Abstract
Drosophila nemo (nmo) is the founding member of the Nemo-like kinase (Nlk) family of serine–threonine kinases. Previous work has characterized nmo's role in planar cell polarity during ommatidial patterning. Here we examine an earlier role for nmo in eye formation through interactions with the retinal determination gene network (RDGN). nmo is dynamically expressed in second and third instar eye imaginal discs, suggesting additional roles in patterning of the eyes, ocelli, and antennae. We utilized genetic approaches to investigate Nmo's role in determining eye fate. nmo genetically interacts with the retinal determination factors Eyeless (Ey), Eyes Absent (Eya), and Dachshund (Dac). Loss of nmo rescues ey and eya mutant phenotypes, and heterozygosity for eya modifies the nmo eye phenotype. Reducing nmo also rescues small-eye defects induced by misexpression of ey and eya in early eye development. nmo can potentiate RDGN-mediated eye formation in ectopic eye induction assays. Moreover, elevated Nmo alone can respecify presumptive head cells to an eye fate by inducing ectopic expression of dac and eya. Together, our genetic analyses reveal that nmo promotes normal and ectopic eye development directed by the RDGN.
THE adult structures of Drosophila melanogaster are patterned during the larval stages in discrete epithelial compartments called imaginal discs. Larval imaginal discs are inherited from the embryo as small groups of progenitor cells (Garcia-Bellido and Merriam 1969). As these cells proliferate, each imaginal disc becomes compartmentalized into fields of cells expressing unique protein sets. Each protein set confers a specific cellular identity. As development progresses, highly complex and integrated signaling networks further refine the fields of cells to achieve the final organ pattern. These signaling networks not only orchestrate cell determination, but also tightly regulate proliferation and cell survival to ensure the proportionality of the resulting adult.
In Drosophila, the adult eyes, antennae, and the majority of head structures are derived from the eye-antennal imaginal discs (Haynie and Bryant 1986). The smaller, anterior region of the disc is fated to become the antenna, and the larger posterior compartment contains the eye and head primordia. In this article, we refer to the anterior and posterior compartments as the antennal and eye discs, respectively (Figure 1B). These discs are composed of two epithelial layers: the main epithelium (ME) and the squamous peripodial epithelium (PE) (Haynie and Bryant 1986). The ME comprises primordia of the compound eye, its surrounding cuticle, and the antennae, while the PE gives rise to the remainder of the head. Studies have revealed a novel role for PE cells in directing cellular events in the ME through cell–cell signaling mediated by lumenal processes (Gibson and Schubiger 2001).
Figure 1.—
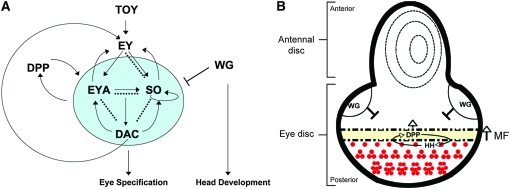
The retinal determination gene network. (A) Regulatory interactions within the RDGN. Solid arrows show direct transcriptional regulation, curved arrows demonstrate feedback loops, and dashed lines indicate physical interactions. So, Eya, and Dac are not required for ey expression during normal eye development, but can activate its expression in ectopic eye assays (Pignoni et al. 1997). Modified from Pappu and Mardon (2004) and Silver and Rebay (2005). (B) Schematic of a third instar eye-antennal imaginal disc. The antennal disc gives rise to the antenna and surrounding head cuticle. In the eye disc, the MF marks the dynamic boundary between the posterior, differentiated eye cells and the anterior head primordia. Hh activates dpp transcription in the furrow, which promotes expression of the RD genes and drives the MF forward. Wg, secreted from the anterior dorsal and ventral lobes, promotes head specification by inhibiting furrow progression and transcription of retinal specification genes. Anterior is up; dorsal is left.
Eye specification is directed in the posterior region of the eye disc by the concerted efforts of the retinal determination gene network (RDGN), a cassette of evolutionarily conserved nuclear factors (Figure 1A; reviewed in Pappu and Mardon 2004; Silver and Rebay 2005; Jemc and Rebay 2006). RDGN mutants are generally characterized by loss of eye tissue (Bonini et al. 1993; Cheyette et al. 1994; Mardon et al. 1994; Quiring et al. 1994). twin-of-eyeless (toy) (Czerny et al. 1999) and eyeless (ey) (Quiring et al. 1994) are Pax-6 genes positioned at the top of the network hierarchy. toy is expressed in the embryonic eye field and activates ey in all cells of the first instar larval imaginal disc (Figure 1A) (Czerny et al. 1999). The primary eye/antennal division of the disc is achieved by downregulation of ey in the anterior-most region of the disc in early second instar, allowing expression of the antennal selector cut (Kenyon et al. 2003). Ey deploys the RDGN by activating sine oculis (so) and eyes absent (eya) expression at the posterior margin (Halder et al. 1998; Kenyon et al. 2003). So is a member of the Six family of homedomain transcription factors (Cheyette et al. 1994). eya encodes a novel nuclear protein with protein tyrosine phosphatase and transactivating activity (Bonini et al. 1993; Rayapureddi et al. 2003; Silver et al. 2003; Tootle et al. 2003). Eya complexes with a variety of cofactors, including So (Pignoni et al. 1997) and Dachshund (Dac) (Chen et al. 1997), to regulate a battery of transcriptional targets. Dac is a nuclear protein required for furrow initiation and ommatidial patterning (Mardon et al. 1994). Dpp is required for expression of eya, so, and dac during the second larval instar (Curtiss and Mlodzik 2000; Kenyon et al. 2003) and early third instar (Chen et al. 1999). So and Eya subsequently maintain dpp expression, thereby forming a positive feedback loop (Pignoni et al. 1997; Hazelett et al. 1998).
Patterning of the retinal field occurs in the posterior region of the third instar eye disc (Figure 1B). The diffusible morphogens Wg and Dpp act antagonistically to promote head and eye fates, respectively (Royet and Finkelstein 1997). The retinal determination (RD) genes so, eya, and dac are key factors in this mutual antagonism. At the onset of third instar, hh alleviates dpp repression along the posterior margin (Royet and Finkelstein 1997; Pappu et al. 2003). Dpp antagonizes wg (Wiersdorff et al. 1996; Chanut and Heberlein 1997; Pignoni and Zipursky 1997; Royet and Finkelstein 1997), allowing initiation and subsequent progression of the morphogenetic furrow (MF) (Dominguez and Hafen 1997; Pignoni and Zipursky 1997). The MF sweeps across the eye disc in a posterior-to-anterior direction, conferring neural identity through induction of atonal (ato) (Jarman et al. 1994, 1995; Zhang et al. 2006). As the furrow traverses the disc, expression of the RD genes so, eya, and dac is maintained in its wake, as well as in the cells immediately anterior to it (Cheyette et al. 1994; Curtiss and Mlodzik 2000; Bessa et al. 2002; Pappu et al. 2003). Wg signaling in the anterior head primordia represses so, eya, and dac transcription (Baonza and Freeman 2002).
A prevalent theme in morphogenesis is the spatial and temporal regulation of specific cofactors to achieve differential interactions and outcomes using common factors. Such combinatorial control is exemplified by the RDGN. Although Ey initiates expression of so, eya, and dac during the second instar, it is restricted to cells anterior to the furrow during the third instar (Halder et al. 1998) where it promotes Wg activity to inhibit furrow progression (Bessa et al. 2002). In cells immediately anterior to the furrow, and thus receiving high levels of Dpp, Ey activates eya expression to repress the Wg target homothorax (hth) (Bessa et al. 2002). Cells more anterior receive a higher dose of Wg than Dpp and are still actively proliferating and adopting head fates. Here Ey complexes with Hth and another Wg effector, Teashirt (Tsh), to repress eya transcription, effectively inhibiting the RDGN and eye determination (Bessa et al. 2002). Thus, Ey can function as both a retinal selector and antagonist depending on its cellular context, an environment specified by the set of available cofactors.
Drosophila nemo (nmo) was first identified as a gene required for ommatidial rotation during establishment of planar cell polarity during eye development (Choi and Benzer 1994). nmo is the founding member of the Nemo-like kinase (Nlk) family of proline-directed serine–threonine kinases (Choi and Benzer 1994). Nlks are highly conserved from worms to mammals and play diverse roles in regulating cell signaling throughout development (Ishitani et al. 1999; Rocheleau et al. 1999; Zeng et al. 2007). Phosphorylation by Nlks has been shown to affect the activity of a number of proteins, including Tcf/Lef family members (Ishitani et al. 1999; Rocheleau et al. 1999) and the Drosophila Smad1 ortholog Mad (Zeng et al. 2007). nmo is an essential gene and loss of both maternal and zygotic nmo results in embryonic lethality (Mirkovic et al. 2002). nmo loss-of-function alleles survive to adulthood through perdurance of maternally supplied gene product and manifest numerous tissue patterning and growth defects (Choi and Benzer 1994; Verheyen et al. 2001; Zeng and Verheyen 2004; Zeng et al. 2007). nmo compound eyes have a distinct morphology; compared to wild type, nmo eyes are long and narrow and display square, rather than hexagonal, packing of ommatidial clusters (Choi and Benzer 1994). In addition to rotation defects, nmo mutants have reduced capacity to specify ommatidia, resulting in smaller eyes (Fiehler and Wolff 2008).
In this study we describe a dynamic pattern of expression for nmo that suggests that it may have previously uncharacterized roles in early division of eye-antennal disc, in eye specification, and in patterning the ocellar region and antennae. We show that nmo is co-expressed with various combinations of the retinal determination genes in the eye disc beginning in the second larval instar during specification of the eye field. Later, nmo not only is expressed within and behind the MF (Choi and Benzer 1994), but also is ubiquitously expressed in the PE, in the presumptive ocelli, and in a discrete pattern in the antennal disc. Loss of nmo modifies ey and eya mutant phenotypes, suggesting that nmo may modulate development mediated by the RDGN. In ectopic eye induction assays, reducing endogenous Nmo represses this effect, suggesting a requirement for nmo in RDGN-mediated fate respecification. Furthermore, Nmo potentiates the ability of Ey, Eya, and Dac to respecify head, wing, and leg tissue to retinal fate. Sufficiently high levels of Nmo induce anterior head-to-eye transformations. These respecified cells show altered transcription of the same genes affected in RD-induced ectopic eyes, supporting a role for Nmo in promoting RDGN activity. Our clonal analysis demonstrates that Nmo does not modify transcription of the canonical RD genes, further suggesting that Nmo may affect output from the RD selector complexes. Reducing endogenous nmo also rescues small-eye defects induced by early misexpression of ey and eya. Moreover, directed co-expression of nmo and ey or eya in this assay severely disrupts eye and head formation, revealing a potent synergy. Together, our data implicate nmo as a positive mediator of RDGN activity in the imaginal eye disc.
MATERIALS AND METHODS
Fly genetics:
All crosses were performed at 25° unless otherwise stated. The following fly strains were used: nmoP, also referred to as nmo-lacZ (Choi and Benzer 1994; Zeng and Verheyen 2004), nmoadk1 and nmoadk2, which express truncated transcripts (Verheyen et al. 1996, 2001), nmoDB24, a molecular null (Zeng and Verheyen 2004), eyR, ci1 (Quiring et al. 1994), eya2 (Zimmerman et al. 2000), dac1/CyO (Mardon et al. 1994), dacE462, FRT40/CyO, dpp-lacZ (Blackman et al. 1991), so-lacZ (Cheyette et al. 1994, provided by U. Waldorf), and ey-lacZ (provided by U. Waldorf). Misexpression analyses were performed using UAS-nmoC5-1e and UAS-nmob27 (Verheyen et al. 2001), UAS-GFP∷nmoII (provided by R. Fiehler, Fiehler and Wolff 2008), dpp-Gal4 (Staehling-Hampton et al. 1994), ey-Gal4 (Hazelett et al. 1998), UAS-ey (Halder et al. 1995), UAS-dac21M5M4(kindly provided by G. Mardon), and UAS-eya1 and UAS-eya2 (Bonini et al. 1998). To examine pharate lethal phenotypes, animals were dissected from pupal cases.
Clonal analysis:
nmo somatic clones were induced using the FLP/FRT method (Xu and Rubin 1993). To induce nmo loss-of-function clones using hs-FLP, embryos from the appropriate crosses were collected for 24 hr and the hatched larvae were heat-shocked at 38° for 90 min at 48 hr of development. The genotypes examined for β-galactosidase staining of dpp-lacZ in nmoDB24 and nmoadk2 clones were dpp-lacZ/ey-FLP; nmo FRT 79D/Ubi-GFP FRT79D, or y hs-FLP22/+; dpp-lacZ/+; nmoFRT 79D/Ubi-GFP FRT79D; for β-galactosidase staining of so-lacZ in nmoDB24 and nmoadk2 clones, so-lacZ/ey-FLP; nmo FRT 79D/Ubi-GFP FRT79D or y hs-FLP22/+; so-lacZ/+; nmo FRT 79D/Ubi-GFP FRT79D; for β-galactosidase staining of ey-lacZ in nmoDB24 and nmoadk2 clones, ey-lacZ/ey-FLP; nmo FRT 79D/Ubi-GFP FRT79D or y hs-FLP22/+; ey-lacZ/+; nmo FRT 79D/Ubi-GFP FRT79D; for all other antibodies, nmoDB24, nmoadk1, nmoadk2, and nmoP alleles were used in the following scheme: ey-FLP/+; nmo FRT 79D/Ubi-GFP FRT79D or y hs-FLP22; +; nmo FRT 79D/Ubi-GFP FRT79D. nmoDB24 somatic clone images in Figure 10 were generated using ey-FLP.
Figure 10.—
Nmo promotes eye development independently of RDGN gene activation. (A, C, E, G, and I) Wild type. (B, D, F, H, and J) nmoDB24 somatic clones are marked by loss of GFP (green). (C–J) nmo loss-of-function clones have no effect on Ey (A and B), Eya (C and D), so-lacZ (E and F), Dac (G and H), or dpp-lacZ expression (I and J). Eye discs are oriented with the anterior up, dorsal left.
dacE462 somatic clones were induced in nmoDB24 and nmoadk1 heterozygotes in the following genotype: y hs-FLP22; dacE462, FRT40/Ubi-GFP, FRT40; nmo/+.
Immunostaining:
Dissection of imaginal discs and antibody staining was performed following standard protocols. The antibodies used were rabbit anti-Atonal (1:1000; gift of Y. N. Jan, Jarman et al. 1994), mouse anti-β-galactosidase (1:500; Promega), rabbit anti β-galactosidase (1:2000; Cappel), mouse anti-β-galactosidase (1:250, Promega), mouse anti-cyclin B [1:20; Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Dac2-3 (1:75; DSHB), mouse anti-Eya10H6 (1:200; DSHB), rabbit anti-Ey (1:1000, gift of U. Walldorf, Halder et al. 1998), rat anti-ELAV (1:100; DSHB), mouse anti-Glass (1:2; DSHB), guinea pig anti-Hth (1:1000, gift of R. Mann, Abu-Shaar et al. 1999), rabbit anti-Hth (1:500, gift of G. Morata, Azpiazu and Morata 2002), and rabbit antiphospho-histone 3 (1:1000, Upstate Biotechnology). Secondary antibodies were used at 1:200 and obtained from Jackson Immunolabs and Molecular Probes.
Microscopy:
Imaginal disc images were acquired with Improvision OpenLab Version 5.0.2 software using a QImaging RETIGA EXi camera mounted to a Zeiss Axioplan 2 microscope unless otherwise stated. Confocal images in Figure 3, H and J, and Figure 8, E–H, were acquired on an inverted Zeiss LSM410 laser-scanning microscope. All images were processed in Adobe Photoshop 6.0.
Figure 3.—
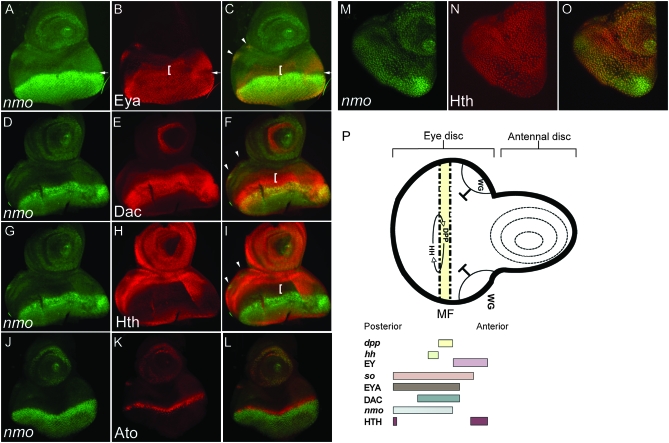
nmo is expressed in multiple cellular contexts in the third instar eye-antennal disc (late third instar:140 hr AEL). All discs are oriented with dorsal left, anterior up. (A–C) nmo-lacZ (green) is coincident with Eya (red) in the MF (arrow) and in the ocellar progenitors (arrowheads) and, to a lesser degree, posterior to the furrow. nmo-lacZ is absent in the PPN domain (bracket). (D–F) nmo-lacZ (green) coincides with Dac (red) in the third antennal disc segment, in addition to the MF and retinal cells. nmo-lacZ overlaps with Dac in the presumptive ocelli (arrowheads in F), although Dac more broadly encompasses the entire dorsal vertex region. (G–I) Hth (red) is absent in eye disc cells expressing nmo-lacZ (red) and reduced in the ocellar primordia (arrowheads in I). (J–L) nmo-lacZ (green) is coincident with Ato (red) in the MF, the ocellar region, and the antennal disc. (M–O) Single confocal section. nmo-lacZ (green) (M) and Hth (red) (N) are expressed in all cells of the PE. (P) Schematic of a third instar eye/antennal disc. The regions of dpp and wg expression and their action on MF progression are shown. The MF moves posterior to anterior. nmo's expression relative to the RD genes and the Wg effector Hth in the eye disc are indicated below, as previously described (Bessa et al. 2002; Silver and Rebay 2005).
Adult flies were preserved in 95% ethanol and photographed using an EOS Rebel 300D digital camera mounted to a Leica MZ6 stereomicroscope. Images were processed in Helicon Focus and Adobe Photoshop 6.0.
RESULTS
nmo is expressed dynamically throughout imaginal eye-antennal disc development:
Analysis of nmo in the eye imaginal disc to date has focused solely on its third instar expression within and posterior to the MF (Choi and Benzer 1994). We have previously described expression of nmo in the wing disc, which is broadly initiated during second instar and is subsequently refined in the third instar (Zeng and Verheyen 2004). Given its dynamic temporal expression during wing development, we considered that nmo may also be expressed in the early eye-antennal imaginal disc. Using the nmoP lacZ strain as a reporter for nmo transcription (Choi and Benzer 1994; Verheyen et al. 2001), we carefully characterized the expression pattern during larval eye development.
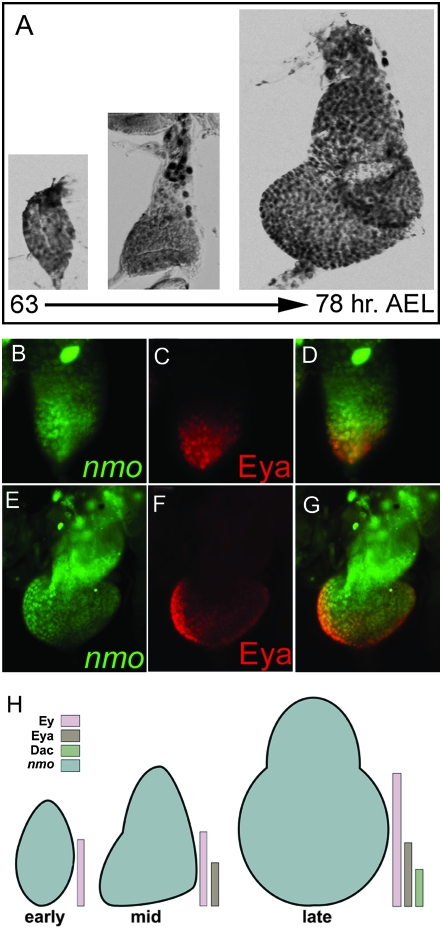
We found that nmo is expressed ubiquitously in the peripodial cells of the second instar eye imaginal disc (Figure 2A), suggesting that Nmo may have an earlier, uncharacterized role in patterning the eye and head. nmo expression coincides with Eya in the posterior eye disc in mid- (Figure 2, B–D) and late (Figure 2, E–G) second instar. During second instar, the eye-antennal imaginal disc is segregated into antennal and eye territories through downregulation of ey in the anterior antennal region (Kenyon et al. 2003). Ey subsequently deploys the retinal determination network in posterior cells, resulting in increasing refinement of eya and dac expression to the posterior margin of the eye disc (Halder et al. 1998; Kenyon et al. 2003). Thus, nmo is co-expressed with different combinations of RD factors in a spatially and temporally regulated manner when the eye territory is initially established (Figure 2H).
Figure 2.—
nmo is co-expressed with the RDGN in the second instar eye disc. Expression of the nmo-lacZ enhancer trap during second instar eye disc development [63–72 hr after egg laying (AEL)], detected with anti-β-gal antibody. (A) nmo-lacZ is expressed in all cells. (B–G) nmo-lacZ (green, B and E) coincides with Eya (red, C and F) in the posterior eye disc in mid (B–D) and late (E–G) second instar. (H) Schematic summarizing nmo's co-expression with the eye-specification genes. Early: nmo and Ey are co-expressed in the posterior eye field. Mid: nmo is co-expressed with Ey in anterior cells of the eye field and with Ey and Eya in posterior cells. Late: same is in mid, except at the posterior margin where nmo is co-expressed with Ey, Eya, and Dac.
As the third larval instar progresses, nmo is expressed in discrete subsets of cells. Posterior co-expression of nmo and Eya in second instar now extends to the anterior edge of the MF (Figure 3, A–C, arrow), but does not extend into the anterior pre-pro-neural (PPN) domain occupied by Eya and Dac (Figure 3, D–F, bracket in F). In late third instar discs, nmo expression is detected in the ocellar primordia. This expression is completely coincident with Eya (Figure 3, A–C, arrowhead) and more refined than Dac, which is more broadly expressed in the dorsal vertex primordia (Figure 3, E and F, arrowhead). Notably, the Wg target Hth is repressed in the ocellar cells co-expressing nmo and Eya (Figure 3, H and I, arrowhead in I). At the posterior margin, nmo-lacZ is repressed in cells expressing Hth (Figure 3I) and Ey (data not shown; Bessa et al. 2002). In the antennal disc, nmo expression is found in the aristal and Johnston's organ progenitors, according to the fate map of Haynie and Bryant (1986). Here, nmo is co-expressed with the pro-neural factor Ato (Figure 3, J–L). Ubiquitous peripodial expression of nmo persists during the third instar as nmo is expressed in all cells of the PE (Figure 3M), coincident with Hth (Figure 3N) and Ey (data not shown; Bessa et al. 2002).
This dynamic pattern of co-expression led us to hypothesize that nmo may contribute to multiple patterning events in the eye-antennal disc, in addition to its characterized role in planar polarity. Consistent with its expression in the ocellar primordia, nmo mutants display defects in the dorsal vertex (L. R. Braid, unpublished results). The antennae of nmo mutants appear normal, but given its co-expression with the neuronal marker ato, more refined analysis may uncover subtle sensory organ defects. nmo's dynamic spatial and temporal co-expression with the eye-specification factors also suggested that it may contribute to early patterning of the eye and antennal fields. In this study, we focused our investigation of nmo's potential novel roles in eye and head development to determine its function in eye specification, specifically by evaluating its ability to modulate the transcription and/or activity of the RDGN.
nmo rescues the ey small-eye phenotype:
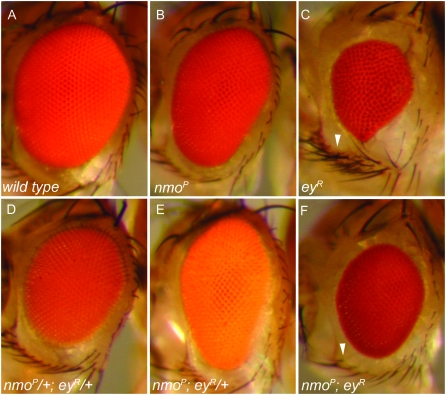
We generated nmo; ey double mutants, using the nmo alleles nmoDB24, nmoadk1, nmoadk2, and nmoP to test whether nmo contributes to RD-mediated eye patterning. Homozygous nmo mutants display narrow eyes and ommatidial rotation defects (Figure 4B; Choi and Benzer 1994). We chose to perform our loss-of-function analysis using the severe hypomorph eyRussian (eyR) (Quiring et al. 1994), which phenocopies the rare square ommatidial array characteristic of nmo mutants (Hartman and Hayes 1971; Ready et al. 1976). ey mutants display variable loss of eye and head tissue (Figure 4C) as a result of large-scale apoptosis early in the third instar (Halder et al. 1998). Flies heterozygous for nmo or eyR appear normal (data not shown). nmo/+; eyR/+ flies displayed slightly smaller eyes (Figure 4D). Heterozygosity for eyR did not significantly modify any aspect of the homozygous nmo eye phenotype (Figure 4E).
Figure 4.—
nmo modifies the ey small-eye phenotype. (A) Wild-type compound eye. (B) nmoP mutants have narrow eyes and a square ommatidial array. (C) eyR compound eyes are small with disorganized ommatidia and uneven eye margins. The ventral row of sensory vibrissae is often duplicated (arrowhead). The most frequent phenotype is shown. (D) nmoP/+; eyR/+ trans-heterozygotes display a slightly smaller eye compared to wild type. (E) nmoP; eyR/+. The nmoP eye phenotype is not modified by reducing a single copy of eyR. (F) nmoP; eyR. The size and periphery of the compound eye are rescued compared to C. A single set of ventral vibrissae is present (arrowhead), as in wild type. Flies are oriented with the anterior left. The same results were obtained using nmoDB24, nmoadk1, and nmoadk2.
Loss of nmo did, however, rescue several aspects of the eyR mutant eye, indicating that Nmo may contribute to some aspects of Ey-mediated eye development. nmo; eyR double mutants had larger eyes than eyR mutants alone (Figure 4F), as the number of ventral ommatidia was increased. eyR mutants frequently display duplicated ventral vibrissae, the set of sensory bristles surrounding the ventral eye margin (Figure 4C, arrowhead). Loss of nmo rescues the bristle duplication to a normal single set (Figure 4F, arrowhead). In addition, the periphery of nmo; eyR compound eyes are restored to wild type, being uniform compared to the irregular eye/head boundary typical of eyR mutants (compare Figure 4, C and F). Interestingly, eyes of nmo;eyR double mutants retained the narrow A–P width characteristic of nmo mutants, although the overall eye is smaller (compare Figure 4, B and F).
nmo and ey are co-expressed in the entire eye disc during second instar (Figure 2), in the third instar PE, and in the ocellar primordia of the ME (Figure 3). nmo is also co-expressed with the eye-specification genes so, eya, and dac during second instar within the furrow itself and the photoreceptor field behind it and in the presumptive ocelli (Figures 2 and 3). We therefore investigated whether nmo also genetically interacts with RD factors downstream of Ey.
Heterozygosity for eya2 enhances the nmo eye phenotype:
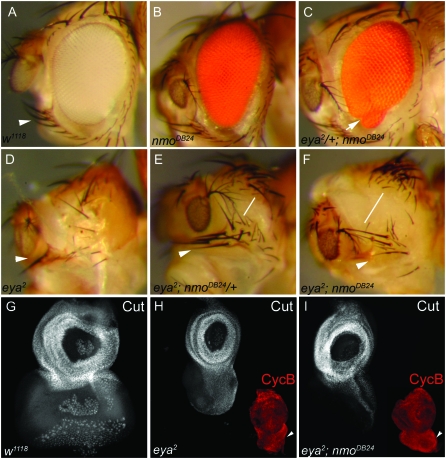
We next characterized flies mutant for both eya and nmo to investigate a possible genetic interaction. The eya2 allele results in specific loss of the compound eye due to complete absence of the type I eya transcript in the retinal progenitors (Bonini et al. 1993; Leiserson et al. 1998; Zimmerman et al. 2000). Flies heterozygous for eya2 or nmo are wild type (not shown), yet we found that heterozygosity for eya2 modified the nmo homozygous mutant phenotype (Figure 5C, arrow). Of these flies, 33.6% (35/104) exhibited ventral defects never observed in nmo mutants. Specifically, 7.7% (8/104) of flies displayed a reduction of the ventral eye, accompanied by a small, secondary eye field (arrow in Figure 5C). An additional 6.7% (7/104) of flies exhibited an eye-to-head transformation, indicated by an ectopic ocellus in the antero-ventral eye field (data not shown). The remaining 19.2% (20/104) of flies displayed ectopic ventral machrochaete bristles, usually accompanied by loss of the ventral eye (data not shown). Similar phenotypes were observed using the nmoadk1 and nmoadk2 alleles, as well as trans-heterozygous combinations of the nmo alleles (data not shown). The manifestation of ectopic cuticle in the eye field, accompanied by inappropriate dorsal head structures, suggests that Eya and Nmo may normally function together in early patterning of the eye and head fields. Indeed, these ventral eye phenotypes were also observed in nmoDB24 and nmoadk2 somatic clones induced during second instar in flies heterozygous for eya2 (data not shown).
Figure 5.—
nmo and eya genetically interact. (A) w1118. Sensory vibrissae surround the ventral eye margin (arrowhead). (B). nmoDB24 compound eyes are elongated and narrow. (C) eya2/+; nmoDB24. A secondary eye field develops at the ventral margin (arrow). This phenotype accounts for 22.9% of total observed ventral eye defects (33.6% of flies; n = 104). (D) eya2 mutants lack eyes and are missing ventral vibrissae (arrowhead). As a result of smaller heads, ventral eye bristles converge with dorsal orbital bristles. (E) eya2; nmoDB24/+. More ventral vibrissae are observed (arrowhead; compare with D), and the distance from the dorsal orbital bristles is increased (line). (F) eya2; nmoDB24. The ventral vibrissae have a nearly wild-type pattern (arrowhead), and their distance from the orbital bristles is further rescued from E (line). (G–I) Imaginal eye discs are oriented with the anterior at the top, dorsal left. White images, anti-Cut; red images in H and I, anti-cyclin B. (G) w1118. Anti-Cut labeling (white) marks the antennal disc and anterior-most eye disc cells, which give rise to head cuticle. Additional posterior staining is observed in the PE and ommatidial clusters. (H) eya2. The eye disc is largely reduced, relative to the antennal disc. (I) eya2; nmoDB24. The ventral eye disc is enlarged compared to eya2 (arrowheads near red images), but proliferation (cyclin B labeling, red) is comparable to eya2 alone (H).
eya2 head defects are suppressed by dose-dependent loss of nmo:
The dorsal perimeter of the eye is flanked by the orbital bristles, while the ventral margin displays a stereotypical set of macrochaetes followed by posterior microchaetes, collectively termed the ventral sensory vibrissae (Figure 5A, arrowhead; Haynie and Bryant 1986). eya2 mutants are eyeless and also display a small head with variably missing ventral vibrissae (Figure 5D, arrowhead). In eya2; nmo/+ flies, we observed an increase in the number of machrochaete-type vibrissae, indicating a rescue in ventral head patterning (Figure 5E, arrowhead). eya2;nmoDB24 double mutants are pharate lethal; however, rare escapers display an expansion of head cuticle indicated by an increase in distance between the orbital bristles and ventral vibrissae (compare Figure 5, D and line in F).
eya mutants are characterized by complete loss of retinal tissue resulting from hyperproliferation during the second instar, followed by massive programmed cell death (PCD) anterior to the MF during third instar (Bonini et al. 1993). Activation of apoptosis in eya2 mutants is an indirect result of the early overproliferation defects (Pignoni et al. 1997) and can also be induced by misexpressing eya (Clark et al. 2002). The ventral eye discs of eya2; nmoDB24 double mutants are larger than eya2 eye discs (Figure 5, H and I, arrowhead), visualized with the sensory organ precursor marker Cut (Blochlinger et al. 1990) and the mitotic marker cyclin B (Knoblich and Lehner 1993). Intriguingly, proliferation appears to occur at the same level in eya2 and eya2; nmoDB24 discs (Figure 5, H and I, red images), while high levels of apoptosis can be detected in both genotypes with acridine orange staining and with an antibody targeted against activated caspase-3 (not shown).
Loss of nmo reduces viability of dac mutants:
We subsequently investigated whether nmo genetically interacts with dac, the most downstream component of the RDGN. We tested interactions between all the nmo alleles and dac1 (Mardon et al. 1994; Shen and Mardon 1997; Martini et al. 2000). dac1 heterozygotes have no external phenotype. As in our loss-of-function analysis with ey, heterozygosity for dac1 did not modify the nmo mutant eye phenotype (not shown). Reducing a single copy of nmo in dac1 mutants induced early larval lethality, precluding our ability to study their potential interaction during eye development. We obtained similar results using the dacE462 allele. We also could not recover heterozygous nmoDB24 or nmoadk1 eye discs in which we had induced dacE462 somatic clones. Moreover, nmo; dac double mutants died as embryos, suggesting a potential interaction for these genes in additional developmental processes that affect viability of the organism.
nmo promotes Ey-mediated ectopic eye induction:
Our loss-of-function analyses suggest that Nmo may interact with the eye-specification factors during eye development. However, these results are difficult to interpret, given the intrinsic positive feedback organization of the RD network. As a result, the network can compensate for simultaneous reduction of two or more of its components, leading to unpredictable phenotypes (Chen et al. 1997; Pignoni et al. 1997). Moreover, ey, so, eya, and dac mutants lack eyes due to hyperactivation of PCD, an indirect result of early overproliferation and patterning defects (Bonini et al. 1993; Cheyette et al. 1994; Pignoni et al. 1997; Halder et al. 1998). Thus, the very nature of these mutants impedes classical genetic analysis as the developing eye field is obliterated before retinal specification is initiated. Epistasis between the canonical RD members has consequently been established using targeted misexpression studies that separate the RD factors under consideration from the feedback loop in the eye disc (Chen et al. 1997, 1999; Shen and Mardon 1997; Halder et al. 1998; Pappu et al. 2003). Therefore, we investigated nmo's potential role in RDGN-mediated eye development using previously established misexpression assays.
A key function of the RDGN is to promote retinal determination. In ectopic eye-induction assays, misexpression of any of the RD factors can deploy the eye-specification program in other tissues, including the head, wing, leg, thorax, and genitals (Halder et al. 1995; Chen et al. 1997; Pignoni et al. 1997; Shen and Mardon 1997; Bonini et al. 1998; Halder et al. 1998; Anderson et al. 2006; Weasner et al. 2007). However, not all cells are able to be respecified to the retinal fate; other endogenous factors, such as Hh and high levels of Dpp, appear to also be required (Pignoni and Zipursky 1997; Halder et al. 1998; Chen et al. 1999; Kango-Singh et al. 2003; Pappu et al. 2003). In these assays, Ey is the most potent inducer of ectopic eyes, causing dramatic induction of organized ommatidial clusters on the ventral head, antennae, leg, and wing (Halder et al. 1995, 1998; Pignoni et al. 1997; Chen et al. 1999; Anderson et al. 2006). We noted that cells able to be respecified as eye cells in the head, wing, and leg frequently correspond to nmo-expressing cells (Chen et al. 1997, 1999; Shen and Mardon 1997; Bonini et al. 1998; Bessa et al. 2002; Anderson et al. 2006; Weasner et al. 2007). Thus, we hypothesized that nmo may be a factor that contributes to eye specification at these remote sites, as well as in the normal eye.
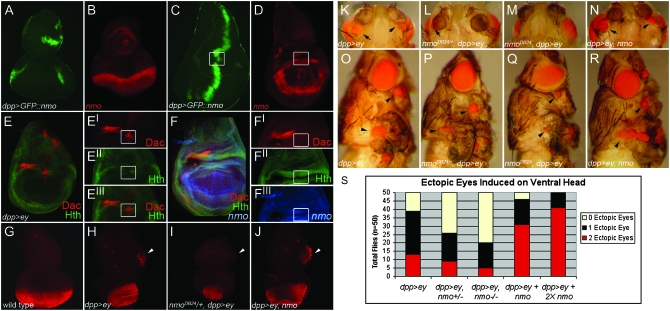
To explore a role for nmo in retinal induction, we modulated nmo levels and assayed the effects on ey-mediated ectopic eye induction utilizing the UAS/GAL4 misexpression system (Brand and Perrimon 1993). dpp-Gal4 drives expression along the posterior and lateral margins of the eye disc and in a ventro-lateral wedge in the antennal disc, which traverses the nmo expression pattern in the MF and antennal segments (Figure 6, A and B). In the wing disc, the A–P stripe of dpp-Gal4 expression bisects nmo-expressing cells where it crosses the dorsal and ventral limits of the presumptive wing hinge (Figure 6, C and D). Ey robustly induces eye development in the dorsal wing hinge primordia (Figure 6O), where loss of Hth is concomitantly observed with ectopic expression of Ey targets, including dac (Figure 6E, Bessa et al. 2002). Nmo is normally co-expressed with Hth in these cells (Figure 6F), suggesting a possible role for nmo in mediating ectopic eye development in these cells.
Figure 6.—
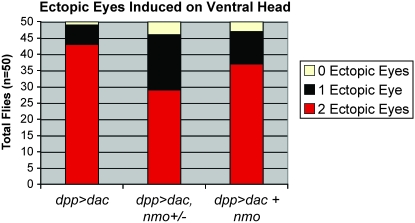
nmo potentiates Ey-mediated ectopic eye induction. (A and C) UAS-GFP∷nmo/+; dpp-Gal4/+ (green). (B and D) nmo-lacZ (red). (A and B) dpp-Gal4 (green) targets expression in the dorsal and ventral poles of the eye disc and in a ventral wedge in the antennal eye disc, which bisects nmo-expressing cells (red, B). (C and D) dpp-Gal4 (green) drives expression along the A–P boundary of the wing disc, which intersects with nmo expression (red) at the dorsal wing hinge (boxes). (E) UAS-ey/+; dpp-Gal4/+ wing disc. E′–E″′ is an enlarged view of the dorsal wing pouch in E. Ectopic eyes are induced in cells ectopically expressing Dac (red, E′) and with reduced Hth (green, E″). E″′ is a composite of E′ and E″. (F) nmo-lacZ (blue) wing disc, Dac (red), and Hth (green). F′–F″′ is an enlarged view of the dorsal wing pouch in F. Dac is not normally expressed in the dorsal wing hinge (F′; compare with E′), although Hth (F″) and nmo (F″′) are normally co-expressed in dorsal wing cells able to be respecified to the eye fate (boxes in C–F). (G–J) Eye discs stained for ELAV (red). (G) w1118. ELAV is normally expressed in the posterior photoreceptors and is absent in the antennal disc. (H) UAS-ey/+; dpp-Gal4/+. ELAV-positive ectopic photoreceptors are detected in the ventral antennal disc (arrowhead). Photoreceptors do not differentiate at the dorsal and ventral boundaries of the eye field, where Dpp targets expression (see A), and the size of the eye disc is reduced compared to the antennal disc. (I) UAS-ey/+; nmoDB24/dppGal4. Ectopic photoreceptors are no longer detected in the antennal disc (arrowhead). The normal photoreceptor field, as well as the overall size of the eye/antennal disc, is further reduced compared to H. (J) UAS-ey/UAS-nmo; dpp-Gal4/+. Large groups of ectopic photoreceptors are detected in the ventral antennal disc (arrowhead). The normal eye field is rescued (compare with H). (K and O) UAS-ey/+; dpp-Gal4/+. Ectopic eyes are induced ventrally to the antennae (arrows, K) and on the legs and wing hinge (arrowheads, O). (L and P) UAS-ey/+; dpp-Gal4/nmoDB24. Ectopic eyes are induced at a lower frequency and are smaller than in K (arrow, L). Ectopic eye fields on the legs and wing hinge are reduced compared to O. (M and Q) UAS-ey/+; dpp-Gal4,nmoDB24/nmoDB24. Ectopic eyes are only rarely induced on the head (M). Ectopic eye fields on the leg and wing hinge are considerably reduced (compare with O). The compound eye has the characteristic nmo morphology. (N and R) UAS-ey/UAS-nmo; dpp-Gal4/+. The size of ectopic eyes induced on the head (N) and on the leg and wing hinge (R) are larger than in K and O, respectively. (S) Quantification of the phenotypes in K–N. The relative frequencies of zero, one, or two ectopic eyes on head cuticle derived from the antennal disc for the indicated genotypes. Loss or co-expression of nmo has a dose-dependent effect on both the frequency and the penetrance of the ectopic eye phenotype. Loss of nmo significantly reduces the penetrance of head-to-eye respecification.
Targeted expression of ey using dpp-Gal4 (dpp>ey) induces ectopic eyes on the anterior head, just below the antennae, at a high frequency (Figure 6, K and S). The normal eye field is also reduced at the dorsal and ventral margins compared to wild type (Figure 6H). Previous studies have shown that induction of ectopic eyes by dpp>ey is effectively abrogated in so, eya, or dac mutant cells (Bonini et al. 1997; Chen et al. 1997; Shen and Mardon 1997; Halder et al. 1998). Similarly, we tested the requirement for nmo in this assay by expressing dpp>ey in nmoDB24 and nmoP heterozygotes and homozygous mutants. We observed a dose-dependent reduction in both size and frequency of ectopic eyes induced on the head (Figure 6, I, L, M, and S), suggesting that Nmo is a positive component of Ey-mediated retinal induction. Moreover, Nmo also contributes to formation of ectopic eye fields in the wing and leg, as we observed a similar dose-dependent reduction in the size of Ey-induced retinal fields in cells heterozygous or mutant for nmo in these tissues (Figure 6, P and Q).
During third instar, ey expression is restricted to anterior cells of the eye disc, where one of its functions is to repress furrow progression by inhibiting eya expression (Bessa et al. 2002). dpp-Gal4 drives expression at the lateral poles of the MF (Figure 6A; Shen and Mardon 1997). The posterior eye field of dpp>ey discs, labeled with an antibody against the nuclear neuronal antigen ELAV (Robinow and White 1991), displays loss of ommatidial clusters from the dorsal and ventral margins and a mild reduction in the size of the eye disc (Figure 6H). In addition, clusters of ectopic ELAV-positive ommatidia are observed in an expanded ventral region of antennal discs (arrowhead in Figure 6H), which give rise to ectopic eyes in the adult (Figure 6K). These cells are fated to give rise to the antero-ventral head cuticle surrounding the eye (Haynie and Bryant 1986). Reducing nmo in dpp>ey flies caused a further decrease in the number of ELAV-labeled photoreceptors from the dorsal and ventral margins of the normal eye field, in addition to reducing the ectopic eye fields in the antennal disc (Figure 6I) We also observed an overall reduction in the size of the eye-antennal disc (Figure 6I). This manifests as a dose-dependent reduction in the compound eye (Figure 6, P and Q). Taken together, these results suggest that nmo promotes eye specification in both normal and misexpression contexts.
We predicted that, if Nmo generally promotes retinal specification, co-expression of nmo with dpp>ey would result in an expansion of both endogenous and ectopic eye fields. Consistent with this hypothesis, the frequency and size of ectopic retinal fields on the anterior head, wing, and leg increased compared to dpp>ey alone (Figure 6, N, R, and S). In dpp>ey, nmo eye discs, large patches of ELAV-positive retinal cells were detected in the antennal disc (Figure 6J, arrowhead). In addition, the normal eye field was expanded along the dorsal–ventral axis, resembling wild type (Figure 6J; compare with Figure 6G). dpp>ey, nmo adults also displayed a variety of inappropriate tissue outgrowths from the ventral head cuticle (data not shown), indicating that Nmo may have additional roles in cell fate decisions.
Nmo promotes Eya-mediated ectopic eye induction:
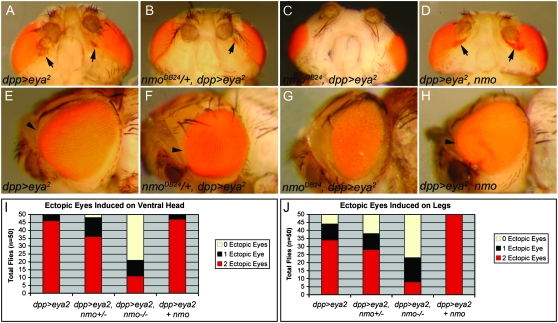
We further tested the requirement for nmo in retinal induction by repeating our misexpression assay using UAS-eya2, which encodes the type II eya cDNA (Bonini et al. 1998). We found that the UAS-eya2 responder line induced ectopic eyes more potently than UAS-eya1. In agreement with a previous study (Bonini et al. 1997), we found that dpp>eya2 produced infrequent small patches of ectopic eyes on the ventral head and only very rare cases of retinal development on the wing and leg when reared at 25° (data not shown). To determine if nmo also promotes Eya-mediated eye formation, we repeated our assay at 29° to increase the penetrance of the dpp>eya2 phenotype (Figure 7, A and I). Again we observed a dose-dependent reduction in the size and number of ectopic retinal fields induced in the head (Figure 7, B, C, and I), wing, and leg (Figure 7J) by expressing dpp>eya2 in nmo heterozygotes (Figure 7B) and homozygotes (Figure 7C), respectively. Notably, nmo contributes more to Ey-mediated ectopic eye induction (Figure 6S) than in assays with exogenous Eya (Figure 7I) or Dac (Figure 8), suggesting that nmo may contribute to Ey-mediated activation of eya and dac in this context.
Figure 7.—
nmo potentiates Eya-mediated ectopic eye formation. (A and E) dppGal4/UAS-eya2. (A) Small fields of ectopic eyes are induced on head cuticle below the antennae (arrows). (B and F) nmoDB24, dppGal4/UAS-eya2. (B) Ectopic eye fields are induced less frequently and are smaller than in A (arrow). (C and G) nmoDB24, dppGal4/nmoDB24, UAS-eya2. (C) Ectopic eyes are rarely induced. (D and H). UAS-nmo/+; dppGal4/UAS-eya2. (D) Large ectopic eye fields frequently merge with the endogenous eye (arrows). (E) The compound eye is overgrown (arrowhead). (F) The compound eye has minimal overgrowth (arrowhead; compare with E). (G) The compound eye is smaller than wild type and resembles nmo mutants. (H) The compound eye is massively overgrown (arrowheads). (I) Quantification of phenotypes in A–D. Loss of nmo dose-dependently reduces the frequency of head-to-eye respecification. (J) Quantification of leg-to-eye transformations for the indicated genotypes. As in the head (I), loss of nmo dose-dependently reduces the frequency of ectopic eyes induced on the leg. Co-expression with nmo increases the penetrance and frequency. Flies were reared at 29°.
Figure 8.—
nmo potentiates Dac-mediated ectopic eye formation. Quantification of the relative frequency of head-to-eye transformations for the genotypes shown. Heterozygosity for nmo reduces the frequency of ectopic eye formation, but less potently than with misexpressed ey (Figure 6S) or eya (Figure 7I).
Eya contributes to propagation of the MF by promoting dpp expression (Pignoni et al. 1997; Hazelett et al. 1998). In dpp>eya2 flies, we observed expansion of the furrow along the lateral margins of the eye disc (L. R. Braid, unpublished results), resulting in an enlarged compound eye (Figure 7E, arrowhead). Consistent with our hypothesis that nmo promotes normal eye development, reducing nmo rescues the overgrowth associated with ectopic eya [Figure 7, F (arrowhead) and G]. In fact, Dpp-driven expression of ey, eya2, or dac is unable to significantly modify the nmo small-eye phenotype (Figure 6Q; Figure 7H; data not shown), indicating that Nmo may function in processes regulating the size of the eye field downstream or independent of the RDGN.
Ectopic eya requires exogenous ey, so, or dac to be a potent inducer of ectopic eyes (Bonini et al. 1997; Chen et al. 1997; Pignoni et al. 1997; Bui et al. 2000). Co-expressing nmo with dpp>eya2 also provides this synergy, as it enhances the frequency and size of ectopic retinal fields in the head, wing, and leg (Figure 7, D, I and J). The ectopic eye fields now frequently merge with the compound eye (Figure 7D, arrows), similar to overexpression of dac (not shown; Chen et al. 1997; Shen and Mardon 1997). We also observed further overgrowth of the compound eye compared to dpp>eya2 alone (Figure 7H, arrowhead), supporting our hypothesis that nmo promotes eye formation. Similar synergy is observed in flies co-expressing nmo and dac (Figure 8).
nmo alone respecifies head precursors as retinal cells:
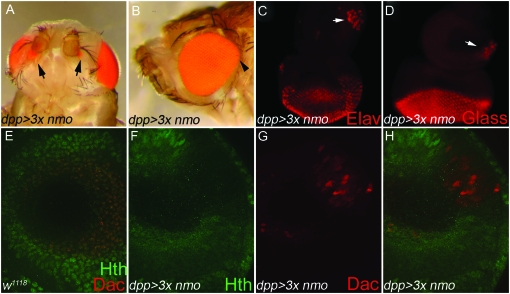
To further characterize nmo's role in ectopic retinal induction, we expressed nmo at high levels using dpp-GAL4. Nmo protein levels appear to be tightly regulated, as elevating expression of UAS-nmo transgenes has minimal effect on raising total Nmo levels in the cell (Fiehler and Wolff 2008; L. R. Braid, unpublished results). Targeted expression of two copies of UAS-nmo with dpp-Gal4 (dpp>2Xnmo) results in mild overgrowth of the dorsal eye (not shown). dpp >3Xnmo flies reared at 29° are pharate lethal, although 15% die as early pupae. Consistent with a role in promoting endogenous eye formation, dpp >3Xnmo pharate adults display dorsal expansion of the compound eye along the A–P axis (Figure 9B). Dorsal overproliferation can be accompanied by ectopic sensory vibrissae along the ventral eye margin, which is occasionally reduced (data not shown). Notably, 16.7% (28/168) display pigmented, ectopic eyes ventral to the antennae (Figure 9A, arrows). dpp>3xnmo pharate adults also have leg, wing, and notum defects (data not shown).
Figure 9.—
Ectopic Nmo induces head-to-eye respecification in the antennal disc. (A–D and F–H) UAS-nmo; dppGal4/UAS-nmo (A) Of pharate adults, 16.7% (28/168) display pigmented retinal cells on the antero-ventral head cuticle (arrows). (B) The dorsal eye is overgrown (arrowhead). (C–H). Imaginal eye/antennal discs. (C) Clusters of ELAV-positive cells (arrow) are detected in the antero-ventral head primordia in 63.6% (28/44) of antennal discs. ELAV is not normally expressed in the antennal disc. (D) Eye-antennal discs labeled with anti-Glass, which is normally absent from the antennal disc, indicate that ventral antennal cells have adopted a retinal fate (arrow). (E–H) Confocal images of the mid-ventral antennal disc taken at ×40. E is a Z-stack of the entire antennal disc. F–H are single confocal planes. (E) w1118. Expression of Hth (green) and Dac (red) in a wild-type disc. Hth is ubiquitously expressed in cells of the outer antennal segments and overlaps with Dac (red) in the third antennal segment. (F) Misexpressed Nmo induces loss of Hth (G) and concomitant ectopic Dac (red) in the outer antennal ring. Endogenous Dac expression is below this focal plane. (H) Composite of F and G. (C–H) Imaginal discs are oriented anterior up, dorsal left.
Each ommatidium of the compound eye comprises 20 cells of various types, including neuronal photoreceptors and non-neuronal accessory cone and pigment cells (Ready et al. 1976; Tomlinson and Ready 1987a,b). Non-neuronal pigment cells are apparent in the dpp>3Xnmo adult ectopic eye phenotype (Figure 9A, arrows). To confirm the presence of photoreceptors, we analyzed dpp >3Xnmo imaginal eye discs using the neural markers ELAV (Robinow and White 1991) and Glass (Ellis et al. 1993), which are specific to the visual system. ELAV and Glass antigens are not present in wild-type antennal discs (Robinow and White 1991; Ellis et al. 1993). Interestingly, we observed clusters of ELAV- and Glass-expressing cells in the ventral antennal disc of 63.6% (28/44) of examined eye discs, which appeared to be forming ommatidia (Figure 9, C and D, arrow). The transformation of presumptive head cells to a retinal fate is therefore more penetrant than the adult phenotype suggests (16.7%). This discrepancy may be the result of early pupal lethality or undetected fate changes in the pharate adults due to the size of the retinal field or the absence of accompanying pigment cells.
Targeted expression of ey, eya, or dac using dpp-Gal4 downregulates Hth at sites of ectopic eye induction in the antennal and wing discs (Figure 6E; Bessa et al. 2002; L. R. Braid, unpublished results). Similarly, we observe concomitant loss of Hth and ectopic expression of dac (Figure 9, F–H) and eya (data not shown) in dpp>3xnmo antennal discs. These cells correspond to the antero-ventral head primordia, and co-analysis with ELAV verified that Hth is repressed and dac is ectopically expressed in cells transformed to an eye fate (not shown). Consistent with previous studies (Anderson et al. 2006; Weasner et al. 2007), we found that only a subset of Dac-positive cells are respecified as photoreceptors (data not shown). Nmo's ability to cause transformations to the eye fate does not extend beyond the antennal disc, although Hth is repressed in dpp>3xnmo wing discs (data not shown). These data imply that elevated Nmo requires endogenous eye factors to deploy the retinal program.
Nmo promotes eye development independently of RDGN gene activation:
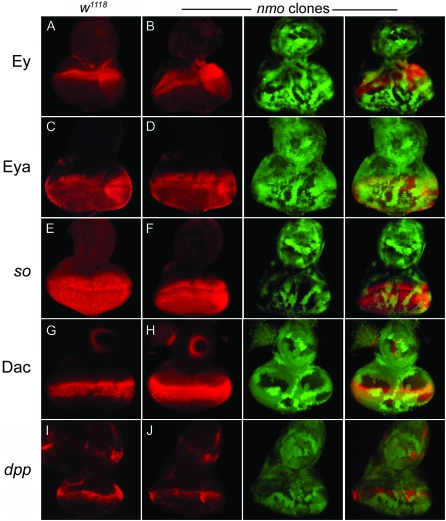
Our study shows that nmo contributes to normal and ectopic eye development mediated by the RDGN. We observed that loss of nmo reduced the ability of Ey, Eya, and Dac to induce ectopic eyes and resulted in smaller compound eyes. We also found that misexpression of nmo with dpp-Gal4 could derepress dac and eya in head cells, causing their respecification to a retinal fate. Thus, we generated nmo somatic clones to examine whether Nmo contributes to eye formation by promoting expression of the RD genes. Surprisingly, we did not observe changes in Ey, Eya, so-lacZ, or Dac expression in nmo loss-of-function clones (Figure 10, B, D, F and H).
The requirement for Dpp in normal and ectopic retinal induction has been well established (Wiersdorff et al. 1996; Chanut and Heberlein 1997; Dominguez and Hafen 1997; Pignoni and Zipursky 1997; Royet and Finkelstein 1997; Halder et al. 1998; Chen et al. 1999; Kango-Singh et al. 2003). dpp expression is promoted by the RDGN downstream of ey (Chen et al. 1997; Hazelett et al. 1998), and high levels of Dpp are required to antagonize wg and promote furrow progression (Wiersdorff et al. 1996; Chanut and Heberlein 1997; Dominguez and Hafen 1997; Pignoni and Zipursky 1997). Therefore, we investigated whether nmo promotes eye formation through regulation of dpp, using dpp-lacZ as a reporter. We found that dpp expression was also unchanged in nmo somatic clones (Figure 10E), indicating that Nmo promotes eye development without directly modulating RD gene expression or levels of Dpp. Using antibodies against cyclin B and phospho-histone3, we also found that proliferation was unaffected in nmo mutant cells (data not shown). These findings suggest that nmo's role in promoting RD-mediated eye development is the result of molecular interactions or the regulation of an as-yet-unidentified common transcriptional target.
nmo mediates defects induced by misexpressed ey:
Ey deploys the RDGN by initiating expression of so and eya in the second instar (Halder et al. 1998; Kenyon et al. 2003) and later complexes with So to activate the neural program in the PPN cells (Zhang et al. 2006). However, Ey also complexes with the anterior Wg effectors Tsh and Hth during third instar to delimit the eye/head boundary (Bessa et al. 2002). In our ectopic eye induction assays, Nmo promoted eye formation by cooperating with Ey in the ectopic eye field and by antagonizing its activity in the normal eye field. nmo and ey are co-expressed in the second instar when Ey initiates the RDGN, but are expressed mainly in complementary domains during third instar. Taken together, we hypothesized that Nmo's interactions with Ey may be context dependent.
Since ectopic eye development requires de novo deployment of the RDGN, we hypothesized that the positive interaction between Nmo and Ey in this context may represent their interaction in early development, when the eye field is initially specified. Our previous misexpression analysis was performed using dpp-Gal4, which drives expression along the posterior and lateral margins of the eye disc in the early instars and in the dorsal and ventral poles of the third instar eye disc. Therefore, we tested this hypothesis using ey-Gal4, which is broadly expressed in the eye disc during all three instars.
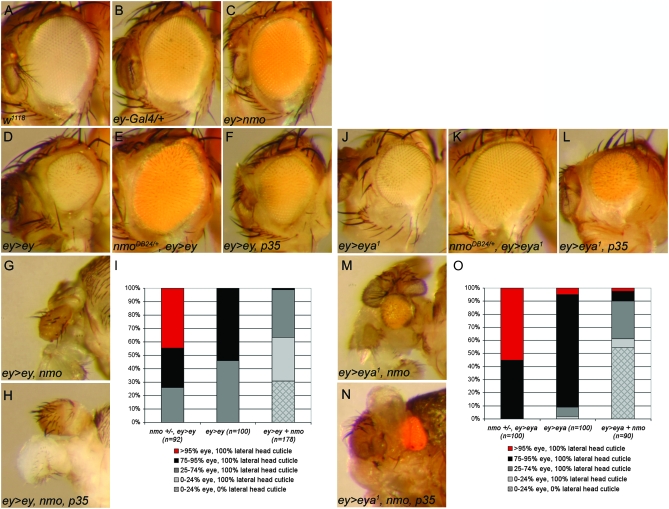
Directed expression of UAS-nmo with ey-Gal4 (ey>nmo) had no effect at 25°, similar to ey-Gal4 alone (Figure 11, B and C). As previously described, ey>ey induced a complete loss of ventral eye and ommatidial disorganization (Figure 11D; Curtiss and Mlodzik 2000; Jiao et al. 2001; Plaza et al. 2001; Curtiss et al. 2007). We misexpressed ey in heterozygous nmoDB24 mutants to test the requirement for nmo in the ey-induced reduced eye. While nmoDB24 heterozygotes appear to be wild type, this background is sufficient to rescue the loss of ventral eye caused by ey>ey (Figure 11, E and I)
Figure 11.—
nmo promotes early eye defects associated with ectopic ey and eya. (A) w1118. (B) ey-Gal4/+ and (C) ey-Gal4/UAS-nmo have no detectable external abnormal phenotype. (D) ey-Gal4/UAS-ey causes a smaller, rough eye. (E) ey-Gal4/UAS-ey; nmoDB24/+. Loss of a single copy of nmoDB24 rescues the small eye induced by UAS-ey (compare with D). (F) ey-Gal4/UAS-ey; UAS-p354/+. Blocking apoptosis does not phenocopy loss of nmo (compare with E). (G) eyGal4/UAS-ey; UAS-nmo/+. Flies frequently lack all eye and most head structures. (H) eyGal4/UAS-ey; UAS-nmo/UAS-p35. Blocking apoptosis does not modify the defects induced by UAS-ey and UAS-nmo (compare with G). (I) Quantification of the phenotypes observed in misexpression analysis with UAS-ey (D–H). (J) ey-Gal4/UAS-eya1 results in smaller eyes with dorsal overproliferation. (K) ey-Gal4/UAS-eya1; nmoDB24/+. Loss of a single copy of nmoDB24 rescues the small eye and dorsal overproliferation induced by eya (compare with J). (L) ey-Gal4/UAS-eya1; UAS-p35/+. Blocking apoptosis does not phenocopy loss of nmo (compare with K). (M) ey-Gal4/UAS-eya1; UAS-nmo/+. Flies display severe reduction of the compound eye and head cuticle. (M) ey-Gal4/UAS-eya1; UAS-nmo/UAS-p35. Blocking apoptosis does not modify the defects induced by UAS-eya1 and UAS-nmo (compare with M).(O) Quantification of phenotypes observed in misexpression analysis with UAS-eya1 (J–N).
Loss or ectopic expression of a single RD factor interferes with normal development, presumably by disrupting the delicate stoichiometry of RD factors (Curtiss and Mlodzik 2000; Curtiss et al. 2007). This abnormal patterning culminates in high levels of cell death and loss of tissue (Shen and Mardon 1997; Halder et al. 1998; Curtiss and Mlodzik 2000; Jiao et al. 2001; Plaza et al. 2001; Curtiss et al. 2007). Since Nmo promotes apoptosis in the pupal eye (Mirkovic et al. 2002), we tested whether the observed rescue upon reducing nmo was an indirect effect of reduced cell death. We reasoned that if eliminating a single copy of nmo rescues the ey>ey small eye by preventing cell death, then blocking apoptosis in ey>ey flies should have a similar effect. Co-expressing the baculovirus caspase inhibitor p35 (Clem et al. 1991) with ey>ey failed to rescue the small-eye phenotype (Figure 11F), indicating that heterozygosity for nmo directly rescues the effects of ectopic ey by restoring eye patterning, rather than by modulating cell death. Consistent with Nmo's role in promoting Ey-mediated ectopic eye formation, this finding suggests that nmo contributes to Ey function in the early eye field.
Although ey>nmo alone had no effect, co-expression of nmo greatly exacerbated the developmental defects induced by ey>ey. ey>ey, nmo animals are pharate lethal and display variable loss of eye and/or head tissue, extending to complete loss of the head and eye, with only the proboscis remaining (Figure 11, G and I). Once again, inhibiting cell death by co-expression of p35 was unable to modify the phenotype (Figure 11H), suggesting that the observed genetic interactions are the direct result of nmo's effects on Ey-mediated patterning.
nmo promotes eya-induced eye phenotypes:
We subsequently tested whether Nmo's effects on Ey activity extend to other RD interactions, as nmo is also highly co-expressed with so, eya, and dac beginning in the second instar (Figures 2 and 3). Eya is another potent RD factor that can disrupt normal patterning when misexpressed (Bui et al. 2000; Hsiao et al. 2001). Driving expression of UAS-eya1, the type I eya transcript (Bonini et al. 1997) with ey-Gal4 caused asymmetric eye defects, with dorsal overproliferation accompanied by loss of the ventral region (Figure 11, J and O). Similar to our genetic analysis with ey, we observed that loss of nmo rescued the reduced ventral eye induced by ey>eya1 (Figure 11, K and O), which could not be phenocopied by co-expression of p35 (Figure 11L). Furthermore, co-expression of nmo enhanced the misexpressed eya1 phenotype, again resulting in gross morphological defects. The retinal field was often still present, although severely reduced and misplaced. Frequently, the majority of head cuticle was absent and malformed although duplicated antennae often remained (Figure 11, M and O). Inhibition of cell death was unable to rescue the ey>eya1, nmo defects (Figure 11N), indicating that nmo's effects on ey>eya1 are not the result of modulating activity of the cell death pathway. We also obtained similar results using the UAS-eya2 transgene (data not shown). Together with our previous analyses, the data imply that Nmo promotes Ey and Eya function during early eye development.
DISCUSSION
In this study we describe novel roles for nmo in early eye patterning that are distinct from its known role in planar polarity during late larval development. The RDGN is composed of a highly complex cascade of positive feedback loops (Figure 1A). The fundamental refinement of this delicate system is apparent from the dramatic defects resulting from reducing or ectopically expressing even a single component. Through loss-of-function and misexpression analyses, we provide genetic evidence that nmo contributes to patterning events orchestrated by the RDGN during eye development.
Co-expression of the RD genes is spatially and temporally regulated and confers cellular identity through the consequential formation of selector complexes (Figure 1A; reviewed in Pappu and Mardon 2004). For example, So and Eya complex to activate dac expression (Chen et al. 1999; Jemc and Rebay 2007). Subsequently, Dac can complex with So or Eya to direct expression of complex-specific gene targets (Chen et al. 1997; Bui et al. 2000). In addition, Ey and So complex to activate ato in cells entering the MF (Zhang et al. 2006). Repression of ey in, and posterior to, the MF limits this interaction to the pro-neural cells (Pignoni et al. 1997; Halder et al. 1998). Spatio-temporal regulation of the RD genes is imperative for normal eye and head development, given the deleterious effects of their misexpression on normal eye development (Bui et al. 2000; Curtiss and Mlodzik 2000; Hsiao et al. 2001; Jiao et al. 2001; Curtiss et al. 2007). It has been proposed that the availability and relative concentrations of these cofactors affect which protein–protein complexes form (Curtiss and Mlodzik 2000; Curtiss et al. 2007). As such, misexpression of the RD genes alters the pool of available cofactors, resulting in mis-specification of cell fate.
Interestingly, reducing any of the eye-specification factors also results in patterning defects, culminating in cell death and loss of tissue (Bonini et al. 1993; Cheyette et al. 1994; Mardon et al. 1994; Quiring et al. 1994; Pignoni et al. 1997; Halder et al. 1998). Thus, reducing an RD factor may be analogous to its misexpression since the relative levels of RD factors are similarly perturbed, leading to abnormal development and hyperactivation of apoptosis. Our data support such a model, since loss of nmo restores eye- and head-patterning defects associated with loss of ey and eya, as it does with early misexpression of these genes. The ey and eya alleles used in this study are not nulls and therefore may retain some level of activity (Bonini et al. 1993; Halder et al. 1998). These interactions imply that reducing nmo can modulate the transcriptional output of RD complexes, restoring developmental integrity. Moreover, inhibiting apoptosis with co-expression of the caspase-inhibitor p35 did not phenocopy this rescue, further supporting our hypothesis that Nmo may contribute to eye development by affecting the activity of RD selector complexes rather than by generally promoting cell death.
Although driving nmo throughout the eye disc in all stages of development with ey-Gal4 has minimal effects on its own, and misexpression of ey or eya causes only small eyes, the combined presence of Nmo and Ey or Nmo and Eya is not compatible with eye and head development. This dramatic synergy, together with the rescue mediated by reducing nmo, is consistent with a model in which Nmo affects the function of one or more of the RD cofactors, thereby affecting the balance of selector factors. We established that Nmo does not regulate Ey, so, Eya, or Dac levels in somatic clones, supporting our hypothesis that the observed genetic interactions occur at the protein level. Whether nmo is itself regulated by the RDGN is yet to be determined.
The context-specific nature of Nmo's role in mediating RD activity was revealed in our ectopic eye induction assay. Misexpression of ey using dpp-Gal4 not only induced ectopic eyes in the antennal, wing, and leg discs, but also interfered with endogenous eye development. Ectopic nmo rescued the dorso-ventral reduction in dpp>ey compound eyes, suggesting that Nmo promotes eye development. It further implies that Nmo may differentially affect Ey activity through cell-specific factors, since early co-expression of nmo with ey>ey had the converse effect, resulting in ablation of the eye and head. Spatial restriction of cofactors to achieve different outcomes is a common developmental strategy. nmo's dynamic pattern of co-expression with Ey, and their complementary expression in the third instar eye and head fields, respectively, supports the hypothesis that Nmo may promote early Ey activity to specify the eye field, while later contributing to patterning of the eye field by antagonizing Ey.
Using ectopic eye induction assays, we investigated Nmo's contribution to eye development in cells expressing exogenous Ey, Eya, and Dac. We demonstrated that endogenous nmo potentiates the induction of ectopic eyes in the antennal disc, as well as in the leg and wing. Interestingly, we find that loss of nmo restricts the ability of Ey, more than Eya or Dac, to induce ectopic eyes. Ey is most potent inducer of ectopic eyes as it can effectively activate transcription of the downstream RD targets (Halder et al. 1995, 1998). Eya, Dac, and So are much less effective in ectopic eye assays (Bonini et al. 1997; Chen et al. 1997; Pignoni et al. 1997; Shen and Mardon 1997; Bui et al. 2000; Weasner et al. 2007) because their transactivating potential is limited by the number of available RD cofactors (Figure 1A). Thus, we expected that misexpressed ey would have the least requirement for nmo in the dpp>ey assay. This finding suggests that Nmo may contribute to deployment of the RDGN by Ey, since cells with exogenous Eya or Dac more readily compensate for loss of endogenous nmo than Ey in the induction of ectopic eyes.
The most convincing evidence for Nmo's role in early eye specification is Nmo's ability to respecify a specific set of head cells as retinal cells when misexpressed alone. Importantly, these are the same subsets of cells able to be transformed by ectopic expression of RD genes (Halder et al. 1995; Chen et al. 1997; Pignoni et al. 1997; Shen and Mardon 1997; Bonini et al. 1998; Halder et al. 1998; Anderson et al. 2006; Weasner et al. 2007) and Tsh, which induces ey expression (Pan and Rubin 1998). Ectopic eyes induced by other factors such as Optix (Weasner et al. 2007) or Eyegone (Eyg) (Jang et al. 2003), which promote eye specification through Ey-independent mechanisms, occur in different subsets of cells. We determined that dac and eya are inappropriately activated in cells transformed by misexpressed nmo. It is tempting to speculate that ectopic Nmo perturbs the basal protein–protein interactions that normally repress them, resulting in deployment of the RDGN in the head primordia. Consistent with this model, we observed loss of Hth in cells ectopically expressing dac.
The ectopic eye induction assay has been utilized to determine epistasis among the RD factors (Chen et al. 1997, 1999; Shen and Mardon 1997; Halder et al. 1998; Pappu et al. 2003). Although we observed loss of Hth in dpp>3xnmo wing discs, this repression does not culminate in activation of any of the retinal genes. This is consistent with our clonal analyses that demonstrate that nmo is not required for expression of the RD genes in the eye disc. Moreover, Nmo antagonizes Dpp and Wg signaling in the wing disc (Zeng and Verheyen 2004; Zeng et al. 2007), both of which contribute to regulation of hth expression in the wing hinge (Azpiazu and Morata 2000). Thus, the observed loss of Hth in dpp>3xnmo eye and wing discs may be the result of different mechanisms. For example, elevated Nmo may promote Eya function to repress hth (Bessa et al. 2002) in the antennal disc. Repression of Hth is not sufficient to deploy the RDGN; therefore Nmo requires the presence of an unidentified factor in the antennal disc to activate eye development.
We showed that nmo is not required for expression of Ey, so, Eya, or Dac or the secreted morphogen dpp. In the eye disc, Wg actively represses eya, so, and dac to antagonize progression of the eye field and promote head development (Baonza and Freeman 2002). We previously showed that nmo is an inducible feedback inhibitor ofWg signaling in the wing imaginal disc (Zeng and Verheyen 2004). Although nmo expression is not coincident with wg in the ME during eye development, we wanted to verify that the observed genetic interactions between Nmo and the RDGN were not due to repression of Wg signaling. Using mutant clonal analysis, we confirmed that, as in the wing, Wg levels are unchanged in both somatic and flp-out nmo clones. Furthermore, we observed no change in Wg activity as assayed by stabilization of cytoplasmic Arm (L. R. Braid, unpublished results). These observations are consistent with a previous study indicating that nmo does not modulate Arm stability in the eye imaginal disc (Freeman and Bienz 2001). It will be interesting to determine what unidentified factors are affected by loss of nmo, and how they contribute to patterning of the eye field.
Novel targets and modes of regulating RDGN activity are rapidly emerging. Recent studies have expanded the repertoire of transcriptional targets regulated by specific RD complexes beyond the scope of the RDGN itself (Ostrin et al. 2006; Zhang et al. 2006; Jemc and Rebay 2007). Moreover, additional proteins have been identified that modify activity of the canonical retinal factors by various mechanisms. For example, Ey acts as a transcriptional activator when bound to So. However, Ey represses the very same target genes when complexed to Tsh and Hth (Bessa et al. 2002). Alternatively, the So–Eya interaction is physically inhibited when So is in complex with the transcriptional corepressor Groucho (Gro) (Silver et al. 2003). In addition, Distal antenna (Dan) and Distal antenna related (Danr) were recently identified as retinal factors that complex with Ey and Dac to promote retinal specification through activation of ato (Curtiss et al. 2007). Whether Nmo directly modulates RDGN output through protein–protein interactions that alter the stoichiometry of available RD cofactors—through post-translational modification of their activity by phosphorylation or indirectly by interactions with noncanonical RDGN regulators—is being investigated. Further characterization of the molecular interactions between Nmo and the RD factors will aid in understanding how cells integrate multiple signals to achieve a specific outcome.
Acknowledgments
We thank Ryan Fiehler, Graeme Mardon, Uwe Waldorf, and the Bloomington Stock Center for fly strains and Lily Y. Jan, Yuh Nung Jan, Richard Mann, Gines Morata, Richard Mann, and Uwe Waldorf for providing antibodies. We also thank Ashraful Anwar for help with dissections and staining, and Nick Harden and Wendy Lee for comments on the manuscript. This research was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
References
- Abu-Shaar, M., H. D. Ryoo and R. S. Mann, 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., C. L. Salzer and J. P. Kumar, 2006. Regulation of the retinal determination gene dachshund in the embryonic head and developing eye of Drosophila. Dev. Biol. 297 536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu, N., and G. Morata, 2000. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development 127 2685–2693. [DOI] [PubMed] [Google Scholar]
- Azpiazu, N., and G. Morata, 2002. Distinct functions of homothorax in leg development in Drosophila. Mech. Dev. 119 55–67. [DOI] [PubMed] [Google Scholar]
- Baonza, A., and M. Freeman, 2002. Control of Drosophila eye specification by Wingless signalling. Development 129 5313–5322. [DOI] [PubMed] [Google Scholar]
- Bessa, J., B. Gebelein, F. Pichaud, F. Casares and R. S. Mann, 2002. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 16 2415–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R. K., M. Sanicola, L. A. Raftery, T. Gillevet and W. M. Gelbart, 1991. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111 657–666. [DOI] [PubMed] [Google Scholar]
- Blochlinger, K., R. Bodmer, L. Y. Jan and Y. N. Jan, 1990. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4 1322–1331. [DOI] [PubMed] [Google Scholar]
- Bonini, N. M., W. M. Leiserson and S. Benzer, 1993. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72 379–395. [DOI] [PubMed] [Google Scholar]
- Bonini, N. M., Q. T. Bui, G. L. Gray-Board and J. M. Warrick, 1997. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124 4819–4826. [DOI] [PubMed] [Google Scholar]
- Bonini, N. M., W. M. Leiserson and S. Benzer, 1998. Multiple roles of the eyes absent gene in Drosophila. Dev. Biol. 196 42–57. [DOI] [PubMed] [Google Scholar]
- Brand, A., and N. Perrimon, 1993. Targetted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Bui, Q. T., J. E. Zimmerman, H. Liu and N. M. Bonini, 2000. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut, F., and U. Heberlein, 1997. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development 124 559–567. [DOI] [PubMed] [Google Scholar]
- Chen, R., M. Amoui, Z. Zhang and G. Mardon, 1997. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91 893–903. [DOI] [PubMed] [Google Scholar]
- Chen, R., G. Halder, Z. Zhang and G. Mardon, 1999. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development 126 935–943. [DOI] [PubMed] [Google Scholar]
- Cheyette, B. N., P. J. Green, K. Martin, H. Garren, V. Hartenstein et al., 1994. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12 977–996. [DOI] [PubMed] [Google Scholar]
- Choi, K.-W., and S. Benzer, 1994. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell 78 125–136. [DOI] [PubMed] [Google Scholar]
- Clark, S. W., B. E. Fee and J. L. Cleveland, 2002. Misexpression of the eyes absent family triggers the apoptotic program. J. Biol. Chem. 277 3560–3567. [DOI] [PubMed] [Google Scholar]
- Clem, R. J., M. Fechheimer and L. K. Miller, 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254 1388–1390. [DOI] [PubMed] [Google Scholar]
- Curtiss, J., and M. Mlodzik, 2000. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127 1325–1336. [DOI] [PubMed] [Google Scholar]
- Curtiss, J., M. Burnett and M. Mlodzik, 2007. Distal antenna and distal antenna-related function in the retinal determination network during eye development in Drosophila. Dev. Biol. 306 685–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny, T., G. Halder, U. Kloter, A. Souabni, W. J. Gehring et al., 1999. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3 297–307. [DOI] [PubMed] [Google Scholar]
- Dominguez, M., and E. Hafen, 1997. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 11 3254–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, M. C., E. M. O'Neill and G. M. Rubin, 1993. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119 855–865. [DOI] [PubMed] [Google Scholar]
- Fiehler, R. W., and T. Wolff, 2008. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev. Biol. 313 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, M., and M. Bienz, 2001. EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep. 2 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido, A., and J. R. Merriam, 1969. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J. Exp. Zool. 170 61–75. [DOI] [PubMed] [Google Scholar]
- Gibson, M. C., and G. Schubiger, 2001. Drosophila peripodial cells: More than meets the eye? BioEssays 23 691–697. [DOI] [PubMed] [Google Scholar]
- Halder, G., P. Callaerts and W. J. Gehring, 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267 1788–1792. [DOI] [PubMed] [Google Scholar]
- Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter et al., 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125 2181–2191. [DOI] [PubMed] [Google Scholar]
- Hartman, H., and T. L. Hayes, 1971. Scanning electron microscopy of Drosophila. J. Hered. 62 41–44. [DOI] [PubMed] [Google Scholar]
- Haynie, J. L., and P. J. Bryant, 1986. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 237 293–308. [DOI] [PubMed] [Google Scholar]
- Hazelett, D. J., M. Bourouis, U. Walldorf and J. E. Treisman, 1998. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125 3741–3751. [DOI] [PubMed] [Google Scholar]
- Hsiao, F. C., A. Williams, E. L. Davies and I. Rebay, 2001. Eyes absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev. Cell 1 51–61. [DOI] [PubMed] [Google Scholar]
- Ishitani, T., J. Ninomiya-Tsuji, S. Nagai, M. Nishita, M. Meneghini et al., 1999. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature 399 798–802. [DOI] [PubMed] [Google Scholar]
- Jang, C. C., J. L. Chao, N. Jones, L. C. Yao, D. A. Bessarab et al., 2003. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development 130 2939–2951. [DOI] [PubMed] [Google Scholar]
- Jarman, A. P., E. H. Grell, L. Ackerman, L. Y. Jan and Y. N. Jan, 1994. Atonal is the proneural gene for Drosophila photoreceptors. Nature 369 398–400. [DOI] [PubMed] [Google Scholar]
- Jarman, A. P., Y. Sun, L. Y. Jan and Y. N. Jan, 1995. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121 2019–2030. [DOI] [PubMed] [Google Scholar]
- Jemc, J., and I. Rebay, 2006. Targeting Drosophila eye development. Genome Biol. 7 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemc, J., and I. Rebay, 2007. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev. Biol. 310 416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, R., M. Daube, H. Duan, Y. Zou, E. Frei et al., 2001. Headless flies generated by developmental pathway interference. Development 128 3307–3319. [DOI] [PubMed] [Google Scholar]
- Kango-Singh, M., A. Singh and Y. Henry Sun, 2003. Eyeless collaborates with Hedgehog and Decapentaplegic signaling in Drosophila eye induction. Dev. Biol. 256 49–60. [DOI] [PubMed] [Google Scholar]
- Kenyon, K. L., S. S. Ranade, J. Curtiss, M. Mlodzik and F. Pignoni, 2003. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell 5 403–414. [DOI] [PubMed] [Google Scholar]
- Knoblich, J. A., and C. F. Lehner, 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 12 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson, W. M., S. Benzer and N. M. Bonini, 1998. Dual functions of the Drosophila eyes absent gene in the eye and embryo. Mech. Dev. 73 193–202. [DOI] [PubMed] [Google Scholar]
- Mardon, G., N. M. Solomon and G. M. Rubin, 1994. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120 3473–3486. [DOI] [PubMed] [Google Scholar]
- Martini, S. R., G. Roman, S. Meuser, G. Mardon and R. L. Davis, 2000. The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development 127 2663–2672. [DOI] [PubMed] [Google Scholar]
- Mirkovic, I., K. Charish, S. M. Gorski, K. McKnight and E. M. Verheyen, 2002. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech. Dev. 119 9–20. [DOI] [PubMed] [Google Scholar]
- Ostrin, E. J., Y. Li, K. Hoffman, J. Liu, K. Wang et al., 2006. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 16 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D., and G. M. Rubin, 1998. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc. Natl. Acad. Sci. USA 95 15508–15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu, K. S., and G. Mardon, 2004. Genetic control of retinal specification and determination in Drosophila. Int. J. Dev. Biol. 48 913–924. [DOI] [PubMed] [Google Scholar]
- Pappu, K. S., R. Chen, B. W. Middlebrooks, C. Woo, U. Heberlein et al., 2003. Mechanism of hedgehog signaling during Drosophila eye development. Development 130 3053–3062. [DOI] [PubMed] [Google Scholar]
- Pignoni, F., and S. L. Zipursky, 1997. Induction of Drosophila eye development by decapentaplegic. Development 124 271–278. [DOI] [PubMed] [Google Scholar]
- Pignoni, F., B. Hu, K. H. Zavitz, J. Xiao, P. A. Garrity et al., 1997. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91 881–891. [DOI] [PubMed] [Google Scholar]
- Plaza, S., F. Prince, J. Jaeger, U. Kloter, S. Flister et al., 2001. Molecular basis for the inhibition of Drosophila eye development by Antennapedia. EMBO J. 20 802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring, R., U. Walldorf, U. Kloter and W. Gehring, 1994. Homology of the eyless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science 265 785–789. [DOI] [PubMed] [Google Scholar]
- Rayapureddi, J. P., C. Kattamuri, B. D. Steinmetz, B. J. Frankfort, E. J. Ostrin et al., 2003. Eyes absent represents a class of protein tyrosine phosphatases. Nature 426 295–298. [DOI] [PubMed] [Google Scholar]
- Ready, D. F., T. E. Hanson and S. Benzer, 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53 217–240. [DOI] [PubMed] [Google Scholar]
- Robinow, S., and K. White, 1991. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22 443–461. [DOI] [PubMed] [Google Scholar]
- Rocheleau, C. E., J. Yasuda, T. H. Shin, R. Lin, H. Sawa et al., 1999. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell 97 717–726. [DOI] [PubMed] [Google Scholar]
- Royet, J., and R. Finkelstein, 1997. Establishing primordia in the Drosophila eye-antennal imaginal disc: the roles of decapentaplegic, wingless and hedgehog. Development 124 4793–4800. [DOI] [PubMed] [Google Scholar]
- Shen, W., and G. Mardon, 1997. Ectopic eye development in Drosophila induced by directed dachshund expression. Development 124 45–52. [DOI] [PubMed] [Google Scholar]
- Silver, S. J., and I. Rebay, 2005. Signaling circuitries in development: insights from the retinal determination gene network. Development 132 3–13. [DOI] [PubMed] [Google Scholar]
- Silver, S. J., E. L. Davies, L. Doyon and I. Rebay, 2003. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 23 5989–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton, K., P. D. Jackson, M. J. Clark, A. H. Brand and F. M. Hoffmann, 1994. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 5 585–593. [PubMed] [Google Scholar]
- Tomlinson, A., and D. Ready, 1987. a Cell fate in the Drosophila ommatidium. Dev. Biol. 123 264–275. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., and D. F. Ready, 1987. b Neuronal differentiation in the Drosophila ommatidium. Dev. Biol. 120 366–376. [DOI] [PubMed] [Google Scholar]
- Tootle, T. L., S. J. Silver, E. L. Davies, V. Newman, R. R. Latek et al., 2003. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature 426 299–302. [DOI] [PubMed] [Google Scholar]
- Verheyen, E. M., K. J. Purcell, M. E. Fortini and S. Artavanis-Tsakonas, 1996. Analysis of dominant enhancers and suppressors of activated Notch in Drosophila. Genetics 144 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen, E. M., I. Mirkovic, S. J. MacLean, C. Langmann, B. C. Andrews et al., 2001. The tissue polarity gene nemo carries out multiple roles in patterning during Drosophila development. Mech. Dev. 101 119–132. [DOI] [PubMed] [Google Scholar]
- Weasner, B., C. Salzer and J. P. Kumar, 2007. Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev. Biol. 303 756–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersdorff, V., T. Lecuit, S. M. Cohen and M. Mlodzik, 1996. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development 122 2153–2162. [DOI] [PubMed] [Google Scholar]
- Xu, T., and G.M. Rubin, 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zeng, Y. A., and E. M. Verheyen, 2004. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development 131 2911–2920. [DOI] [PubMed] [Google Scholar]
- Zeng, Y. A., M. Rahnama, S. Wang, W. Sosu-Sedzorme and E. M. Verheyen, 2007. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development 134 2061–2071. [DOI] [PubMed] [Google Scholar]
- Zhang, T., S. Ranade, C. Q. Cai, C. Clouser and F. Pignoni, 2006. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133 4881–4889. [DOI] [PubMed] [Google Scholar]
- Zimmerman, J. E., Q. T. Bui, H. Liu and N. M. Bonini, 2000. Molecular genetic analysis of Drosophila eyes absent mutants reveals an eye enhancer element. Genetics 154 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]