Abstract
The steroid hormone ecdysone triggers the rapid and massive destruction of larval tissues through transcriptional cascades that culminate in rpr and hid expression and caspase activation. Here we describe the use of genetic screens to further our understanding of this steroid-triggered programmed cell death response. Pupal lethal mutants were screened for specific defects in larval salivary gland destruction. A pilot screen using existing P-element collections resulted in the identification of mutations in known cell death regulators, E74 and hid, as well as multiple alleles in CBP (nejire) and dTrf2. A large-scale EMS mutagenesis screen on the third chromosome resulted in the recovery of 48 mutants. These include seven multiallelic complementation groups, at least five of which do not map to regions or genes previously associated with cell death. Five mutants display defects in the transcriptional induction of rpr and hid, and all display a penetrant block in caspase activation. Three were mapped to specific genes: CG5146, which encodes a protein of unknown function, Med24, which encodes a component of the RNA polymerase II mediator complex, and CG7998, which encodes a putative mitochondrial malate dehydrogenase. These genetic screens provide new directions for understanding the regulation of programmed cell death during development.
PROGRAMMED cell death plays a central role in animal development, eliminating obsolete tissues, controlling cell numbers, and sculpting complex structures. Genetic screens in Caenorhabditis elegans identified three genes that provide the framework for our understanding of how cell death is appropriately restricted during development: ced-3, ced-4, and ced-9 (Ellis and Horvitz 1986; Metzstein et al. 1998). ced-3 encodes a caspase, a member of a conserved family of cysteine proteases that are present in all cells as inactive zymogens. Binding of CED-3 to the CED-4 adaptor facilitates its activation by proteolytic cleavage and initiates a cascade of caspase activation that directs cell death. Unlike CED-3 and CED-4, which promote cell death, CED-9 interacts with CED-4 to block caspase activation and the death response. These three factors form the core cell death machinery that has been conserved through evolution, from nematodes to humans (Shi 2002; Danial and Korsmeyer 2004). The vertebrate ortholog of CED-4, APAF-1, performs a similar function, mediating caspase activation in the presence of cytochrome c, via a structure referred to as the apoptosome (Schafer and Kornbluth 2006). CED-9 represents the founding member of the Bcl-2 family of death regulators, with multiple mammalian homologs that can act in either a proapoptotic or antiapoptotic manner. Likewise, activation of mammalian caspase-9 leads to cleavage of caspase-3 and caspase-7, which drive cell death (Shi 2002).
Genetic studies in Drosophila have revealed an additional level for controlling programmed cell death through inhibitor of apoptosis (IAP) proteins. A key member of this family, DIAP1, is expressed widely and is essential for preventing cell death during normal development (Wang et al. 1999; Goyal et al. 2000; Lisi et al. 2000). DIAP1 blocks death by binding to caspases and maintaining them in an inactive state. Expression of death activators, encoded by reaper (rpr), head involution defective (hid), or grim, overcome the inhibitory effect of DIAP1 (White et al. 1994; Grether et al. 1995; Chen et al. 1996). The Rpr, Hid, and Grim proteins bind to DIAP1, disrupting its interaction with caspases and targeting it for degradation, allowing caspase activation and cell death (for a review, see Martin 2002). Elimination of all three death activator genes blocks most programmed cell death in Drosophila, while ectopic expression of any is sufficient to trigger a death response (White et al. 1994; Grether et al. 1995; Chen et al. 1996). The mammalian death activators Smac/Diablo and Omi/HtrA2 appear to function in a similar manner as rpr, hid, and grim, inhibiting death repressors such as Survivin and XIAP, demonstrating that this pathway has been conserved through evolution (Riedl and Shi 2004).
Upstream signaling pathways dictate the appropriate temporal and spatial patterns of programmed cell death during development, ensuring that this response is restricted to cells that are fated to die. Key among these signals are small lipophilic hormones that act through members of the nuclear receptor family of transcription factors. In frogs, thyroid hormone signals the destruction of the tadpole tail and remodeling of the intestine as the animal progresses from a juvenile to adult form (Shi et al. 2001). Similarly, steroid hormones regulate mammalian apoptotic pathways, including the glucocorticoid-induced apoptosis of immature thymocytes and mature T cells (Winoto and Littman 2002).
Although relatively little is known about the mechanisms of hormone-regulated programmed cell death in vertebrates, significant insights into this pathway have been gained in Drosophila, where the steroid hormone ecdysone directs the massive and rapid destruction of larval tissues during metamorphosis. A high titer pulse of ecdysone at the end of larval development acts through the EcR/USP nuclear receptor heterodimer to signal puparium formation and the destruction of several larval tissues, including the midgut (Baehrecke 2003; Yin and Thummel 2005). A second ecdysone pulse, ∼10 hr after puparium formation (APF), triggers adult head eversion, marking the prepupal-to-pupal transition and signaling the rapid destruction of the larval salivary glands (Robertson 1936; Jiang et al. 1997). Destruction of the larval midguts and salivary glands is accompanied by classic hallmarks of apoptosis, including acridine orange staining, TUNEL staining, and caspase activation (Jiang et al. 1997). Ecdysone triggers cell death through a transcriptional cascade that converges on rpr and hid induction, overcoming the inhibitory effect of DIAP1 and inducing the apical caspase DRONC (Jiang et al. 1997, 2000; Lee et al. 2002; Daish et al. 2004; Yin and Thummel 2004). Ecdysone directly regulates rpr transcription through at least one response element in its promoter (Jiang et al. 2000). This effect is augmented through the ecdysone induction of several transcription factor-encoding genes, including the Broad-Complex (BR-C), E74, and E93, which are required for proper rpr and hid expression as well as larval tissue cell death (Baehrecke 2003; Yin and Thummel 2005). The larval salivary glands also display hallmarks of autophagy, characterized by the formation of intracellular autophagic vesicles (Lee and Baehrecke 2001; Neufeld and Baehrecke 2008). Autophagy is induced just prior to salivary gland cell death and appears to act in parallel with caspases to drive tissue destruction (Berry and Baehrecke 2007).
Although the core cell death machinery was discovered through genetic screens in C. elegans, relatively few efforts have been made to exploit genetics for furthering our understanding of cell death control. For example, the only screen in Drosophila for loss-of-function mutations that affect cell death was restricted to 129 chromosomal deletions, resulting in identification of the H99 deficiency that removes rpr, hid, and grim (White et al. 1994). The Drosophila eye has also been used as a context for genetic screens to identify cell death regulators (Hay et al. 1995, 1997; Bergmann et al. 1998; Goyal et al. 2000; Lisi et al. 2000). These studies screen for enhancement or suppression of ectopically expressed cell death regulators, and led to the discovery of several key regulators of cell death, including Ras signaling and DIAP1 (Hay et al. 1995; Bergmann et al. 1998). Although the adult eye provides an easy platform for conducting genetic screens, these efforts have a distinct limitation in that they are based on ectopic expression and a nonnatural death response. In large part, this paucity of genetic approaches is due to the difficulty of screening for defects in cell death responses, which are usually restricted to isolated clusters of cells that die at specific times during development.
We have exploited the massive destruction of larval tissues during metamorphosis as a context for identifying new regulators of programmed cell death. By expressing GFP specifically in larval salivary glands, we can rapidly and easily screen for mutants that affect this death response (Ward et al. 2003). Our understanding of the molecular mechanisms by which ecdysone triggers cell death also provides an ideal framework for integrating new cell death regulators into this pathway.
Here, we describe open-ended genetic screens for loss-of-function mutations that affect ecdysone-triggered salivary gland cell death. A pilot screen using P-element insertions identified five genes as essential for this response, including genes that encode the CBP transcriptional cofactor encoded by nejire and the TATA box-binding protein (TBP)-related factor 2 (dTRF2). We also identified two known regulators of salivary gland cell death, E74 and hid, validating our approach and indicating that our screens have the potential to reveal new factors that directly impact the core cell death machinery. We expanded our effort by conducting a large-scale open-ended screen on the third chromosome for pupal lethal mutations that display defects in salivary gland cell death. Through this work, we identify seven multiallelic complementation groups and map three of these to specific genes: CG5146, which encodes a protein of unknown function, Med24, which encodes a component of the RNA polymerase II mediator complex, and CG7998, which encodes a putative mitochondrial malate dehydrogenase. These studies provide a new basis for characterizing the regulation of programmed cell death in Drosophila and have implications for understanding how this response is controlled in all higher organisms.
MATERIALS AND METHODS
Drosophila stocks:
All crosses were carried out at 25° on standard cornmeal molasses medium. P-element induced lethal mutations on the X and third chromosomes were obtained from the Bloomington Drosophila Stock Center. The X chromosome collection consisted of 398 stocks that included most lethal alleles generated in the original P-element mutagenesis screen (Peter et al. 2002). The third chromosome collection consisted of 467 lethal P-element insertions characterized and reduced to a unigene set by the Berkeley Drosophila Genome Project (Bellen et al. 2004). Two different GFP reporters were used to follow larval salivary gland cell death. For the pilot screen on the X chromosome, we used SG>GFP (w1118; {UAS-GFP}; {SG-GAL4}) kindly provided by A. Andres (University of Nevada, Las Vegas). For the third chromosome screens, we used fkh-GAL4 and UAS-GFP transgenes recombined onto a w1118 chromosome (w1118, {fkh-GAL4}, {UAS-GFP}). Dominant marker stocks and deficiency stocks used for mapping were obtained from the Bloomington Drosophila Stock Center.
EMS screen for pupal lethal mutants:
Several w1118 lines were isogenized for the third chromosome, and the healthiest of these strains was used for the screen. Newly eclosed w1118 males were aged for 3–4 days, starved for 8–12 hr, and then fed 10 mm EMS in 5% sucrose for 12–24 hr. Males were allowed to recover on regular food for several hours before mating with virgin females that carry the appropriate reporter transgene, phenotypic markers, and balancer chromosome (Figure 2). The F0 crosses were set up en masse with 25 males and 50 virgin females per bottle. Each F0 bottle was transferred to fresh bottles every day for 4 days. Individual F1 males were collected and crossed to 3–5 multiply marked virgin females. Each F2 stock established from these F1 crosses carried a single mutagenized third chromosome; these were given a unique number and scored for lethality. Absence of the dominant pupal marker Tubby on the TM6B, Hu Tb balancer chromosome was used to identify homozygous mutant pupae (Figure 2). Thus, F2 stocks carrying a lethal mutation on the third chromosome were scored by the absence of empty non-Tubby pupal cases on the wall of the vial. We then selected stocks in which the mutants arrest primarily during metamorphosis, with ≥75% of the expected non-Tubby progeny dying after puparium formation. Among these mutants, only those that arrested during or after head eversion were considered pupal lethals and examined for salivary gland cell death defects. For the pilot screens, we first selected stocks that appeared to have a significant number of pupae that never eclosed. These stocks were crossed to appropriate balancer chromosomes (FM7i, Act-GFP for the X chromosome lethal mutations and TM6B, Hu Tb for the third chromosome lethal mutations) to select stocks in which ≥75% of the mutant progeny die as pupae.
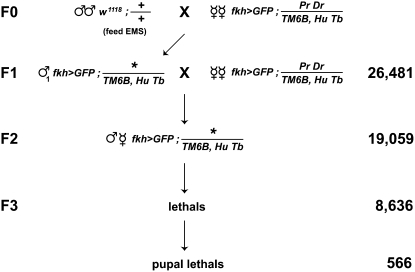
Figure 2.—
A genetic screen for pupal lethal mutations. Males fed 10 mM EMS were mated en masse to females carrying a salivary gland-specific GFP marker on the X chromosome (fkh>GFP = w1118, {fkh-GAL4}, {UAS-GFP}) and dominant markers Pr Dr along with a Tubby-marked balancer (TM6B, HuTb) on the third chromosome. Individual F1 males were crossed to the same female stock used in the F0 generation, allowing the establishment of F2 stocks, each of which carries a unique mutagenized third chromosome. We established 19,059 F2 stocks from 26,481 F1 crosses and identified 8636 lethal mutations in the F3 generation. Of these, 566 had a primary lethal phase during pupal stages of development.
Screening for persistent larval salivary glands:
Persistent salivary glands (PSGs) were visualized by tissue specific GFP expression in intact animals (Ward et al. 2003). For the pilot screen on the X chromosome, the SG>GFP reporter was crossed to each pupal lethal stock and the F1 mutant progeny examined for PSGs at ∼12 hr after head eversion (AHE), when glands in wild-type pupae are no longer detectable. For the large-scale EMS mutagenesis screen, all F2 stocks carried a salivary gland-specific GFP reporter on the X chromosome (w1118, {fkh-GAL4}, {UAS-GFP}), allowing non-Tubby pupae to be scored for PSGs at ∼12 hr AHE. The selection of mutations with PSGs was done in two steps. First, pupal lethal stocks were subjected to a rapid screen in which culture vials were rotated under a dissecting microscope equipped with a UV light source to identify PSGs. Stocks that consistently contained pupae that displayed PSGs for at least 1 day were selected and subjected to a second, more thorough, examination. For the detailed PSG screen, embryos were collected for 2 days from each mutant stock and allowed to develop. All pupae were removed from these vials, transferred to moist black filter paper in a petri dish, and scored at 12 hr AHE for two phenotypes: PSGs (by examining GFP expression) and morphogenesis of adult structures (by examining external pupal morphology, primarily head eversion and leg elongation). We developed criteria to identify pupae that had undergone the normal morphogenetic movements associated with pupation (see results). In this manner, we were able to document the percentage of total PSGs and the percentage of normal pupae with PSGs for each pupal lethal stock. These data were tabulated and used to select mutations in potential regulators of programmed cell death.
Immunohistochemistry and microscopy:
Larval salivary glands were dissected at the appropriate stage, fixed, and stained with antibodies directed against the cleaved/active form of caspase-3 (Cell Signaling Technologies) and Cy3-labeled donkey anti-rabbit secondary antibodies (Jackson Laboratories), both at 1:200 dilution, as described (Boyd et al. 1991). Samples were mounted in VECTAShield and imaged using a Zeiss Axioskop 2 microscope. All figures were processed in parallel with Photoshop CS3.
RNA isolation and Northern blot hybridizations:
Larval salivary glands were dissected from staged prepupae or pupae. Equal amounts of total RNA, isolated using Tripure (Roche), were fractionated on 1% formaldehyde gels and transferred to nylon membranes for Northern blot hybridization. Probes were prepared as previously described (Andres et al. 1993).
RESULTS
A pilot screen for mutants with defects in salivary gland cell death:
Previous studies have identified a number of mutations in regulators or effectors of cell death that result in both pupal lethality and defects in the destruction of larval salivary glands, including BR-C, E74A, βFTZ-F1, E93, and dronc (Jiang et al. 2000; Lee et al. 2002; Daish et al. 2004). This observation suggested that we could identify new genes that contribute to steroid-triggered cell death by conducting an open-ended genetic screen in two steps, first selecting for pupal lethality and then selecting for specific defects in salivary gland destruction. To test this idea, we conducted a small-scale screen taking advantage of the ability to follow the fate of salivary glands in living animals with GFP, where a block in cell death results in persistent salivary glands (PSGs) (Figure 1, A–D; Ward et al. 2003). We started with a collection of 398 lethal P-element insertions on the X chromosome. Of these, 94 lines were selected as pupal lethal mutants, in which ≥75% of mutant animals died during metamorphosis (see materials and methods). For each pupal lethal mutant stock, virgin females carrying the lethal mutation and a GFP-marked balancer chromosome (FM7i, Act-GFP) were crossed to males carrying the SG>GFP salivary gland reporter. This allowed us to examine salivary gland cell death in each mutant pupa (non-FM7i, Act-GFP males) using a dissecting microscope equipped with a UV light source. Surprisingly, 51 of the 94 pupal lethals displayed significant defects in salivary gland destruction. Most of these mutants, however, also displayed defects in other ecdysone-triggered processes like head eversion and leg elongation, indicating that the corresponding mutations were general regulators of ecdysone signaling and not specific to the death pathway. By eliminating mutations with pleiotropic ecdysone signaling defects, we identified nine mutant lines with the desired phenotype: normal overall pupal morphology, relatively normal head eversion, and proper adult leg elongation, but clear defects in salivary gland cell death (Figure 1F and Table 1). These nine P-element insertions corresponded to three genes: three mutations mapped to CBP/nejire [l(1)G0350, l(1)G0112, l(1)G0470], four mutations mapped to dTrf2 [l(1)G0039, l(1)G0071, l(1)G0295, l(1)G0425], and one mutation mapped to Rala [l(1)G0501]. The remaining P-element insertion resides within a yoyo transposable element and was not mapped.
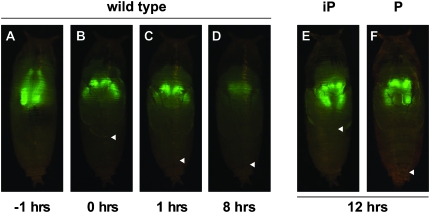
Figure 1.—
Salivary gland-specific GFP expression provides a marker to follow cell death in wild-type and mutant pupae. Salivary gland-specific GFP expression is clearly visible in wild-type late prepupae ∼1 hour before adult head eversion (AHE) (A), as well as a few minutes (B) or 1 hr after the onset of head eversion (C), but is significantly reduced by ∼8 hr AHE (D) due to cell death. This GFP expression persists at 12 hr AHE in mutant pupae in which salivary gland cell death is disrupted (E and F). The adult legs normally extend to full length at head eversion (marked by arrowheads in B and C) and maintain this length through pupal stages (arrowhead, D). Mutations in general regulators of ecdysone responses, like rbp5, block leg elongation as well as salivary gland cell death (E), resulting in incomplete pupation (iP), while mutations recovered from the pilot screen, such as CBP, show normal leg elongation and head eversion despite the disruption of salivary gland cell death (F), resulting in relatively normal pupation (P).
TABLE 1.
Summary of pilot screen for persistent larval salivary glands
| Lethals (total) | Pupal lethals | PSG alleles | PSG loci | |
|---|---|---|---|---|
| X chromosome | 398 | 94 | 9a | 3 |
| Third chromosome | 467 | 131 | 2 | 2 |
| Total | 865 | 225 | 11 | 5 |
P-element induced lethal mutations on the X or third chromosomes were first screened for pupal lethality. A salivary gland-specific GFP marker was then crossed into these pupal lethals and alleles were selected that displayed a persistent salivary gland (PSG) phenotype. The numbers corresponding to each class are listed.
One P-element mutation could not be mapped to a gene because it resides within a yoyo transposable element.
We attempted to expand this pilot screen to the autosomes by examining a collection of 467 lethal P-element mutations on the third chromosome. This effort, however, was less successful because relatively few stocks had a high penetrance of pupal lethality, most likely due to an accumulation of second-site mutations over time. Despite the reduced number of mutant stocks that could be screened, we recovered two additional mutations from this effort: E74 (Eip74neo24) and hid (W05014). In total, 865 P-element-induced lethal mutations were screened on the X and third chromosomes, and five mutations were identified that led to specific defects in the destruction of larval salivary glands in otherwise normal-looking pupae (Table 1). Two of these mutations reside in known regulators of salivary gland cell death, E74 and hid (Jiang et al. 2000; Yin and Thummel 2004). In addition, subsequent characterization of CBP and dTrf2 revealed roles for these factors in regulating the salivary gland death response (Bashirullah et al. 2007; Yin et al. 2007). The identification of these new cell death regulators thus validated our screening parameters and established a foundation for conducting a large scale EMS screen to identify other factors that contribute to steroid-triggered programmed cell death.
An EMS screen for pupal lethal mutants:
Given the low penetrance of pupal lethality in most available collections of lethal mutations, we set out to generate a new collection of pupal lethal mutants that could serve as a starting point for our screen. By using a relatively low dose of EMS (10 mm) we tried to minimize the frequency of multiple lethal hits per chromosome and thus increase the probability of recovering late lethal mutations. After 9 months of weekly mutageneses, we established 19,059 F2 stocks from 26,481 F1 crosses, with each F2 stock derived from a single mutagenized third chromosome and carrying a TM6B, Hu Tb balancer chromosome (Figure 2). We used the absence of the dominant marker Tubby (Tb) on the balancer chromosome to follow the homozygous progeny in each F2 stock that carry the mutagenized third chromosome and selected 8636 stocks with lethal mutations. Based on a Poisson distribution of the number of lethal stocks recovered (45% of all F2's), we expect that our mutagenesis protocol generated <1 lethal hit per chromosome (data not shown). We then selected stocks in which at least three-quarters of the mutant animals die after puparium formation (see materials and methods). After repeating this selection four times in consecutive generations, we identified 637 stocks with a consistent lethal phase during metamorphosis. Of these stocks, 71 had a lethal phase during the prepupal stage, prior to head eversion and the onset of larval salivary gland cell death. These prepupal lethal mutants were not examined further, leaving us with 566 stocks that carry a pupal lethal mutation on the third chromosome.
Large-scale screen for persistent salivary glands:
The 566 pupal lethal mutants were subjected to a two-step selection process to identify those mutants that display a high frequency of PSGs in an otherwise normal pupa. All F2 stocks from the screen carry a salivary gland-specific GFP reporter on the X chromosome and a Tubby-marked balancer chromosome maintaining the mutagenized third chromosome (Figure 2). For an initial rapid screen to identify PSGs, vials of each F2 pupal lethal stock were examined under a dissecting microscope to visualize the larval salivary glands in mutant (non-Tubby) pupae. Salivary glands with a complete block in cell death remain visible for several days, facilitating the identification of stocks that display even a low frequency of PSGs (Ward et al. 2003). In this manner, 266 stocks with apparent PSGs were selected for further characterization. In this more detailed analysis, all mutant pupae from each lethal stock were removed from the culture vial and followed over the course of several days, scoring them individually for PSGs and overall adult morphology. The goal of this effort was to identify stocks that display a high penetrance of PSGs with little or no effect on other ecdysone responses. Two key indicators of successful ecdysone-triggered pupation were scored in each mutant line, adult head eversion and adult leg elongation, both of which are easy to visualize in intact living animals. The head everts rapidly in an anterior direction to differentiate in the adult head of the fly (0 hr AHE; Figure 1B) (Chadfield and Sparrow 1985). This is followed by leg elongation, which is completed ∼1 hr AHE (see arrowheads in Figure 1, B and C) (Ward et al. 2003). Mutant pupae without any signs of morphogenesis of adult structures were scored as “PP” to indicate an arrest during “prepupal” stages (resembling the animal depicted in Figure 1A). Mutant pupae with defects in adult structures were scored as “iP” to indicate “incomplete pupation” (Figure 1E). Mutant pupae with normal adult head and leg morphology were scored as “P” to indicate the formation of a normal pupa (Figure 1F). These phenotypes were scored for each mutant animal to identify stocks with the most specific defects in larval salivary gland cell death.
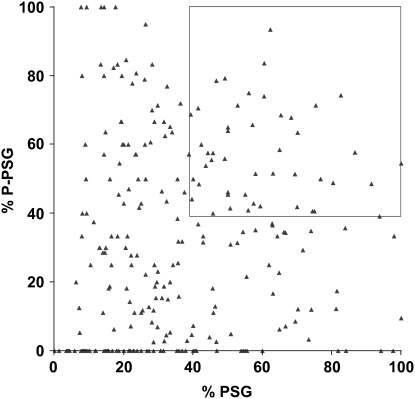
On average, 47 mutant pupae were examined from each of the 266 pupal lethal stocks that display PSGs. Each mutant pupa was first scored for the presence or absence of PSGs and then subdivided into phenotypic classes on the basis of the extent of pupation events: prepupal lethality (PP), incomplete pupation (iP), or normally formed pupae (P). An example of this breakdown is shown in Table 2 for the seven multiallelic complementation groups that are described in more detail below. The fraction of mutant pupae that display PSGs in each stock—percentage of PSG—was calculated by dividing the number of mutants in the PSG(iP) and PSG(P) classes by the total number of mutant pupae analyzed. Prepupa with PSGs [PSG(PP)] were not included in this calculation because salivary glands are not destroyed until after the prepupal stage. Although the fraction of mutant pupae that display PSGs (a high percentage of PSG) is important, we added a second parameter to identify those stocks in which most animals with PSGs also form relatively normal adult morphology—percentage of P-PSG. This number was calculated by dividing the number of PSG(P) animals by the total number of pupae with PSGs. A plot of the percentage of PSG and percentage of P-PSG for each of the 266 pupal lethal mutants is shown in Figure 3. Although a high percentage of P-PSG is the ideal phenotype, many of these mutant stocks have a low percentage of PSG. For example, among the seven mutations with ≥90% P-PSG, only one has >35% PSG. To restrict our initial studies to the most promising PSG mutations, we arbitrarily selected a cutoff of ≥39% PSG and ≥39% P-PSG (box, Figure 3). Among the 266 pupal lethal mutants, 102 stocks have ≥39% P-PSG and 98 stocks have ≥ 39% PSG, but only 48 fulfilled both criteria. These 48 stocks represent a collection of pupal lethal mutations that appear to specifically disrupt the ecdysone-triggered destruction of larval salivary glands and provide a good starting point for more detailed phenotypic characterization.
TABLE 2.
Phenotypic analysis of l(3)psg alleles recovered from the EMS screen
| PSG
|
Non-PSG
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci | Allele | n | PP | iP | P | Sum | PP | iP | P | Sum | % PSG | % P-PSG |
| l(3)psg1 | 1305 | 43 | 1 | 16 | 17 | 34 | 0 | 1 | 8 | 9 | 77 | 50 |
| 3908 | 49 | 3 | 8 | 24 | 35 | 0 | 0 | 14 | 14 | 65 | 69 | |
| 4584 | 43 | 1 | 6 | 20 | 27 | 2 | 0 | 14 | 16 | 61 | 74 | |
| l(3)psg2 | 2300 | 30 | 0 | 11 | 15 | 26 | 0 | 0 | 4 | 4 | 87 | 58 |
| 15203 | 49 | 1 | 16 | 18 | 35 | 0 | 2 | 12 | 14 | 69 | 51 | |
| l(3)psg3 | 3469 | 43 | 0 | 19 | 13 | 32 | 3 | 0 | 8 | 11 | 74 | 41 |
| 4672 | 50 | 4 | 12 | 17 | 33 | 0 | 6 | 11 | 17 | 58 | 52 | |
| l(3)psg4 | 3629 | 55 | 5 | 12 | 15 | 32 | 0 | 4 | 19 | 23 | 49 | 47 |
| 5780 | 48 | 1 | 27 | 18 | 46 | 0 | 0 | 2 | 2 | 96 | 39 | |
| l(3)psg5 | 5307 | 41 | 0 | 9 | 19 | 28 | 0 | 3 | 10 | 13 | 68 | 68 |
| 9293 | 64 | 7 | 9 | 20 | 36 | 0 | 3 | 25 | 28 | 45 | 56 | |
| l(3)psg6 | 10202 | 40 | 2 | 17 | 13 | 32 | 0 | 5 | 3 | 8 | 75 | 41 |
| 14416 | 59 | 0 | 10 | 15 | 25 | 0 | 17 | 17 | 34 | 42 | 60 | |
| l(3)psg7 | 6753 | 112 | 4 | 11 | 33 | 48 | 0 | 5 | 59 | 64 | 39 | 69 |
| 14144 | 95 | 0 | 13 | 32 | 45 | 0 | 2 | 48 | 50 | 47 | 71 | |
| 16677 | 47 | 1 | 15 | 12 | 28 | 0 | 7 | 12 | 19 | 57 | 43 | |
Alleles from each of the seven lethal complementation groups are listed, along with the total number of mutant pupae screened for each (n). These animals were divided into two groups, those with persistent salivary glands at 12–36 hr AHE (PSG) and those without (non-PSG). Each group was subdivided into phenotypic classes representing mutants that arrest as prepupae (PP), mutants that undergo incomplete pupation (iP), or mutants that display essentially normal pupation (P). % PSG = [(PSG(iP) + PSG(P))/n]. % P-PSG = [(PSG(P)/Sum(PSG)].
Figure 3.—
A graphical depiction of the persistant salivary gland phenotypes for 266 pupal lethal stocks. The percentage of PSG (x-axis) and percentage of P-PSG (y-axis) for all 266 pupal lethal stocks are plotted. Many stocks that have a high percentage of PSG do not also have a high percentage of P-PSG, suggesting that they are separable measures that can be used to select the best candidates for further study. The 48 stocks defined by the cutoff of ≥39% PSG and ≥39% P-PSG are marked by a box.
Characterization of seven loci with persistent salivary glands:
Inter se complementation tests among the top 48 pupal lethal mutants that display PSGs identified seven multiallelic loci [referred to as l(3)psg1-7] represented by 16 mutations (listed in Table 2). The remaining 32 monoallelic loci are not pursued further here. As a first step toward characterizing the PSG defects associated with these seven loci, we asked whether caspases are activated in the salivary glands of mutant pupae. As expected, caspase activation is evident in wild-type salivary glands by 1.5 hr AHE (Figure 4, A and B). No caspase activation, however, is seen in salivary glands dissected from representatives of each of the seven l(3)psg loci (Figure 4, C–W). In addition, the morphology of the larval salivary glands in these mutants showed few signs of tissue breakdown by 6 hr AHE, a time when wild-type salivary glands are no longer intact (data not shown). Thus, each of the seven l(3)psg loci regulate critical aspects of the cell death machinery that result in caspase activation and subsequent destruction of larval salivary glands.
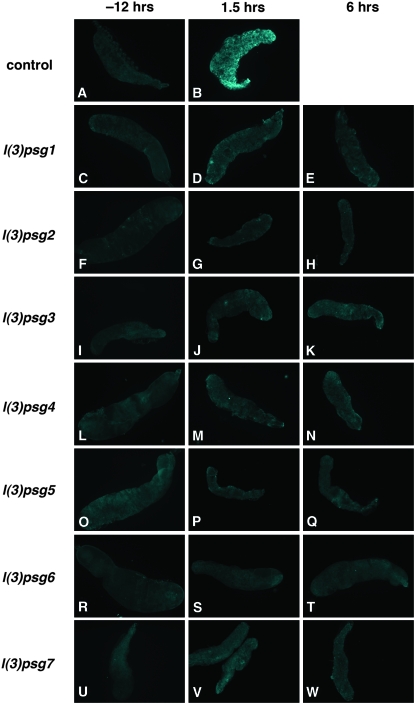
Figure 4.—
Caspases are not activated in salivary glands from staged mutant pupae. Salivary glands dissected from white prepupae (−12 hr AHE, left column) or after head eversion (1.5 hr AHE, middle column or 6 hr AHE, right column) were stained with antibodies directed against cleaved active caspase-3. Although caspases are activated soon after head eversion in wild-type (w1118) salivary glands (B), no caspase activation is detected in salivary glands from representative mutants of each of the seven l(3)psg loci isolated in the screen (C-W).
The l(3)psg loci have different effects on rpr and hid expression:
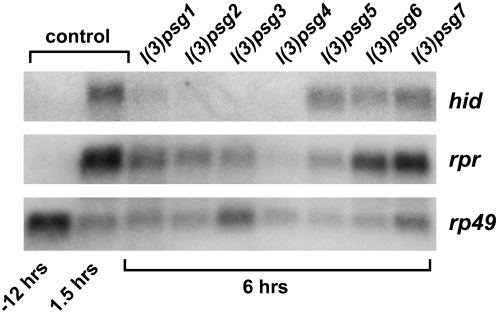
The ecdysone-induced transcriptional induction of rpr and hid initiates caspase activation and larval salivary gland cell death. To determine if mutations in any of the seven l(3)psg loci affect this ecdysone-triggered genetic cascade, we examined rpr and hid expression in mutant salivary glands. RNA was extracted from salivary glands of control pupae at head eversion or 1.5 hr later and at 6 hr AHE from representatives of each of the seven l(3)psg loci. Levels of rpr and hid mRNA were determined by Northern blot hybridization (Figure 5). As expected, rpr and hid are induced in control salivary glands by the prepupal ecdysone pulse, with high levels of mRNA accumulation by 1.5 hr AHE (Figure 5). Salivary glands from each of the seven l(3)psg mutants, however, show different effects on rpr and hid expression (Figure 5). Mutations in four loci, l(3)psg1-4, result in little or no detectable hid mRNA and reduced levels of rpr expression at 6 hr AHE, with almost no rpr or hid induction in l(3)psg4 mutants. Salivary glands from l(3)psg5 mutants also show effects on death activator expression, but with more significant effects on rpr expression than that of hid. These five l(3)psg loci appear to represent genes that act upstream of the death activators in the ecdysone-triggered transcriptional cascade that leads to salivary gland cell death. In contrast, l(3)psg6 and l(3)psg7 appear to have no significant effects on rpr or hid induction in spite of their penetrant defects in larval salivary gland cell death, suggesting that these genes act in parallel with, or downstream from, the induction of death activators.
Figure 5.—
Most l(3)psg mutants display defects in rpr and hid expression. RNA was extracted from salivary glands of staged control animals at either puparium formation (−12 hr) or 1.5 hr AHE (1.5 hr) and from salivary glands of each l(3)psg mutant staged at 6 hr AHE. RNA was analyzed by Northern blot hybridization for levels of rpr and hid transcript. Both rpr and hid are induced by 1.5 hr AHE in control salivary glands, in response to the prepupal pulse of ecdysone that triggers pupation. Mutants l(3)psg1, l(3)psg2, l(3)psg3, l(3)psg4, and l(3)psg5 show defects in induction in one or both death activators, while l(3)psg6 and l(3)psg7 do not show significant changes in death activator expression. Hybridization to detect rp49 mRNA was used as a control for loading and transfer.
Recombination mapping of the l(3)psg loci using dominant markers:
More detailed functional characterization of the l(3)psg loci requires the identification of their corresponding genes. As a first step toward this goal, we used recombination mapping and crosses with mapped deficiencies to position each l(3)psg locus on the third chromosome. Rather than using recessive phenotypic markers or molecularly defined SNPs as is commonly done, we chose to use dominant phenotypic markers for recombination mapping. Although there are a few classic examples of using dominant phenotypic markers for recombination mapping (e.g., Shearn 1974), their use had fallen out of favor because there are not enough dominant markers to provide a high-density genetic map. The recent availability, however, of small molecularly defined deficiencies that cover large regions of the genome provide a rapid and simple means of refining the relatively broad genetic intervals defined by dominant marker mapping. We estimated the location of each l(3)psg locus using three sets of dominant markers: Roughened (1.4) and Dichaete (40.7); Glued (41.4), Stubble (58.2), and Hairless (69.5); and Hairless (69.5) and Prickly (90.0). Virgin females carrying the l(3)psg mutation and a set of dominant markers were crossed to males carrying a different allele of the l(3)psg locus being mapped along with a balancer chromosome. The most informative crosses were those in which the l(3)psg locus mapped between the dominant markers. In those crosses, the recombination frequency of the dominant markers among the viable nonbalancer progeny provided a recombination map position for each l(3)psg locus. These map positions were used to select large cytologically defined deficiencies from the Bloomington deficiency collection (DK3) to perform complementation tests. Deficiencies that failed to complement the l(3)psg mutations were, in turn, used to select overlapping molecularly defined deficiencies to further refine the location of each l(3)psg locus. Only one large cytologically defined deficiency was identified that failed to complement l(3)psg1 mutations [Tp(3:Y)ry506, 85C], providing an approximate chromosomal location for this locus at 87F. In addition, all deficiencies tested complemented l(3)psg4 mutations, which thus appears to lie within one of the gaps that still exist in the deficiency coverage of the third chromosome, near 83E. Precise locations were determined for the remaining five l(3)psg loci, to relatively small intervals on the third chromosome (Figure 6). An example of the mapping results for l(3)psg2, l(3)psg5, and l(3)psg7 is presented in Table 3. Although we used lethality to follow the l(3)psg loci during the mapping process, we found that both lethality and the PSG phenotype map to the same final small deficiency for each mapped l(3)psg locus (data not shown).
Figure 6.—
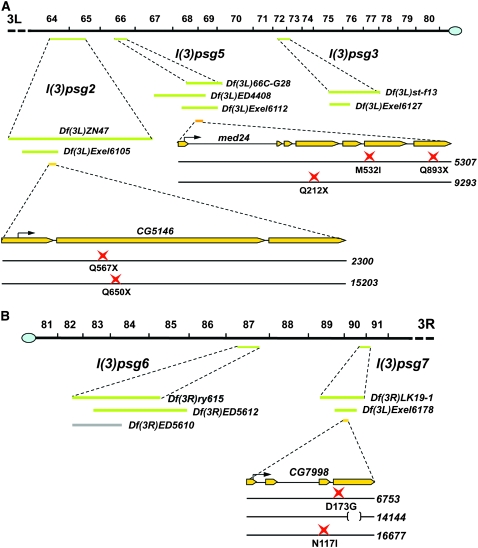
Map locations and lesions associated with the mapped l(3)psg loci. (A) Left arm of the third chromosome, from position 64 to the centromere. (B) Right arm of the third chromosome from the centromere to postion 91. Cytological map positions are shown on top. The green bars below the cytological map indicate the location of noncomplementing overlapping deficiencies for each mapped l(3)psg locus. Gray bars indicate deficiencies that complemented the l(3)psg mutations and helped to restrict their location. The yellow bars indicate the structure of mapped genes, with the start codon depicted by a black arrow. The point mutation(s) associated with each mutant allele is shown at the bottom, represented by a red star. A deletion is represented by a bracket. Df(3L)ZP1, which fails to complement l(3)psg5, is not depicted because it has essentially the same coverage as Df(3L)ED4408, which is shown. The l(3)psg22300 allele also carries seven missense mutations: E158D, L198P, P382S, H515P, A542V, D1007E, and P1160S. The l(3)psg714144 mutation is a 236-bp deletion that shifts the reading frame after codon 242, changing amino acids 243–253 from GAGSATLSMAY to EGQEEHPEGHX. Deficiency maps and gene structures are derived from FlyBase (Wilson et al. 2008).
TABLE 3.
Mapping of l(3)psg2, l(3)psg5, and l(3)psg7
| PSG loci | Alleles | Dominant marker mapping | Estimated position | Cytological deficiencies tested | Defined deficiencies tested |
|---|---|---|---|---|---|
| l(3)psg2 | 2300 | (Recombination within R and D) | ∼25 | Df(3L)BSC23 | Df(3L)ED4342 |
| 15203 | 1 ++, 28 R+, 42 + D | 66C–66D | Df(3L)HR119 | Df(3L)ED210 | |
| Df(3L)GN24 | Df(3L)Exel6104 | ||||
| Df(3L)ZN47 | Df(3L)Exel6105 | ||||
| Df(3L)XDI98 | Df(3L)Exel6106 | ||||
| Df(3L)BSC27 | Df(3L)Exel6107 | ||||
| Df(3L)BSC33 | Df(3L)Exel7210 | ||||
| Df(3L)pbl-X1 | Df(3L)ED212 | ||||
| Df(3L)ZP1 | |||||
| Df(3L)66C-G28 | |||||
| Df(3L)h-i22 | |||||
| Df(3L)BSC35 | |||||
| Df(3L)AC1 | |||||
| l(3)psg5 | 5307 | (Recombination within R and D) | ∼34 | Df(3L)BSC27 | Df(3L)Exel6112 |
| 9293 | 1 ++, 20 R+, 91 + D | 67E-F | Df(3L)BSC33 | Df(3L)ED4408 | |
| Df(3L)pbl-X1 | Df(3L)Exel6279 | ||||
| Df(3L)ZP1 | Df(3L)Exel8104 | ||||
| Df(3L)66C-G28 | |||||
| Df(3L)BSC13 | |||||
| Df(3L)h-i22 | |||||
| Df(3L)BSC35 | |||||
| Df(3L)AC1 | |||||
| Df(3L)BSC14 | |||||
| Df(3L)vin5 | |||||
| Df(3L)vin7 | |||||
| l(3)psg7 | 6753 | (Recombination within Sb and H) | ∼66 | Df(3R)Cha7 | Df(3R)Exel6178 |
| 14144 | 1 ++, 9 Sb+, 23 + H | 91F | Df(3R)DG4 | ||
| 16677 | Df(3R)LK19-1 |
The three mapped l(3)psg loci are listed on the left along with the alleles that define each lethal complementation group. One example of dominant marker mapping is provided, showing the number of recombinants recovered along with the estimated map position. All cytological and defined deficiencies used to map each locus are listed, with the noncomplementing deficiencies underlined.
Most l(3)psg loci correspond to novel death regulators:
Finally, we set out to determine if any of the five l(3)psg loci mapped by molecularly defined deficiencies correspond to known lethal mutations. For this purpose, we crossed the l(3)psg alleles to all available lethal mutations in the Bloomington Drosophila Stock Center collection that map within the smallest genetic interval defined for that locus. Two loci—l(3)psg3 and l(3)psg6—complemented all available lethal mutations, suggesting that they may represent the first lethal alleles in their respective genes. The l(3)psg3 alleles map very close to diap1, but complement diap1 lethal alleles (th4 or thj5c8) and Df(3)brm11, which removes the diap1 locus, indicating that they do not correspond to loss-of-function mutations in this known death inhibitor. In contrast, the remaining three loci—l(3)psg2, l(3)psg5, and l(3)psg7—failed to complement available lethal mutations (Table 4). These three loci correspond to previously uncharacterized Drosophila genes. The l(3)psg2 locus maps to CG5146, a gene of unknown function. The l(3)psg5 locus maps to Med24, a component of the RNA polymerase II mediator complex, and l(3)psg7 maps to CG7998, a gene that encodes a predicted mitochondrial malate dehydrogenase (Figure 6). Sequencing of genomic DNA from the original isogenized w1118 parental stock and genomic DNA from the l(3)psg2, l(3)psg5, and l(3)psg7 alleles confirmed these molecular assignments and revealed the specific lesion associated with each mutation (Figure 6). None of the five mapped l(3)psg loci corresponds to genes or chromosomal regions associated with known regulators of programmed cell death and thus provide an opportunity to extend our understanding of this biological pathway in new directions.
TABLE 4.
Complementation analysis of l(3)psg loci using available lethal mutations in mapped intervals
| PSG loci | Known lethals | Stock no. | Complementation results |
|---|---|---|---|
| l(3)psg2 | spo | BL-3276 | Complement |
| sinu | BL-8577 | Complement | |
| sinu | BL-11690 | Complement | |
| CG10635 | BL-10064 | Complement | |
| CG10630 | BL-13914 | Complement | |
| Unknown | BL-16520 | Complement | |
| CG5146 | BL-17332 | Fail to complement | |
| l(3)psg5 | mus301 | BL-916 | Complement |
| l(3)L0139 | BL-10169 | Complement | |
| nmt | BL-12071 | Complement | |
| atg18 | BL-13945 | Complement | |
| Unknown | BL-17117 | Complement | |
| arp66B | BL-17149 | Complement | |
| Ect4 | BL-18166 | Complement | |
| med24 | BL-12847 | Fail to complement | |
| l(3)psg7 | Med17 | BL-10307 | Complement |
| repo | BL-11604 | Complement | |
| sr | BL-11618 | Complement | |
| l(3)05697 | BL-11668 | Complement | |
| ssdp | BL-13020 | Complement | |
| dlc90F | BL-14912 | Complement | |
| Unknown | BL-15475 | Complement | |
| 14-3-3epsilon | BL-17142 | Complement | |
| eIF-1A | BL-17203 | Complement | |
| CG8064 | BL-17732 | Complement | |
| CG7998 | BL-15383 | Fail to complement |
Each l(3)psg mutant was crossed to all lethal mutations in the Bloomington Drosophila Stock Center collection that are located within the region defined by the smallest deficiency that fails to complement the allele. Each lethal mutant stock is listed along with its Bloomington stock number and whether or not it complemented the l(3)psg allele.
DISCUSSION
A genetic screen for naturally occurring cell death in Drosophila:
Our understanding of the molecular mechanisms of programmed cell death traces back to classic genetic screens in C. elegans, defining the core death machinery shared by all higher organisms (Metzstein et al. 1998). In contrast, relatively little has been done to exploit open-ended genetic screens in Drosophila as a means of characterizing cell death. This is due largely to the difficulty of scoring for cell death defects in small clusters of cells during development. We have overcome this problem by scoring for defects in programmed cell death during metamorphosis, when GFP markers can be used to visualize the destruction of entire larval tissues in living animals. Moreover, the trigger that initiates this death response has been identified, the steroid ecdysone, and its downstream transcriptional cascades have been well defined, providing a molecular context for characterizing new cell death regulators. This study represents an initial effort to use the power of open-ended genetic screens to extend our understanding of cell death regulation in Drosophila.
There are three steps to our screening strategy: identification of pupal lethal mutants, identification of mutants with defects in salivary gland cell death (i.e., the presence of a PSG phenotype), and elimination of PSG mutants that show more global defects in ecdysone responses. Although we could, in principle, examine viable mutants for defects in larval tissue cell death, we scored for pupal lethality as the first step in our screen. This is because all known mutations that affect larval tissue destruction lead to lethality, with most arresting development during metamorphosis (Jiang et al. 2000; Lee et al. 2002; Daish et al. 2004). Selecting for pupal lethality should thus increase our frequency of identifying mutants of interest. Given that most lethal mutations result in arrest during embryogenesis or immediately thereafter, it was critical to ensure that we generated ∼1 lethal hit per chromosome to not mask mutations that lead to later lethality, during metamorphosis. The standard mutagenesis protocol using 25 mm EMS generates about 3 lethal hits per chromosome (Ashburner et al. 2004). We used 10 mm EMS, which should generate, on average, 1.23 lethal hits per chromosome (Ashburner et al. 2004). This is consistent with our recovery of lethal mutant stocks in 45% of the F2 progeny, corresponding to <1 lethal hit per chromosome.
By screening through multiple rounds for mutations that lead to lethality primarily during pupal stages, we seem to have selected against hypomorphic mutations in essential genes required earlier in development. This is indicated by the observation that all mutations we recovered in the seven loci act as null (amorphic) alleles, in which homozygous mutants display the same lethal phase as hemizygous mutants (in trans over a deletion for the region; data not shown). In addition, our stringent screening resulted in recovering fewer pupal lethal mutants compared to the traditional proportion of ∼25% (Ashburner et al. 2004). Of the 8636 lethal mutations recovered in the mutagenesis screen, we identified 637 (7%) with a primary lethal phase after puparium formation. We further restricted our collection by removing the 71 mutants that arrest development prior to head eversion and the onset of salivary gland cell death, leaving us with 566 pupal lethal mutants that were screened for defects in cell death.
Interestingly, a significant proportion of animals carrying pupal lethal mutations also appear to have PSGs as evidenced in both the pilot screen on the X chromosome (51/94 or 54%) and the EMS screen on the third chromosome (266/566 or 47%). To enrich for stocks that display specific defects in salivary gland cell death, we imposed additional rounds of selection on these mutant stocks. Each stock that displayed PSGs was scored for the number of mutant animals that arrest prior to salivary gland cell death (the PP class), those that display other defects in ecdysone responses (the iP class), and those that formed relatively normal pupae (the P class). This data allowed us to calculate the percentage of mutant pupae that display PSGs as well as the percentage of animals with PSGs that had normal-looking adult morphology. By comparing these two numbers, it became clear that the stocks that display a high penetrance of PSGs do not correlate with stocks that have a high penetrance of PSGs in normal-looking pupae (Figure 3). In other words, PSGs are often associated with defects in the overall process of pupation, an observation consistent with pupation being a major developmental transition coordinated by a large gene regulatory network triggered by the prepupal pulse of ecdysone. This observation also explains the high proportion of pupal lethal mutations with a PSG phenotype. It is possible that these loci represent functional targets of ecdysone-regulated transcription factors such as crol or BR-C, that direct the complex biological pathways associated with metamorphosis (Restifo and Merrill 1994; D'Avino and Thummel 1998). As a starting point for identifying mutants that display selective defects in ecdysone-triggered cell death, we chose an arbitrary cutoff of stocks that display at least 39% PSG and 39% P-PSG, resulting in a final collection of 48 pupal lethal mutants. This represents a >10-fold enrichment of potential regulators of cell death among the pupal lethal mutants (48/566). Inter se complementation tests between these 48 mutants revealed that 16 mutations represent 7 multiallelic complementation groups, leaving 32 monoalleleic complementation groups that represent other genes. The fact that a significant proportion (33%) of the PSG mutations selected by our 39% cutoff correspond to multiple hits in a small number of genes, suggests that the screening parameters are specific for loci that impact the destruction of larval salivary glands.
The mapped l(3)psg loci represent novel regulators of steroid-triggered cell death:
Taken together, the pilot screen and large-scale EMS mutagenesis screen identified 12 genes that are required for larval salivary gland cell death. Two of these genes are known to play a role in this response, E74 and hid, demonstrating that the screen can identify mutations in both trans-acting regulators as well as effectors that control the death response (Jiang et al. 2000; Yin and Thummel 2004). The P-element screens also identified mutations in genes that had not been previously associated with cell death, including multiple alleles of CBP and dTrf2. More detailed characterization of these loci defined a central role for CBP in establishing the competence to initiate salivary gland cell death (Yin et al. 2007). CBP is both necessary and sufficient in the mid-third instar for downregulation of the DIAP1 death inhibitor. In the absence of this downregulation, high levels of DIAP1 persist into the prepupal stages and block the ability of ecdysone-induced rpr and hid to trigger cell death. Interestingly, this early function of CBP appears to be in response to a low titer mid-third instar ecdysone pulse, suggesting that the hormone first provides the competence to die, through DIAP1 downregulation, and then directs tissue destruction via rpr and hid-mediated cell death. Functional studies of dTrf2 provided a less complete understanding of its role in regulating salivary gland destruction, although a number of genes that encode key ecdysone-regulated transcription factors are expressed at a reduced level and delayed in dTrf2 mutants (Bashirullah et al. 2007). The large-scale EMS screen also identified seven multiallelic complementation groups, at least five of which map to loci or genes not previously associated with programmed cell death. These screens thus provide a new direction for defining the regulation of this critical biological pathway.
Each of the seven l(3)psg loci appears to be required for caspase activation, a critical final step in programmed cell death (Figure 4). As a first step toward understanding the mechanisms by which these loci control salivary gland cell death, we examined the transcription of rpr and hid at 6 hr AHE, when salivary glands are no longer present in wild-type pupae (Figure 5). Two loci, l(3)psg6 and l(3)psg7, have no significant effect on rpr or hid mRNA levels. This observation suggests that these genes, like CBP, act downstream from, or in parallel with, the rpr and hid death activators. The identification of one of these loci, l(3)psg7, as encoding a predicted malate dehydrogenase, a nuclear-encoded, mitochondrial-targeted enzyme that acts in the citric acid cycle, is consistent with this proposal. Several recent papers have shown that Rpr and Hid are rapidly localized to mitochondria where they can regulate the dynamics of DIAP1 degradation and the cell death response (Abdelwahid et al. 2007; Goyal et al. 2007; Freel et al. 2008). It is thus possible that CG7998 mutations could lead to a disruption of mitochondrial function or integrity that, in turn, inhibits Rpr or Hid activity. Detailed characterization of CG7998 and its role in salivary gland cell death is in progress.
The remaining five l(3)psg mutants display reduced levels of death activator expression. Salivary glands from l(3)psg5 mutants show a significant reduction in rpr mRNA levels and a modest reduction of hid expression, while l(3)psg1, l(3)psg2, and l(3)psg3 mutants display more significant effects on hid expression, and l(3)psg4 mutants have very low levels of both rpr and hid mRNA. It is thus possible that these genes act upstream from rpr and hid to direct the proper levels of death activator expression required for caspase activation and a cell death response. Previous genetic studies have shown that rpr and hid regulation can be uncoupled. For example, E74A mutant salivary glands display significantly reduced levels of hid expression with no effect on rpr (Jiang et al. 2000). Similarly, the Ras/MAPK pathway selectively regulates hid expression (Bergmann et al. 1998; Kurada and White 1998). It is thus possible that these l(3)psg loci encode factors that regulate distinct subsets of the transcriptional cascade required for salivary gland cell death. Our identification of l(3)psg5 as corresponding to Med24, which encodes a component of the RNA polymerase II mediator complex, is consistent with this model. Med24 may play a critical role in rpr transcriptional induction with little or no effect on hid expression. This effect on rpr, however, is not sufficient to explain the penetrant block in salivary gland cell death seen in Med24 mutants, since loss of rpr alone has no effect on this response (Peterson et al. 2002). Rather, it is likely that additional death regulators depend on Med24 for their proper expression. As with CG7998, functional studies of Med24 are in progress to define its role in cell death control.
In conclusion, this study represents, to our knowledge, the first large-scale genetic screen for mutations in a naturally occurring programmed cell death response in Drosophila. We have identified a total of forty-four loci in these screens that selectively impact the destruction of larval salivary glands. Here we report on 12 of these loci, including seven multiallelic complementation groups that correspond to genes or genetic intervals not previously associated with cell death regulation. Further study of these genes should advance our understanding of the molecular mechanisms by which steroids control programmed cell death during development.
Acknowledgments
We thank S. Moayedi for preparing the fly food for these studies, the Bloomington Drosophila Stock Center for providing fly stocks, A. Andres for the SG>GFP stock, and U. Schäfer for help with the P-element lethal collection on the X chromosome. This work was supported by the National Institutes of Health (1R01 GM073670).
References
- Abdelwahid, E., T. Yokokura, R. J. Krieser, S. Balasundaram, W. H. Fowle et al., 2007. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell 12 793–806. [DOI] [PubMed] [Google Scholar]
- Andres, A. J., J. C. Fletcher, F. D. Karim and C. S. Thummel, 1993. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 160 388–404. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., K. G. Golic and R. S. Hawley, 2004. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Baehrecke, E. H., 2003. Autophagic programmed cell death in Drosophila. Cell Death Differ. 10 940–945. [DOI] [PubMed] [Google Scholar]
- Bashirullah, A., G. Lam, V. P. Yin and C. S. Thummel, 2007. dTrf2 is required for transcriptional and developmental responses to ecdysone during Drosophila metamorphosis. Dev. Dyn. 236 3173–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, A., J. Agapite, K. McCall and H. Steller, 1998. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95 331–341. [DOI] [PubMed] [Google Scholar]
- Berry, D. L., and E. H. Baehrecke, 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, L., E. O'Toole and C. S. Thummel, 1991. Patterns of E74A RNA and protein expression at the onset of metamorphosis in Drosophila. Development 112 981–995. [DOI] [PubMed] [Google Scholar]
- Chadfield, C. G., and J. C. Sparrow, 1985. Pupation in Drosophila melanogaster and the effect of the lethalcryptocephal mutation. Dev. Genet. 5 103–114. [Google Scholar]
- Chen, P., W. Nordstrom, B. Gish and J. M. Abrams, 1996. grim, a novel cell death gene in Drosophila. Genes Dev. 10 1773–1782. [DOI] [PubMed] [Google Scholar]
- D'Avino, P. P., and C. S. Thummel, 1998. crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development 125 1733–1745. [DOI] [PubMed] [Google Scholar]
- Daish, T. J., K. Mills and S. Kumar, 2004. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell 7 909–915. [DOI] [PubMed] [Google Scholar]
- Danial, N. N., and S. J. Korsmeyer, 2004. Cell death: critical control points. Cell 116 205–219. [DOI] [PubMed] [Google Scholar]
- Ellis, H. M., and H. R. Horvitz, 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44 817–829. [DOI] [PubMed] [Google Scholar]
- Freel, C. D., D. A. Richardson, M. J. Thomenius, E. C. Gan, S. R. Horn et al., 2008. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J. Biol. Chem. 283 367–379. [DOI] [PubMed] [Google Scholar]
- Goyal, L., K. McCall, J. Agapite, E. Hartwieg and H. Steller, 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, G., B. Fell, A. Sarin, R. J. Youle and V. Sriram, 2007. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev. Cell 12 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether, M. E., J. M. Abrams, J. Agapite, K. White and H. Steller, 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9 1694–1708. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., D. A. Wassarman and G. M. Rubin, 1995. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83 1253–1262. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., R. Maile and G. M. Rubin, 1997. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 94 5195–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., E. H. Baehrecke and C. S. Thummel, 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124 4673–4683. [DOI] [PubMed] [Google Scholar]
- Jiang, C., A. F. Lamblin, H. Steller and C. S. Thummel, 2000. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell 5 445–455. [DOI] [PubMed] [Google Scholar]
- Kurada, P., and K. White, 1998. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95 319–329. [DOI] [PubMed] [Google Scholar]
- Lee, C. Y., and E. H. Baehrecke, 2001. Steroid regulation of autophagic programmed cell death during development. Development 128 1443–1455. [DOI] [PubMed] [Google Scholar]
- Lee, C. Y., C. R. Simon, C. T. Woodard and E. H. Baehrecke, 2002. Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 252 138–148. [DOI] [PubMed] [Google Scholar]
- Lisi, S., I. Mazzon and K. White, 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. J., 2002. Destabilizing influences in apoptosis: sowing the seeds of IAP destruction. Cell 109 793–796. [DOI] [PubMed] [Google Scholar]
- Metzstein, M. M., G. M. Stanfield and H. R. Horvitz, 1998. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 14 410–416. [DOI] [PubMed] [Google Scholar]
- Neufeld, T. P., and E. H. Baehrecke, 2008. Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy 4 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, A., P. Schottler, M. Werner, N. Beinert, G. Dowe et al., 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C., G. E. Carney, B. J. Taylor and K. White, 2002. reaper is required for neuroblast apoptosis during Drosophila development. Development 129 1467–1476. [DOI] [PubMed] [Google Scholar]
- Restifo, L. L., and V. K. Merrill, 1994. Two Drosophila regulatory genes, deformed and the Broad-Complex, share common functions in development of adult CNS, head, and salivary glands. Dev. Biol. 162 465–485. [DOI] [PubMed] [Google Scholar]
- Riedl, S. J., and Y. Shi, 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5 897–907. [DOI] [PubMed] [Google Scholar]
- Robertson, C. W., 1936. The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. J. Morphol. 59 351–399. [Google Scholar]
- Schafer, Z. T., and S. Kornbluth, 2006. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10 549–561. [DOI] [PubMed] [Google Scholar]
- Shearn, A., 1974. Complementation analysis of late lethal mutants of Drosophila melanogaster. Genetics 77 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9 459–470. [DOI] [PubMed] [Google Scholar]
- Shi, Y. B., L. Fu, S. C. Hsia, A. Tomita and D. Buchholz, 2001. Thyroid hormone regulation of apoptotic tissue remodeling during anuran metamorphosis. Cell Res. 11 245–252. [DOI] [PubMed] [Google Scholar]
- Wang, S. L., C. J. Hawkins, S. J. Yoo, H. A. Muller and B. A. Hay, 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98 453–463. [DOI] [PubMed] [Google Scholar]
- Ward, R. E., P. Reid, A. Bashirullah, P. P. D'Avino and C. S. Thummel, 2003. GFP in living animals reveals dynamic developmental responses to ecdysone during Drosophila metamorphosis. Dev. Biol. 256 389–402. [DOI] [PubMed] [Google Scholar]
- White, K., M. E. Grether, J. M. Abrams, L. Young, K. Farrell et al., 1994. Genetic control of programmed cell death in Drosophila. Science 264 677–683. [DOI] [PubMed] [Google Scholar]
- Wilson, R. J., J. L. Goodman and V. B. Strelets, 2008. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 36 D588–D593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winoto, A., and D. R. Littman, 2002. Nuclear hormone receptors in T lymphocytes. Cell 109(Suppl): S57–S66. [DOI] [PubMed] [Google Scholar]
- Yin, V. P., and C. S. Thummel, 2004. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc. Natl. Acad. Sci. USA 101 8022–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, V. P., and C. S. Thummel, 2005. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell Dev. Biol. 16 237–243. [DOI] [PubMed] [Google Scholar]
- Yin, V. P., C. S. Thummel and A. Bashirullah, 2007. Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J. Cell Biol. 178 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]