Abstract
The first step of bacteriophage (phage) infection is the attachment of the phage virion onto a susceptible host cell. This adsorption process is usually described by mass-action kinetics, which implicitly assume an equal influence of host density and adsorption rate on the adsorption process. Therefore, an environment with high host density can be considered as equivalent to a phage endowed with a high adsorption rate, and vice versa. On the basis of this assumption, the effect of adsorption rate on the evolution of phage optimal lysis time can be reinterpreted from previous optimality models on the evolution of optimal lysis time. That is, phage strains with a higher adsorption rate would have a shorter optimal lysis time and vice versa. Isogenic phage λ-strains with different combinations of six different lysis times (ranging from 29.3 to 68 min), two adsorption rates (9.9 × 10−9 and 1.3 × 10−9 phage−1 cell−1 ml−1 min−1), and two markers (resulting in “blue” or “white” plaques) were constructed. Various pairwise competitions among these strains were conducted to test the model prediction. As predicted by the reinterpreted model, the results showed that the optimal lysis time is shorter for phage strains with a high adsorption rate and vice versa. Competition between high- and low-adsorption strains also showed that, under current conditions and phenotype configurations, the adsorption rate has a much larger impact on phage relative fitness than the lysis time.
THE life cycle of a generic virus can be divided into three successive stages: (1) “searching” for a susceptible host cell to initiate an infection, (2) producing viral progeny inside the infected host, and (3) exiting current host cell to start a new infection cycle. From the point of view of a lytic bacteriophage (phage), the three stages of its life cycle correspond to the processes of adsorption, maturation, and lysis. The rates and timing of these processes can thus be seen as a phage's life-history traits.
The genetic and molecular bases for many of these traits have been known for quite some time and the study of these processes has formed the foundation of modern molecular biology (Stent 1965). To initiate an infection, a phage virion has to first adsorb onto the surface of a susceptible host cell. This is accomplished by the recognition of receptors on the host cell surface by phage tail fiber (or attachment protein) and various other appendages (Katsura 1983; Goldberg et al. 1994). Only a few phage gene products, usually in the range of one to three, are involved in the adsorption process. The initial step of host recognition is commonly seen as a ligand-receptor binding problem; therefore, the rate of “finding” a host is usually assumed to follow the mass-action kinetics (Schlesinger 1932), as described in Stent (1965). Consequently, the search time (tS) would in part be determined by the host density in the environment and the adsorption rate constant (r) of the phage.
Once the phage genetic material is injected into the host cell, a series of molecular events would be initiated and host synthesis machinery is then hijacked and intracellular resources diverted to produce materials needed for phage progeny. Typically, the first infectious phage progeny is not assembled until sometime after adsorption and injection of phage genetic materials. This period of time, traditionally called the eclipse period (e), is required to synthesize enough phage-encoded materials for progeny production. For many larges phages, the process of phage morphogenesis (or assembly) is complex and involves many proteins. Even for small phages that are encapsulated with only few capsid proteins, the interaction between the genetic materials and phage proteins is also complex. However, it can be inferred that the phage components are being continuously synthesized and maintained at a certain level at the same time that the phage progeny are assembled. This is because for at least a few known phages (Hutchison and Sinsheimer 1966; Josslin 1970; Wang et al. 1996; Wang 2006), the progeny are accumulated linearly before host lysis.
While the progeny is being assembled, the phage-encoded lysis proteins are also expressed to prepare for the eventual release of the accumulated progeny from the infected host cells. In many phages, host lysis is achieved by a single-gene lysis protein or a holin–endolysin system (Young 1992; Wang et al. 2000; Young et al. 2000). The molecular mechanisms of phage-mediated host lysis have been studied extensively (Wang et al. 2000; Young et al. 2000). Under most conditions, only one to three gene products are needed for host lysis. The length of time it takes for a phage to complete the intracellular phase of its life cycle constitutes the lysis time tL, traditionally called the latent period.
The number of phage progeny released from each infected host is the phage fecundity (or burst size, b), which is jointly determined by three phage life-history traits: the eclipse period, the lysis time, and the maturation (assembly) rate (m). Empirically, the relationship can be expressed as b = m(tL − e), at least to a certain extent (Wang 2006). Clearly, the progeny phage cannot be accumulated indefinitely. The burst size reaches a certain maximum after a while (Wang 2006). Generally, for a phage, it takes tS + tL amount of time to complete its life cycle. Therefore, tS + tL can be seen as the generation time. At the end of each generation, b amount of the progeny is produced. As a result, the phage growth rate can be expressed as ln(b)/(tS + tL) (Wang et al. 1996; Bull 2006).
The simplicity of the phage life cycle makes it relatively easy to be analyzed with optimality models (Abedon 1989; Wang et al. 1996; Bull et al. 2004). The wealth of phage biology also allows us to hypothesize trade-off relationships among various phage life-history traits (De Paepe and Taddei 2006; Caraco and Wang 2008). So far, among these traits, the problem of optimal lysis time has received the most theoretical and experimental attention (Abedon 1989; Wang et al. 1996; Abedon et al. 2001, 2003; Bull 2006; Wang 2006; Heineman and Bull 2007). The existence of an optimal lysis time can be understood as the consequence of the trade-off between generation time and burst size. This is because to maximize the phage growth rate is to shorten the generation time and/or to increase the burst size. For the phage, these two traits have been empirically shown to be positively linked; i.e., the longer the generation time (due to a longer lysis time), the larger the burst size. Consequently, a shorter lysis time (hence a shorter generation time) would inevitably lead to a smaller burst size, and vice versa. As has been shown previously (Wang et al. 1996), a phage with too long a lysis time would lose the opportunity of initiating many new infections, though its burst size would increase linearly as a result. On the other hand, a phage with too short a lysis time would be able to infect other host cells in the environment earlier, but with a much reduced gain because of reduced burst size. On balance, the phage with the maximal growth rate (fitness) would be the one that has the intermediate (optimal) lysis time.
The influence of host density on the evolution of phage optimal lysis time is mainly mediated through its effect on the length of search time. That is, a phage in an environment containing high host density will experience a shorter search time (and, by extension, a shorter generation time) and vice versa. As the analysis showed, the optimal lysis time is shorter when the host density is high and vice versa (Wang et al. 1996). Several empirical studies using different phage systems have given some support to the utility of this optimality model in describing the effect of host density on the evolution of phage lysis time (Abedon et al. 2003; Wang 2006; Heineman and Bull 2007); although the original model tends to break down under conditions of low host density (Abedon et al. 2001; Heineman and Bull 2007). This is because when the host density (N) is high, the average search time for a phage to be adsorbed onto the host cell surface can be approximated by tS = 1/rN, again assuming the mass-action kinetics. However, when N is small, the stochastic nature of phage adsorption becomes dominant. That is, the times for individual adsorption events would be exponentially distributed (McAdams and Arkin 1997). In fact, ∼63% of individual search times would be shorter than the average estimated by tS = 1/rN and ∼5% >3/rN (McAdams and Arkin 1997). That is, these earlier-than-average adsorption events would contribute to an effectively shorter search time than would be expected. Therefore, when the host density is low, the expected optimal lysis time would also be shorter than the one predicted by using tS = 1/rN as the average search time (Abedon et al. 2001). Interestingly, using an optimality model based on phage population dynamics in a chemostat [or more precisely cellstat (Husimi et al. 1982)], Bull (2006) showed that, when the phage population is at the dynamic equilibrium, the relationship between the optimal lysis time and maximal growth rate can be expressed simply as 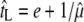 , where a circumflex (∧) indicates a value at the optimum, μ the phage growth rate, and e the eclipse period. Simulation analyses showed that the above formula performed well even under conditions of low host density (Bull 2006).
, where a circumflex (∧) indicates a value at the optimum, μ the phage growth rate, and e the eclipse period. Simulation analyses showed that the above formula performed well even under conditions of low host density (Bull 2006).
Even though previous studies focused mainly on the effects of ecological factors on the evolution of phage lysis time, the optimality approach can also be used to explore the evolution of other phage life-history traits, such as adsorption rate, maturation rate, eclipse period, etc. (Bull et al. 2006). Among these many possibilities, the effect of adsorption rate on the evolution of optimal lysis time would be a natural first choice. This is possible because of an implicit equivalency between adsorption rate and host density. That is, an environment with high host density is equivalent to a phage endowed with a high adsorption rate, for which both conditions would result in a shortened search time for the phage. Therefore, the previous optimality model can now be reinterpreted to make predictions on the joint effects of life-history traits on phage fitness. For instance, the model would predict that the optimal lysis time would be shorter for phages with a high adsorption rate and vice versa.
In this study, we competed various marked isogenic phage λ-strains that differ only in their adsorption rate and lysis time and demonstrated that, as predicted by the model, phage strains with a high adsorption rate have a shorter optimal lysis time than strains with a low adsorption rate. However, as discussed later, these predictions are more qualitative than quantitative. We further showed that, under most conditions, adsorption rate has a much larger impact on phage fitness than lysis time.
MATERIALS AND METHODS
Bacterial and phage strains, plasmids, and primers:
All bacterial and phage strains, plasmids, and primers used in this study are listed in Table 1. Bacteria cultures were grown in LB media with various antibiotics when appropriate. The concentrations of antibiotics were as follows: 100 μg/ml for ampicillin and 10 μg/ml for chloramphenicol.
TABLE 1.
Bacterial and phage strains, plasmids, and primers
| Name | Genotype and relevant features | Reference |
|---|---|---|
| Bacterial strains | ||
| IN25 | Originally MC4100 | Wang (2006) |
| IN56 | MC4100(λcI857) | Wang (2006) |
| SYP045 | MC4100(λcI857 Swt R∷lacZα+ stf−) | This study |
| SYP046 | MC4100(λcI857 Swt R∷lacZα+ stf+) | This study |
| SYP047 | MC4100(λcI857 ΔS∷Cam R∷lacZα+ stf−) | This study |
| SYP049 | MC4100(λcI857 Swt R∷lacZα− stf−) | This study |
| SYP052 | MC4100(λcI857 Swt R Δ(J-orf401)∷Cam | This study |
| SYP053 | MC4100(λcI857 Swt R stf+) | This study |
| SYP056 | MC4100(λcI857 Swt R∷lacZα− stf+) | This study |
| SYP061 | MC4100(λcI857 ΔS∷Cam R∷lacZα+ stf+) | This study |
| SYP062 | MC4100(λcI857 ΔS∷Cam R∷lacZα− stf+) | This study |
| SYP085 | MC4100(λcI857 SS68C R∷lacZα+ stf−) | This study |
| SYP086 | MC4100(λcI857 SM1L R∷lacZα+ stf−) | This study |
| SYP087 | MC4100(λcI857 SM1L/C51S R∷lacZα+ stf−) | This study |
| SYP088 | MC4100(λcI857 SM1L/C51S/S76C R∷lacZα+ stf−) | This study |
| SYP093 | MC4100(λcI857 SS68C R∷lacZα+ stf+) | This study |
| SYP094 | MC4100(λcI857 SM1L R∷lacZα+ stf+) | This study |
| SYP095 | MC4100(λcI857 SM1L/C51S R∷lacZα+ stf+) | This study |
| SYP096 | MC4100(λcI857 SM1L/C51S/S76C R∷lacZα+ stf+) | This study |
| SYP107 | MC4100(λcI857 SM1L/V77G R∷lacZα+ stf−) | This study |
| SYP111 | MC4100(λcI857 SM1L/V77G R∷lacZα+ stf+) | This study |
| SYP119 | MC4100(λcI857 SM1L/C51S R∷lacZα− stf+) | This study |
| SYP120 | MC4100(λcI857 SM1L/C51S/S76C R∷lacZα− stf+) | This study |
| Phage strains | ||
| All phage strains were thermally induced (see materialsandmethods) from the lysogens above. | ||
| Plasmids | ||
| pSwt | Contains the wt λ-lysis genes: S, R, and Rz/Rz1 | Wang (2006) |
| pS105 | The same as pSwt, but with an M1L mutation at the S gene | Wang (2006) |
| pS105/C51S | The same as pS105, but with an additional C51S mutation at the S gene | Wang (2006) |
| pS105/C51S/S76C | The same as pS105/C51S, but with an additional S76C mutations at the S gene | Wang (2006) |
| pS105/V77G | The same as pS105, but with an additional V77G mutation at the S gene | This study |
| pSwtRlacZblueRz | The same as pSwt, but with an R∷lacZα+ insertion | This study |
| pSwtRlacZwhiteRz | The same as pSwt, but with an R∷lacZα− insertion | This study |
| pZE1-J-stf | Contains the genomic region encompassing part of J, all of lom and orf401, and part of orf314 | This study |
| pZE1-J-stf+ | The same as pZE1-J-stf, but with a cytocine (C) insertion in orf401, resulting in fusion of orf401 and orf314 to become stf | This study |
| pZE1-J-Cam-stf | The same as pZE1-J-stf, but with cam replacing the region between J and orf314 | This study |
| pSwtdelS_Cam | same as pSwt, but with cam replacing part of S | This study |
| Primers | ||
| RlacZα_For | GGAACGGTCAGAGAGATTGATatgaccatgattacggattcac | This study |
| RlacZα_Rev | AATCGCGGTGACTCTGCTCATACgatgggcgcatcgtaaccgtg | This study |
| white_α_For | GCCGTCGTTTTACAAGCTGCTGCCTGGGCAGCCCCTGGCGTTACCC | This study |
| white_α_Rev | GGGTAACGCCAGGGGCTGCCCAGGCAGCAGCTTGTAAAACGACGGC | This study |
| J_to_stf_For | GCGCGACGTCAGGTTGAAACCAGCACGCG | This study |
| J_to_stf_Rev | GCGCTCTAGACAGCACGACCGCTGGCGGG | This study |
| del_stf_For | CACCGCTTTTGTACTGGCCG | This study |
| del_J_Rev | CACATTACTGAGCGTCCCGG | This study |
| Lam_Ur_For | CGAACAGAAAAGCCCCACTGGACAGTCCGGC | This study |
| Lam_Ur_Rev | GCCGGACTGTCCAGTGGGGCTTTTCTGTTCG | This study |
| delS16-92_For | CGCTTCGCTGCTAAAAAAGCCGGA | This study |
| delS16-92_Rev | CGCGAGAATGGCGGCCAACAGGTC | This study |
Phage strain constructions:
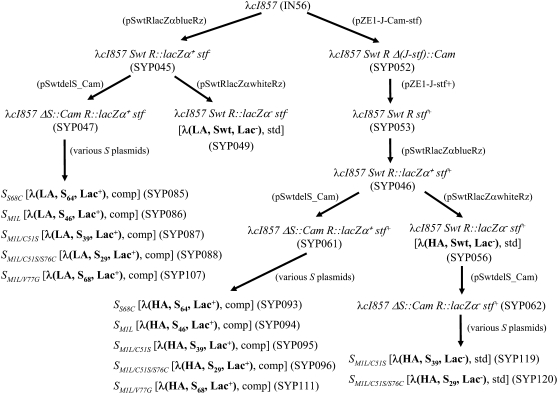
The goal of phage strain construction is to construct isogenic phage strains that differ in three traits: (1) adsorption rate, (2) lysis time, and (3) marker state. The high and low adsorption rates are achieved by the presence or the absence of the side tail fiber, encoded by the stf gene. The different lysis times are results of different S alleles, most of which have been described previously (Wang 2006). The two marker states are the results of wild-type and mutant α-fragments of Escherichia coli's β-galactosidase (β-gal). Due to numerous steps involved, the details of phage strain construction are presented in the supplemental data. The flowchart for strain construction is shown in Figure 1. The identities of various strain constructs were confirmed by DNA sequencing.
Figure 1.—
Flowchart of phage λ-strain construction. Relevant genotypes of each λ-phage strain are shown. Names of λ-lysogens and plasmids used in construction are shown in parentheses. The names of the standard (std) and competing (comp) λ-strains are represented by the phage phenotype with the format of λ(X, Y, Z) in boldface type within brackets. X is the state of the adsorption rate (LA, low adsorption; HA, high adsorption), Y the approximate lysis time (Swt, the wt S allele with a lysis time of ∼52 min), and Z the marker state [Lac+ produces blue plaques; Lac− produces clear (“white”) plaques]. Please refer to the supplemental data section for details of strain construction.
Standard PCR and DNA sequencing:
Standard PCR reactions were performed using the following condition: one cycle of 95° for 1 min, followed by 30 cycles of 95° for 30 sec, 50° for 30 sec, and 72° for several minutes, depending on the size of the template (using 1 min/kb). A high-fidelity thermal-stable DNA polymerase, PfuUltra (Stratagene, La Jolla, CA), was used for amplification. The BigDye Terminator cycle sequencing kit (v.3.1; ABI, Columbia, MD) was used for DNA sequencing reactions, following the vendor's recommendation. DNA sequencing was performed at the Molecular Core Facility of the Life Sciences Research Building located in the State University of New York at Albany.
Determination of adsorption rate:
A similar protocol (Hendrix and Duda 1992) was adopted for determining the adsorption rate of λ-phages. Briefly, in 10 ml LB containing 5 mm MgCl2, ∼4 × 106 cells/ml of exponentially grown IN25 were mixed with ∼5 × 103 plaque-forming units (pfu)/ml of λcI857 SWT R∷lacZα− stf− or λcI857 SWT R∷lacZα− stf+ and incubated in a water bath at 37° with constant shaking. Samples were withdrawn and filtered using an AcroPrep 96 filter plate (Pall, Ann Arbor, MI) every 4 min. The filtrate was plated on IN25 for plaque counting. The adsorption rates were estimated by fitting the data with the following model of 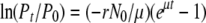 , where Pt and P0 are phage concentrations at times t and 0, respectively; r the adsorption rate to be estimated; N0 the bacteria concentration at time 0; and μ the bacteria growth rate (see appendix a for equation derivation). The nonlinear fit was performed using the statistical package JMP v.5.0.1a for MacOS. Three replicate experiments were conducted to estimate adsorption rate. The cell concentrations at the beginning and the end of each experiment were determined and were used separately for the estimation of each replicate adsorption rate.
, where Pt and P0 are phage concentrations at times t and 0, respectively; r the adsorption rate to be estimated; N0 the bacteria concentration at time 0; and μ the bacteria growth rate (see appendix a for equation derivation). The nonlinear fit was performed using the statistical package JMP v.5.0.1a for MacOS. Three replicate experiments were conducted to estimate adsorption rate. The cell concentrations at the beginning and the end of each experiment were determined and were used separately for the estimation of each replicate adsorption rate.
Determination of lysis time:
The previously described procedure (Wang 2006) was used for the determination of phage lysis time. Three replicates were conducted for each lysis time determination.
Phage competition experiments and determination of phage growth rate and relative fitness:
Unless otherwise stated, all competition experiments were conducted by inoculating a total of ∼2 × 104 pfu/ml into a 125-ml flask containing 10 ml of prewarmed (to 37°) LB plus 5 mm MgSO4 and ∼106 cells/ml of MC4100 cells that have been grown exponentially to A550 ≈ 0.25. The culture was incubated at 37° in a water bath shaker (New Brunswick Scientific, Edison, NJ) with constant shaking of 225 rpm for 4 hr. In most cases, the initial ratio of blue and white phages was kept 1:1 when competition was conducted within each stf background. When competition was between stf+ and stf− phages, the ratio was adjusted (usually 1:10) so that at the end of the competition both blue and white plaques could be reasonably counted. Standardized protocol and precautions (Wang 2006) were followed for phage plating. To differentiate the standard and the competing strains, XL-1 Blue was used as the plating host with the same IPTG and X-gal conditions as described above. Emerging blue and clear plaques were counted separately.
The growth rates for the competing (μC) and the standard (μS) strains were calculated as μC = ln(PC4/PC0)/4 and μS = ln(PS4/PS0)/4, where PC and PS are the concentrations for the competing and standard strains, respectively, and the subscripts 0 and 4 are the times 0 and 4 hr after infection, respectively. The relative fitness of the competing phage strain was defined as w = μC/μS. Three replicates were conducted for each pairwise competition.
Statistical analyses:
Since most data were collected with few replicate experiments (usually three), the calculated standard errors have all been corrected for small sample sizes according to Sokal and Rohlf (1995, p. 53). The correction would result in a larger standard deviation and standard error than the typically calculated ones.
RESULTS
Effect of side tail fiber on phage λ's adsorption rate:
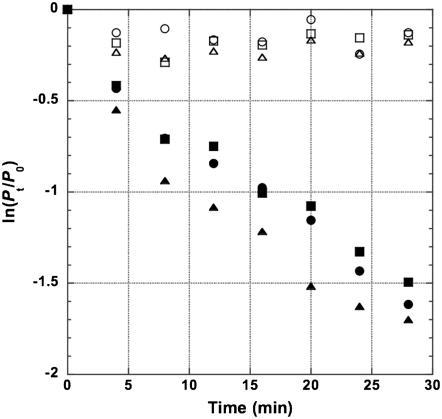
The adsorption rate of the laboratory wild-type (wt) λ (λPaPa) is much lower than that of the original λ-strain (Ur-λ) (Hendrix and Duda 1992). The reduced adsorption rate is due to a frameshift mutation (a deletion of cytosine) in the side tail fiber (stf) gene that resulted in the loss of side tail fibers (Hendrix and Duda 1992). By using site-directed mutagenesis (see materials and methods), we successfully restored the functional stf gene back into the genome of laboratory wt λ. As shown in Figure 2, the relative concentrations of the stf+ phage declined appreciably during the assay period while the decline of the stf− phage was barely noticeable. A similar pattern was also shown elsewhere (Hendrix and Duda 1992). Clearly, the presence of the side tail fibers greatly increases the rate of adsorbing onto the cell surface.
Figure 2.—
The adsorption kinetics of high- and low-adsorption λ-phages. After mixing with exponentially grown E. coli cells, cultures containing λ(HA, Swt, Lac−) (solid symbols) or λ(LA, Swt, Lac−) (open symbols) phages were withdrawn and filtered every 4 min for 28 min. Ratios of unadsorbed phages were logarithmically transformed and plotted against time. Different symbols represent different replicate assays. Adsorption rates were estimated using the equation 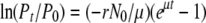 , where Pt and P0 are phage concentrations at times t and 0, respectively; r is the adsorption rate to be estimated; N0 the bacteria concentration at time 0; and μ the bacteria growth rate (see materials and methods and appendix a for equation derivation).
, where Pt and P0 are phage concentrations at times t and 0, respectively; r is the adsorption rate to be estimated; N0 the bacteria concentration at time 0; and μ the bacteria growth rate (see materials and methods and appendix a for equation derivation).
Traditionally, the adsorption rate is estimated by fitting the logarithmically transformed relative phage concentration data with a linear regression line. The slope of the line is the product of adsorption rate and bacteria cell concentration. Assuming constant host density during the assay period, the adsorption rate can thus be calculated by dividing the value of the slope by the host density. Since the length of time used for adsorption rate determination (28 min in this study) is about the generation time of bacterial growth in a rich medium, the host density is not kept constant. To account for the changing host density, thus affecting the estimated adsorption rate, a nonlinear regression model was used to estimate the adsorption rates (see appendix a for equation derivation). The estimated adsorption rates for the stf+ and stf− phages are 9.90 ± 3.00 × 10−9 and 1.33 ± 0.383 × 10−9 phage−1 cell−1 ml−1 min−1, with mean ± 95% confidence limits shown. That is, when compared to the stf− phages, the stf+ phages would be ∼7.4 times more likely to “find” a host. Therefore, in this study, phages with the stf+ genotype would have the high-adsorption (HA) phenotype and would be referred to as the HA phages and phages with the stf− genotype would have the low-adsorption (LA) phenotype, referred to as the LA phages.
Range of the lysis time spectrum:
Six different S alleles, each resulting in different lysis times, were used in this study. Table 2 lists the genotypes and the estimated lysis times for the phage strains that carry these alleles. In this study, the lysis times ranged from the shortest of 29.3 min to the longest of 68.0 min.
TABLE 2.
The growth rates of the standard and competing strains
| Competing strain namea | S allele | stf status | Phenotype designation | Lysis time (min ± 95% confidence intervals) | Competing strain growth rate (hr−1 ± 95% confidence intervals) | Standard strain growth rateb (hr−1 ± 95% confidence intervals) | Relative fitness (mean ± 95% confidence intervalsc) |
|---|---|---|---|---|---|---|---|
| SYP088 | SM1L/C51S/S76C | — | λ(LA, S29) | 29.3 ± 1.47 | 0.929 ± 0.052 | 2.761 ± 0.073 | 0.336 ± 0.012 |
| SYP087 | SM1L/C51S | — | λ(LA, S39) | 38.7 ± 1.47 | 2.298 ± 0.028 | 2.771 ± 0.023 | 0.829 ± 0.004 |
| SYP086 | SM1L | — | λ(LA, S46) | 46.0 ± 0.00 | 2.611 ± 0.046 | 2.783 ± 0.067 | 0.938 ± 0.019 |
| SYP045 | Swt | — | λ(LA, S52) | 52.3 ± 1.27 | 2.821 ± 0.022 | 2.800 ± 0.030 | 1.001 ± 0.017 |
| SYP085 | SS68C | — | λ(LA, S64) | 64.0 ± 0.00 | 2.967 ± 0.065 | 2.761 ± 0.075 | 1.075 ± 0.031 |
| SYP107 | SM1L/V77G | — | λ(LA, S68) | 68.0 ± 0.00 | 2.338 ± 0.230 | 2.776 ± 0.072 | 0.842 ± 0.067 |
| SYP096 | SM1L/C51S/S76C | + | λ(HA, S29) | 29.3 ± 1.47 | 0.447 ± 0.073 | 3.777 ± 0.455 | 0.118 ± 0.010 |
| SYP095 | SM1L/C51S | + | λ(HA, S39) | 38.7 ± 1.47 | 2.053 ± 0.274 | 3.160 ± 0.179 | 0.649 ± 0.062 |
| SYP094 | SM1L | + | λ(HA, S46) | 46.0 ± 0.00 | 3.062 ± 0.065 | 2.985 ± 0.084 | 1.026 ± 0.001 |
| SYP046 | Swt | + | λ(HA, S52) | 52.3 ± 1.27 | 3.110 ± 0.108 | 3.058 ± 0.099 | 1.017 ± 0.002 |
| SYP093 | SS68C | + | λ(HA, S64) | 64.0 ± 0.00 | 3.097 ± 0.131 | 3.188 ± 0.089 | 0.971 ± 0.016 |
| SYP111 | SM1L/V77G | + | λ(HA, S68) | 68.0 ± 0.00 | 2.896 ± 0.144 | 2.973 ± 0.226 | 0.975 ± 0.032 |
The bacterial lysogen strains that have each λ-strain residing in them.
The growth rates for each standard strain used in pairwise competitions.
The 95% confidence intervals were calculated from individual relative fitness values.
For simplicity of strain notation, each S allele is referred to by its phenotypic effect, namely the lysis times. For example, the Swt allele is named Swt, which has a lysis time of 52.3 min, and SM1L/C51S is named S39, which has a lysis time of 38.7 min.
Effect of the lacZα marker on phage fitness:
To differentiate between the standard and the competing strains, the genetic marker lacZα, which encodes the first 100 amino acid residues of the lacZ gene from E. coli, was inserted at the 3′ end of λ's R gene (Wang et al. 2003). Two versions of the marker were constructed: (1) lacZα+, the functional version that could complement the ω-fragment, thus restoring the β-gal activity, and (2) lacZα−, the inactive version that, due to various introduced mutations (see materials and methods), would fail to form the required tetramer that is necessary for β-gal activity. The marker state is differentiated by a common colorimetric method, with the lacZα+-carrying strain producing blue plaques and the lacZα−-carrying strain clear ones. In this study, strains carrying the lacZα+ allele are represented as Lac+ and those carrying the lacZα− allele as Lac−.
Strain designation:
To reduce confusion and facilitate communication, the phage strain name from this point on is represented by λ(X, Y, Z), where X is the state of the adsorption rate, HA or LA; Y is the lysis time, S29 to S68; and Z is the marker state, Lac+ for blue plaque and Lac− for clear (white) plaque. For example, the strain with the genotype of λcI857 SM1L R∷lacZα+ stf+ is referred as λ(HA, S46, Lac+).
To assess the potential marker effect on phage relative fitness, two pairwise competitions were conducted, using λ(HA, Swt, Lac+) vs. λ(HA, Swt, Lac−) and λ(LA, Swt, Lac+) vs. λ(LA, Swt, Lac−) strains. As shown in Figure 3, neither the HA nor the LA strains showed discernible marker effect on phage relative fitness. That is, the relative fitness in both cases is ∼1.000 (1.017 ± 0.002 for stf+ and 1.001 ± 0.017 for stf−, with mean ± 95% confidence limits shown).
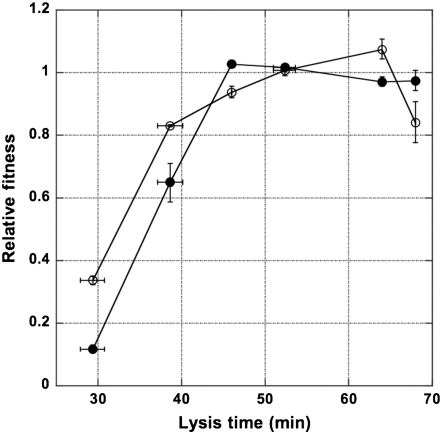
Figure 3.—
Effects of adsorption rates on optimal lysis times. Relative fitnesses of the high-adsorption (solid circle) and low-adsorption (open circle) λ-phages were estimated with pairwise competitions and plotted against the lysis time of the competing strains. In this plot, only phage strains with the same adsorption rate were competed against each other. Phage strains used in competitions are shown in Figure 1. The standard strains for the low- and high-adsorption phages are λ(LA, Swt, Lac−) and λ(HA, Swt, Lac−), respectively. The lysis times and relative fitness for each pairwise competition are listed in Table 2. Error bars show the 95% confidence intervals of both the relative fitness and the lysis time. Some values are too small to appear in the plot.
Experimental conditions and phage competitions:
All the competition experiments were conducted by competing the standard strains against the competing strains. In this study, the standard strains are λ(HA, Swt, Lac−) and λ(LA, Swt, Lac−). Since the standard strains are the same in each pairwise competition, the standard strains can be seen as an internal control for the culture environments. If the growth rates of the standard strains are similar across all pairwise competitions, then it can be argued that all phage strains that participated in pairwise competitions have experienced a similar culture environment.
Table 2 lists the growth rates of the standard and the competing strains for various pairwise competitions. To test to see if the growth rates of the standard strains are different from each other, unplanned comparisons are conducted using a Tukey–Kramer honestly significant difference (HSD) test at the α = 0.05 level, as implemented in JMP (v.5.0.1a for MacOS). For competitions among the LA phages, there is no significant difference among the growth rates of the standard strain. With one exception [i.e., λ(HA, S29, Lac+) vs. λ(HA, Swt, Lac−)], all the other HA standard strain growth rates are not significantly different from each other either. This result suggests that the culture environment, especially the host density, is quite similar across these pairwise competition experiments.
Effect of adsorption rate on optimal lysis time:
The phage strain with a higher adsorption rate would on average have a shorter time in encountering and attacking a host; therefore, based on the reinterpreted optimality model, it would have a shorter optimal lysis time when compared to the phage strain with a lower adsorption rate. To test this prediction, several pairwise competition experiments were conducted between various isogenic λ-strains that differ in their lysis times and adsorption rates.
Figure 3 shows the relative fitness of isogenic λ-strains with different lysis times and adsorption rates. Note that each pairwise competition was conducted among the HA or LA phages. As shown in Figure 3, the relative fitness curves peaked at different lysis times, depending on phage adsorption rates. As predicted by the model, the HA phages have a shorter optimal lysis time (with the fitness curve peaking at 46.0 min) than that of the LA phages (which peaked at 64.0 min).
Interestingly, the relative fitness curves are not symmetrical around either optimum. In the case of HA phages, there is a steep decline of relative fitness when the lysis time is below the optimum, but a very shallow decline, almost plateau-like, when the lysis time is longer than the optimum. On the other hand, for the LA phages, the optimum peak is more prominent, with notable declines on relative fitness when the lysis times are suboptimal. It is interesting to note that simulation studies (Bull 2006; Wang 2006) also tend to find sharp declines on the left side of the fitness curves.
Combined effects of adsorption rate and lysis time on phage relative fitness:
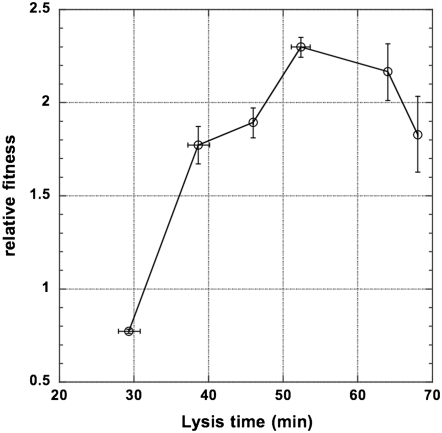
The results shown in Figure 3 may give an erroneous impression that both phage traits—adsorption rate and lysis time—have a similar impact on relative fitness. In reality, under the current assay conditions, adsorption rate has a much larger effect on phage relative fitness than the lysis time (our unpublished results). As a result, regardless of the lysis times, the competition outcome for the most part can be predicted by the state of the stf allele alone, with the stf+ strains outcompeting the stf− ones. The exceptions are when the stf+ allele is associated with suboptimal lysis time alleles that resulted in much too short a lysis time. Figure 4 shows the outcomes of pairwise competition experiments, using the λ(HA, S29, Lac−) phage as the standard strain to compete against LA strains with various lysis times. As shown in Figure 4, only under such a disparate configuration can we observe the fitness advantage conferred by high adsorption rate being overcome by the disadvantage incurred by suboptimal lysis times. However, if the standard strain is replaced with a strain with a slightly longer lysis time [i.e., λ(HA, S39, Lac−)], the standard HA strain outcompetes all other competing LA strains, regardless of the lysis times (data not shown).
Figure 4.—
Competition between the low- and high-adsorption λ-phages. Relative fitness was determined and plotted against the lysis time of competing strains. In this plot the standard strain is the high-adsorption λ(HA, S29, Lac−) and the competing strains are low-adsorption (LA) strains with various lysis times. Error bars show the 95% confidence intervals of both lysis time and relative fitness. Some values are too small to appear in the plot.
Even though the presence of stf+ is in general advantageous in liquid culture conditions, the relative fitness gain conferred by stf+ still depends on the state of the S allele. As shown in Figure 4, when compared to λ(HA, S29, Lac−), the relative fitness of λ(LA, S29, Lac+) is 0.774 ± 0.011 (with a 95% confidence limit shown). However, when compared to λ(HA, Swt, Lac−) (lysis time 52.3 min), the relative fitness of λ(LA, Swt, Lac+) was estimated as 0.329 ± 0.0183. That is, as the lysis time approaches the optimum, the effect of the adsorption rate becomes more important.
DISCUSSION
The pros and cons of the lacZα marker system:
Genetic markers are routinely used to differentiate between different bacterial or viral strains in competition experiments. Typically, the methods for differentiation are based on selection, e.g., resistance to phage T5 infection in E. coli competition experiments (Dykhuizen et al. 1987), the ability to utilize arabinose (Lenski 1988), the host range marker in phage φ6 (Chao 1990), and resistance to monoclonal antibodies in vesicular stomatitis virus (Duarte et al. 1992) and foot-and-mouth disease virus (Martinez et al. 1991). With few exceptions [e.g., T5 resistance in E. coli (Dykhuizen et al. 1987) and the ability to utilize arabinose (Lenski 1988)], the most common problem with the selection-based marker system is the fitness cost imposed by the marker. The marker effect is especially frequent when viruses are used as the study organisms; although the fitness cost is usually taken into account when estimating relative fitness.
We adopted a screening-based marker system, specifically color screening, which is unlike the selection-based marker system. As we demonstrated in this study, the LacZα-based screening has virtually no marker effect on phage fitness. Although the marker does impose a slight fitness cost when compared to the phage strain without the marker. (The relative fitness of the LacZα-marked strain is ∼0.91.) To eliminate the potential marker effect, it is important to construct isogenic markers with minimal differences; in our case, the difference is five amino acid residues. Also, the screening-based system has its own limit as well. In our case, the limit of detection of the LacZα marker is set by the ratio of the two phage strains and the total number of plaques that can be counted (manually, in this case) on a typical petri dish, which is ∼500 plaques. Because of this, the screening-based method would have a smaller dynamic range in detecting phage concentration differences. Consequently, the ability to determine large fitness differences is more limited in the screening-based than the selection-based methods.
Experimental conditions and their effects on phage life-history traits and fitness:
Since all the λ-genes required for host lysis and progeny virion assembly are coexpressed as one long late transcript (Friedman and Gottesman 1983 and references therein), it is conceivable that insertion of the lacZα gene at the end of the endolysin R gene (see materials and methods) may have interfered with the expression pattern of these genes, thus altering lysis time and burst size and consequently phage fitness. In our case, it is unclear which trait(s) is affected and hence responsible for the fitness cost. All lysis times in our study are determined using lysogens carrying lacZα-marked λ-prophages, while five of the current six S alleles were also determined in a previous study but without the marker (Wang 2006). Comparison between these two sets of results showed that the lysis time was slightly affected by the marker insertion. But there is no consistent direction of bias in lysis time. Furthermore, the timing usually differs within 2–3 min. Since we have not determined the burst size for the marked phage strains, we do not know the impact of marker insertion. However, it is not unreasonable to speculate that both the lysis time and the burst size have been somewhat affected by the marker insertion. This may explain why the observed optimum lysis times (under the stf− background) are different between these two studies.
Optimality models and the quantitative prediction of optimal lysis times:
Currently, there are two seemingly different theoretical approaches to the question of optimal lysis time. The first one, as described in this study, explicitly incorporates phage life-history traits into the commonly used formula for growth rate calculation. That is, the phage growth rate is expressed as 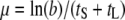 , where b is the burst size, tS the search time, and tL the lysis time (Wang et al. 1996). Maximization of μ is through optimization of tL. The other approach is derived from comprehensive phage population dynamics in a chemostat culture (Bull 2006; Bull et al. 2006). The phage growth rate is found to be
, where b is the burst size, tS the search time, and tL the lysis time (Wang et al. 1996). Maximization of μ is through optimization of tL. The other approach is derived from comprehensive phage population dynamics in a chemostat culture (Bull 2006; Bull et al. 2006). The phage growth rate is found to be 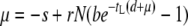 , where s is the free phage death rate, r the adsorption rate, N the host concentration, and d the washout rate of the chemostat (Bull 2006, Equation 2b). By assuming s = 0 and d = 0, and the empirical relationship of b = m(tL − e), the optimal lysis time is found to be
, where s is the free phage death rate, r the adsorption rate, N the host concentration, and d the washout rate of the chemostat (Bull 2006, Equation 2b). By assuming s = 0 and d = 0, and the empirical relationship of b = m(tL − e), the optimal lysis time is found to be 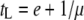 . In fact, as shown in appendix b, the same solution for the optimal lysis time can also be derived from the first theoretical approach of calculating phage growth rate. Although different in perspectives, both approaches arrived at the same solution for the optimal lysis time.
. In fact, as shown in appendix b, the same solution for the optimal lysis time can also be derived from the first theoretical approach of calculating phage growth rate. Although different in perspectives, both approaches arrived at the same solution for the optimal lysis time.
On the basis of the above solution, it is possible to test the predicted optimal lysis times against the empirical ones (Heineman and Bull 2007), by plugging in the independently estimated eclipse period e [∼28 min for phage λ (Wang 2006)] and empirically determined growth rate μ (see Table 2). In the case of LA phages, the highest relative fitness was attained by λSS68C, which has the maximum growth rate of 0.0494 min−1 and observed optimal lysis time of 64 min. Among the HA phages, it is the strain λSM1L that has the highest relative fitness with a maximum growth rate of 0.0510 min−1 and the observed optimal lysis time of 46 min. According to the above equation, a maximum growth rate of 0.0494 min−1 (the LA phages) should have a predicted optimal lysis time of 48.2 min and a maximum growth rate of 0.0510 min−1 (the HA phages) a predicted optimal lysis times of 47.6 min. At present, it seems that the solution based on the current optimality models is able to predict the optimal lysis time for the HA phages with reasonable accuracy (predicted 47.6 min vs. observed 46 min), but not that for the LA phages (predicted 48.2 min vs. observed 64 min).
But the failure in prediction is not due to the LA phages per se. Analysis of the data from a previous study (Wang 2006) showed a somewhat different conclusion. Based on the maximum growth rate of 0.047 min−1 (2.83 hr−1, Table 2 of Wang 2006), the predicted optimal lysis time is 49.2 min, a reasonably close prediction to the empirically determined 51.0 min. It seems that the discrepancy is partly due to the higher growth rate of the LA phages in the current study, which may be caused by the higher initial host concentration (∼106 cells/ml) when compared to the previous study (∼105 cells/ml) (Wang 2006). That is, the search time (thus the generation time) of the LA phages would be shorter, and consequently the growth rate higher, in this study.
But the main reason for such an uncertainty in predicting the optimal lysis time may be because of the nonequilibrium nature of the batch culture. For both optimality models, the phage growth rate, and thus the optimal lysis time, should be evaluated under the condition of constant host density. However, for most batch culture experiments, the phage growth rate is usually determined under the condition of exponential host growth, resulting in a progressively shortened search time and thus a progressively higher instantaneous growth rate. The empirically determined growth rate is simply an average over the assay period. When the growth rate μ is larger, then the optimal lysis time becomes shorter (cf. 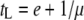 ). Therefore, it seems that caution is needed in interpreting quantitative predictions of the optimal lysis time when the study is conducted under the batch culture condition. This conclusion does not deny the utility of the batch culture. Besides its easy setup, studies conducted with the batch culture can nevertheless provide us the qualitative predictions of optimality models, as is demonstrated in this study.
). Therefore, it seems that caution is needed in interpreting quantitative predictions of the optimal lysis time when the study is conducted under the batch culture condition. This conclusion does not deny the utility of the batch culture. Besides its easy setup, studies conducted with the batch culture can nevertheless provide us the qualitative predictions of optimality models, as is demonstrated in this study.
Fitness landscape and evolutionary pathways:
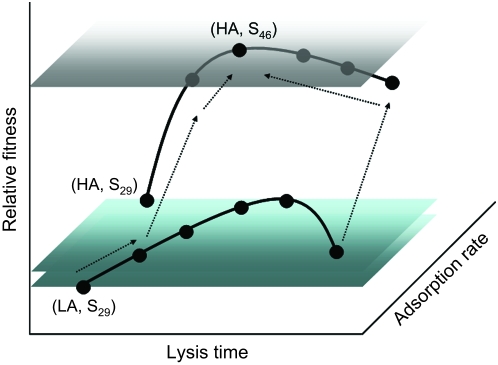
Even though results from this study are relatively sparse and are not enough to allow us to reconstruct a detailed fitness landscape, we can nevertheless infer what a fitness landscape might be like for phage λ (Figure 5). Implicitly, this study presumes a Fisherian fitness landscape with one global maximum. That is, for a given environment, there is one best combination of life-history traits that results in maximal fitness. Given a long enough time, we also assume that each part of the phage genome will coadapt to a particular environment, resulting in the highest fitness possible. In this discussion, we presume that all strains will eventually arrive at the combined traits of (HA, S46) or one close to it. Results from this study allow us to speculate on the possible evolutionary pathways to achieve this maximum fitness.
Figure 5.—
A hypothetical fitness landscape in which the phage relative fitness is expressed as a function of adsorption rate and lysis time. The two solid curves are redrawn from Figure 3. Solid dots represent λ-strains used in the competition experiments. The three horizontal planes indicates the relative fitnesses of, from low to high, λ(LA, S29), λ(HA, S29), and λ(HA, S46), respectively. The positions of adsorption rates and lysis times are not to scale and are for reference only. The dotted lines with arrowheads indicate the speculated evolutionary sequences from suboptimal strains to the optimal strain. Except for λ(LA, S29), which is hypothesized to first evolve a longer lysis time, followed by a higher adsorption rate, all the others should evolve to a higher adsorption rate and then an optimal lysis time.
For example, in the case of λ(LA, S29), it has the lowest fitness among all strains. To climb up the fitness peak, the first step for λ(LA, S29) could be to increase its adsorption rate or lysis time. As shown in Figure 4, the fitness gain for λ(LA, S29) to evolve to HA (while retaining its lysis time) is not as large as if it retains its LA status but evolves to other longer lysis times. (This because when compared to λ(HA, S29), all the other low-adsorption, but longer lysis phages have a >1.0 relative fitness.) That is, starting from λ(LA, S29), the evolutionarily profitable pathway would be to evolve to a longer lysis time first. An observation from this study provides circumstantial support for the above speculation. It was found that during competition the λ(LA, S29, Lac+) strain consistently showed polymorphism of plaque morphology, from entirely small irregular shapes at the beginning of the competition to a mix of small irregular and medium-sized round shapes at the end. An independent study showed that the slightly increased plaque size is consistent with a lengthened lysis time (our unpublished results). DNA sequencing of round λ(LA, S29, Lac+) showed that four of six random plaques have, besides the original mutations, acquired extra mutations in the S gene, presumably resulting in lengthened lysis time (our unpublished results). In fact, the evolution of this particular S allele is so consistent that all competition results were discarded if the total λ(LA, S29, Lac+) plaque count had >10% of medium-sized round plaques.
On the other hand, results showed that λ(HA, S39) outcompeted all the other LA phages with a different lysis time (our unpublished results, see the results section), suggesting that the evolutionarily profitable pathway for λ(HA, S39) would be to evolve to a higher adsorption rate first, followed by evolution to the optimal lysis time. Since all the other LA phages have a relative fitness similar to or higher than that of λ(LA, S39) (see Figure 3), it is expected that the fitness advantage of evolving a high adsorption rate would be similar. Therefore, except for λ(LA, S29), it is expected that the other suboptimal strains should evolve with the sequence of “high adsorption rate” → “optimal lysis time.” Interestingly, simulation studies by Bull (2006) showed a similar conclusion. In Figure 3 of the study (Bull 2006), the relationship between phage growth rate and lysis time is shown with three curves, some representing different host densities. Since the effect of host density and adsorption rate on phage growth rate can be treated as equivalent, these curves can be interpreted as representing phages with different adsorption rates. Closer inspection of these curves suggests that, regardless of the lysis time, it is always evolutionarily profitable for phages to evolve to a higher adsorption rate first.
So far, this analysis suggests that most suboptimal strains would follow the evolutionary sequence of high adsorption rate → optimal lysis time, though under some restricted conditions, say certain combinations of low adsorption rate and suboptimal lysis times, the sequence would be longer (or shorter) lysis time → high adsorption rate. It is not clear how general this conclusion is. One problem is that the ideas of a high or a low adsorption rate and a long or a short lysis time are context dependent and should be evaluated in the presence of other life-history traits and environmental conditions. After all, this study was conducted using phage λ. It is likely that this simple conclusion may be limited to phages with a similar range of life-history trait values to that of phage λ. Nevertheless, it would be interesting to develop a theoretical basis for understanding the nature of the phage fitness landscape such that we would be able to predict the evolutionary sequences of climbing up the fitness peak.
Mutations and evolutionary steps:
For the above discussion, we have solely focused on the phenotypic imperatives derived from optimality analysis, i.e., which trait to evolve first to extract the most fitness gain. Whether the evolution of the suboptimal strain would follow the most profitable pathway would depend on the genetic basis underlying these traits. The evolution of phenotype is always constrained by the underlying genotype.
In the case of adsorption rate, there are at least two mechanisms to achieve a high adsorption rate: mutations at the tail fiber gene J (our unpublished data) or reacquisition of the side tail fiber through a revertant, or possibly a compensatory, mutation. In the discussion above, we did not assume that the current adsorption rate is the highest rate phage λ can achieve. In fact, we recently obtained λ-strains with even higher adsorption rates through combinations of mutations at the J gene and reacquisition of the stf gene. However, note that the genetic basis for the current phenotypic difference between the LA and HA phages is only a single point mutation. Therefore, it is biologically possible for an LA phage to become an HA phage in one single mutational step.
A similar case can also be made with the evolution of lysis time. A preliminary study on the lysis time distribution within the one-step mutational neighborhood of the λ S holin gene showed that it is relatively easy to access biologically relevant lysis times through single missense mutations (our unpublished data). This initial result suggests that the holin gene is quite malleable. Therefore, it is also possible for a suboptimal phage to climb up the fitness peak by improving its lysis time.
On the basis of various anecdotal evidence, it is highly likely that both evolutionary pathways (through changing lysis time or increasing adsorption rate) of increasing fitness are readily accessible; although it may be easier to change the lysis time than the adsorption rate. Nonetheless, it would be interesting to subject suboptimal strains with different genetic backgrounds to the protocol of a long-term evolution experiment to see if the first evolutionary step confirms the hypothesis presented above.
A coadapted genome in the presence of mosaic genomes:
One of the most striking features of the phage genome is the pattern of the “mosaic genome,” in which pairwise comparisons of phage genomes often yield different stretches of highly homologous sequences juxtaposed with entirely unrelated ones (Highton et al. 1990; Casjens et al. 1992; Juhala et al. 2000). This mosaic pattern suggests that phage genomes in the wild are constantly experiencing rampant genetic recombination. The demarcation of the genetic exchange is usually at borders of various functional modules (Juhala et al. 2000). The implication of genetic recombination is that alleles responsible for different adsorption rates, lysis times, and assembly rates are likely to be shuffled in and out of any phage genome. Analogous to the effect of mutation on fitness, genetic recombination can break up an already coadapted genome (like a deleterious mutation), recreate a combination of alleles that results in a higher fitness (like a beneficial mutation), or have no effect at all on fitness (like a neutral mutation). Since we do not know the magnitude of the recombination rate, it is difficult to evaluate its effect on the dynamics of phage adaptation. Nevertheless, it is an interesting and important topic worthy of further study.
Acknowledgments
The authors thank Kuangnang Xiong for constructing the pS105/V77G plasmid and Jim Bull for his helpful and thoughtful comments. This study is sponsored by National Institutes of Health grant GM 072815.
APPENDIX A
The adsorption assay is simply a truncated infection process. Therefore, a previously described model for the population dynamics of phage infection (Wang 2006), after some simplifications, can also be used to describe the adsorption process.
The main difference between the full-scale infection and a simple adsorption is that the assay period is generally shorter than the lysis time; therefore, there is no production of phage progeny. That is, during the assay period the host and phage dynamics can be simplified as
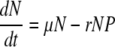 |
(A1) |
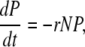 |
(A2) |
where N and P are the bacteria and phage concentrations, μ is the bacterial growth rate, and r the adsorption rate.
Since the adsorption experiments were always conducted with an excess of host cells (MOI ≈ 0.001), the decline of cell concentration due to phage adsorption in Equation A1 can be ignored. That is, Equation A1 can be simplified as
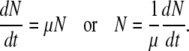 |
(A3) |
By combining Equations A2 and A3, it can be shown that
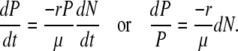 |
(A4) |
The solution for Equation A4 is obtained as 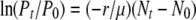 , where Pt and P0 and Nt and N0 are phage and bacteria concentrations at times t and 0, respectively. Since
, where Pt and P0 and Nt and N0 are phage and bacteria concentrations at times t and 0, respectively. Since 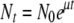 , it can be shown that
, it can be shown that 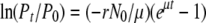 .
.
APPENDIX B
Using a population dynamics model that is based on chemostat culture condition, Bull (2006; Bull et al. 2006) derived the optimal lysis time as 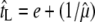 , where tL is the lysis time, e the eclipse period, and μ the phage growth rate, with the circumflexes indicating values at optimum. However, this relationship can also be derived using phage growth rate expressed as
, where tL is the lysis time, e the eclipse period, and μ the phage growth rate, with the circumflexes indicating values at optimum. However, this relationship can also be derived using phage growth rate expressed as 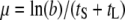 , where tS is the search time for the bacterial host and b is the burst size, which can be expressed as b = m(tL − e), where m is the maturation rate. The optimal lysis time is the time when the phage growth rate μ is maximum with respect to the lysis time tL, i.e., when
, where tS is the search time for the bacterial host and b is the burst size, which can be expressed as b = m(tL − e), where m is the maturation rate. The optimal lysis time is the time when the phage growth rate μ is maximum with respect to the lysis time tL, i.e., when 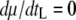 . It can then be shown that when
. It can then be shown that when 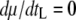 ,
, 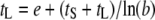 , or
, or 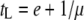 .
.
References
- Abedon, S. T., 1989. Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microb. Ecol. 18 79–88. [DOI] [PubMed] [Google Scholar]
- Abedon, S. T., T. D. Herschler and D. Stopar, 2001. Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 67 4233–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedon, S. T., P. Hyman and C. Thomas, 2003. Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl. Environ. Microbiol. 69 7499–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J., D. W. Pfennig and I.-N. Wang, 2004. Genetics details, optimization and phage life histories. Trends Ecol. Evol. 19 76–82. [DOI] [PubMed] [Google Scholar]
- Bull, J. J., 2006. Optimality models of phage life history and parallels in disease evolution. J. Theor. Biol. 241 928–938. [DOI] [PubMed] [Google Scholar]
- Bull, J. J., J. Millstein, J. Orcutt and H. A. Wichman, 2006. Evolutionary feedback mediated through population density, illustrated with viruses in chemostats. Am. Nat. 167 E39–E51. [DOI] [PubMed] [Google Scholar]
- Caraco, T., and I. N. Wang, 2008. Free-living pathogens: life-history constraints and strain competition. J. Theor. Biol. 250 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens, S. R., G. F. Hatfull and R. W. Hendrix, 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin. Virol. 3 383–397. [Google Scholar]
- Chao, L., 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348 454–455. [DOI] [PubMed] [Google Scholar]
- De Paepe, M., and F. Taddei, 2006. Viruses' life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4 e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, E., D. Clarke, A. Moya, E. Domingo and J. Holland, 1992. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc. Natl. Acad. Sci. USA 89 6015–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen, D. E., A. M. Dean and D. L. Hartl, 1987. Metabolic flux and fitness. Genetics 115 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, D. I., and M. Gottesman, 1983. Lytic mode of lambda development, pp. 21–51 in Lambda II, edited by R. W. Hendrix, J. W. Roberts, F. W. Stahl and R. A. Weisberg. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Goldberg, E., L. Grinius and L. Letellier, 1994. Recognition, attachment, and injection, pp. 347–356 in Molecular Biology of Bacteriophage T4, edited by J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling et al. American Society for Microbiology, Washington, DC.
- Heineman, R. H., and J. J. Bull, 2007. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution 61 1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix, R. W., and R. L. Duda, 1992. Bacteriophage λPaPa: not the mother of all λ phages. Science 258 1145–1148. [DOI] [PubMed] [Google Scholar]
- Highton, P. J., Y. Chang and R. J. Myers, 1990. Evidence for the exchange of segments between genomes during the evolution of lambdoid bacteriophages. Mol. Microbiol. 4 1329–1340. [DOI] [PubMed] [Google Scholar]
- Husimi, Y., K. Nishigaki, Y. Kinoshita and T. Tanaka, 1982. Cellstat—a continuous culture system of a bacteriophage for the study of the mutation rate and the selection process of the DNA level. Rev. Sci. Instrum. 53 517–522. [DOI] [PubMed] [Google Scholar]
- Hutchison, 3rd, C. A., and R. L. Sinsheimer, 1966. The process of infection with bacteriophage φX174. X. Mutations in a φX lysis gene. J. Mol. Biol. 18 429–447. [DOI] [PubMed] [Google Scholar]
- Josslin, R., 1970. The lysis mechanism of phage T4: mutants affecting lysis. Virology 40 719–726. [DOI] [PubMed] [Google Scholar]
- Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull et al., 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299 27–51. [DOI] [PubMed] [Google Scholar]
- Katsura, I., 1983. Tail assembly and injection, pp. 331–346 in Lambda II, edited by R. W. Hendrix, J. W. Roberts, F. W. Stahl and R. A. Weisberg. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Lenski, R. E., 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution 42 425–432. [DOI] [PubMed] [Google Scholar]
- Martinez, M. A., C. Carrillo, F. Gonzalez-Candelas, A. Moya, E. Domingo et al., 1991. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J. Virol. 65 3954–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H. H., and A. Arkin, 1997. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA 94 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger, M., 1932. Adsorption of bacteriophages to homologous bacteria. II. Quantitative investigation of adsorption velocity and saturation. Estimation of the particle size of the bacteriophage. Immunitaetsforschung 114 149–160. [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry. W. H. Freeman, New York.
- Stent, G. S., 1965. Papers on Bacterial Viruses. Little, Brown, Boston.
- Wang, I. N., 2006. Lysis timing and bacteriophage fitness. Genetics 172 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.-N., D. E. Dykhuizen and L. B. Slobodkin, 1996. The evolution of phage lysis timing. Evol. Ecol. 10 545–558. [Google Scholar]
- Wang, I.-N., D. L. Smith and R. Young, 2000. HOLINS: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54 799–825. [DOI] [PubMed] [Google Scholar]
- Wang, I.-N., J. Deaton and R. Young, 2003. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J. Bacteriol. 185 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, R., 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56 430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, R., I.-N. Wang and W. D. Roof, 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8 120–128. [DOI] [PubMed] [Google Scholar]