several other behaviors and properties, such as locomotion, taste preference, longevity, and fertility; none of them were affected in sxe167 homozygous mutant males (Table 1). In summary, the genetic analysis of males lacking a functional sxe1 gene confirmed the mating phenotypes observed with sxe1 RNAi, and demonstrate that sxe1 is necessary in males for efficient mating.
Quantitative analysis of lipids and fatty acids in the head of sxe1 mutant male:
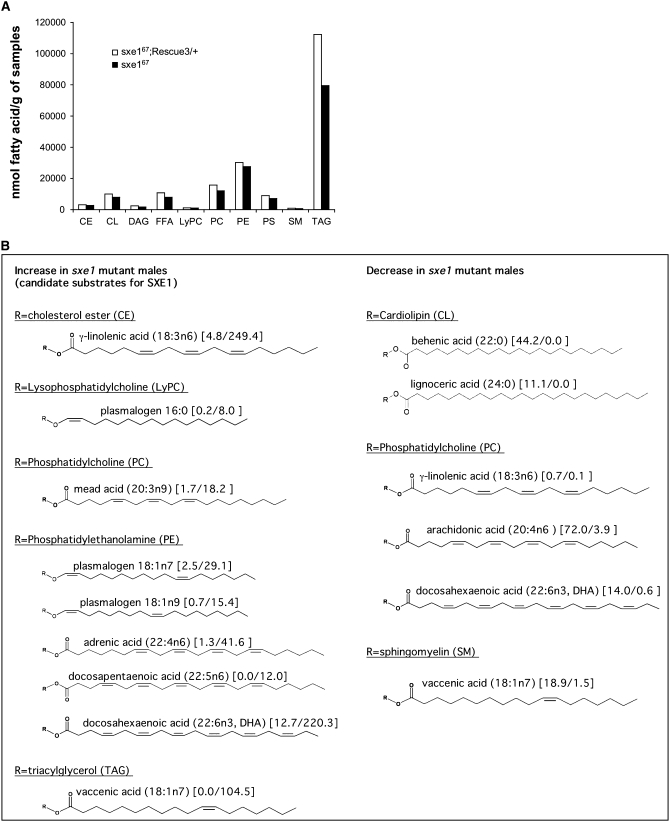
The cytochrome p450 enzymes can be subdivided into several protein subfamilies. In Drosophila, the CYP4 subfamily is one of the largest and includes 22 members, and several insect and mammalian CYP4 enzymes were shown to function as ω-hydroxylases. For example, human CYP4A11 and CYP4F2 are fatty acid ω-hydroxylases for lauric acids (Powell et al. 1996) and leukotriene B4, a potent mediator of inflammation (Kikuta et al. 1993), respectively. In Drosophila, CYP4G1 functions as an ω-hydroxylase for regulating triacylglycerol (TAG) levels in oenocytes (Gutierrez et al. 2007). To test whether SXE1/CYP4D21 may have a role in fatty acid metabolism, we analyzed and quantified lipids in the head of flies in the presence and absence of a functional sxe1 gene (Figure 5 and supplemental Table 1). Overall, the concentration of all lipid classes is similar, regardless of whether a functional sxe1 gene is present or not, with the possible exception of TAGs (Figure 5A). Interestingly, however, the concentration of plasmalogen 18:1n7, plasmalogen 18:1n9, adrenic acid (22:4n6), docosapentaenoic acid (22:5n6), docosahexaenoic acid (DHA, 22:6n3) of phosphatidylethanolamine (PE) are dramatically elevated in the head of sxe1 mutant males (Figure 5B). In addition, the concentrations of γ-linolenic acid (18:3n6) of cholesterol ester (CE), plasmalogen 16:0 of lysophosphatidylcholine (LyPC), mead acid (20:3n9) of phosphatidylcholine (PC), and vaccenic acid (18:1n7) of TAG are elevated, whereas the concentrations of two saturated fatty acids, behenic acid (22:0) and lignoceric acid (24:0) of cardiolipin (CL), γ-linolenic acid (18:3n6), adrenic acid (22:4n6), and DHA (22:6n3) of phosphatidylcholine (PC), and vaccenic acid (18:1n7) of sphingomyelin (SM) are reduced in the head of sxe1 mutant males (Figure 5B and supplemental Table 1). These results strongly suggest that SXE1 is involved in fatty acid metabolism of a substantial number of lipids, presumably as an ω-hydroxylase.
Figure 5.—
The concentration of all lipid classes in the head of sxe1 mutant and control males is similar. Quantitative lipid concentration in the head of files with (sxe167/sxe167;Rescue3/+) and without (sxe167/sxe167) a functional sxe1 gene. (A) No large differences were observed for any of the various lipid classes, with the possible exception of triacylglycerol. CE, cholesterol ester; CL, cardiolipin; DAG, diacylglycerol; FFA, free fatty acid; LyPC, lysophosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; SM, sphingomyelin; and TAG, triacylglycerol. (B) Some specific compounds are drastically increased or reduced between the two genotypes (for detail, see supplemental Table 1). All chemical structures are referred from http://www.lipomics.com/fatty_acids. Values in brackets indicate nanomoles of fatty acid per gram of sample in the heads of sxe167/sxe167; Rescue 3/+ and sxe167/sxe167mutant males, respectively.
DISCUSSION
Hundreds of putative, sex-specific and sex-biased effector genes thought to be collectively responsible for sex-specific differentiation during development and sex-specific physiological processes in males and females have been identified in several large-scale expression studies (Arbeitman et al. 2002, 2004; Dauwalder et al. 2002; Fujii and Amrein 2002; Parisi et al. 2004). The main criteria of a sex-specific effector gene are (i) sex-specific or -biased expression outside the germ line, (ii) regulation by the sex determination genes and (iii) functional requirement in a sex-specific process in males or females. Despite the relatively large number of putative effector genes, virtually none of these genes have been studied in any detail. A main difficulty in uncovering their function is probably the subtle phenotypes that animals mutant for any of these genes display. One effector gene that has been functionally characterized is to, which is expressed in fat cells of the head in males and which was shown to be necessary for male sexual behavior (Lazareva et al. 2007). Fat cells are known to have endocrine roles, and TO, which is a member of the odorant/pheromone binding protein family is thought to be secreted from these cells and may function as a neuromodulator (Lazareva et al. 2007). Interestingly, fat cells in the adult fly are known to express several other putative sex-specific/biased effector genes, including the yolk protein (yp) genes and female-specific independent of transformer (fit) in females and turn on sex-specificity (tsx) in males, which also encodes a odorant/pheromone binding protein (Fujii and Amrein 2002).
Sxe1 is only the second male-specific effector gene with a role in male sexual behavior. Expression studies revealed that SXE1 protein is found in nonneuronal cells associated with chemo- and mechanosensory bristles of the head and thorax in males. Albeit SXE1 is also detected in the labial palps of females, both Northern and Western analysis revealed that the RNA/protein levels are severely reduced when compared to males. We also showed that both sxe1 mRNA and protein oscillate in males during the day, with peak values at night and separated by ∼4 hr.
While sxe1 is necessary for efficient reproductive behavior of the male, demonstrated both in our RNAi studies as well as with the sxe1 mutant fly strain, only sxe1 RNAi males, but not sxe1 homozygous mutant males, exhibited a significant decrease of the CI. The most likely explanation for this difference is that RNAi of sxe1 causes off-target effects and downregulates other members of the many and highly conserved cytochrome p450 genes. The observation that sxe1 mutant males show relatively normal levels of courtship, yet copulate less efficiently than control males suggests that they are slow maters. For example, it is possible that sxe1 is necessary for aggressive pursuit of the female, when she exhibits coyness toward the end of courtship. However, once sxe1 mutant males have copulated, they produce similar numbers of progeny as control males (Table 1), further supporting the notion that sxe1 is indeed a behavioral, but not a fertility gene.
How does SXE1 function in male mating success, given its nonneural expression in sensory bristles? To address this, we reflected on the biochemical role of this enzyme. While the specific chemical reaction carried out by SXE1 remains to be characterized, the lack of SXE1 activity has severe consequences on the abundance of various metabolites in the male's head (Figure 5B and supplemental Table 1). Like other CYP4 enzymes, SXE1 may modify arachidonic acid derivatives such as leukotriene B4 and prostaglandins. Strikingly, sxe1 mutant males accumulate plasmalogens (fatty acids with the dm prefix in supplemental Table 1) in their heads. Plasmalogens serve as a starting point for the release of arachidonic acid by phospholipase A2 (PLA2) and the synthesis of signaling molecules derived from arachidonic acid such as prostaglandins in mammals (Farooqui et al. 1995). In sxe1 mutant males, accumulation of a SXE1 substrate may decrease PLA2 activity and prevent release of arachidonic acid from plasmalogens, thereby increasing the concentration of plasmalogens. Given the SXE1-dependent metabolites are small and soluble, they can easily reach other cells, not only within the vicinity of their production, but possibly in remote regions in the head and beyond via transport through the hemolymph. Thus, we propose that SXE1 expressed in sensory bristles acts in an endocrine fashion via a diffusible metabolite on cells within the sensilla or possibly on neurons located elsewhere, including the brain.
Expression of SXE1 (and TO; see above) in nonneuronal cells of tissues not associated with sex-specific behaviors is intriguing, especially in light of the fact that several other sex-specific/biased genes have been found to be expressed in the same tissues. For example, the three yp genes and fit were all shown to be expressed in the fat cells in the female head, while tsx was found to be expressed in the same cells of males, and yet another gene, sxe2 is expressed in nonneuronal cells in the head of males in a fashion similar to sxe1 (Fujii and Amrein 2002) (S. Fujii and H. Amrein, unpublished data). A common feature of all these genes is that they either encode secreted proteins (TO, TSX, YP1, 2, and 3) or enzymes associated with the production of small metabolites (SXE1 and SXE2); hence, we propose that these tissues have endocrine properties and may be the source of various, hormone-like molecules involved in sexual behavior and other sex-specific physiological processes by acting on remote cells or neurons in the CNS.
Interestingly, sxe1 and a few other sex-specific/biased genes (to and tsx), are regulated by the clock genes (Dauwalder et al. 2002; Fujii and Amrein 2002), and both to and sxe1 expression oscillate in diurnal fashion. Therefore, one may expect that mating success in sxe1 mutant males may be affected differently at various Zeitgeber times; instead, we observed a reduction throughout the day. Similarly, no circadian-dependent courtship phenotype has been reported in to mutant males, albeit a second function associated with this gene, food intake, has been reported to exhibit a circadian phenotype in to mutants. Regardless, the functional relevance of sxe1 oscillation remains to be investigated in more detail in the future.
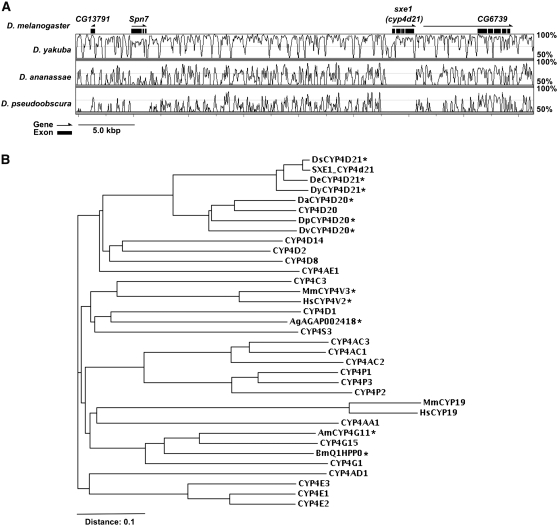
At last, we note that sxe1 is a gene that emerged recently in evolution. Comparison of the genome sequence of various Drosophila species revealed that sxe1 orthologs are only found in members of the melanogaster subgroup (Figure 6, A and B), which include D. simulans, D. melanogaster, D. erecta, D. sechellia, and D. yakuba. Drosophila species of the melanogaster group (D. ananassae) or the obscura group (D. pseudoobscura) lack sxe1 in the syntenic region or elsewhere in the genome (Figure 6, A and B), as do more distant diptera such as the housefly or mosquitos. This suggests that the function of sxe1 may be limited to a few, closely related Drosophila species, perhaps as a gene involved in speciation. However, many other sex-specific/biased effector genes such as tsx (obp99b), fit, sxe2, to, and the yp genes are conserved in all Drosophila species for which sequence data are available (data not shown).
Figure 6.—
The sxe1 gene is conserved only in the melanogaster subgroup, but not in other Drosophila species. (A) The sxe1 region (chr2L: 7,576,849–7,618,455) on the D. melanogaster genome (April 2004, the BDGP, Release 4), D. yakuba genome (April 2004, the Washington University School of Medicine, St. Louis, Release 1.0, SLAGAN), D. ananassae genome (July 2004, the Institute for Genomic Research, SLAGAN), and D. pseudoobscura genome (July 2003, the Human Genome Sequencing Center at Baylor College of Medicine, Freeze 1, SLAGAN) displayed by VISTA Browser (version 2.2.31 at http://pipeline.lbl.gov/cgi-bin/gateway2). Plots for D. yakuba/D. melanogaster (top), D. ananassae/D. melanogaster (middle), and D. pseudoobscura/D. melanogaster (bottom) are displayed on the scale of D. melanogaster sequence. Conserved regions above the level of 50% in a 100-bp window are plotted by the curve. (B) Neighbor-joining phylogenetic tree of CYP4-type chytochrome P450 in D. melanogaster with some SXE1 homologs in other Drosophila species. Proteins were aligned using ClustalW (EMBL-EBI, http://www.ebi.ac.uk) with default parameters. Prefixes Ds, De, Dy, Da, Dp, Dv, Mm, Hs, Ag, Am, and Bm indicate D. simulans, D. erecta, D. yakuba, D. ananassae, D. pseudoobscura, D. virilis, Mus musculus (mouse), Homo sapiens (human), Anopheles gambiae (mosquito), Apis mellifera (honey bee), and Bombyx mori (silkworm), respectively. D. melanogaster, D. simulans, D. erecta, and D. yakuba are melanogaster subgroup species of the genus Drosophila. Asterisks indicate the most similar protein of SXE1 in each species determined by tblastn (FlyBase and NCBI).
Acknowledgments
We thank Jeff Hall for providing ClkJRK, cyc01, per01, tim 01, Rodolfo Costa for UAS-y_ds, and Kent Golic for w1118; 70FLP, 70I-SceI, Sco/CyO and w1118;70FLP. This work was supported by grants from National Institutes of Health (DC-005606) and National Science Foundation (IBN-03-49671) to H.A.
References
- Arbeitman, M. N., A. A. Fleming, M. L. Siegal, B. H. Null and B. S. Baker, 2004. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131 2007–2021. [DOI] [PubMed] [Google Scholar]
- Arbeitman, M. N., E. E. Furlong, F. Imam, E. Johnson, B. H. Null et al., 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297 2270–2275. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Bray, S., and H. Amrein, 2003. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron 39 1019–1029. [DOI] [PubMed] [Google Scholar]
- Burtis, K. C., and B. S. Baker, 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56 997–1010. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang, A., H. Wijnen, F. Naef, C. Boothroyd, N. Rajewsky et al., 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32 657–671. [DOI] [PubMed] [Google Scholar]
- Coschigano, K. T., and P. C. Wensink, 1993. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 7 42–54. [DOI] [PubMed] [Google Scholar]
- Dauwalder, B., S. Tsujimoto, J. Moss and W. Mattox, 2002. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 16 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir, E., and B. J. Dickson, 2005. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121 785–794. [DOI] [PubMed] [Google Scholar]
- Dunipace, L., S. Meister, C. McNealy and H. Amrein, 2001. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 11 822–835. [DOI] [PubMed] [Google Scholar]
- Erdman, S. E., and K. C. Burtis, 1993. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui, A. A., H. C. Yang and L. A. Horrocks, 1995. Plasmalogens, phospholipases A2 and signal transduction. Brain Res. Brain Res. Rev. 21 152–161. [DOI] [PubMed] [Google Scholar]
- Fujii, S., and H. Amrein, 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 21 5353–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S., P. Krishnan, P. Hardin and H. Amrein, 2007. Nocturnal male sex drive in Drosophila. Curr. Biol. 17 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, E., D. Wiggins, B. Fielding and A. P. Gould, 2007. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445 275–280. [DOI] [PubMed] [Google Scholar]
- Hall, J. C., 1994. The mating of a fly. Science 264 1702–1714. [DOI] [PubMed] [Google Scholar]
- Hardeland, R., 1972. Species differences in the diurnal rhythmicity of courtship behaviour within the melanogaster group of the genus Drosophila. Anim. Behav. 20 170–174. [DOI] [PubMed] [Google Scholar]
- Hardin, P. E., 2005. The circadian timekeeping system of Drosophila. Curr. Biol. 15 R714–R722. [DOI] [PubMed] [Google Scholar]
- Ito, H., K. Fujitani, K. Usui, K. Shimizu-Nishikawa, S. Tanaka et al., 1996. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. USA 93 9687–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta, Y., E. Kusunose, K. Endo, S. Yamamoto, K. Sogawa et al., 1993. A novel form of cytochrome P-450 family 4 in human polymorphonuclear leukocytes. cDNA cloning and expression of leukotriene B4 omega-hydroxylase. J. Biol. Chem. 268 9376–9380. [PubMed] [Google Scholar]
- Lazareva, A. A., G. Roman, W. Mattox, P. E. Hardin and B. Dauwalder, 2007. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 3 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., M. Han, B. Shimada, L. Wang, T. M. Gibler et al., 2002. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99 9562–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, M. J., and M. Rosbash, 2001. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107 567–578. [DOI] [PubMed] [Google Scholar]
- Nagoshi, R. N., and B. S. Baker, 1990. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 4 89–97. [DOI] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, P. Edwards, J. Minor, D. Naiman et al., 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5 R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, P. K., I. Wolf and J. M. Lasker, 1996. Identification of CYP4A11 as the major lauric acid omega-hydroxylase in human liver microsomes. Arch. Biochem. Biophys. 335 219–226. [DOI] [PubMed] [Google Scholar]
- Ryner, L. C., S. F. Goodwin, D. H. Castrillon, A. Anand, A. Villella et al., 1996. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87 1079–1089. [DOI] [PubMed] [Google Scholar]
- Sakai, T., and N. Ishida, 2001. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc. Natl. Acad. Sci. USA 98 9221–9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov-Blat, L., W. V. So, L. Liu and M. Rosbash, 2000. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 101 647–656. [DOI] [PubMed] [Google Scholar]
- Shirangi, T. R., B. J. Taylor and M. McKeown, 2006. A double-switch system regulates male courtship behavior in male and female Drosophila melanogaster. Nat. Genet. 38 1435–1439. [DOI] [PubMed] [Google Scholar]
- Simpson, A. E., 1997. The cytochrome P450 4 (CYP4) family. Gen. Pharmacol. 28 351–359. [DOI] [PubMed] [Google Scholar]
- Tauber, E., H. Roe, R. Costa, J. M. Hennessy and C. P. Kyriacou, 2003. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr. Biol. 13 140–145. [DOI] [PubMed] [Google Scholar]
- Thorne, N., C. Chromey, S. Bray and H. Amrein, 2004. Taste perception and coding in Drosophila. Curr. Biol. 14 1065–1079. [DOI] [PubMed] [Google Scholar]