Abstract
The Arabidopsis mutant Atubp26 initiates autonomous endosperm at a frequency of ∼1% in the absence of fertilization and develops arrested seeds at a frequency of ∼65% when self-pollinated. These phenotypes are similar to those of the FERTILIZATION INDEPENDENT SEED (FIS) class mutants, mea, fis2, fie, and Atmsi1, which also show development of the central cell into endosperm in the absence of fertilization and arrest of the embryo following fertilization. Atubp26 results from a T-DNA insertion in the UBIQUITIN-SPECIFIC PROTEASE gene AtUBP26, which catalyzes deubiquitination of histone H2B and is required for heterochromatin silencing. The paternal copy of AtUBP26 is able to complement the loss of function of the maternal copy in postfertilization seed development. This contrasts to the fis class mutants where the paternal FIS copy does not rescue aborted seeds. As in the fis class mutants, the Polycomb group (PcG) complex target gene PHERES1 (PHE1) is expressed at higher levels in Atubp26 ovules than in wild type; there is a lower level of H3K27me3 at the PHE1 locus. The phenotypes suggest that AtUBP26 is required for normal seed development and the repression of PHE1.
IN angiosperms, fertilization of the egg and central cell nuclei in the embryo sac by the two sperm nuclei is essential for the formation of embryo and endosperm. The development of the embryo, endosperm, and seed coat is coordinated and controlled by both genetic and epigenetic processes (reviewed by Berger et al. 2006). The breakdown of coordinated development in some mutant lines is shown by endosperm development in the absence of fertilization when any of the four genes, MEA, FIS2, FIE, and AtMSI1 are not functional in Arabidopsis. The FIS genes encode homologs of the Drosophila Polycomb group (PcG) proteins, members of an epigenetic repressive complex PRC2 (Grossniklaus et al. 1998; Luo et al. 1999; Ohad et al. 1999; Kohler et al. 2003a). Loss of function in any of the genes leads to the formation of diploid autonomous endosperm derived from the central cell nucleus in the absence of fertilization. When the mutants are fertilized, the maternal genotype determines the fate of the seed irrespective of the paternal genotype. An ovule carrying a fis mutant allele develops into an arrested seed with an arrested embryo and noncellularized endosperm whereas an ovule carrying a wild-type allele develops a viable seed.
The FIS PcG complex mediates the addition of the repressive mark H3K27me3 to the target gene PHE1 through the histone methyltransferase activity of MEA (Makarevich et al. 2006). A mutation in the MEA gene reduces the level of H3K27me3 at PHE1 and leads to increased expression of PHE1 (Makarevich et al. 2006). PHE1 expression is also elevated in other mutants of the fis PcG genes (Kohler et al. 2003b). It has been speculated that the FIS complex may have functions before and after fertilization as fis mutants have phenotypes in both nonfertilized and fertilized ovules (Sørensen et al. 2001; Makarevich et al. 2006).
To obtain other mutants with autonomous endosperm development, we screened for arrested embryo and noncellularized endosperm in a small collection of T-DNA insertion lines (http://www.arabidopsis.org) and checked for autonomous seed development following emasculation. We isolated one mutant that had a weak autonomous endosperm phenotype and arrested seed development. The T-DNA insertion caused the loss of function of a H2B deubiquitinating enzyme, AtUBP26 (Sridhar et al. 2007) and resulted in mutant seed phenotypes. PHE1 was upregulated in the mutant ovules and there was a reduced level of H3K27me3 at the PHE1 locus suggesting that AtUBP26 is required for addition of H3K27me3 at PHE1 and that AtUBP26 may be necessary for FIS PcG action in seeds.
MATERIALS AND METHODS
Mutant identification, materials, and growth condition:
A small collection of Salk lines from the Arabidopsis Stock Center was screened for shriveled seeds that resembled fis fertilized seeds (Chaudhury et al. 1997) (http://www.arabidopsis.org). A putative mutant which produced shriveled seeds was backcrossed to Columbia (Col) to cross out any other T-DNA insertions in the same line. An allele of mea (fis1, Chaudhury et al. 1997), was emasculated to compare the autonomous endosperm development with that of the putative mutant.
The Arabidopsis ecotypes used in this study were Landsberg erecta (Ler) and Col. Two T-DNA insertion lines, Salk_024392 and CS826614 (SAIL_621_F01), which have T-DNA insertions in At3g49600, were obtained from the Arabidopsis Biological Resource Center and confirmed by PCR.
Plants were grown in pots with compost soil under continuous artificial light at 20° in growth chambers. The artificial light was achieved by using an incandescent light source producing a fluence of 180 micro-E for most of the experiments.
Cloning of AtUBP26 and expression analysis:
For mapping, the homozygous mutant plant was crossed as a pollen donor to Ler. The following SSLP markers located on different part of the chromosomes were used to test F2 plants: nga63, nga111, nga361, nga172, CIW4, CIW6, and NGA151 (http://www.arabidopsis.org). The recombination frequency between markers and seed phenotypes was scored. Further detailed mapping was done by using the following insertion-deletion (indel) markers on chromosome III on the basis of sequence differences between Col and Ler (http://www.arabidopsis.org). They are T32N15 FR with primers 5′-AGCTAGGGTTTGGCAACATT-3′ and 5′-CTTTTGTGCACGACGGAATCT-3′, T21J18 FR with primers 5′-GCTTTCAAAATTGCTGGAAAA-3′ and 5′-TCAGGCTGCATTAGGCTCTT-3′, F2K15FR with primers 5′-GAATCAGGTTTGAATAAAATGGAA-3′ and 5′-TTCAATCCTCCAAAATTATCAACA-3′, and T16K5FR with primers 5′-GCGTGATCATGGAAATATTGG-3′ and 5′-AGCAAGGTTATGCATTTCAGA-3′. The genotypes of each plant for the SSLP and indel markers were visualized on 4% agarose gel. The plant DNA preparation and the PCR reactions were carried out as described by Bell and Ecker (1994).
For isolating T-DNA border sequences, DNA was extracted using a QIAGEN DNeasy mini kit. T-DNA border isolation and sequencing were carried out as described (http://signal.salk.edu/). Primers used to amplify T-DNA junctions were LBb1, 5′-GCGTGGACCGCTTGCTGCAAC-3′ and AtUBP26 TF2, 5′-TCGTTAATCGGAATCAAAATTG-3′. Individual plants of the mapping F2 population were PCR amplified with LBb1 and AtUBP26 TF2.
For complementation, a genomic fragment including promoter and coding region was generated by PCR and fused to FLAG in vector pCH252 (Helliwell et al. 2006) using the Gateway recombination system. The primer sequences used were GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATCTGGTGTGATGGAATTGATC and GGGGACCACTTTGTACAAGAAAGCTGGGTAGCAGGCTTCAGAAGAGATATTGGG. Wild-type Col was transformed by floral dip (Clough and Bent 1998). Stable transgenic plants were screened by Western blots with anti-FLAG (Sigma). Homozygous progeny of three independent transgenic plants, which produced the hybrid protein of expected size, were selected to cross to the Atubp26-2 homozygote. The arrested seeds were scored in the F1 plants between Atupb26-2 and the transgenic plants. The arrested seeds were also scored in the F1 plants of a control cross between Atubp26-2 and Col.
RT–PCR was performed on RNA isolated from buds, cauline leaves, rosette leaves, young seedlings, seeds 3 days postfertilization, embryos 7 days postfertilization, endosperm 7 days postfertilization, young siliques 1–2 days postfertilization, and stems using a Promega Access RT–PCR system kit. Primers used for AtUBP26 are 5′-CATTGCAGCTTCCACAAAAA-3′ and 5′-TGCTTTTGTCAAGACGTTGG-3′. For the control gene FORMALDEHYDE DEHYDROGENASE (FDH), primers 5′-TGGGAAACCCATTTATCACTTCA-3′ and 5′-CAGCAAGTCCAACAGTGCCAAG-3′ were used (Luo et al. 2005).
Microscopy:
Fertilized and unfertilized siliques were dissected under a dissecting microscope. Developing seeds were cleared (Boisnard-Lorig et al. 2001). Specimens were examined with a Leica (Deerfield, IL) microscope using differential interference contrast optics. Mature seeds of Col, Atubp26-2, Atubp26-3, and Atubp26-4 were photographed and captured as digital images under a dissection microscope.
Real-time PCR:
RNA was isolated from young siliques 36, 60, and 84 hr postfertilization from both Atubp26-2 and Col, using a QIAGEN RNeasy plant mini kit with Dnase I treatment according to the manufacturer's protocol. RNAs were also harvested from rosette leaves and buds from both Atubp26-2 and Col. Silique RNAs 84 hr postfertilization from Ler and mea (fis1, Chaudhury et al. 1997) were isolated. Approximately 3 μg total RNA was used for cDNA synthesis using an Invitrogen Superscript III reverse transcriptase kit. Real-time PCR was performed using PHE1 primers by following Kohler et al. (2003b). FDH primers were used as internal control (Luo et al. 2005). For each stage, at least three biological repeats were conducted.
Real-time PCRs were also performed on cDNAs from young silique 84 hr postfertilization for both Atubp26-2 and Col with MEA, FIS2, FIE, and AtMSI1.
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (Johnson et al. 2002) was carried out using an antibody against histone H3K27me3 (Upstate). Buds (including all the tips without open flowers) and siliques 60–84 hr postfertilization were harvested for chromatin preparation. After chromatin immunoprecipitation, DNA was used for real-time PCR using primers amplifying region 2 of the PHE1 locus described by Makarevich et al. (2006). For an internal control, we used primers 5′-CCCAAAGATTTTAGTGCCTCA-3′ and 5′-GGTTCAAGAGGGCAATCAC-3′ from the 5′-UTR of AG described by Finnegan and Dennis (2007). This region has been shown to be enriched by the H3K27me3 antibody (Schubert et al. 2006). AG gene expression was not affected in the Atubp26 mutant at bud or silique stage. Thus the AG 5′-UTR is a suitable control in our experiment. Three pull-down experiments have been carried out for both stages. DNA precipitated by H3K27me3 antibody was normalized with the 5′-UTR of AG. We calculated the relative enrichment and statistical analysis as described by Finnegan and Dennis (2007).
RESULTS
T-DNA insertions in Atubp26, a ubiquitin-specific protease gene, cause arrested seed phenotypes:
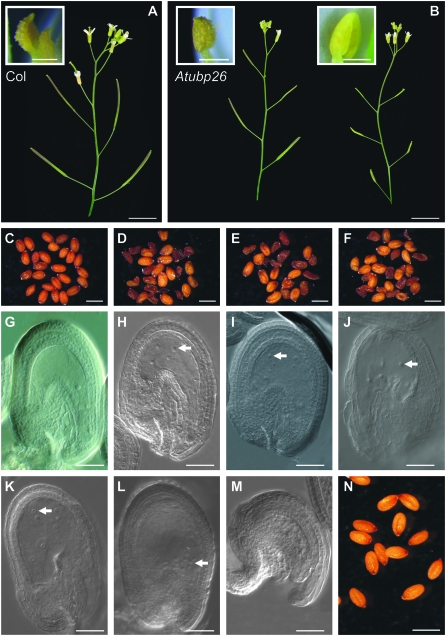
In a screen of ∼200 T-DNA insertion lines, 20 lines with a proportion of shriveled seeds were identified. Two shriveled seeds coming from one line, Salk_014298, germinated on an MS plate while the shriveled seeds from other lines failed to germinate. The resulting plants displayed short siliques and produced ∼60% shriveled seeds with arrested embryos (Figure 1, A–D). These seed phenotypes are similar to those of the FERTILIZATION INDEPENDENT SEED (FIS) class mutants, mea, fis2, fie, and Atmsi1 (Chaudhury et al. 1997; Grossniklaus et al. 1998; Ohad et al. 1999; Kohler et al. 2003a). As the Salk_014298 line is known to have a T-DNA insertion in the gene ITB1 (At2g35110) and itb1 does not display any seed phenotype (Zhang et al. 2005), the seed phenotype segregated away from the itb mutation in a backcross to Col.
Figure 1.—
Phenotypes of Atubp26. (A) Col wild type with fertile flowers and elongated siliques. Inset is an anther with pollen grains. (B) Siliques on an early-emerged branch (left) are longer than that of a late-emerged branch (right) in Atubp26-2 due to the lack of dehiscent anthers. Right inset shows a nondehiscent anther from an open flower in a late-emerging branch. Left inset shows a dehiscent anther from a Atubp26-2 homozygote from an open flower in an early-emerging branch. (C) Col mature seeds. (D) Atubp26-2 homozygote seeds showing both full and shriveled seeds. (E) Atubp26-3 homozygote seeds. (F) Atubp26-4 homozygote seeds. (G) An unfertilized ovule 7 days postemasculation in Col showing a fused central-cell nucleus. (H) An Atubp26-2 unfertilized ovule with six endosperm nuclei (two out of focus). (I) An Atubp26-3 unfertilized ovule with four endosperm nuclei. (J) An Atubp26-4 unfertilized ovule with endosperm nuclei (some out of focus). (K) An unfertilized ovule with endosperm nuclei of an Atubp26-2 heterozygote in the ap3/ap3 background. (L) An unfertilized ovule with endosperm nuclei of an Atubp26-2 heterozygote in the ap3/ap3 background. Note that the nuclei are concentrated at the micropylar region. (M) An Atubp26-2 aborted ovule with no developed embryo sac. (N) An Atubp26-2 homozygote pollinated with wild-type pollen showing viable seeds. Arrows in H–L indicate the endosperm nuclei. Bars in A and B, 10 mm; C–F and N, 0.5 mm; G–M, 0.1 mm.
We established a F2 mapping population between Ler and the mutant. F1 plants did not display 50% shriveled F2 seeds as do the fis class mutants mea, fis2, fie, and Atmsi1; instead they produced 14/120 shriveled F2 seeds. The selfed progeny from the two original plants did not contain any wild-type plants but displayed a uniform arrested seed phenotype, indicating that the original plants were homozygous mutants. The mutation was closely linked to the SSLP marker CIW4 and located on the bottom of chromosome 3 in a 240-kb region in which no fis class mutations have been found (Figure 2A).
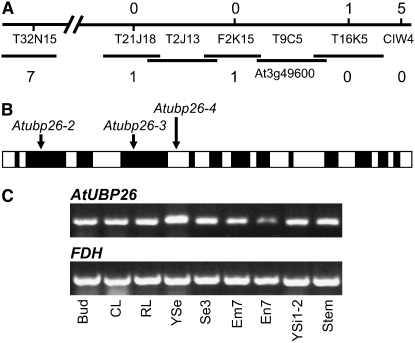
Figure 2.—
Mapping and expression of AtUBP26. (A) The mutant gene was mapped to a contig containing At3g49600/AtUBP26. Markers CIW4 and T16K5 FR were used to identify five and one recombinants, respectively, which showed crossovers between the mutant locus and the markers. Markers T32N15 FR, T21J18 FR, and F2K15 FR identified seven, one, and one recombinants, respectively. (B) Schematic of AtUBP26 showing the T-DNA insertions in three alleles. Solid boxes represent exons and open boxes, introns. (C) RT–PCR showing that AtUBP26 is expressed in buds, cauline leaves (CL), rosette leaves (RL), young seedlings (YSe), seeds 3 days postfertilization (Se3), embryos 7 days postfertilization (Em7), endosperm 7 days postfertilization (En7), young siliques 1–2 days postfertilization (YSi1-2), and stems.
We isolated the junction sequences of T-DNA insertions from 14 plants derived from the original mutant plants. T-DNAs were inserted in three different genes, At2g35110, At3g49600, and At1g69870. At2g35110 is the ITB1 gene; At3g49600 is located in the same region as the new mutant (Figure 2A), suggesting that At3g49600 may be the locus mutated. PCR primers that specifically amplified the T-DNA junction in At3g49600 were used to test a total of 260 plants heterozygous for the mutation and 20 wild-type plants in the F2 of Col crossed with the original mutant. The T-DNA insertion cosegregated with the shriveled seed phenotype. To confirm that the T-DNA insertion in At3g49600 causes the mutant phenotype, we obtained additional T-DNA insertion lines, Salk_024392 and CS826614, in which At3g49600 is interrupted (Figure 2B). Both alleles gave shriveled seeds and autonomous endosperm development (Figure 1, E, F, I, and J). These mutant alleles failed to complement each other, suggesting that the T-DNA insertions in At3g49600 cause the mutant seed phenotype. At3g49600 encodes a UBIQUITIN-SPECIFIC PROTEASE and has previously been designated AtUBP26 (Yan et al. 2000). Atubp26-2 has a T-DNA insertion in the second exon, Atubp26-3 in the fourth exon, and Atubp26-4 in the fifth intron (Figure 2B). The positions of the T-DNA insertions suggest that all the alleles are null mutants.
We further confirmed by complementation that the AtUBP26 is the gene responsible for the phenotype. Three independent transgenic homozygotes carrying a wild-type allele-FLAG fusion were crossed to Atubp26-2. Five mature siliques were scored to determine the frequency of arrested seeds in each plant. Five plants were investigated for each F1 population. F1 plants with the transgene showed a reduced frequency of arrested seeds (Table 1) in contrast to the control F1 plants, which produced ∼13% arrested seed. This result showed that the transgene complemented the lesion caused by the Atubp26-2 mutation.
TABLE 1.
The frequency of arrested seeds in F1 plants
| Atubp26 × Col | Atubp26 × tra*1 | Atubp26 × tra 2 | Atubp26 × tra 3 | |
|---|---|---|---|---|
| Plant1 | 40/305 (13) | 4/310 (1.3) | 5/275 (1.8) | 10/312 (3.2) |
| Plant2 | 40/300 (13) | 7/305 (2.2) | 8/285 (2.8) | 8/313 (2.6) |
| Plant3 | 40/295 (13) | 6/290 (2.1) | 9/295 (3.1) | 7/250 (3.4) |
| Plant4 | 38/280 (14) | 4/275 (1.5) | 8/300 (2.6) | 6/270 (2.9) |
| Plant5 | 35/275 (12) | 6/305 (2) | 7/311 (2.3) | 4/290 (1.4) |
| Average % | 13 | 1.82 | 2.52 | 2.7 |
tra*, Col carrying a construct of wild-type allele of AtUBP26-FLAG. Numbers in parentheses are percentages.
AtUBP26 is expressed in buds, cauline leaves, rosette leaves, young seedlings, seeds 3 days postfertilization, embryos 7 days postfertilization, endosperm 7 days postfertilization, young siliques 1–2 days postfertilization, and stems (Figure 2C) consistent with the results of Sridhar et al. (2007). MPSS data also suggest that AtUBP26 is expressed in inflorescences, siliques, roots, and leaves (http://mpss.udel.edu/at/).
Loss of Atubp26 leads to endosperm initiation at low frequency in the absence of fertilization:
Both homozygous Atubp26-2 and Col siliques were slightly elongated after emasculation. In wild-type Col, no ovules showed division of the central cell nucleus (of a total of 1200 ovules checked) (Figure 1G). Approximately 1% (16/1200) of the ovules in Atubp26-2 became slightly enlarged and showed autonomous division of the central cell nucleus (Figure 1H). Most cases of autonomous endosperm development in Atubp26-2 only reached the 4- to 8-nuclei stage (n = 13) but in a few ovules the endosperm developed beyond 8 nuclei (n = 3). Atubp26-3 and Atubp26-4 showed similar autonomous seed phenotypes (Figure 1, I and J; Table 2). In contrast to Atubp26, 10–15% of ovules of a mea (fis1) allele were enlarged and most developed autonomous endosperm with >20 nuclei.
TABLE 2.
Ovule and seed phenotypes of Atubp26-2
| Multiple nuclei after emasculation | Aborted seed ratio
|
|||||
|---|---|---|---|---|---|---|
| Aborted ovules | Globular | Globular heart | Torpedo | Full seeds | ||
| Wild type | 0/1200 | 2/310 (0.65) | None | 308/310 (99.35) | ||
| Atubp26-2 homozygote | 16/1200 (1.3) | 48/240 (20) | 15/240 (6.3) | 62/240 (25.8) | 35/240 (14.6) | 80/240 (33.3) |
| Atubp26-2 heterozygote | 4/900 (0.44) | 4/450 (0.89) | 9/450 (2) | 35/450 (7.8) | 17/450 (3.8) | 385/450 (85.6) |
| Atubp26-3 homozygote | 3/350 (0.86) | 17/100 (17) | 5/100 (5) | 30/100 (30) | 12/100 (12) | 36/100 (36) |
| Atubp26-4 homozygote | 2/270 (0.74) | 16/105 (15.2) | 5/105 (4.8) | 34/105 (32.4) | 10/105 (9.5) | 39/105 (37.1) |
Numbers in parentheses are percentages.
To investigate whether the autonomous phenotype is determined by the genotype of the embryo sac or the maternal somatic tissue, we emasculated heterozygous Atubp26-2 plants (in a Col background). Autonomous development of the central cell was observed. We reconfirmed that the autonomous endosperm initiation occurred in heterozygous Atubp26-2 using an ap3/ap3 male sterile plant (Figure 1, K and L; Table 2). Sometimes, the autonomous endosperm nuclei congregated at the micropylar end (Figure 1L) where the earliest endosperm divisions take place.
AtUBP26-2 has an arrested embryo phenotype following fertilization:
In homozygous Atubp26-2 plants, 20% (48/240) of ovules failed to form seeds upon self-pollination, resulting in slightly shorter siliques than in wild type (Figure 1, A and B). These ovules did not develop an embyo sac (Figure 1M). Late-developing branches set even fewer seeds (Figure 1B) and gave short siliques. In newly opened flowers on branches with short siliques most of the anthers had not opened, in contrast to wild-type flowers at the same stage in which released pollen grains could be seen (Figure 1, A and B insets). Flowers on late branches responded to pollination with wild-type pollen as did some of the flowers in early branches. Thus the partial sterility could be caused by either or both the lack of pollen from nondehiscent anthers and the lack of embryo sacs in Atubp26-2. We tested whether transmission of the mutant gene by pollen had been affected by pollinating wild-type with pollen from a heterozygote plant. There were 55 heterozygote mutants and 48 wild-type plants among the progeny, indicating that mutation in AtUBP26 does not affect pollen viability.
While 20% of ovules failed to develop embryo sacs, 80% (192/240) of ovules give seeds upon fertilization. In self-fertilized Atubp26-2 seeds (240 − 48 = 192), 58.4% (112/192) of the resulting seeds were arrested at various stages of development and 41.6% (80/192) of seeds developed fully (Table 2; Figure 1D). When Atubp26-2 was pollinated with wild-type pollen, all the seeds developed fully (Figure 1N). Thus, the paternal wild-type AtUBP26 can fully complement the loss of function of the maternal Atubp26 allele. In fis class mutants the maternal gametophyte genotype determines the seed phenotype irrespective of the paternal contribution (Chaudhury et al. 1997; Grossniklaus et al. 1998; Ohad et al. 1999; Kohler et al. 2003a).
We did not see any other abnormal morphology in Atubp26 homozygotes. The heterozygote of Atubp26-2 set 61/450 (15%) arrested seeds upon self-pollination and did not have the unopened anthers seen in the Atubp26-2 homozygote. There were some aborted ovules but at a level comparable to wild type (Table 2). In contrast, heterozygous mea, fis2, fie, and Atmsi1 displayed 50% arrested seeds when self-pollinated or pollinated with wild-type pollen as expected for a locus operating in the gametophyte.
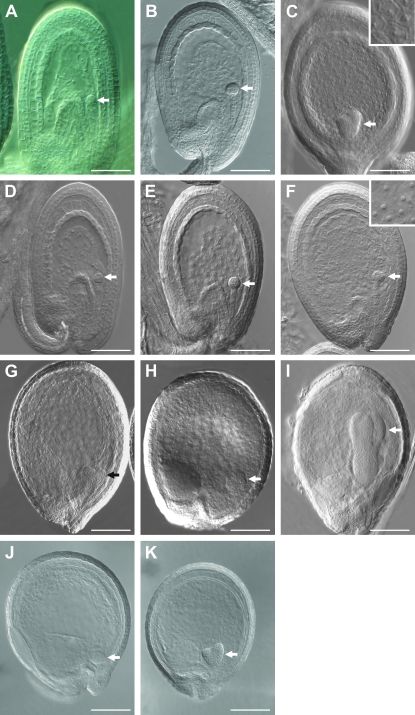
We characterized the defects in the self-pollinated Atubp26-2 seeds by analyzing the early seed morphology of the mutant and wild type using cleared whole seeds (Boisnard-Lorig et al. 2001). Seed arrested 32.2% (62/192) at heart stage (Figure 3, G and H), 7.8% (15/192) at the globular stage (Figure 3F), 18.2% (35/192) beyond heart stage (Figure 3I), while 41.6% (80/192) of embryos developed fully. Fully developed seeds showed 100% (n = 80) germination. Early embryo development up to the globular stage is similar in Atubp26-2 and wild type (Figure 3, A–E). The wild-type endosperm was cellularized at heart stage (Figure 3C), whereas all the arrested embryos were associated with un-cellularized endosperm (n = 112; Figure 3, F–H). Seeds in Figure 3, G and H, showed overproliferated endosperm with 710 and 750 nuclei, respectively, while the same stage wild-type endosperm had only 410 ± 30 nuclei (n = 2). We also observed an enlarged chalazal cyst in some mutant seeds (Figure 3H). The two other Atubp26 alleles showed similar seed abortion and embryo arrest frequencies (Table 2). The seed-arrested phenotype in the heterozygote is similar to that in the homozygote, indicating that there is no maternal sporophytic effect on seed development in the mutant (Figure 3, J and K; Table 2).
Figure 3.—
Atubp26 aborted seed phenotypes. (A–C) Embryo and endosperm development in wild-type seeds at 48, 72, and 96 hr postfertilization. In A and B, globular embryos and syncytial endosperm were observed. In C, heart-stage embryo and cellularized endosperm were observed. (Inset) Cell wall of endosperm. (D and E) Atubp26 seeds at 48 and 72 hr showing similar development to wild type of the same age. (F) An Atubp26 seed at 96 hr containing an arrested globular embryo and syncytial endosperm. (Inset) Free endosperm nuclei. (G and H) Atubp26 seeds at 120 hr containing arrested heart or globular embryo and syncytial endosperm. (I) An Atubp26 seed at 120 hr containing a torpedo stage embryo. (J) Seed with arrested globular embryo from a heterozygote. (K) Seed with arrested heart embryo from a heterozygote. Arrows indicate the embryos. Bars in A, B, D, and E are 50 μm; in C, F, and G–I are 0.1 mm.
AtUBP26 regulates PHE1 expression:
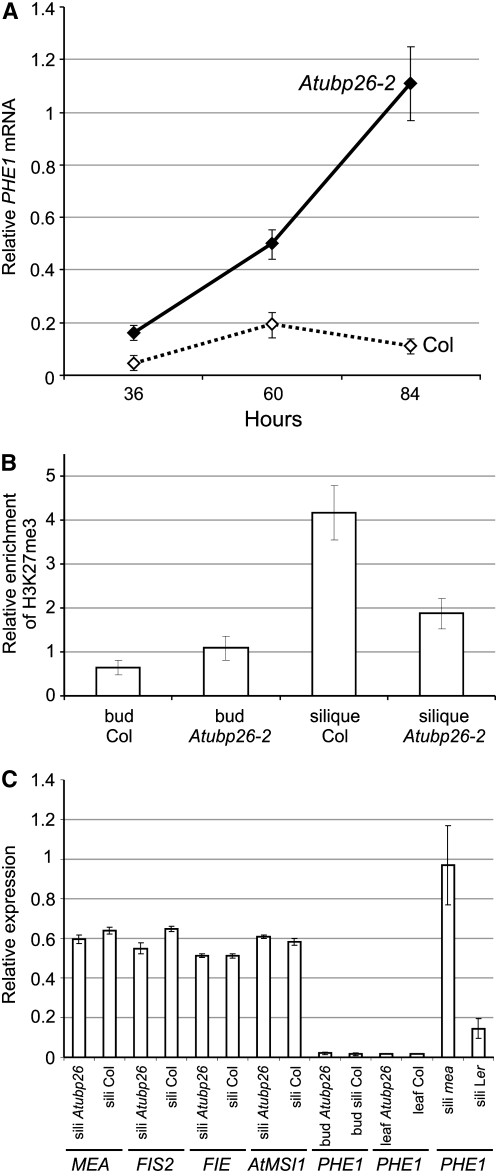
PHE1, a type I MADS-box gene was upregulated relative to wild type (Kohler et al. 2003b) in mea, fis2, and fie young siliques. Given that Atubp26-2 displayed phenotypes similar to the fis mutants, we tested whether PHE1 was upregulated in developing siliques. Because PHE1 has a similar sequence to PHE2 (Kohler et al. 2003b), we confirmed that the RT–PCR products were specific for PHE1 by sequencing 18 subclones. PHE1 expression was significantly increased compared to the control Col in RNA extracted from 36-hr- (mainly containing early globular embryos), 60-hr- (globular embryos), and 84-hr- (late globular and heart-stage embryo) old siliques of Atubp26-2 and Col (Figure 4A). We also observed increased expression of PHE1 in mea. PHE1 expression in buds and leaves is very low and was not elevated in the mutant (Figure 4C). At late globular and heart stage, the PHE1 mRNA level was up to ninefold higher in Atubp26-2 siliques than in wild type (Figure 4A). In early stages, there was a two- to threefold increase of PHE1 expression in Atubp26-2 siliques compared with wild-type siliques. The upregulation of PHE1 could be caused indirectly by a reduction in expression of FIS class genes if AtUBP26 were a positive regulator of FIS genes. We did not find any reduction of expression of MEA, FIS2, FIE, and AtMSI1 in 60-hr-old siliques of Atubp26-2 compared to Col (Figure 4C). We conclude that AtUBP26 regulates PHE1 in developing seed but does not regulate the FIS class genes.
Figure 4.—
PHE1 expression is elevated in Atubp26 siliques. (A) Real-time PCR showing PHE1 expression in Atubp26 and wild-type siliques at 36, 60, and 84 hr postfertilization. (B) Chromatin immunoprecipitation analysis of H3K27me3 at the PHE1 locus. There was a higher enrichment of the PHE1 region in Col than in Atubp26 siliques (60–84 hr old) after chromatin immunoprecipitation using a H3K27me3 antibody but no difference in buds. (C) MEA, FIS2, FIE, AtMSI1, and FWA expression in Atubp26 and wild-type siliques of 2–3 days; PHE1 expression in Atubp26 and wild-type leaves; PHE1 expression in mea and wild-type siliques of 2–3 days.
H3K27me3 is reduced at PHE1 chromatin in Atubp26 young siliques:
Repression of PHE1 expression is associated with increased H3K27me3 at the PHE1 locus, which is dependent on the histone methyltranferase activity of MEA (Kohler et al. 2003b; Makarevich et al. 2006). As shown above, PHE1 expression was elevated in Atubp26-2 siliques. In these siliques, H3K27me3 was reduced to half the wild-type level at the PHE1 locus, suggesting that AtUBP26 is required to maintain H3K27me3 at the PHE1 locus (Figure 4B).
We did not observe any difference in the level of H3K27me3 at the PHE1 locus in buds of Atubp26-2 and Col wild type (Figure 4B).
DISCUSSION
Lack of activity of AtUBP26, which catalyses the deubiquitination of H2B (Sridhar et al. 2007), causes low frequency autonomous endosperm growth without fertilization and embryo arrest if self-pollinated. The mutant seed phenotypes of Atubp26 are similar to those of the fis class mutants mea, fis2, fie, and Atmsi1 suggesting that AtUBP26 may act, at least in part, in the same pathway as the FIS genes. We consistently observed a lower frequency of autonomous endosperm development in the three alleles of Atubp26 than in fis class mutants. Autonomous endosperm rarely develops beyond 16 nuclei in Atubp26, while in the mutant mea, autonomous endosperm may produce >20 nuclei. The observation of low-frequency autonomous development of the central cell in Atubp26 heterozygotes suggests that this phenotype is under gametophytic control with low penetrance.
The weaker autonomous endosperm phenotype in Atubp26 is paralleled by its weaker postfertilization phenotype. When homozygous, all three alleles of Atubp26 formed ∼40% viable seeds following self-pollination while mea and fis2 homozygotes produce <1% viable seed. The low penetrance of the autonomous endosperm phenotype in Atubp26 is unlikely to be due to a partially functioning Atubp26 gene because all three alleles are likely to be null mutations as the T-DNA insertions are in exons. AtUBP26 may be redundant with other UBP genes as there are 27 genes identified encoding UBPs in the Arabidopsis genome. AtUBP26 is the only one in its class (Yan et al. 2000) but other genes may be able to partially complement the loss of AtUBP26. In the Atubp26 mutant, the level of ubiquitinated histone H2B is only slightly increased over that in wild type (Sridhar et al. 2007), suggesting that other UBPs may be involved. An alternative explanation of the weak “fis” phenotype in Atubp26 is that the target loci of H2B deubiquitination by AtUBP26 do not completely overlap with the target loci of H3K27 trimethylation by the FIS Polycomb complex; the misregulated genes in the Atubp26 ovules may not be exactly the same set of genes misregulated in fis mutants. We speculate that there are some common target genes, including PHE1, which, when derepressed in Atubp26, cause a fis phenotype. There are at least three genes PHE1, PHE2, and MEIDOS that are upregulated in the mea mutant siliques with only PHE1 having been characterized for its status of histone H3K27 trimethylation in siliques (Kohler et al. 2003b; Makarevich et al. 2006). In this study, we focused on PHE1 and showed that PHE1 expression is elevated in mutant siliques relative to wild type and is accompanied by the loss of H3K27me3 as in a mea mutant (Makarevich et al. 2006). It remains to be determined whether the other two genes PHE2 and MEIDOS are upregulated and histone-demethylated in the Atubp26 mutant. In Drosophila, the ubiquitin-specific protease USP7 catalyzes the deubiquitination of H2B and contributes to epigenetic silencing of homeotic genes by PcG proteins. The silencing of USP7 by RNAi causes global reduction of H3K27me3 (van der Knaap et al. 2005). Although the mechanism causing this reduction of H3K27me3 remains unknown, one hypothesis is that ubiquitinated H2B prevents the trimethylation of H3K27 by PcG proteins. Our results and the finding in Drosophila suggest that deubiquitination of H2B mediated by UBPs is required for H3K27me3 addition.
Atubp26 and fis mutants both have embryo-arrested seeds but display different modes of transmission. When a homozygous Atubp26 mutant was self-pollinated most seeds were aborted with embryos arrested at the globular or heart stage and the endosperm remained noncellularized as in fis class mutants. When pollinated with wild-type pollen, all the seeds developed normally, indicating that the paternal copy of AtUBP26 fully complements the loss of function of the maternal copy in controlling seed development and that the arrested seed phenotype is under sporophytic control. In contrast to the Atubp26 mutants, the arrested seed phenotype of the fis mutants, which cannot be reversed by wild-type pollen, is completely under gametophytic control (Chaudhury et al. 1997; Grossniklaus et al. 1998; Ohad et al. 1999; Kohler et al. 2003a).
It is likely that AtUBP26 has a number of functions in development. The abnormal development of Atubp26 is not restricted to endosperm and embryo. We observed that pollen dehiscence was affected especially in late branches. There was a proportion of aborted embryo sacs in the mutant with both factors contributing to the partial sterility of the mutant.
Ubiquitination plays a critical role in target protein degradation but also participates in gene regulation by modifying histones (Hershko 2005). Recent reports indicate that ubiquitinated H2A might be involved in gene silencing and ubiquitinated H2B in gene activation (Henry et al. 2003; Kao et al. 2004; Wang et al. 2004). AtUBP26 acts to remove ubiquitin from ubiquitinated H2B (Sridhar et al. 2007). Thus AtUBP26 may act as a repressor, causing gene silencing via chromatin remodeling. It has been reported that, in the Atubp26 mutant, H3K9 dimethylation is reduced, siRNA-directed DNA methylation is suppressed and heterochromatic silencing is released. H2B deubiquitination by AtUBP26 is thought to play an early and crucial role in heterochromatin formation (Sridhar et al. 2007). Here we show that the loss of function of AtUBP26 has effects on seed development similar to the loss of function of the seed development repressors, the FIS PcG proteins, leading to both the autonomous growth of endosperm in the absence of fertilization and arrested embryos with noncellularized endosperm. This suggests that AtUBP26 may have a broad role in chromatin remodeling affecting different aspects of silencing processes such as siRNA-mediated heterochromation formation and the FIS Polycomb repression.
References
- Bell, C. J., and J. R. Ecker, 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Berger, F., P. E. Grini and A. Schnittger, 2006. Endosperm: an integrator of seed growth and development. Curr. Opin. Plant Biol. 9 664–670. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., A. Colon-Carmona, M. Bauch, S. Hodge, P. Doerner et al., 2001. Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A. M., L. Ming, C. Miller, S. Craig, E. S. Dennis et al., 1997. Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Finnegan, E. J., and E. S. Dennis, 2007. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17 1978–1983. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., J. P. Vielle-Calzada, M. A. Hoeppner and W. B. Gagliano, 1998. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Helliwell, C. A., C. Wood, M. Robertson, W. J. Peacock and E. S. Dennis, 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular weight protein complex. Plant J. 46 183–192. [DOI] [PubMed] [Google Scholar]
- Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre et al., 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., 2005. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 12 1191–1197. [DOI] [PubMed] [Google Scholar]
- Johnson, L., X. Cao and S. Jacobsen, 2002. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kao, C. F., C. Hillyer, T. Tsukuda, K. Henry, S. Berger et al., 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., L. Hennig, R. Bouveret, J. Ghevselinck, U. Grossniklaus et al., 2003. a Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J. 22 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., L. Hennig, C. Spillane, S. Pien, W. Gruissem et al., 2003. b The polycomb group protein MEDEA regulates seed development by controlling expression of the MADS box gene PHERES1. Genes Dev. 17 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., P. Bilodeau, A. Koltunow, E. S. Dennis, W. J. Peacock et al., 1999. Genes controlling fertilization independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., E. S. Dennis, F. Berger, W. J. Peacock and A. Chaudhury, 2005. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich, G., O. Leroy, U. Akinci, D. Schubert, O. Clarenzl et al., 2006. Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., R. Yadegari, I. Margossian, M. Hannon, D. Michaeli et al., 1999. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, D., L. Primavesi, A. Bishopp, G. Roberts, J. Doonan et al., 2006. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25 4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, M. B., A. M. Chaudhury, H. Robert, E. Bancharel and F. Berger, 2001. Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11 277–281. [DOI] [PubMed] [Google Scholar]
- Sridhar, V. V., A. Kapoor, K. Zhang, J. Zhu, T. Zhou et al., 2007. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447 735–738. [DOI] [PubMed] [Google Scholar]
- van der Knaap, J. A., B. R. P. Kumar, Y. M. Moshkinoshkin, K. Langenberg, J. Krijgsveld et al., 2005. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17 695–707. [DOI] [PubMed] [Google Scholar]
- Wang, H., L. Wang, H. M. Erdjument-Bromage, M. Vidal, P. Tempst et al., 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431 873–878. [DOI] [PubMed] [Google Scholar]
- Yan, N., J. H. Doelling, T. G. Falbel, A. M. Durski and R. D. Vierstra, 2000. The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124 1828–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., J. Dyachok, S. Krishnakumar, L. G. Smith and D. G. Oppenheimer, 2005. IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex Activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]