Abstract
Unlike animals, whose gametes are direct products of meiosis, plant meiotic products undergo additional rounds of mitosis, developing into multicellular haploid gametophytes that produce egg or sperm cells. The complex development of gametophytes requires extensive expression of the genome, with DNA-dependent RNA polymerases I, II, and III being the key enzymes for nuclear gene expression. We show that loss-of-function mutations in genes encoding key subunits of RNA polymerases I, II, or III are not transmitted maternally due to the failure of female megaspores to complete the three rounds of mitosis required for the development of mature gametophytes. However, male microspores bearing defective polymerase alleles develop into mature gametophytes (pollen) that germinate, grow pollen tubes, fertilize wild-type female gametophytes, and transmit the mutant genes to the next generation at moderate frequency. These results indicate that female gametophytes are autonomous with regard to gene expression, relying on transcription machinery encoded by their haploid nuclei. By contrast, male gametophytes make extensive use of transcription machinery that is synthesized by the diploid parent plant (sporophyte) and persists in mature pollen. As a result, the expected stringent selection against nonfunctional essential genes in the haploid state occurs in the female lineage but is relaxed in the male lineage.
IN flowering plants, three rounds of postmeiotic mitosis and development give rise to an eight-nucleate female gametophyte, one cell of which is the egg cell (Schneitz et al. 1995; Grossniklaus and Schneitz 1998; Drews and Yadegari 2002). Pollen, the male gametophyte, consists of three haploid cells, two of which are sperm cells. The three pollen cells are clonally related and are all descended from a single haploid meiotic product of a pollen mother cell (McCormick 1993, 2004). The male gametophyte can survive independent of the sporophyte (the parent plant) and upon landing on a receptive flower, the pollen germinates and develops a pollen tube that elongates through the transmitting tract of the pistil, the female floral organ, to reach the ovary. Within the ovary, the pollen tube grows toward chemical signals emanating from the two synergid cells of the female gametophyte (Higashiyama 2002; Higashiyama et al. 2001, 2003; Johnson and Preuss 2002). Upon reaching a synergid cell, adjacent to the egg, the pollen tube ruptures, releasing the sperm. One sperm cell fuses with the egg to give rise to the diploid embryo. The second sperm cell fuses with the female gametophyte's central cell, giving rise to the endosperm. Proper development of both embryo and endosperm as a result of double fertilization is required for seed maturation (Russell 1993; Grossniklaus and Schneitz 1998; Yadegari et al. 2000).
Large-scale analyses of cDNA libraries generated from mRNAs purified from maize and wheat female gametophytes have shown that thousands of genes are expressed in female gametophytes (Sprunck et al. 2005; Yang et al. 2006). Comparative microarray-based transcript profiling analyses using ovules of Arabidopsis wild-type plants and mutants lacking embryo sacs have similarly identified large numbers of female gametophyte-specific genes (Yu et al. 2005; Johnston et al. 2007; Jones-Rhoades et al. 2007; Steffen et al. 2007). Collectively, expression-profiling studies combined with analyses of female gametophytic mutants (Pagnussat et al. 2005) provide evidence for extensive transcriptional regulatory networks that are critical for the proper development of female gametophytes.
In Arabidopsis, ∼62% of all genes in the genome are expressed during at least one stage of male gametophyte development, with ∼10% of these transcripts being pollen specific (Honys and Twell 2003, 2004). Moreover, labeled UTP is incorporated into RNA in pollen and the transcription inhibitor, actinomycin D inhibits pollen tube growth (Mascarenhas 1989, 1993; Honys and Twell 2004). These observations indicate that male gametophytes are actively engaged in the transcription of their haploid genomes.
The enzymes central to nuclear gene expression are DNA-dependent RNA polymerases I, II, and III (Pol I, Pol II, and Pol III), each of which is composed of between 12 and 17 subunits. Pol I is responsible for transcribing the 45S preribosomal RNAs (rRNAs) that are then processed into the 18S, 5.8S, and 25–28S (the latter size depends on the species) rRNAs that form the catalytic core of ribosomes. Pol II transcribes messenger RNAs (mRNAs) as well as RNAs that do not encode proteins, such as micro RNAs and small nuclear RNAs that guide mRNA and rRNA processing events. Pol III is primarily responsible for transcribing transfer RNAs (tRNAs) and repetitive 5S rRNA genes (Kassavetis et al. 1994; Paule and White 2000).
For purposes of gene and subunit nomenclature, Arabidopsis Pol I is denoted as nuclear RNA polymerase A (NRPA), Pol II is denoted as NRPB, and Pol III is denoted as NRPC. Their second-largest subunits, denoted as NRPA2, NRPB2, and NRPC2, respectively, are homologs of the β-subunits of eubacterial RNA polymerase. Together with the largest subunits, the β-like second-largest subunits help form the active sites of the enzymes and are essential for RNA synthesis. In Arabidopsis thaliana, the Pol I, Pol II, and Pol III second-largest subunits are encoded by single-copy genes located on chromosomes 1, 4, and 5, respectively (Larkin and Guilfoyle 1993; Onodera et al. 2005); see also phylogenetic analyses by Craig S. Pikaard and Jonathan Eisen discussed in Arabidopsis Genome Initiative (2000).
Contrary to our expectation that loss-of-function mutations in NRPA2, NRPB2, or NRPC2 genes would be unrecoverable due to lethality in both the haploid male and female gametophytes, transgenic lines hemizygous for T-DNA disruptions of each gene can be identified and maintained. Detailed analysis of these lines revealed that the mutant RNA polymerase alleles are not transmitted through the female lineage due to the failure of mutant female gametophytes to complete their development. By contrast, the mutant alleles are transmitted to subsequent generations through the male gametophyte at moderate efficiency compared to wild type. Our data indicate that pollen can develop to maturity, grow pollen tubes, and carry out fertilization in the absence of functional RNA polymerase genes, apparently by utilizing transcription machinery synthesized premeiotically in pollen mother cells. By contrast, female gametophyte development is autonomous and requires transcription machinery generated de novo in the haploid state.
MATERIALS AND METHODS
Plant strains and growth conditions:
Arabidopsis thaliana wild-type and T-DNA insertion mutants (ecotype Columbia in both cases) were grown at 22° with a 16-hr photoperiod. Gene locus identifiers for NRPA2, NRPB2, and NRPC2 are At1g29940, At4g21710, and At5g45140, respectively. The T-DNA insertion alleles we named nrpa2-1, nrpa2-2, nrpb2-1, and nrpb2-2 are carried within Torrey Mesa Research Institute (San Diego) transgenic lines: GARLIC_726_H01, GARLIC_918_C10, GARLIC_859_B04, and GARLIC_110_G08, respectively [GARLIC is the former name of the Syngenta Biotechnology's SAIL collection of T-DNA lines, available from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University]. The parental line for GARLIC_110_G08 was homozygous for the qrt1-2 allele of the QUARTET gene (ecotype Columbia) (Preuss et al. 1994); other GARLIC lines are wild type at the QRT locus. The T-DNA allele nrpc2-1 is present in Salk line 007865 (Alonso et al. 2003) obtained from the ABRC. Seeds of plants bearing the nrpc2-2 (GABI_131_B09) allele were obtained from GABI-Kat (Rosso et al. 2003). The transgenic Arabidopsis line (SAIL _100_H07) carrying a LAT52∷GUS reporter gene(s) inserted in an intergenic region was obtained from ABRC.
Genotyping:
To identify T-DNA disrupted alleles in segregating families, PCR was carried out using primers complementary to the T-DNA left border (5′-GCATCTGAATTTCATAACCAATCTC-3′, 5′-CGTCCGCAATGTGTTATTAAG-3′, or 5′-CCCATTTGGACGTGAATGTAGACAC-3′) and primers specific for NRPA2 (5′-AGAGAGGTAGAGAAACTCACG-3′ or 5′-ATAAACAGTTAGGCAAGCGAA-3′), NRPB2 (5′-CGATTTGAGCTTCTACCGTTT-3′ or 5′-CCTAGAACATACCATGCGAAA-3′) or NRPC2 (5′-CTCGCACAATGAAGGATGTTT-3′ or 5′-TAATTCTTGCCGCAAATTGAC-3′). Wild-type alleles of NRPA2, NRPB2, and NRPC2 were identified using the gene-specific primers above in combination with 5′-GATGAGTTGGATAACACGAAC-3′ or 5′-AGCACCCTTTAAGCTACAAAG-3′ for NRPA2; 5′-CCATCAGACTCTGTCATCATA-3′ or 5′-ACGAAGGGTAAGCATGCAGTT-3′ for NRPB2; and 5′-AGCTACTCCAGGGGAGATTAT-3′ or 5′-GGCAAGTACTATAGCCCCCTG-3′ for NRPC2.
The unique genomic DNA/T-DNA junction sequences at both ends of the single T-DNA loci in nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 alleles were amplified by PCR and verified by sequencing.
Production of transgenic plants:
Genomic sequences for NRPA2 (positions −1433 to +7346 relative to the translation start site), NRPB2 (positions −338 to +6514), or NRPC2 (positions −1947 to +10295) were amplified by PCR. Amplified gene sequences included promoter regions and all introns and exons. Resulting PCR products were captured in pENTR/D-TOPO and recombined into the Gateway recombination (Invitrogen)-compatible expression vector pEarleyGate 302 (Earley et al. 2006). Resulting NRPA2, NRPB2, or NRPC2 full-length transgenes were introduced into hemizygous plants bearing a corresponding mutant allele (+/nrpa2-1, +/nrpb2-1, or +/nrpc2-1). Progeny of transgenic plants that were homozygous for the nrpa2-1, nrpb2-1, or nrpc2-1 mutations and were rescued by the full-length transgenes were identified by PCR genotyping.
Confocal laser scanning microscopy:
Examination of specimens was carried out using a Zeiss LSM confocal microscope system equipped with a Helium/Neon laser. Images were processed using Adobe Photoshop 7.0 software. Floral stages were defined according to Bowman (1994). Developmental stages of female gametophytes were defined according to Christensen et al. (1997).
Cytological and histochemical analysis of pollen:
In vitro pollen germination was carried out as described by Hashida et al. (2007). Pollen were stained with 1 μg/ml DAPI in 20 mm Tris-HCl pH 7.65, 0.5 mm EDTA, 1.2 mm spermidine, 7 mm 2-mercaptoethanol, 0.4 mm phenylmethylsulfonyl fluoride, 0.1 mg/ml FDA in 0.5 m sucrose, or Alexander solution (ftp://ftp.arabidopsis.org/home/tair/Protocols/EMBOmanual/ch1.pdf). Pollen and self-pollinated pistils were incubated at 37° for 12 hr in GUS staining solution (50 mm sodium phosphate pH 7.2, 0.2% Triton X-100, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, and 1 mg/ml X-Gluc).
RESULTS
Sex-biased defects in the transmission of mutant alleles encoding RNA polymerase I, II, and III second-largest subunits:
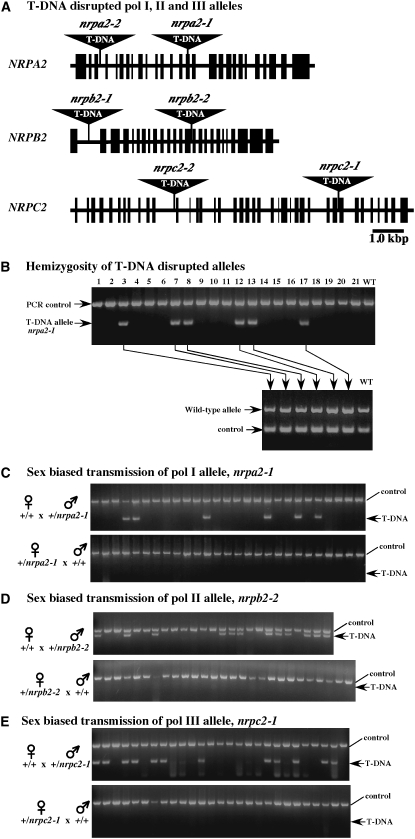
We used a PCR-based strategy to verify the existence of T-DNA-disrupted alleles for the catalytic second-largest subunits of RNA polymerase I (alleles nrpa2-1 and nrpa2-2), RNA polymerase II (alleles nrpb2-1 and nrpb2-2), or RNA polymerase III (alleles nrpc2-1 and nrpc2-2) (Figure 1A). We then genotyped the progeny resulting from self-fertilization of plants bearing these alleles. In all cases, individuals that carried a mutant RNA polymerase allele also carried a corresponding wild-type allele (Figure 1B and data not shown), indicating that these plants were hemizygous for the mutations. No plants homozygous for the Pol I (nrpa2-1, nrpa2-2), Pol II (nrpb2-1, nrpb2-2), or Pol III (nrpc2-1 or nrpc2-2) mutant alleles were recovered, indicating that the alleles are all severe loss-of-function mutations in essential genes, consistent with the essential roles of Pol I, Pol II, and Pol III in nuclear gene expression.
Figure 1.—
Sex-biased transmission of disrupted alleles for second-largest subunits of RNA polymerases I, II, and III (NRPA2, NRPB2, and NRPC2, respectively). (A) Structures of the NRPA2, NRPB2, and NRPC2 genes showing the positions of nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 T-DNA insertions. Solid boxes represent exons. (B) PCR-based genotyping of progeny of a self-fertilized +/nrpa2-1 hemizygote. Disrupted alleles were detected using a T-DNA-specific primer in conjunction with a gene-specific primer. Wild-type alleles were detected using primers that flank the T-DNA insertion site. (C–E) PCR-based detection of T-DNA disrupted alleles in progeny generated from reciprocal crosses between wild-type (+/+) and +/nrpa2-1, +/nrpb2-2, and +/nrpc2-1 hemizygotes.
Hemizygotes should outnumber homozygous wild-type siblings 67%:33% (2:1) among the progeny of a hemizygous parent bearing one copy of a defective essential gene, assuming that the homozygous mutant is inviable. However, as shown in Table 1, PCR-based genotyping revealed that only 8–38% of the progeny were hemizygous for Pol I (nrpa2-1 or nrpa2-2), Pol II (nrpb2-1 or nrpb2-2), or Pol III (nrpc2-1 or nrpc2-2) mutant alleles (Table 1). Instead, the majority of the progeny possessed only wild-type alleles, indicating a defect in the transmission of the mutant RNA polymerase alleles.
TABLE 1.
Genotypes of progeny of Pol I, II, and III hemizygotes
| Parental genotype | % homozygous wt (+/+) | % hemizygous (+/−) | % homozygous mutant |
|---|---|---|---|
| +/nrpa2-1 | 76 (62/82) | 24 (20/82) | 0 (0/82) |
| +/nrpa2-2 | 63 (32/51) | 37 (19/51) | 0 (0/51) |
| +/nrpb2-1 | 86 (18/21) | 14 (3/21) | 0 (0/21) |
| qrt1-2, +/nrpb2-2 | 80 (67/84) | 20 (17/84) | 0 (0/84) |
| +/nrpc2-1 | 62 (39/63) | 38 (24/63) | 0 (0/63) |
| +/nrpc2-2 | 92 (45/49) | 8 (4/49) | 0 (0/49) |
Mutant alleles nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 are underrepresented among the progeny of self-fertilized hemizygotes. Numbers in parentheses represent the number of individuals displaying a given genotype and the total number of individuals examined. wt, wild type.
To test for sex-biased defects in the transmission of the mutant alleles through the male or female gametophytes, Pol I hemizygotes (+/nrpa2-1 or +/nrpa2-2), Pol II hemizygotes (+/nrpb2-1 or +/nrpb2-2, qrt1-2; the latter is a Pol II mutant hemizygote in a homozygous quartet mutant background), or Pol III hemizygotes (+/nrpc2-1 or +/nrpc2-2) were reciprocally crossed with wild-type (+/+) plants by hand-pollinating emasculated flowers. Resulting progeny were then genotyped by PCR. None of the mutant polymerase alleles were found to be transmitted to the progeny via the maternal parent (Figure 1, C–E; Table 2); instead all progeny of hemizygous (+/−) female plants crossed with wild-type (+/+) males were homozygous wild type (+/+). By contrast, the nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 alleles were all pollen transmissible, such that 13–38% of the progeny inherited a mutant allele from the hemizygous paternal parent when crossed with a wild-type female (Table 2). Note, however, that equal numbers of hemizygous (+/−) and homozygous (+/+) progeny are expected from a (+/+) × (+/−) cross if the wild-type and mutant alleles are transmitted with equal efficiency; the male-transmitted Pol I, II, and III mutant alleles were not inherited at such high levels.
TABLE 2.
Male-specific transmission of Pol I, II, and III mutant alleles
| Parental genotype
|
Genotypes of progeny
|
||
|---|---|---|---|
| Female parent | Male parent | % homozygous wt (+/+) | % hemizygous (+/−) |
| +/nrpa2-1 | +/+ | 100 (55/55) | 0 (0/55) |
| +/nrpa2-2 | +/+ | 100 (46/46) | 0 (0/46) |
| +/nrpb2-1 | +/+ | 100 (52/52) | 0 (0/52) |
| qrt1-2, +/nrpb2-2 | +/+ | 100 (42/42) | 0 (0/42) |
| +/nrpc2-1 | +/+ | 100 (56/56) | 0 (0/56) |
| +/nrpc2-2 | +/+ | 100 (47/47) | 0 (0/47) |
| +/+ | +/nrpa2-1 | 75 (42/56) | 25 (14/56) |
| +/+ | +/nrpa2-2 | 62 (24/39) | 38 (15/39) |
| +/+ | +/nrpb2-1 | 79 (38/48) | 21 (10/48) |
| +/+ | qrt1-2, +/nrpb2-2 | 70 (19/27) | 30 (8/27) |
| +/+ | +/nrpc2-1 | 67 (36/54) | 33 (18/54) |
| +/+ | +/nrpc2-2 | 87 (45/52) | 13 (7/52) |
Paternally biased transmission of nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 alleles. Wild-type (+/+) plants were reciprocally crossed with +/nrpa2-1, +/nrpa2-2, +/nrpb2-1 (in qrt1-2 mutant background); +/nrpb2-2, +/nrpc2-1, and +/nrpc2-2 and resulting progeny were genotyped. Numbers in parentheses are the number of progeny displaying the specified genotype out of the total number of progeny examined.
The reciprocal crossing data summarized in Tables 1 and 2 indicate a lack of transmission of the mutant polymerase second-largest subunit alleles through female gametophytes and a partial defect in their transmission through the male gametophyte. Similar allele transmission behavior was observed for the RNA polymerase subunit mutant nrpb12a (supplemental Table S1). The homolog of NRPB12a in yeast is a single-copy gene whose encoded protein is incorporated into all three nuclear polymerases (Pol I, II, and III). As was the case for the second-largest subunit mutants, homozygous nrpb12a mutants were not recoverable. Moreover, nrpb12a mutant alleles were transmitted via pollen but not through the female gametophytes. Collectively, our results indicate that male-specific transmissibility of defective RNA polymerase alleles is a general characteristic of RNA polymerase subunit genes and not a peculiarity of second-largest subunit genes.
Defective RNA polymerase alleles cause female gametophyte developmental arrest:
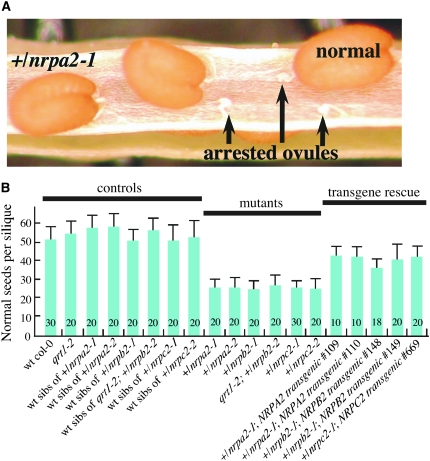
Lack of maternal transmission of the Pol I (nrpa2-1 or nrpa2-2), Pol II (nrpb2-1 or nrpb2-2), or Pol III (nrpc2-1 or nrpc2-2) alleles prompted an examination of siliques (seed pods) of self-pollinated hemizygous +/nrpa2-1, +/nrpa2-2, +/nrpb2-1, +/nrpc2-1, +/nrpc2-2, or +/nrpb2-2, qrt1-2 plants. Siliques of these plants contain small unfertilized ovules interspersed with an equal number of normal seeds; as an example, a silique from a +/nrpa2-1 plant is shown in Figure 2A. Whereas wild-type plants produce 51–58 seeds per silique, siliques of Pol I (nrpa2-1 or nrpa2-2), Pol II (nrpb2-1 or nrpb2-2), or Pol III (nrpc2-1 or nrpc2-2) mutant hemizygotes contain only 25–27 mature seeds (Figure 2B).
Figure 2.—
Failed seed development in siliques of nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 hemizygotes. (A) A silique of a hemizygous +/nrpa2-1 plant. Normal seeds and undeveloped (arrested) ovules occur in a silique of a hemizygous plant. (B) Average amounts of normal seeds per silique from wild-type and hemizygous plants. Numbers of siliques examined are indicated.
Defects in seed set caused by the polymerase mutations were rescued by transforming Pol I, Pol II, or Pol III hemizygotes with full-length NRPA2, NRPB2, or NRPC2 genomic clone transgenes expressed from their endogenous promoters (Figure 2B). Southern blot and segregation analyses showed that the transgenes in each case were integrated in multiple copies at a single locus (data not shown) such that the plants tested in Figure 2B were hemizygous for the polymerase mutant alleles as well as being hemizygous for the rescuing transgene loci. As a result, seed set is rescued by the transgenes to a level intermediate between the mutant and wild-type phenotypes. This is due to the independent segregation of the transgenes and polymerase alleles such that only half of the gametophytes bearing a mutant polymerase allele inherit a rescuing transgene. Collectively, our data indicate that functional RNA polymerases are essential for one or more critical aspects of female gametophyte development, fertilization, or seed development.
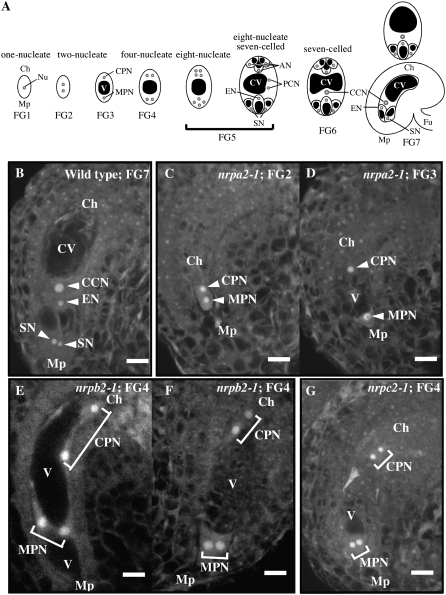
To further investigate the defects in ovule development and female transmission of mutant alleles (Figure 1, C–E; Table 2), ovaries of flowers at floral stage 13 (Bowman 1994), a stage just prior to flower opening, were examined by confocal laser scanning microscopy (CLSM). Female gametophytes develop relatively synchronously (Christensen et al. 1997) such that gametophytes that have undergone all three rounds of mitosis (female gametophyte stages FG5–FG7; see Figure 3A) are observed at floral stage 13 in wild-type pistils (Figure 3B and Table 3). By contrast, in floral stage 13 pistils of hemizygous plants segregating mutant alleles for Pol I (+/nrpa2-1 or +/nrpa2-2), Pol II (+/nrpb2-1 or +/nrpb2-2, qrt1-2), or Pol III (+/nrpc2-1 or +/nrpc2-2), ∼50% of the female gametophytes arrest after only one or two rounds of mitosis (2–4 nuclei), at developmental stages FG2–FG4 (Table 3, Figure 3, C–G, and supplemental Figure S1). The other ∼50% of the gametophytes in these ovaries display normal development, as in wild-type plants, consistent with the 1:1 segregation of wild-type and mutant alleles within the siliques of plants hemizygous for the mutations.
Figure 3.—
Developmental arrest of mutant female gametophytes in flowers just prior to anthesis was visualized by confocal fluorescence microscopy. (A) Stages of female gametophyte development (FG1–FG7), according to Christensen et al. (1997). Mp, micropylar pole; Ch, chalazal pole; Nu, nucleus; V, vacuole; CPN, chalazal pole nucleus; MPN, micropylar nucleus; AN, antipodal cell nucleus; CV, central cell vacuole; EN, egg cell nucleus; PCN, polar cell nucleus; CCN, central cell nucleus; SN, synergid cell nucleus; Fu, funiculus. (B) A wild-type female gametophyte, at floral stage 13, that is fully developed (FG7). The nuclei and vacuoles for the 2N central cell, the egg cell, and two synergid cells are apparent. (C and D) nrpa2-1 female gametophytes arrested at the two-nucleate stage (FG2 and FG3). (E and F) nrpb2-1 female gametophytes arrested at the four-nucleate stage. (G) A nrpc2-1 female gametophyte arrested at the four-nucleate stage. Scale bars, 10 μm.
TABLE 3.
Female gametophyte development in polymerase mutants
| No. of female gametophytes at specified developmental stages
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plant genotype | Pistil identification no. | FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 | Total |
| wt col-0 | 1 | 7 | 2 | 8 | 17 | ||||
| 2 | 1 | 2 | 11 | 14 | |||||
| 3 | 3 | 1 | 12 | 16 | |||||
| 4 | 1 | 14 | 15 | ||||||
| qrt1-2 | 1 | 1 | 12 | 13 | |||||
| 2 | 5 | 3 | 9 | 17 | |||||
| 3 | 13 | 13 | |||||||
| +/nrpa2-1 | 1 | 4 | 4 | 1 | 2 | 4 | 15 | ||
| 2 | 2 | 4 | 2 | 1 | 3 | 12 | |||
| 3 | 4 | 4 | 2 | 7 | 2 | 4 | 23 | ||
| 4 | 1 | 6 | 1 | 9 | 17 | ||||
| +/nrpa2-2 | 1 | 3 | 6 | 1 | 2 | 1 | 7 | 20 | |
| 2 | 2 | 7 | 1 | 4 | 1 | 1 | 16 | ||
| 3 | 1 | 4 | 1 | 7 | 3 | 1 | 17 | ||
| 4 | 1 | 6 | 4 | 2 | 1 | 7 | 21 | ||
| +/nrpb2-1 | 1 | 2 | 8 | 1 | 4 | 15 | |||
| 2 | 1 | 4 | 2 | 2 | 9 | ||||
| 3 | 3 | 3 | 1 | 1 | 7 | 15 | |||
| 4 | 9 | 2 | 8 | 19 | |||||
| 5 | 1 | 10 | 2 | 9 | 22 | ||||
| 6 | 4 | 10 | 8 | 22 | |||||
| qrt1-2, +/nrpb2-2 | 1 | 2 | 6 | 2 | 1 | 4 | 15 | ||
| 2 | 5 | 8 | 3 | 2 | 3 | 21 | |||
| 3 | 1 | 3 | 8 | 12 | |||||
| +/nrpc2-1 | 1 | 4 | 12 | 2 | 1 | 7 | 26 | ||
| 2 | 2 | 7 | 2 | 1 | 5 | 17 | |||
| 3 | 2 | 3 | 3 | 1 | 7 | 16 | |||
| 4 | 4 | 4 | 1 | 2 | 6 | 17 | |||
| +/nrpc2-2 | 1 | 3 | 10 | 1 | 3 | 1 | 6 | 24 | |
| 2 | 2 | 10 | 1 | 8 | 21 | ||||
| 3 | 3 | 6 | 1 | 3 | 6 | 19 | |||
| 4 | 1 | 8 | 1 | 9 | 19 | ||||
| 5 | 2 | 7 | 1 | 1 | 8 | 19 | |||
Developmentally arrested nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 female gametophytes. Pistils from flowers just prior to anthesis (flower opening) were fixed, and female gametophytes within these pistils were classified according to their developmental stage (FG1–FG7). wt, wild type.
Detailed examination of ovules within +/nrpa2-1 plants indicated that female gametophytes lacking functional Pol I arrest most frequently at the two-nucleus stage (FG2 and FG3; Figure 3, C and D and Table 3) and were not observed to progress beyond the four-nucleus stage. Similar results were observed for hemizygous plants bearing the nrpa2-2 Pol I mutant allele (Table 3 and supplemental Figure S1, A and B).
As shown in Table 3, Figure 3, E–G, and supplemental Figure S1, most of the nrpb2-1, nrpb2-2, and nrpc2-1 female gametophytes arrested after the second mitotic division (FG4), at the four-nucleus stage, whereas the majority of nrpc2-2 female gametophytes displayed developmental arrest at the two-nucleus stage (FG2 and FG3). The difference in the severity of the nrpc2-1 and nrpc2-2 alleles is presumably due to the relative locations of the T-DNA insertions, with the T-DNA in the stronger nrpc2-2 allele occurring in an earlier intron (see Figure 1).
Collectively, the microscopic analyses suggest that female gametophytes carrying defective alleles for RNA polymerases I, II, or III arrest early in development, at or prior to the four-nucleus stage, FG4.
Certation explains reduced male transmissibility of defective polymerase alleles:
As shown in Table 2 and Figure 1, C–E, nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 alleles are all transmitted via the male gametophyte. However, homozygous wild-type individuals outnumber hemizygous individuals among the progeny of self-fertilized hemizygotes or among the progeny of wild-type females outcrossed with a hemizygous male (Tables 1 and 2). These data indicate that male gametophytes bearing wild-type RNA polymerase alleles are either more viable or more successful at fertilization than are male gametophytes bearing mutant polymerase alleles.
To investigate the influence of defective RNA polymerase alleles on pollen development and viability using tetrad analysis, we generated lines that carry a Pol I (nrpa2-1), Pol II, (nrpb2-1), or Pol III (nrpc2-1) mutant allele in the quartet (qrt) mutant background. The quartet mutation causes the four pollen that develop from the four meiotic products (microspores) to remain associated with one another, rather than dissociating into individual pollen grains. Thus, pollen tetrads of plants hemizygous for the polymerase mutants include two pollen-bearing mutant polymerase alleles and two bearing wild-type polymerase alleles.
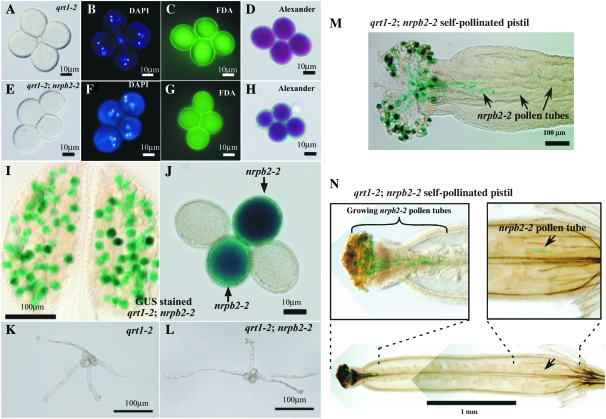
Pollen tetrads were examined by DAPI (4′,6-diamidino-2-phenylindole), FDA (fluorescein diacetate), or Alexander staining (Figure 4, A–H). DAPI staining of chromatin in pollen of quartet (qrt1-2) mutant plants; Pol I hemizygote quartet (+/nrpa2-1; qrt1-2), Pol II hemizygote quartet (+/nrpb2-1; qrt1-2 as well as +/nrpb2-2; qrt1-2), or Pol III hemizygote quartet (+/nrpc2-1; qrt1-2) plants revealed the normal pattern of one diffuse vegetative cell nucleus and two compact sperm cell nuclei in each of the four attached pollen (Figure 4, B and F, and data not shown). FDA and Alexander staining detected no differences in viability among the individual pollen in tetrads of wild-type or mutant plants (Figure 4, C, D, G, and H, and data not shown).
Figure 4.—
Development and early tube elongation of pollen are unaffected by defects in RNA polymerases. (A–H) Cytological examination of mature pollen from qrt1-2 (a–d) and qrt1-2; +/nrpb2-2 (e–h). (a and e) Bright-field microscopy; (b and f) DAPI staining test; (c and g) FDA staining test; (d and h) Alexander staining test. (I and J) LAT52∷GUS expression in pollen defective for the Pol II subunit (nrpb2-2 pollen). (K and L) Germinating qrt1-2 (k) and qrt1-2; nrpb2-2 (l) pollen. Pollen was incubated for 18 hr at 22 ° in a germination medium and its images were captured. Note that four tubes of quartet pollen from wild-type (k, qrt1-2) and mutant (l, qrt1-2; +/ nrpb2-2) plants grew equally in this assay, to a length of ∼100–150 mm. (M and N) Self-pollinated pistils from qrt1-2; +/ nrpb2-2 plants. LAT52∷GUS was expressed during pollen tube growth in the absence of the functional allele of a catalytic subunit of Pol II. A considerable number of nrpb2-2 pollen tubes (blue stained) was present in the top portions of the pistils. Note that a tube from nrpb2-2 pollen grew into ∼2.0 mm in length (N).
Two of the pollen in each tetrad of a polymerase mutant hemizygote carry defective RNA polymerase alleles and lack wild-type alleles. In the case of the nrpb2-2 hemizygotes, the mutant alleles are tagged by a LAT52∷GUS reporter gene that is present within the T-DNA inserted into the Pol II NRPB2 gene (Figure 4, I and J). The LAT52 promoter is specifically expressed in mature pollen and pollen tubes, thereby allowing the pollen bearing the mutant nrpb2-2 alleles to be visualized by GUS staining. Equal numbers of GUS-positive (blue) and GUS-negative pollen are present in nrpb2-2/+ pollen quartets, indicating that wild-type and mutant pollen develop in equal abundance and that the nrpb2-2 mutant allele segregates normally (Figure 4, I and J).
It is noteworthy that mRNA-encoded proteins, such as the GUS enzyme, are synthesized by RNA polymerase II and require the distinctive 5′7-methylguanosine caps and poly A tails of Pol II transcripts to be translated. Pol I and Pol III transcripts lack these features and are not translated. Despite the disruption of the gene encoding the essential Pol II second-largest subunit (NRPB2), the GUS enzyme is clearly expressed from the LAT52 promoter in nrpb2 mutant pollen (Figure 4, I and J). Expression of the GUS gene cannot be attributed to stored GUS mRNA transcribed premeiotically; if so, it would be present in all four pollen of the tetrad. Moreover, the LAT52 promoter has previously been shown to be expressed only postmeiotically, making it a useful male-gametophyte-specific marker (Eady et al. 1994; Twell et al. 1990). We conclude that Pol II transcription takes place in nrpb2-2 mutant pollen despite the lack of a functional NRPB2 allele.
Examination of pollen germination and pollen tube growth in vitro revealed no differences among pollen tubes that grew from pollen quartets consisting of two pollen-bearing defective RNA polymerase alleles and two pollen-bearing wild-type alleles, at least up to a pollen tube length of 100–150 μm (Figure 4, K and L, and data not shown). Self-pollinated pistils of qrt1-2; +/nrpb2-2 plants stained for GUS also reveal pollen tube growth from pollen bearing the disrupted allele in vivo (Figure 4, M and N). Most of the GUS-stained tubes from nrpb2-2 pollen are observed at the stigma and upper portions of the ovary (Figure 4, M and N; Figure 5C). However, in rare cases, tubes from nrpb2-2 pollen are observed in the distal portion of the ovary (Figure 4N, images at top right and bottom). Collectively, these observations suggest that in pollen that do not encode endogenous functional RNA polymerase II, Pol II-dependent GUS activity is sustained during pollen development and early pollen tube growth.
Figure 5.—
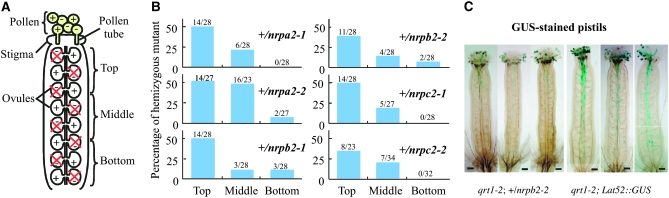
Reduced paternal transmission of nrpa2-1, nrpa2-2, nrpb2-1, nrpb2-2, nrpc2-1, and nrpc2-2 alleles relative to wild-type alleles in self-fertilized hemizygotes is due to decreased, competitive fertilization of ovules farthest from the stigma. (A) A diagram of the female floral organ (the pistil), whose surface (the stigma) is the site where a pollen grain germinates and initiates formation of a pollen tube. Half of the pollen of a hemizygote has wild-type (+) RNA polymerase alleles and half are mutant (−), but all develop and mature. Likewise, within the ovary of a hemizygote, half of the ovules are wild type and half are mutant with respect to the RNA polymerase alleles. However, the latter fail to develop (denoted with an “X”) such that mutant alleles in fertilized ovules and seeds are derived from the male gametophyte. (B) Seeds collected from the top, middle, and bottom portions of siliques of the hemizygotes were germinated and resultant plants were genotyped. The numbers of plants of each genotype are indicated. Note that mutant alleles are more abundant in seeds developing nearest the stigma, at the top of the siliques, where the shortest pollen tubes would be needed to reach the ovules. (C) Self-pollinated pistils from qrt1-2; +/ nrpb2-2 plants and transformants hemizygously carrying a LAT52∷GUS reporter gene(s) inserted in an intergenic region (qrt1-2; LAT52∷GUS). Pollen tubes from qrt1-2; nrpb2-2 pollen (blue stained) were present in top portions of the pistils, while control pollen tubes (qrt1-2; LAT52∷GUS) were observed all the way from the tops to the bottoms of the pistils.
To test the hypothesis that pollen bearing Pol I (nrpa2-1 or nrpa2-2), Pol II (nrpb2-1 or nrpb2-2), or Pol III (nrpc2-1 or nrpc2-2) mutant alleles are at a competitive disadvantage compared to wild-type pollen, we determined the distribution of seeds bearing mutant alleles within the siliques of self-pollinated hemizygous plants. Due to the previously demonstrated lethality of the 50% of female gametophytes that inherit a mutant polymerase allele (depicted as ovules with an “X” through them in Figure 5A), only the 50% of female gametophytes that bear wild-type alleles are available to be fertilized. Therefore, any mutant alleles detected in the seeds are inherited via the male gametophytes (refer to Figure 1, C–E, and Table 2). Seeds were collected from the top one-third of the silique, which is nearest to the stigma where the pollen germinates to initiate the growth of pollen tubes, or from the middle or bottom one-third of the silique. Following germination of the seeds, resulting plants were genotyped (Figure 5B). This test revealed that mutant alleles were found most frequently among seeds that developed within the top one-third of the siliques; 35–50% of these seeds develop as hemizygotes (note that a frequency of 50% is expected if there is no difference in the fitness of wild-type and mutant pollen). The frequency of hemizygous seeds within the middle portions of the siliques were significantly reduced (11–21%) in comparison with the top one-third, except for the nrpa2-2 allele that was detected in 16 of the 23 sibs examined. In the bottom one-third of the siliques, where fertilization of the ovules would require the growth of the longest pollen tubes, hemizygotes represented only a small proportion of the seeds (0–11%).
The extent of mutant pollen tube growth fits with the distribution of hemizygous seeds following fertilization. A nonmutant transgenic line in which a T-DNA bearing the LAT52∷GUS reporter gene inserted into an intergenic region was used as a control for comparison to nrpb2-2. Whereas GUS-stained nrpb2-2 pollen tubes are rarely observed deeper than the top one-third of the pistil, GUS-stained control pollen tubes are easily detected throughout the top and middle one-thirds of the pistils and can be observed all the way to the base of the pistil (Figure 5C). Taken together, our results suggest that pollen germination, early pollen tube elongation, and fertilization are not severely affected by the lack of functional alleles for the RNA polymerase I, II, or III subunits. However, sustained pollen tube growth presumably requires de novo synthesis of essential RNA polymerase genes such that mutant pollen are at a competitive disadvantage compared to wild-type pollen, the phenomenon known as certation (Heribert-Nilsson 1920).
DISCUSSION
Genetic analyses have identified a large number of female gametophytic mutants in Arabidopsis, a significant fraction of which correspond to mutant alleles of transcription factors (Pagnussat et al. 2005). Our demonstration that mutations in RNA polymerases I, II, and III cause female gametophyte lethality are generally consistent with these findings and indicate that the female gametophyte is dependent on endogenous transcription machinery synthesized de novo during gametophyte development. In the absence of functional RNA polymerase subunits, female gametophytes can often progress to the two-nucleate stage, but typically arrest before, or shortly after, the second of the three mitotic divisions required for development of mature gametophytes. It is noteworthy that the SeedGenes Project database (http://www.seedgenes.org/index.html) (Tzafrir et al. 2003, 2004) includes information for two T-DNA insertion alleles of nrpb2, named emb 1989-1 and emb 1989-2. Embryos fail to develop in 90–94% of ovules bearing these mutant alleles, consistent with the female gametophytic lethal phenotype we describe in this article. However, 6–10% of emb 1989-1 and emb 1989-2 ovules are reported to arrest as preglobular embryos, indicating that the female gametophytes in these cases had completed development and had been fertilized, but produced embryos that were then unable to complete development. Cloning and sequencing of the region that defines the junction between the NRPB2 gene and the T-DNA revealed that the T-DNA in emb 1989-1 inserted 34 nucleotides upstream from the translation start site (Y. Onodera, data not shown). Because the protein coding region is not disrupted, it is possible that the emb 1989-1 allele is partially functional, which may explain how development can sometimes proceed to stages beyond what we have observed for the nrpb2-1 and nrpb2-2 alleles. We currently lack analogous data concerning the precise location of the T-DNA in the emb 1989-2 allele.
A recent study of developing and mature pollen showed that 61.9% of all Arabidopsis genes are expressed during at least one stage of male gametophyte development, with 9.7% of the transcripts being pollen specific (Honys and Twell 2004). A large number of transcription factors are expressed during pollen development, suggesting that orchestrated waves of transcription are essential for pollen maturation. Mature pollen is also known to contain proteins, ribosomes, mRNAs, rRNAs, and tRNAs that are synthesized postmeiotically during pollen maturation or pollen tube growth (Mascarenhas 1975, 1989). Therefore, we were surprised to find that functional alleles of RNA polymerases I, II, and III are not absolutely required in the haploid pollen genome to complete pollen development, germination, pollen tube growth, or fertilization. The simplest explanation is that transcription in pollen-bearing defective polymerase alleles is conducted using RNA polymerases, or stored mRNAs encoding RNA polymerase subunits, that are synthesized premeiotically in the hemizygous microspore mother cell and are then partitioned into the microspores following meiosis. The one functional allele is apparently sufficient for microspore mother cells to load microspores with enough polymerase to support subsequent pollen development and postgermination pollen functions, including pollen tube growth and fertilization.
Transcript profiling using DNA microarray technology has shown that mRNAs encoding the core subunits for nuclear RNA polymerases are present within unicellular microspores at similar or greater abundance than in sporophytic tissues (Honys and Twell 2004). However, in mature pollen, mRNAs encoding transcription factors, RNA processing proteins, and translation machineries are less abundant than in vegetative tissues of the plant (Honys and Twell 2003; Pina et al. 2005; Grennan 2007). This holds true for transcripts encoding the core subunits for nuclear RNA polymerases I, II, and III, which either are not detected in mature pollen or are present at very low levels (Honys and Twell 2003; Pina et al. 2005). The idea that maternally derived polymerase subunit mRNAs are stored for translation late in pollen development is not readily supported by these observations, but the possibility cannot be ruled out. An alternative hypothesis is that polymerase proteins derived from the microspore mother cell, or translated from mRNAs partitioned into the unicellular microspores, persist in mature pollen. Plants hemizygous for a single-copy transgene expressing a polymerase subunit-GFP fusion protein would be useful for testing this hypothesis. If the transgene were capable of rescuing plants that were homozygous for null alleles of the corresponding endogenous genes, one would expect the GFP marker to segregate 2:2 among the pollen. If GFP were observed in all pollen, this would indicate maternal loading of the polymerase subunit. Regardless of whether stored mRNA or stored protein is responsible for allowing the transmission of mutant polymerase alleles through the pollen, there are enough of the stored molecules to complete pollen development, germination, and fertilization. These developmental events are thought to span a period of at least 90 hr (Bowman 1994). However, additional de novo synthesis of Pol I, II, and III is apparently needed for full pollen vigor and for growth of pollen tubes long enough to reach the ovules farthest from the stigma.
Given the reduced fitness of mutant pollen relative to wild-type pollen, deleterious mutant polymerase alleles are unlikely to become widespread among a population. However, some gene evolution phenomena would seem to be favored by allowing mutant alleles to persist in the population for some period of time. For instance, a characteristic of the RNA polymerase I transcription system is that it evolves rapidly, such that the transcription machinery of one species cannot transcribe the rRNA genes of an unrelated species (Grummt et al. 1982; Miesfeld and Arnheim 1984; Doelling and Pikaard 1996). Species specificity appears to be explained by the rapid evolution of rRNA gene sequences and the corresponding coevolution of the transcription machinery, such that changes in gene sequences can be tolerated as a result of compensatory changes in the proteins that bind these sequences (or vice versa). Because haploid selection against defective alleles is less stringent in the male gametophyte than in the female gametophyte, at least for subunits of RNA polymerases I, II, and III, it is tempting to speculate that the male lineage could be the conduit for transmitting mutations that might initially be deleterious but could be tolerated if a compensatory mutation in an interacting protein or DNA sequence were to occur. Transmitting mutations at moderate frequency via the pollen would presumably buy time for such compensatory mutations to occur. However, the null hypothesis is that the capacity to transmit mutations in essential housekeeping genes such as RNA polymerases via pollen has no evolutionary advantage and is merely an unintended consequence of pollen development.
Acknowledgments
Research at Hokkaido University was supported in part by a grant in aid for scientific research from the Ministry of Education, Science and Culture and grants from the Akiyama Foundation and Grant Program of The Sumitomo Foundation. Pikaard lab research was supported by National Institutes of Health grants GM-060380 and GM-077590. The content of this article is solely the responsibility of the authors and does not necessarily reflect the views of the funding agencies that supported the work.
References
- Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen et al., 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bowman, J. (Editor), 1994. Arabidopsis: An Atlas of Morphology and Development. Springer-Verlag, New York.
- Christensen, C., E. King, J. R. Jordan and G. N. Drews, 1997. Megagametogenesis in Arabdopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Doelling, J. H., and C. S. Pikaard, 1996. Species-specificity of rRNA gene transcription in plants manifested as a switch in polymerase-specificity. Nucleic Acids Res. 24 4725–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G., and R. Yadegari, 2002. Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36 99–124. [DOI] [PubMed] [Google Scholar]
- Eady, C., K. Lindsey and D. Twell, 1994. Differential activation and conserved vegetative cell-specific activity of a late pollen promoter in species with bicellular and tricellular pollen. Plant J. 5 543–550. [Google Scholar]
- Earley, K., J. Haag, O. Pontes, K. Opper, T. Juehne et al., 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Grennan, A., 2007. An analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 145 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., and K. Schneitz, 1998. The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9 227–238. [DOI] [PubMed] [Google Scholar]
- Grummt, I., E. Roth and M. R. Paule, 1982. rRNA transcription in vitro is species-specific. Nature 296 173–174. [DOI] [PubMed] [Google Scholar]
- Hashida, S., H. Takahashi, M. Kawai-Yamada and H. Uchimiya, 2007. Arabidopsis thaliana nicotinate/nicotinamide mononucleotide adenyltransferase (AtNMNAT) is required for pollen tube growth. Plant J. 49 694–703. [DOI] [PubMed] [Google Scholar]
- Heribert-Nilsson, N., 1920. Zuwachsgeschwindigkeit der Pollenschläuche und gestörte Mendelzahlen bei Oenothera Lamarckiana. Hereditas 1 41–67. [Google Scholar]
- Higashiyama, T., 2002. The synergid cell: attractor and acceptor of the pollen tube for double fertilization. J. Plant Res. 115 149–160. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., S. Yabe, N. Sasaki, Y. Nishimura, S. Miyagishima et al., 2001. Pollen tube attraction by the synergid cell. Science 293 1480–1483. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., H. Kuroiwa and T. Kuroiwa, 2003. Pollen-tube guidance: beacons from the female gametophyte. Curr. Opin. Plant Biol. 6 36–41. [DOI] [PubMed] [Google Scholar]
- Honys, D., and D. Twell, 2003. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys, D., and D. Twell, 2004. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M., and D. Preuss, 2002. Plotting a course: multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. [DOI] [PubMed] [Google Scholar]
- Johnston, A., P. Meier, J. Gheyselinck, S. Wuest, M. Federer et al., 2007. Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8 R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M., J. Borevitz and D. Preuss, 2007. Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 3 1848–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis, G. A., C. Bardeleben, B. Bartholomew, B. R. Braun, C. A. P. Joazeiro et al., 1994. Transcription by RNA polymerase III, pp. 107–126 in Transcription Mechanisms and Regulation, edited by R. C. Conaway and J. W. Conaway. Raven Press, New York.
- Larkin, R., and T. Guilfoyle, 1993. The second largest subunit of RNA polymerase II from Arabidopsis thaliana. Nucleic Acids Res. 21 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas, J. P., 1975. The Biochemistry of angiosperm pollen development. Bot. Rev. 41 259–314. [Google Scholar]
- Mascarenhas, J. P., 1989. The Male gametophyte of flowering plants. Plant Cell 1 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas, J. P., 1993. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S., 1993. Male gametophyte development. Plant Cell 5 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S., 2004. Control of male gametophyte development. Plant Cell 16 S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld, R., and N. Arnheim, 1984. Species-specific rDNA transcription is due to promoter-specific binding factors. Mol. Cell. Biol. 4 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera, Y., J. R. Haag, T. Ream, P. C. Nunes, O. Pontes et al., 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 613–622. [DOI] [PubMed] [Google Scholar]
- Pagnussat, G., H. Yu, Q. Ngo, S. Rajani, S. Mayalagu et al., 2005. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Paule, M. R., and R. J. White, 2000. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina, C., F. Pinto, J. Feijó and J. Becker, 2005. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., S. Rhee and R. Davis, 1994. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264 1458–1460. [DOI] [PubMed] [Google Scholar]
- Rosso, M., Y. Li, N. Strizhov, B. Reiss, K. Dekker et al., 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Russell, S., 1993. The egg cell: development and role in fertilization and early embryogenesis. Plant Cell 5 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., M. Hülskamp and R. E. Pruitt, 1995. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7 731–749. [Google Scholar]
- Sprunck, S., U. Baumann, K. Edwards, P. Langridge and T. Dresselhaus, 2005. The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J. 41 660–672. [DOI] [PubMed] [Google Scholar]
- Steffen, J., I. Kang, J. MacFarlane and G. Drews, 2007. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51 281–292. [DOI] [PubMed] [Google Scholar]
- Twell, D., J. Yamaguchi and S. McCormick, 1990. Pollen-specific gene-expression in transgenic plants: coordinate regulation of 2 different tomato gene promoters during microsporogenesis. Development 109 705. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., A. Dickerman, O. Brazhnik, Q. Nguyen, J. McElver et al., 2003. The Arabidopsis SeedGenes Project. Nucleic Acids Res. 31 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir, I., R. Pena-Muralla, A. Dickerman, M. Berg, R. Rogers et al., 2004. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135 1206–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari, R., T. Kinoshita, O. Lotan, G. Cohen, A. Katz et al., 2000. Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., N. Kaur, S. Kiriakopolos and S. McCormick, 2006. EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta 224 1004–1014. [DOI] [PubMed] [Google Scholar]
- Yu, H., P. Hogan and V. Sundaresan, 2005. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 139 1853–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]